Soil Bacterial Communities and Diversity in Alpine Grasslands on the Tibetan Plateau Based on 16S rRNA Gene Sequencing
- 1Key Laboratory of Mountain Surface Processes and Ecological Regulation, Institute of Mountain Hazards and Environment, Chinese Academy of Sciences, Chengdu, China
- 2University of the Chinese Academy of Sciences, Chinese Academy of Sciences, Beijing, China
- 3Key Laboratory of Aquatic Botany and Watershed Ecology, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
The Tibetan Plateau, widely known as the world’s “Third Pole,” has gained extensive attention due to its susceptibility to climate change. Alpine grasslands are the dominant ecosystem on the Tibetan Plateau, albeit little is known about the microbial community and diversity among different alpine grassland types. Here, soil bacterial composition and diversity in the upper soils of five alpine grassland ecosystems, alpine meadow (AM), alpine steppe (AS), alpine meadow steppe (AMS), alpine desert (AD), and alpine desert steppe (ADS), were investigated based on the 16S rRNA gene sequencing technology. Actinobacteria (46.12%) and Proteobacteria (29.67%) were the two dominant soil bacteria at the phylum level in alpine grasslands. There were significant differences in the relative abundance at the genus level among the five different grassland types, especially for the Rubrobacter, Solirubrobacter, Pseudonocardia, Gaiella, Haliangium, and Geodermatophilus. Six alpha diversity indices were calculated based on the operational taxonomic units (OTUs), including Good’s coverage index, phylogenetic diversity (PD) whole tree index, Chao1 index, observed species index, Shannon index, and Simpson index. The Good’s coverage index value was around 0.97 for all the grassland types in the study area, meaning the soil bacteria samplings sequenced sufficiently. No statistically significant difference was shown in other diversity indices’ value, indicating the similar richness and evenness of soil bacteria in these alpine grasslands. The beta diversity, represented by Bray–Curtis dissimilarity and the non-metric multidimensional scaling (NMDS), showed that OTUs were clustered within alpine grasslands, indicating a clear separation of soil bacterial communities. In addition, soil organic matter (SOM), total nitrogen (TN), total phosphorus (TP), pH, and soil water content (SWC) were closely related to the variations in soil bacterial compositions. These results indicated that soil bacterial taxonomic compositions were similar, while soil bacterial community structures were different among the five alpine grassland types. The environmental conditions, including SOM, TN, TP, pH, and SWC, might influence the soil bacterial communities on the Tibetan Plateau.
Introduction
The Tibetan Plateau, which is considered the world’s “Third Pole,” has attracted extensive attention because of its high altitude and harsh environment (Chen et al., 2013). Environmental factors on the plateau, such as altitude (Cui et al., 2019), precipitation (Zhang K. et al., 2016), and topography (Frindte et al., 2019), have a large impact on soils of alpine ecosystems, resulting in marked changes in microbial diversity and function (Donhauser and Frey, 2018). Alpine grasslands are the representative landscape of the Tibetan Plateau, occupying more than 70% of the total area (Lu et al., 2018; Yu et al., 2019). Based on geographical, climatic, and vegetation characteristics, alpine grasslands on the Tibetan Plateau can be classified as alpine meadow (AM), alpine steppe (AS), alpine meadow steppe (AMS), alpine desert (AD), alpine desert steppe (ADS), etc. (Shen et al., 2015; Lu et al., 2018). Extensive studies have been carried out to examine vegetation and soil biogeochemical cycling within these grasslands (Tian et al., 2017; Che et al., 2018; Wu et al., 2019). However, soil microbial communities, which are the key drivers of many below-ground processes, remain poorly understood on the Tibetan Plateau.
