- Laboratory of Molecular Developmental Biology, Faculty of Biology-Oriented Science and Technology, Kindai University, Kinokawa-shi, Japan
Actin is a highly abundant protein in eukaryotic cells and dynamically changes its polymerized states with the help of actin-binding proteins. Its critical function as a constituent of cytoskeleton has been well-documented. Growing evidence demonstrates that actin is also present in nuclei, referred to as nuclear actin, and is involved in a number of nuclear processes, including transcriptional regulation and chromatin remodeling. The contribution of nuclear actin to transcriptional regulation can be explained by its direct interaction with transcription machineries and chromatin remodeling factors and by controlling the activities of transcription factors. In both cases, polymerized states of nuclear actin affect the transcriptional outcome. Nuclear actin also plays an important role in activating strongly silenced genes in somatic cells for transcriptional reprogramming. When these nuclear functions of actin are considered, it is plausible to speculate that nuclear actin is also implicated in embryonic development, in which numerous genes need to be activated in a well-coordinated manner. In this review, we especially focus on nuclear actin’s roles in transcriptional activation, reprogramming and development, including stem cell differentiation and we discuss how nuclear actin can be an important player in development and cell differentiation.
Introduction
Eukaryotic cells have a globular, multifunctional protein called actin. Actin plays key roles in the cytoplasm by controlling a dynamic equilibrium between the monomeric and filamentous states for a variety of cellular processes, such as cell motility and adhesion. The amount of actin in nuclei is lower than that in the cytoplasm in most of cell types. Nevertheless, actin has also been found in the nucleus of various types of cells (Lestourgeon et al., 1975; Clark and Merriam, 1977; Clark and Rosenbaum, 1979; Pederson and Aebi, 2002). Nuclear actin has attracted a lot of attention, although its function has often been regarded as a controversial topic due to the technical difficulties of discriminating nuclear actin from cytoplasmic actin, which is highly abundant in cells. With the development of experimental tools, convincing evidence has been obtained that nuclear actin indeed does exist and plays crucial roles in many nuclear processes. For example, actin is found in all kinds of RNA polymerase complexes (Visa and Percipalle, 2010) and in a number of chromatin remodeling complexes (Zhao et al., 1998; Kapoor and Shen, 2014) (Figure 1A). It also binds to transcription regulators to control their activities (Grosse and Vartiainen, 2013).
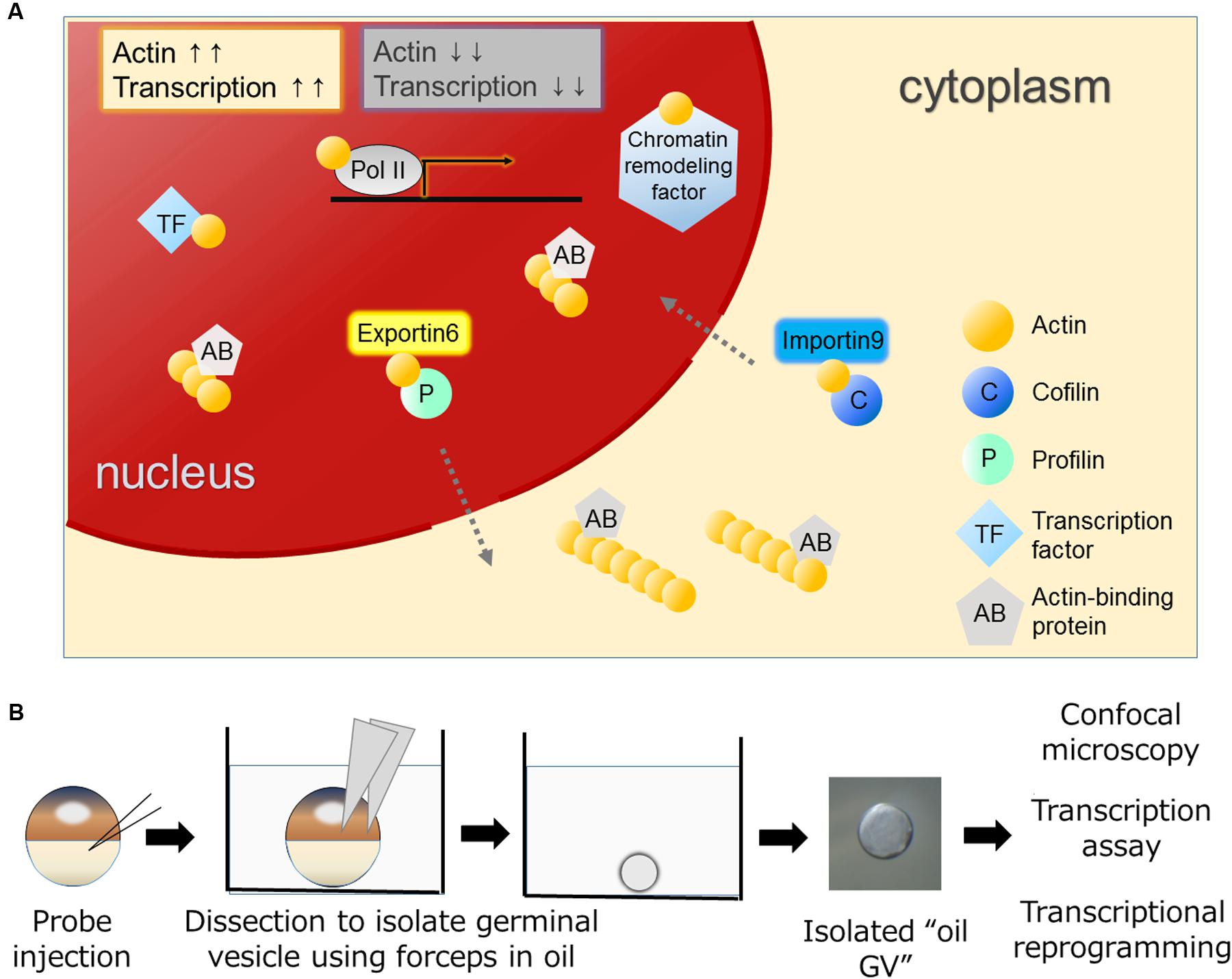
FIGURE 1. Various functions of actin in the nucleus and the oil GV system to study nuclear actin. (A) Nuclear actin binds to many nuclear proteins including transcription machineries, transcription factors, and chromatin remodeling factors. Nuclear actin-binding proteins such as actin nucleation promoting factors regulate nuclear actin polymerization. Nuclear concentrations of actin are regulated by IMPORTIN9 and EXPORTIN6 and high concentrations of nuclear actin are generally associated with high transcriptional activities. (B) Actin probes such as UtrCH-GFP mRNA are injected into Xenopus oocytes (Miyamoto et al., 2011). Germinal vesicles (GVs) filled with actin probes are isolated in mineral oil and these oil GVs can be observed by confocal microscopy to monitor nuclear actin dynamics. Oil GVs retain transcriptional activities, and when somatic nuclei are transplanted into the GVs, the injected nuclei undergo transcriptional reprogramming.