Soil microorganisms are crucial for maintaining soil fertility and sustainability because of their role in regulating organic matter and nutrient cycling (Fierer, 2017). For example, Trichoderma, associated with plant roots, can trigger systemic resistance and improve plant nutrient uptake (Contreras-Cornejo et al., 2016). Additionally, genomically divergent microorganisms were reported as mediators of soil carbon (C) and nitrogen (N) compound turnovers in Mediterranean grasslands (Diamond et al., 2019). Therefore, understanding the differences in microbial communities among multiple ecosystems and their underlying mechanisms is essential for improving ecosystem function predictions and their responses to climate change and human activities (Zhang Y. et al., 2016; Karlowsky et al., 2018). Equally important are the microbial co-occurrence and network analysis to reveal keystone taxa that significantly impact the microbiome structure and functioning regardless of their abundance across space and time (Banerjee et al., 2018). Recent studies have also found that both competition and environmental filtering affect the abundance, composition, and encoded gene functions of bacterial and fungal communities (Bahram et al., 2018). In a tropical mountain ecosystem on Mount Kilimanjaro, interactions between climate and land use explained the soil microbial composition satisfactorily, indicating that climate and land use can modulate soil biodiversity and ecosystem functions (Peters et al., 2019). Steidinger et al. (2019) analyzed the global distribution of the dominant root-associated microbial symbionts and found that climate variables are the primary drivers of the global distribution of major symbionts. Even in other ecosystems, such as deserts (Evans et al., 2019) and farmlands (Jiao et al., 2020), environmental factors also considerably influenced the soil microbial community, but these effects may differ among different ecosystem types.
On the Tibetan Plateau, the spatial distribution of alpine grassland vegetation communities is strongly related to soil properties (Lu et al., 2015), climatic conditions (Wu et al., 2019), and elevation (Ren et al., 2018; Wu et al., 2019). The variations in these factors could also explain soil microbe distribution (Ciccazzo et al., 2016; Ren et al., 2018). For example, Chu et al. (2016) found that the microbial community structure was distinct between the surface and subsurface soil layers, strongly correlating with variation in the total carbon and carbon to nitrogen ratio (C:N) in the western Tibetan Plateau. Chen et al. (2016) found that soil microbial community variations in Tibetan alpine grasslands were mainly explained by edaphic factors, such as soil organic carbon (SOC), C:N ratio, pH, and soil texture, followed by biotic factors, including aboveground biomass and plant species richness, and further by climatic factors, such as mean annual precipitation. Xu et al. (2014) found that soil pH was a major factor affecting microbial communities, and the impact of soil pH was closely correlated with temperature and vegetation changes along the elevational gradient of Mount Segrila on the Tibetan Plateau. Therefore, the main controlling factors may vary at different regional scales because the spatial regulators of soil microbes vary at different spatial scales and in different ecosystems (White et al., 2020).