Dynamic changes between globular (G-) and filamentous (F-) forms of nuclear actin affect these nuclear events. Nuclear actin polymerization is regulated by multiple mechanisms; more than 30 of actin-binding proteins that affect its polymerized states have been discovered in nuclei (Gieni and Hendzel, 2009; Castano et al., 2010; Kristo et al., 2016). It is of great interest to reveal how monomeric/polymeric actin is orchestrated by these actin-binding proteins during the above mentioned nuclear events. The regulation of the total amount of nuclear actin is another critical factor that affects activities of actin-mediated nuclear events. Nuclear actin is actively exported to the cytoplasm by EXPORTIN6 as the profilin-actin heterodimer (Stuven et al., 2003). Conversely, cytoplasmic actin together with cofilin is imported to the nucleus by IMPORTIN9 (Dopie et al., 2012). The active maintenance of nuclear actin levels is needed to support transcription (Dopie et al., 2012) (Figure 1A). In addition to these functions of actin to support fundamental nuclear processes, nuclear actin influences cellular phenotypes through transcriptional regulation and chromatin alternation, exemplified by transcriptional activation of the osteogenic genes in differentiating mesenchymal stem cells (MSCs; Sen et al., 2015) or by reorganization of heterochromatin during lineage commitment of epidermal stem cells (Le et al., 2016) (Figure 2). In this review, we summarize recent findings of functional roles of nuclear actin in transcriptional activation and chromatin remodeling. Currently, discovered functions of nuclear actin are executed in many different crucial cellular events such as transcription (de Lanerolle and Serebryannyy, 2011; Percipalle, 2013), transcriptional regulation (Grosse and Vartiainen, 2013), RNA processing (Percipalle et al., 2001; Visa and Percipalle, 2010), nuclear export (Percipalle et al., 2002), chromatin remodeling (Fenn et al., 2011; Kapoor et al., 2013; Kapoor and Shen, 2014), and nuclear reprogramming (Miyamoto and Gurdon, 2011; Miyamoto et al., 2011). We discuss how the nuclear functions of actin are related to development and cellular differentiation/dedifferentiation.
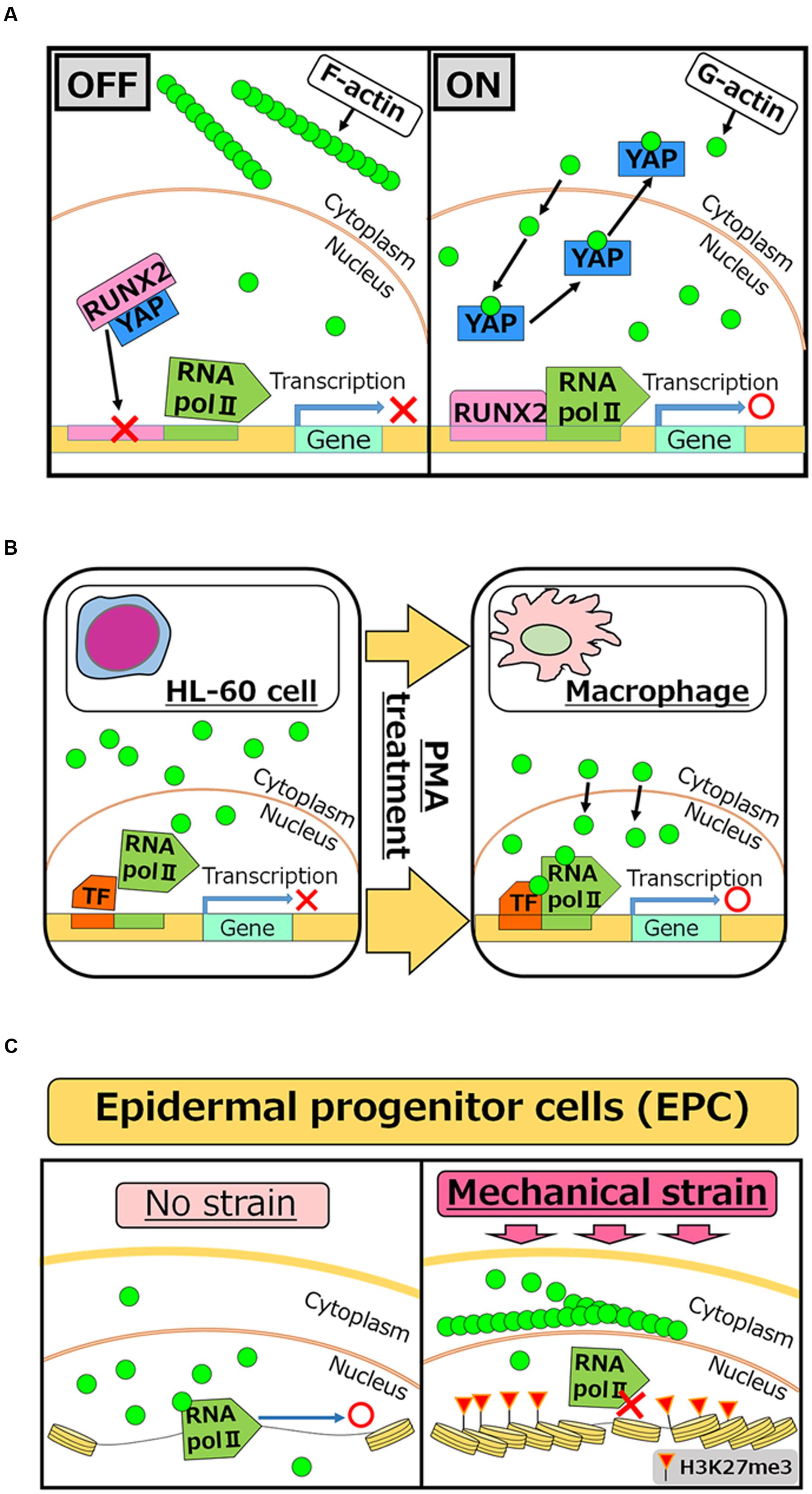
FIGURE 2. Roles of nuclear actin in cell differentiation and lineage commitment. (A) RUNX2 is a transcription factor which is required for activating the osteogenic gene, while interaction of RUNX2 with YAP protein inhibits its function as an activator (OFF). Increased concentrations of nuclear actin induce nuclear export of YAP, and therefore freed RUNX2 activates target genes for osteogenesis (ON) (Sen et al., 2015). (B) PMA treatment induces differentiation of HL-60 cells to macrophages. After the treatment, nuclear actin accumulates and induces transcription of genes related to macrophage activation (Xu et al., 2010). (C) When human EPCs are exposed to mechanical strain, actin polymerization is enhanced at the outer nuclear membrane, which in turn reduces the levels of nuclear actin. Reduced nuclear actin is associated with attenuated Pol II activities and the acquisition of H3K27me3, a repressive histone mark (Le et al., 2016).
Nuclear Actin and Transcriptional Activity
The implication of nuclear actin in transcription was first reported in the 1980s using amphibian oocytes (Scheer et al., 1984). Subsequently molecular evidence suggested that actin is contained in the transcription initiation complex of RNA polymerase II (Pol II) (Hofmann et al., 2004), bound to the C terminus domain of Pol II (Kukalev et al., 2005), and also bound to heterogeneous ribonucleoproteins (hnRNPs) and histone acetyltransferase PCAF during transcriptional elongation (Percipalle et al., 2003; Obrdlik et al., 2008). Furthermore, actin’s binding partners in the nucleus include not only the transcription machineries per se, but also the complexes of chromatin-remodeling factors. A variety of chromatin remodeling complexes contain actin, such as BAF (Zhao et al., 1998; Nishimoto et al., 2012), INO80 (Fenn et al., 2011; Kapoor et al., 2013), and SWR1 complexes (Kapoor and Shen, 2014). Additionally, it is also contained in histone-modifying complexes like NuA4 (Galarneau et al., 2000) and interacts with histone deacetylases (HDACs) (Serebryannyy et al., 2016). Examples of how nuclear actin plays a role in the chromatin remodeling factors have been shown in the BAF and INO80 complexes. The BAF complex, which belongs to the SWI/SNF family, slides and ejects nucleosomes in an ATP-dependent manner (Cairns, 2009). It can function as both transcriptional activators and repressors and its essential roles in development and reprogramming have been shown (Ho and Crabtree, 2010). Actin has been found as a monomer in the BAF complex and binding of β-actin to actin-related protein 4 (ARP4) modulates the activity of the complex (Nishimoto et al., 2012). The INO80 complex can evict and/or slide nucleosomes and is involved in telomere regulation, chromosome segregation, and DNA replication (Ho and Crabtree, 2010). In the INO80 complex, a monomer of nuclear actin is included in the INO80 subcomplex together with ARP4 and ARP8 (Fenn et al., 2011; Kapoor et al., 2013). Actin cannot bind to DNA directly, and instead actin-ARPs modules enable their binding to DNA (Gerhold et al., 2012). Requirement of monomeric actin for INO80-mediated chromatin remodeling has been demonstrated (Kapoor et al., 2013). Moreover, an increased nuclear actin concentration inhibits HDAC activities (Serebryannyy et al., 2016). Taken together, nuclear actin is involved in enhanced transcriptional states and active chromatin remodeling in general (Figure 1A). As a consequence, the amount of nuclear actin, regulated by IMPORTIN 9 (Dopie et al., 2012) and EXPORTIN 6 (Stuven et al., 2003), is crucial for transcription.