To date, most of the soil bacterial community studies have been conducted in tropical and temperate forests (Selmants et al., 2016; Wei et al., 2018) and grasslands (Wang et al., 2019; Yu et al., 2020). Information on the soil bacterial communities in alpine ecosystems is relatively scarce. In this study, a field investigation was conducted to explore the soil bacterial compositions and diversity in the upper soils from five different alpine grassland ecosystem types, including AM, AS, AMS, AD, and ADS, on the Tibetan Plateau using the 16S rRNA gene sequencing technology. We aimed to answer the following two questions: (1) do soil bacterial compositions and diversity show variations among different alpine grassland types? If so, (2) what are the determining factors that could explain these variations?
Materials and Methods
Sampling Sites
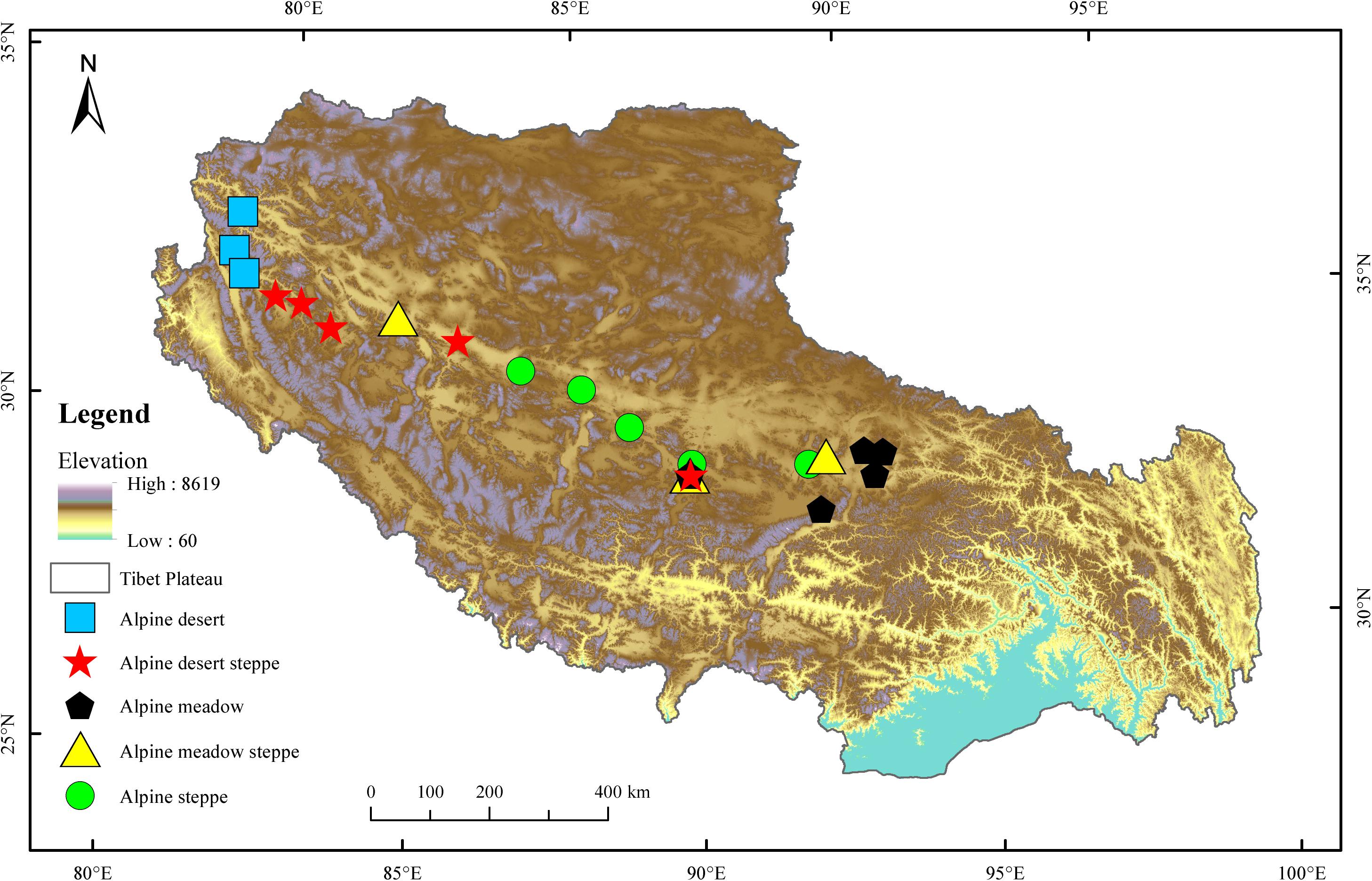
The Tibetan Plateau covers approximately one-eighth of China’s total area, with an average altitude higher than 4,000 m. This region shows wide ranges of temperature and vegetation due to its extensive territory and highly dissected topography. The mean annual temperature is relatively low, ranging from −4 to 18°C, with an annual precipitation of less than 1,000 mm. The radiation is intense, with annual sunshine ranging from 1,800 to 3,200 h. Alpine grasslands are not only the most important and largest ecosystem in this area but are also a key resource that supports the subsistence of the local population. Five typical alpine grassland types (AM, AS, AMS, AD, and ADS) were chosen to investigate the soil bacterial compositions and diversity in the present study (Figure 1). AM is composed of perennial mesic and mesoxeric herbs under cold and wet climatic conditions, occupying approximately 31.3% of the total grassland area of the Tibetan Plateau. AS is composed of drought-tolerant perennial herbs or small shrubs under cold and arid/semiarid climatic conditions, representing approximately 38.9% of the total grassland area. AD is a grassland type developed and controlled by cold and extreme drought conditions, covering 6.71% of the total grassland area. AMS is a transitional type of alpine grassland from the meadow to the steppe and ADS is a transitional type of alpine grassland from steppe to desert, covering 7.32 and 10.7%, respectively, of the total grassland area in the Tibetan Plateau (Lu et al., 2018).
Twenty-one sampling sites were set from east to west across the Tibetan Plateau according to the distribution of alpine grassland types (Supplementary Table S1). Five sampling sites were selected for each of the three main natural grassland types, AM, AS, and ADS, and three sampling sites were selected at the relatively smaller natural grassland areas, AMS and AD. At each sample site, three random quadrats (1 m × 1 m) were laid out at intervals of approximately 50 m during July and August 2019. In total, 63 quadrats of alpine grasslands on the Tibetan Plateau were sampled with 45 quadrats (15 sites × 3 quadrats) for AM, AS, and ADS, and 18 quadrats (6 sites × 3 quadrats) for AMS and AD. Five soil samples were collected from each quadrat at the depths of 0–15 cm and then composited into a single soil sample. Rocks and visible roots were removed from the soil, and it was passed through a 2 mm mesh. All samples were shipped in a refrigerator (−21°C) and immediately transported to the laboratory. All the soil samples were divided into two parts: one part was used to analyze the soil physiochemical properties, and the other was used to analyze soil bacteria based on 16S rRNA gene sequencing. The samples for genomic deoxyribonucleic acid (DNA) extraction and molecular analysis were stored at −80°C.