On top of these direct interactions between nuclear actin and chromatin proteins, actin regulates transcription levels by sequestering transcriptional activators (Kang et al., 2004) or repressors (Huang et al., 2011). A well-documented case is represented by serum response factor (SRF). SRF activity is regulated by binding of MAL, a co-activator of SRF transcription factor, to target genes (Vartiainen et al., 2007). However, if actin monomer binds to MAL, this MAL/actin binding induces not only inhibition of interaction between MAL and Pol II, but also nuclear export of the MAL/actin complex (Vartiainen et al., 2007). Increased actin polymerization results in reduced monomeric actin and consequently activates SRF target genes. Interestingly, nuclear actin polymerization is sufficient to activate SRF targets (Baarlink et al., 2013). In summary, multiple mechanisms related to nuclear actin lead to transcriptional activation and changes in polymerized states of nuclear actin often influence the transcriptional outcome.
Nuclear Actin in Transcriptional Reprogramming
Differentiated cell states of somatic cells can be reversed to the undifferentiated one by nuclear reprogramming. Nuclear reprogramming of somatic cells entails global changes in transcriptional programs. This transcriptional reprogramming is achieved by nuclear transplantation of differentiated somatic cells to oocytes (Jullien et al., 2011). Especially when somatic nuclei are transplanted into the germinal vesicle (GV) – a giant nucleus – of the Xenopus laevis oocyte, the transplanted nuclei undergo widespread transcriptional reprogramming within 2 days without the need for cell divisions and new protein synthesis (Jullien et al., 2014). These characteristic features of the Xenopus oocyte nuclear transfer system enabled us to reveal several important factors and mechanisms underlying transcriptional reprogramming (Jullien et al., 2010, 2014; Miyamoto et al., 2011, 2013). Of note, nuclear actin and actin-binding protein play important roles during transcriptional reprogramming in oocytes (Miyamoto et al., 2011, 2013). Xenopus oocytes have advantages for studying nuclear actin for two main reasons. Firstly, GVs of Xenopus oocytes contain a large amount of nuclear actin due to the lack of EXPORTIN 6 (Bohnsack et al., 2006). This large amount of nuclear actin makes actin polymerization in nuclei easily visible. Secondly, GVs can be isolated in non-aqueous liquid, like mineral oil, without disrupting their transcriptional activities (Figure 1B) (Gall and Wu, 2010). Hereafter, we name these isolated GVs in mineral oil as “oil GV.” Oil GVs represent live purified nuclei and hence provide an optimal platform for nuclear actin research, since we do not have to consider the cytoplasmic function of actin (Figure 1B). Even transcriptional reprogramming can be induced in the oil GV system after nuclear transplantation of somatic nuclei into oil GVs (Miyamoto et al., 2011). Using the oil GV system, we have shown that perturbation of polymerized states of nuclear actin affects transcriptional reprogramming of Oct4, a key pluripotency gene; namely decreased actin polymerization results in failed activation of Oct4 and increased actin polymerization is associated with enhanced Oct4 expression (Miyamoto et al., 2011). Furthermore, we found WASF1 (also known as WAVE1), an actin-binding protein that can regulate actin polymerization, in GVs (Miyamoto et al., 2013). The knockdown of nuclear WAVE1 causes defects in transcriptional reprogramming (Miyamoto et al., 2013). These results indicate that both nuclear actin and nuclear actin-binding protein play roles in transcriptional reprogramming. Importantly, recent studies confirm nuclear actin’s contributions to transcriptional reprogramming using cultured cells. Forced expression of nuclear actin in Hela cells results in altered expression of approximately 2000 genes, including Oct4 (Yamazaki et al., 2015). Moreover, nuclear F-actin, but not nuclear G-actin, plays a role in recruiting β-catenin to the Oct4 locus for its activation (Yamazaki et al., 2016). Sadhukhan et al. (2014) revealed a nuclear role of Wiskott-Aldrich Syndrome Protein (WASP), an actin nucleation-promoting factor, in transcriptional activation during human TH cell differentiation. They also found that WASP accelerates epigenetic modification, namely histone H3 lysine 4 trimethylation, independent of actin polymerization activities (Sadhukhan et al., 2014), in good agreement with WAVE1’s function in the frog system. Taken together, functional roles of actin and actin nucleation-promoting factor in the nucleus seem well-conserved across different species and different cell types although the amount of nuclear actin varies. It is also noteworthy that when cells are exposed to stimuli to change their stable states, nuclear actin tends to be formed. This is exemplified by transcriptional reprogramming (Miyamoto et al., 2011), DNA damage (Belin et al., 2015), mechanical strain (Le et al., 2016), and serum stimulation (Vartiainen et al., 2007; Baarlink et al., 2013). It would be therefore intriguing to observe dynamics of nuclear actin in vivo when cells are exposed to such stimuli during differentiation and development.
Nuclear Actin in Development and Cell Differentiation
Considering nuclear roles of actin in transcriptional regulation, it is reasonable to speculate that nuclear actin is involved in cellular differentiation and development, which entail changes in transcriptional programs. Accumulating evidence indicates that this is indeed true at least in several different cell types. Induced accumulation of nuclear actin in MSCs causes transcriptional activation of the osteogenic genes osterix and osteocalcin, leading to the osteogenic phenotype. Mechanistically, YAP protein binds to and inhibits the osteogenic transcription factor RUNX2. The accumulation of nuclear actin induces nuclear export of YAP protein. As a result, RUNX2 is relieved from its repressive interaction with YAP and osteogenesis is enhanced in a RUNX2-dependent manner (Sen et al., 2015) (Figure 2A). Translocation of actin to nuclei is also observed when HL-60 cells differentiate toward macrophages after phorbol 12-myristate 13-acetate (PMA) treatment (Xu et al., 2010). In this case, nuclear β-actin is involved in transcriptional activation of target genes important for macrophage differentiation (Figure 2B). Another recent study shows that nuclear actin is involved in lineage commitment of epidermal progenitor cells (EPCs) through the regulation of the transcriptional activity (Le et al., 2016). Upon induction of mechanical strain on EPCs, enhanced actin polymerization at the outer nuclear membrane is observed, which in turn reduces the amount of nuclear actin, leading to suppression of RNA Pol II activity and the increase of histone H3 lysine 27 trimethylation (H3K27me3). This increased level of H3K27me3 is crucial for suppressing the expression of lineage-commitment genes (Figure 2C). In addition to these roles of nuclear actin in cell differentiation and lineage commitment, nuclear actin-binding proteins also serve as a key player for cell differentiation. In hematopoietic cells, WASP is localized in nuclei during T cell differentiation (Taylor et al., 2010). WASP supports histone H3K4 methyltransferase activity for transcriptional activation of TH1 genes, such as IFNG and TBX21, and is important for differentiation (Sadhukhan et al., 2014). It is noteworthy that nuclear roles of WASP seem uncoupled from its actin polymerization activity (Sadhukhan et al., 2014), although the actin-related protein 2/3 (ARP2/3) complex, which binds to WASP through the VCA domain, exists in nuclei (Yoo et al., 2007). Differential functions of other actin-binding proteins between the nucleus and the cytoplasm need to be determined in future studies.
Nuclear actin is found in female germ cells of Xenopus laevis (Clark and Merriam, 1977; Clark and Rosenbaum, 1979), Drosophila (Kelpsch et al., 2016), and avian species (Maslova and Krasikova, 2012). In the case of Drosophila, nurse cells and GVs contain nuclear actin during oogenesis and nuclear actin rod formation is regulated by Fascin (Kelpsch et al., 2016). In Xenopus, a large amount of nuclear actin in GVs contribute to mechanical stability of the giant nucleus (Bohnsack et al., 2006), stabilization of nuclear structure (Maslova and Krasikova, 2012; Feric and Brangwynne, 2013) and transcription (Scheer et al., 1984; Miyamoto et al., 2011). The amount of nuclear actin seems to decrease after oocyte maturation and fertilization since EXPORTIN 6 protein starts to be expressed after GV breakdown (Bohnsack et al., 2006). The abundance of nuclear actin and its functional roles in early embryonic development need further investigation. We have shown that an actin nucleation-promoting factor WAVE1, stored in the nuclei of Xenopus oocytes, is important for early embryonic development. Knockdown of maternal WAVE1 protein induces downregulation of Hox genes in gastrula embryos, which is rescued by nuclear WAVE1 expression (Miyamoto et al., 2013). In good agreement with our study, the requirement of nuclear N-WASP (neuronal Wiskott-Aldrich Syndrome Protein), another actin nucleation-promoting factor, and nuclear actin polymerization for HoxB expression upon retinoic acid treatment has been shown in cultured cells (Ferrai et al., 2009). However, it remains unknown to what extent WAVE1-regulated nuclear actin polymerization contributes to embryonic gene activation and early embryonic development since an isoform of WAVE1, which lacks the ARP2/3-binding VCA domain, can rescue Hox gene expression and embryonic development (Miyamoto et al., 2013). It is possible that nuclear roles of actin nucleation-promoting factor might be separated from their cytoplasmic functions, as is the case in WASP (Sadhukhan et al., 2014). Further research will reveal mechanisms of selecting actin-dependent/independent actions.
Conclusion
Recent studies have uncovered the implication of nuclear actin in a variety of nuclear processes, including basal transcription (Percipalle, 2013), transcriptional activation (Grosse and Vartiainen, 2013; Miyamoto and Gurdon, 2013), chromosome movement (Chuang et al., 2006; Dundr et al., 2007; Mehta et al., 2010), structural integrity (Feric and Brangwynne, 2013), chromatin remodeling (Szerlong et al., 2008; Fenn et al., 2011; Nishimoto et al., 2012; Kapoor et al., 2013), apoptosis (Grzanka et al., 2014; Grzanka et al., 2015), cell cycle progression (Goyal et al., 2011; Spencer et al., 2011; Kalendova et al., 2014), DNA repair (Andrin et al., 2012; Belin et al., 2015) and nuclear assembly (Krauss et al., 2003). These various features of nuclear actin could be related to cellular phenotypes during or after differentiation. It is likely that roles of nuclear actin will expand even more in the near future. For detailed understanding of nuclear actin, especially in the context of actin polymerization, it is critically important to develop new probes and systems to monitor nuclear actin dynamics (Belin et al., 2013; Plessner et al., 2015) and to distinguish nuclear and cytoplasmic actin. It is equally important to use genome-wide or a large-scale approaches to identify interactome of nuclear actin (Rohn et al., 2011; Samwer et al., 2013; Dopie et al., 2015). Structural insights into nuclear actin (Cao et al., 2016) will also accelerate our understanding. Finally, biological relevance of nuclear actin formation in vivo cells and/or in the context of pathogenesis warrants further investigation.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
KM is supported by Human Frontier Science Program (RGP0021/2016), by JSPS KAKENHI Grant Numbers JP16H01321, JP16H01222, by Grant for Basic Science Research Projects from The Sumitomo Foundation (150810), and by Kindai University Research Grant (15-I-2).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms. N. Backes Kamimura and Mr. J. Horvat for proof reading.