Analyses of Soil Properties
The soil water content (SWC) was measured gravimetrically after 10 h of desiccation at 105°C. Soil pH was determined at a fresh soil to water ratio of 1:2.5 using a pH monitor (Mettler-Toledo, Switzerland). Above-ground plant biomass (APB) and underground plant biomass (UPB) were measured gravimetrically after drying at 65°C for 48 h. Soil organic matter (SOM) and soil total nitrogen (TN) concentrations were determined with an element analyzer (Elementar, LiquiTOC, Hanau, Germany). The soil total phosphorus (TP) was determined using the NaHCO3 alkali digestion method, and the molybdenum antimony colorimetric method.
16S rRNA Gene Sequencing Technology
Total genomic DNA was extracted using DNeasy PowerSoil Kit # 12888-100 (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instructions. DNA quality and quantity were verified using NanoDrop and agarose gel electrophoresis. Extracted DNA was diluted to a concentration of 1 ng/μL and stored at −20°C until further processing. The diluted DNA was used as a template for polymerase chain reaction (PCR) amplification of bacterial 16S rRNA genes with the barcoded primers and Takara Ex Taq (Takara Biochemicals, Beijing, China).
For bacterial diversity analysis, V3–V4 variable regions of 16S rRNA genes were amplified with the universal primers 343F and 798R in PCR amplification (580BR10905, Bio-Rad, CA, United States). The PCR was performed according to previously described PCR methods (Wu et al., 2015; Scarlett et al., 2020). The first thermocycling involved an initial denaturation at 94°C for 5 min followed by 26 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 5 min, which included a combined annealing and extension time. Amplicon quality was visualized using gel electrophoresis, purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, United States), and amplified for another round of PCR (seven cycles); the second thermocycling was the same as in the first round. After purification with the AMPure XP beads again, the final amplicon was quantified using the Qubit dsDNA assay kit (Q328520, Life Technologies, Grand Island, NY, United States). Equal amounts of purified amplicon were pooled for subsequent sequencing on a Miseq Machine (Illumina, San Diego, CA, United States).
Raw sequencing data were in FASTQ format. Paired-end reads were then preprocessed using Trimmomatic software (Bolger et al., 2014) to detect and cut off ambiguous bases. Low-quality sequences with an average quality score below 20 were cut off using a sliding window trimming approach. After trimming, paired-end reads were assembled using FLASH software (Reyon et al., 2012). The assembly parameters were: 10 bp of minimal overlapping, 200 bp of maximum overlapping, and 20% of maximum mismatch rate. Sequences were subjected to further denoising as follows: reads with ambiguous, homologous sequences or below 200 bp were removed. Reads with 75% of bases above Q20 were retained. Then, reads with chimera were detected and removed. These two steps were performed using QIIME software (Quantitative Insights Into Microbial Ecology, version 1.8.0) (Caporaso et al., 2010). Clean reads were subjected to primer sequence removal and clustering to generate operational taxonomic units (OTUs) using Vsearch software with a 97% similarity cutoff (Edgar et al., 2011). The representative read of each OTU was selected using the QIIME package. All representative reads were annotated and blasted against the SILVA database (Version 132) using the RDP classifier (confidence threshold of 70%) (Wang et al., 2007).
Diversity Estimates
Six commonly used alpha diversity indices of the bacterial community were calculated. The Good’s coverage index represents the sample coverage detected. A higher Good’s coverage index means that the result approximates to the actual situation (Yuan et al., 2018). The phylogenetic diversity (PD) whole tree index is defined as the sum of the branch lengths of a phylogenetic tree connecting all species in the target assemblage, which can be regarded as a phylogenetic generalization of species richness (Chao et al., 2016). Chao1 is an index of species richness, which is sensitive to rare species, but does not represent abundance or uniformity (Chao and Bunge, 2002). The observed species index is the number of species in the sample (Yuan et al., 2018). The Shannon index is positively correlated with species richness and evenness, and that gives more weight per individual to rare than common species (Hill et al., 2003). The Simpson index focuses on the uniformity of the community (Zhong et al., 2010). Beta diversity was represented by Bray–Curtis dissimilarity between each type of grassland pair (Chen et al., 2019), and then non-metric multidimensional scaling (NMDS) was used to explain the pairwise dissimilarity between objects in a low-dimensional space. Additionally, the analysis of similarities (ANOSIM) was used to determine the significance of separation along alpine grassland types (Ren et al., 2018).
Statistical Analyses
All statistical analyses were performed in the R environment (v4.0.01). The relative abundance of the top 15 species at the phylum and genus levels was used to compare the distribution of soil bacterial species in the five alpine grassland ecosystems. Before conducting a one-way analysis of variance (ANOVA), the Shapiro function was used to test for the normality of residuals, and the Bartlett function was used to test the homogeneity of variance. The ANOVA was used when the application conditions were satisfied; otherwise, the non-parametric statistics (Kruskal–Wallis test) was chosen. The least significant difference (LSD) comparison of bacteria among different alpine grasslands was performed using the “agricolae” package (Version 1.3-3) (Felipe, 2020); then the indicator bacteria were determined using the random forest model (“randomForest” package, Version 4.6-14) (Liaw and Wiene, 2002).
The alpha diversity indices of the soil bacteria community were calculated based on OTUs using the QIIME software (Version 1.80) (Caporaso et al., 2010). Beta diversity was estimated based on Bray–Curtis dissimilarities between samples, where the distance was computed using OTU tables. The “vegan” package (Version 2.5-6) (Jari Oksanen et al., 2019) was used to compute Bray–Curtis dissimilarity, NMDS, and ANOSIM, which were used to assess the beta diversity of soil bacterial communities. Correlations between the soil bacterial compositions and environmental factors were determined using redundancy analysis (RDA) in the vegan’ package. The selected environmental factors were APB, UPB, SOM, TN, TP, pH, SWC, and altitude. The significance of the RDA results was tested using the Monte Carlo permutation test. Graphs were drawn by the “ggplot2” package (Version 3.3.2) (Wickham, 2016).
Results
Soil Bacterial Community Structure in Alpine Grasslands
A total of 48,366–51,566 clean reads with high-quality 16S rRNA sequences were identified from the 21 samples from the Tibetan Plateau. After chimera detection and removal, about 37,532–47,699 valid tags with an average read length of 412.52–416.3 bp were obtained. Accordingly, an average of 1,799–3,089 OTUs at a 97% similarity cutoff were generated from 16S rRNA sequencing analysis. The number of different OUTs was 855, covering 94 different genera and eight different phyla.
In these five alpine grassland ecosystems, Actinobacteria (46.12%) and Proteobacteria (29.67%) were the two dominant bacteria at the phylum level, followed by Gemmatimonadetes (8.30%), Bacteroidetes (7.25%), Firmicutes (4.23%), and Acidobacteria (2.35%) (Figure 2A). At the genus level, the dominant bacteria were inhomogeneously distributed in different alpine grasslands, accounting for 20% of all relative abundance of taxonomic, where each dominant genus was between 0.7 and 2.4%. The relative abundance from highest to lowest was Rubrobacter (2.38%), Solirubrobacter (2.25%), Blastococcus (1.65%), Pseudonocardia (1.43%), Crossiella (1.29%), Gaiella (1.17%), Bacteroides (1.17%), Nocardioides (1.17%), Sphingomonas (1.12%), Gemmatirosa (1.10%), Escherichia-Shigella (1.07%), MND1 (0.90%), Haliangium (0.78%), Geodermatophilus (0.77%), and Microvirga (0.73%) (Figure 2B). The relative abundance of Rubrobacter was statistically similar in AD, ADS, and AS; the relative abundance of Geodermatophilus in AD was only higher than in AMS. The relative abundance of Geodermatophilus in the soils of AD and ADS was 0.65 and 1.45%, respectively, 9.29 and 20.71 times higher than that of the AMS (0.07%), respectively. Moreover, the relative abundance of Solirubrobacter, Pseudonocardia, Gaiella, and Haliangium were also the least observed genera in AMS soils. Nevertheless, these bacteria were the most abundant in the AM soils, where their content was two to four times that in AMS soils (Figure 3). Furthermore, the relative abundance of Gaiella showed obvious differentiation in alpine grasslands and it was most abundant in the AM soils.