References
Andrin, C., McDonald, D., Attwood, K. M., Rodrigue, A., Ghosh, S., Mirzayans, R., et al. (2012). A requirement for polymerized actin in DNA double-strand break repair. Nucleus 3, 384–395. doi: 10.4161/nucl.21055
Baarlink, C., Wang, H., and Grosse, R. (2013). Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340, 864–867. doi: 10.1126/science.1235038
Belin, B. J., Cimini, B. A., Blackburn, E. H., and Mullins, R. D. (2013). Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell 24, 982–994. doi: 10.1091/mbc.E12-09-0685
Belin, B. J., Lee, T., and Mullins, R. D. (2015). DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-(1/2) that promotes efficient DNA repair. [corrected]. Elife 4:e07735. doi: 10.7554/eLife.07735
Bohnsack, M. T., Stuven, T., Kuhn, C., Cordes, V. C., and Gorlich, D. (2006). A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat. Cell Biol. 8, 257–263. doi: 10.1038/ncb1357
Cairns, B. R. (2009). The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198. doi: 10.1038/nature08450
Cao, T., Sun, L., Jiang, Y., Huang, S., Wang, J., and Chen, Z. (2016). Crystal structure of a nuclear actin ternary complex. Proc. Natl. Acad. Sci. U.S.A. 113, 8985–8990. doi: 10.1073/pnas.1602818113
Castano, E., Philimonenko, V. V., Kahle, M., Fukalova, J., Kalendova, A., Yildirim, S., et al. (2010). Actin complexes in the cell nucleus: new stones in an old field. Histochem. Cell Biol. 133, 607–626. doi: 10.1007/s00418-010-0701-2
Chuang, C. H., Carpenter, A. E., Fuchsova, B., Johnson, T., de Lanerolle, P., and Belmont, A. S. (2006). Long-range directional movement of an interphase chromosome site. Curr. Biol. 16, 825–831. doi: 10.1016/j.cub.2006.03.059
Clark, T. G., and Merriam, R. W. (1977). Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell 12, 883–891. doi: 10.1016/0092-8674(77)90152-0
Clark, T. G., and Rosenbaum, J. L. (1979). An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell 18, 1101–1108. doi: 10.1016/0092-8674(79)90223-X
de Lanerolle, P., and Serebryannyy, L. (2011). Nuclear actin and myosins: life without filaments. Nat. Cell Biol. 13, 1282–1288. doi: 10.1038/ncb2364
Dopie, J., Rajakyla, E. K., Joensuu, M. S., Huet, G., Ferrantelli, E., Xie, T., et al. (2015). Genome-wide RNAi screen for nuclear actin reveals a network of cofilin regulators. J. Cell Sci. 128, 2388–2400. doi: 10.1242/jcs.169441
Dopie, J., Skarp, K. P., Rajakyla, E. K., Tanhuanpaa, K., and Vartiainen, M. K. (2012). Active maintenance of nuclear actin by importin 9 supports transcription. Proc. Natl. Acad. Sci. U.S.A. 109, E544–E552. doi: 10.1073/pnas.1118880109
Dundr, M., Ospina, J. K., Sung, M. H., John, S., Upender, M., Ried, T., et al. (2007). Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179, 1095–1103. doi: 10.1083/jcb.200710058
Fenn, S., Breitsprecher, D., Gerhold, C. B., Witte, G., Faix, J., and Hopfner, K. P. (2011). Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J. 30, 2153–2166. doi: 10.1038/emboj.2011.118
Feric, M., and Brangwynne, C. P. (2013). A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 15, 1253–1259. doi: 10.1038/ncb2830
Ferrai, C., Naum-Ongania, G., Longobardi, E., Palazzolo, M., Disanza, A., Diaz, V. M., et al. (2009). Induction of HoxB transcription by retinoic acid requires actin polymerization. Mol. Biol. Cell 20, 3543–3551. doi: 10.1091/mbc.E09-02-0114
Galarneau, L., Nourani, A., Boudreault, A. A., Zhang, Y., Heliot, L., Allard, S., et al. (2000). Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell. 5, 927–937. doi: 10.1016/S1097-2765(00)80258-0
Gall, J. G., and Wu, Z. (2010). Examining the contents of isolated Xenopus germinal vesicles. Methods 51, 45–51. doi: 10.1016/j.ymeth.2009.12.010
Gerhold, C. B., Winkler, D. D., Lakomek, K., Seifert, F. U., Fenn, S., Kessler, B., et al. (2012). Structure of Actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res. 40, 11036–11046. doi: 10.1093/nar/gks842
Gieni, R. S., and Hendzel, M. J. (2009). Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem. Cell Biol. 87, 283–306. doi: 10.1139/O08-133
Goyal, P., Behring, A., Kumar, A., and Siess, W. (2011). STK35L1 associates with nuclear actin and regulates cell cycle and migration of endothelial cells. PLoS ONE 6:e16249. doi: 10.1371/journal.pone.0016249
Grosse, R., and Vartiainen, M. K. (2013). To be or not to be assembled: progressing into nuclear actin filaments. Nat. Rev. Mol. Cell Biol. 14, 693–697. doi: 10.1038/nrm3681
Grzanka, D., Gagat, M., and Izdebska, M. (2014). Involvement of the SATB1/F-actin complex in chromatin reorganization during active cell death. Int. J. Mol. Med. 33, 1441–1450. doi: 10.3892/ijmm.2014.1710
Grzanka, D., Kowalczyk, A. E., Izdebska, M., Klimaszewska-Wisniewska, A., and Gagat, M. (2015). The interactions between SATB1 and F-actin are important for mechanisms of active cell death. Folia Histochem. Cytobiol. 53, 152–161. doi: 10.5603/fhc.a2015.0018
Ho, L., and Crabtree, G. R. (2010). Chromatin remodelling during development. Nature 463, 474–484. doi: 10.1038/nature08911
Hofmann, W. A., Stojiljkovic, L., Fuchsova, B., Vargas, G. M., Mavrommatis, E., Philimonenko, V., et al. (2004). Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 6, 1094–1101. doi: 10.1038/ncb1182
Huang, W., Ghisletti, S., Saijo, K., Gandhi, M., Aouadi, M., Tesz, G. J., et al. (2011). Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature 470, 414–418. doi: 10.1038/nature09703
Jullien, J., Astrand, C., Halley-Stott, R. P., Garrett, N., and Gurdon, J. B. (2010). Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl. Acad. Sci. U.S.A. 107, 5483–5488. doi: 10.1073/pnas.1000599107
Jullien, J., Miyamoto, K., Pasque, V., Allen, G. E., Bradshaw, C. R., Garrett, N. J., et al. (2014). Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming. Mol. Cell. 55, 524–536. doi: 10.1016/j.molcel.2014.06.024
Jullien, J., Pasque, V., Halley-Stott, R. P., Miyamoto, K., and Gurdon, J. B. (2011). Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat. Rev. Mol. Cell Biol. 12, 453–459. doi: 10.1038/nrm3140
Kalendova, A., Kalasova, I., Yamazaki, S., Ulicna, L., Harata, M., and Hozak, P. (2014). Nuclear actin filaments recruit cofilin and actin-related protein 3, and their formation is connected with a mitotic block. Histochem. Cell Biol. 142, 139–152. doi: 10.1007/s00418-014-1243-9
Kang, M. I., Kobayashi, A., Wakabayashi, N., Kim, S. G., and Yamamoto, M. (2004). Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. U.S.A. 101, 2046–2051. doi: 10.1073/pnas.0308347100
Kapoor, P., Chen, M., Winkler, D. D., Luger, K., and Shen, X. (2013). Evidence for monomeric actin function in INO80 chromatin remodeling. Nat. Struct. Mol. Biol. 20, 426–432. doi: 10.1038/nsmb.2529
Kapoor, P., and Shen, X. (2014). Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 24, 238–246. doi: 10.1016/j.tcb.2013.10.007
Kelpsch, D. J., Groen, C. M., Fagan, T. N., Sudhir, S., and Tootle, T. L. (2016). Fascin regulates nuclear actin during Drosophila oogenesis. Mol. Biol. Cell 27, 2965–2979. doi: 10.1091/mbc.E15-09-0634
Krauss, S. W., Chen, C., Penman, S., and Heald, R. (2003). Nuclear actin and protein 4.1: essential interactions during nuclear assembly in vitro. Proc Natl Acad Sci U.S.A. 100, 10752–10757. doi: 10.1073/pnas.1934680100
Kristo, I., Bajusz, I., Bajusz, C., Borkuti, P., and Vilmos, P. (2016). Actin, actin-binding proteins, and actin-related proteins in the nucleus. Histochem. Cell Biol. 145, 373–388. doi: 10.1007/s00418-015-1400-9
Kukalev, A., Nord, Y., Palmberg, C., Bergman, T., and Percipalle, P. (2005). Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 12, 238–244. doi: 10.1038/nsmb904
Le, H. Q., Ghatak, S., Yeung, C. Y., Tellkamp, F., Gunschmann, C., Dieterich, C., et al. (2016). Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18, 864–875. doi: 10.1038/ncb3387
Lestourgeon, W. M., Forer, A., Yang, Y. Z., Bertram, J. S., and Pusch, H. P. (1975). Contractile proteins. Major components of nuclear and chromosome non-histone proteins. Biochim. Biophys. Acta 379, 529–552. doi: 10.1016/0005-2795(75)90160-9
Maslova, A., and Krasikova, A. (2012). Nuclear actin depolymerization in transcriptionally active avian and amphibian oocytes leads to collapse of intranuclear structures. Nucleus 3, 300–311. doi: 10.4161/nucl.20393
Mehta, I. S., Amira, M., Harvey, A. J., and Bridger, J. M. (2010). Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 11:R5. doi: 10.1186/gb-2010-11-1-r5
Miyamoto, K., and Gurdon, J. B. (2011). Nuclear actin and transcriptional activation. Commun. Integr. Biol. 4, 582–583. doi: 10.4161/cib.4.5.16491
Miyamoto, K., and Gurdon, J. B. (2013). Transcriptional regulation and nuclear reprogramming: roles of nuclear actin and actin-binding proteins. Cell Mol. Life Sci. 70, 3289–3302. doi: 10.1007/s00018-012-1235-7
Miyamoto, K., Pasque, V., Jullien, J., and Gurdon, J. B. (2011). Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 25, 946–958. doi: 10.1101/gad.615211
Miyamoto, K., Teperek, M., Yusa, K., Allen, G. E., Bradshaw, C. R., and Gurdon, J. B. (2013). Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science 341, 1002–1005. doi: 10.1126/science.1240376
Nishimoto, N., Watanabe, M., Watanabe, S., Sugimoto, N., Yugawa, T., Ikura, T., et al. (2012). Heterocomplex formation by Arp4 and beta-actin is involved in the integrity of the Brg1 chromatin remodeling complex. J. Cell Sci. 125, 3870–3882. doi: 10.1242/jcs.104349
Obrdlik, A., Kukalev, A., Louvet, E., Farrants, A. K., Caputo, L., and Percipalle, P. (2008). The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol. Cell. Biol. 28, 6342–6357. doi: 10.1128/MCB.00766-08
Pederson, T., and Aebi, U. (2002). Actin in the nucleus: What form and what for? J. Struct. Biol. 140, 3–9. doi: 10.1016/S1047-8477(02)00528-2
Percipalle, P. (2013). Co-transcriptional nuclear actin dynamics. Nucleus 4, 43–52. doi: 10.4161/nucl.22798
Percipalle, P., Fomproix, N., Kylberg, K., Miralles, F., Bjorkroth, B., Daneholt, B., et al. (2003). An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 100, 6475–6480. doi: 10.1073/pnas.1131933100
Percipalle, P., Jonsson, A., Nashchekin, D., Karlsson, C., Bergman, T., Guialis, A., et al. (2002). Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 30, 1725–1734. doi: 10.1093/nar/gks292
Percipalle, P., Zhao, J., Pope, B., Weeds, A., Lindberg, U., and Daneholt, B. (2001). Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 153, 229–236.
Plessner, M., Melak, M., Chinchilla, P., Baarlink, C., and Grosse, R. (2015). Nuclear F-actin formation and reorganization upon cell spreading. J. Biol. Chem. 290, 11209–11216. doi: 10.1074/jbc.M114.627166
Rohn, J. L., Sims, D., Liu, T., Fedorova, M., Schock, F., Dopie, J., et al. (2011). Comparative RNAi screening identifies a conserved core metazoan actinome by phenotype. J. Cell Biol. 194, 789–805. doi: 10.1083/jcb.201103168
Sadhukhan, S., Sarkar, K., Taylor, M., Candotti, F., and Vyas, Y. M. (2014). Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J. Immunol. 193, 150–160. doi: 10.4049/jimmunol.1302923
Samwer, M., Dehne, H. J., Spira, F., Kollmar, M., Gerlich, D. W., Urlaub, H., et al. (2013). The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. EMBO J. 32, 1886–1902. doi: 10.1038/emboj.2013.108
Scheer, U., Hinssen, H., Franke, W. W., and Jockusch, B. M. (1984). Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell 39, 111–122. doi: 10.1016/0092-8674(84)90196-X
Sen, B., Xie, Z., Uzer, G., Thompson, W. R., Styner, M., Wu, X., et al. (2015). Intranuclear actin regulates osteogenesis. Stem Cells 33, 3065–3076. doi: 10.1002/stem.2090
Serebryannyy, L. A., Cruz, C. M., and de Lanerolle, P. (2016). A role for nuclear actin in HDAC 1 and 2 regulation. Sci. Rep. 6:28460. doi: 10.1038/srep28460
Spencer, V. A., Costes, S., Inman, J. L., Xu, R., Chen, J., Hendzel, M. J., et al. (2011). Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J. Cell Sci. 124, 123–132. doi: 10.1242/jcs.073197
Stuven, T., Hartmann, E., and Gorlich, D. (2003). Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 22, 5928–5940. doi: 10.1093/emboj/cdg565
Szerlong, H., Hinata, K., Viswanathan, R., Erdjument-Bromage, H., Tempst, P., and Cairns, B. R. (2008). The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 15, 469–476. doi: 10.1038/nsmb.1403
Taylor, M. D., Sadhukhan, S., Kottangada, P., Ramgopal, A., Sarkar, K., D’Silva, S., et al. (2010). Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci. Transl. Med. 2:37ra44. doi: 10.1126/scitranslmed.3000813
Vartiainen, M. K., Guettler, S., Larijani, B., and Treisman, R. (2007). Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752. doi: 10.1126/science.1141084
Visa, N., and Percipalle, P. (2010). Nuclear functions of actin. Cold Spring Harb. Perspect. Biol. 2:a000620. doi: 10.1101/cshperspect.a000620
Xu, Y. Z., Thuraisingam, T., Morais, D. A., Rola-Pleszczynski, M., and Radzioch, D. (2010). Nuclear translocation of beta-actin is involved in transcriptional regulation during macrophage differentiation of HL-60 cells. Mol. Biol. Cell 21, 811–820. doi: 10.1091/mbc.E09-06-0534
Yamazaki, S., Yamamoto, K., de Lanerolle, P., and Harata, M. (2016). Nuclear F-actin enhances the transcriptional activity of beta-catenin by increasing its nuclear localization and binding to chromatin. Histochem. Cell Biol. 145, 389–399. doi: 10.1007/s00418-016-1416-9
Yamazaki, S., Yamamoto, K., Tokunaga, M., Sakata-Sogawa, K., and Harata, M. (2015). Nuclear actin activates human transcription factor genes including the OCT4 gene. Biosci. Biotechnol. Biochem. 79, 242–246. doi: 10.1080/09168451.2014.972332
Yoo, Y., Wu, X., and Guan, J. L. (2007). A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J. Biol. Chem. 282, 7616–7623. doi: 10.1074/jbc.M607596200
Keywords: nuclear actin, actin polymerization, transcriptional reprogramming, chromatin remodeling, regulation of gene expression, actin-binding protein, stem cell, differentiation
Citation: Misu S, Takebayashi M and Miyamoto K (2017) Nuclear Actin in Development and Transcriptional Reprogramming. Front. Genet. 8:27. doi: 10.3389/fgene.2017.00027
Received: 26 December 2016; Accepted: 20 February 2017;
Published: 07 March 2017.
Edited by:
Piergiorgio Percipalle, New York University Abu Dhabi, UAEReviewed by:
Cinzia Allegrucci, University of Nottingham, UKHolger Richly, Institute of Molecular Biology, Germany
Copyright © 2017 Misu, Takebayashi and Miyamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kei Miyamoto, kmiyamo@waka.kindai.ac.jp
 Shinji Misu
Shinji Misu Marina Takebayashi
Marina Takebayashi Kei Miyamoto
Kei Miyamoto