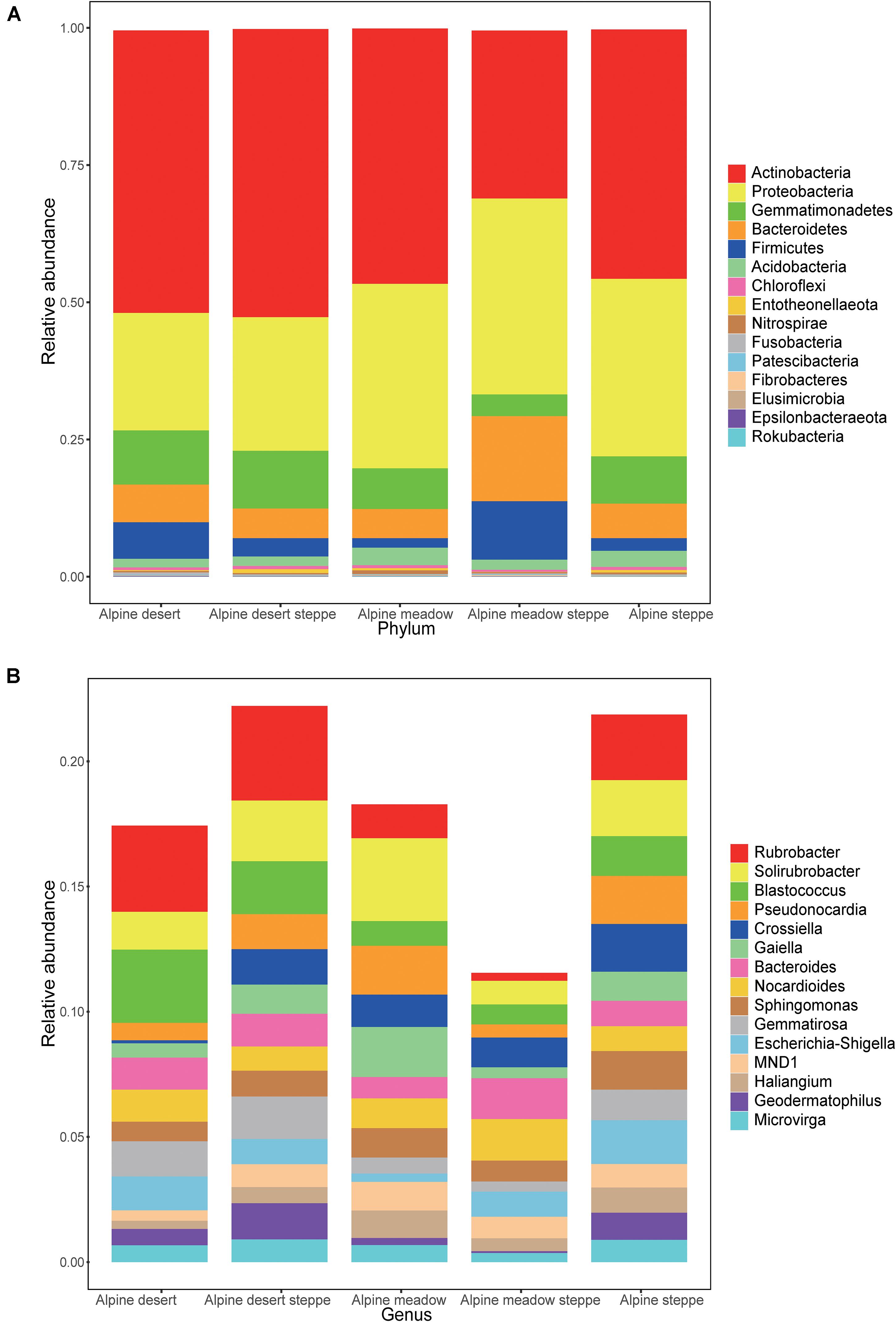
Figure 2. Relative abundance of bacteria in different grasslands at the phylum and genus levels on the Tibetan Plateau. The bacteria whose relative abundance are in the top 15 at phylum (A) and genus (B) levels. AD, alpine desert; ADS, alpine desert steppe; AM, alpine meadow; AMS, alpine meadow steppe; AS, alpine steppe.
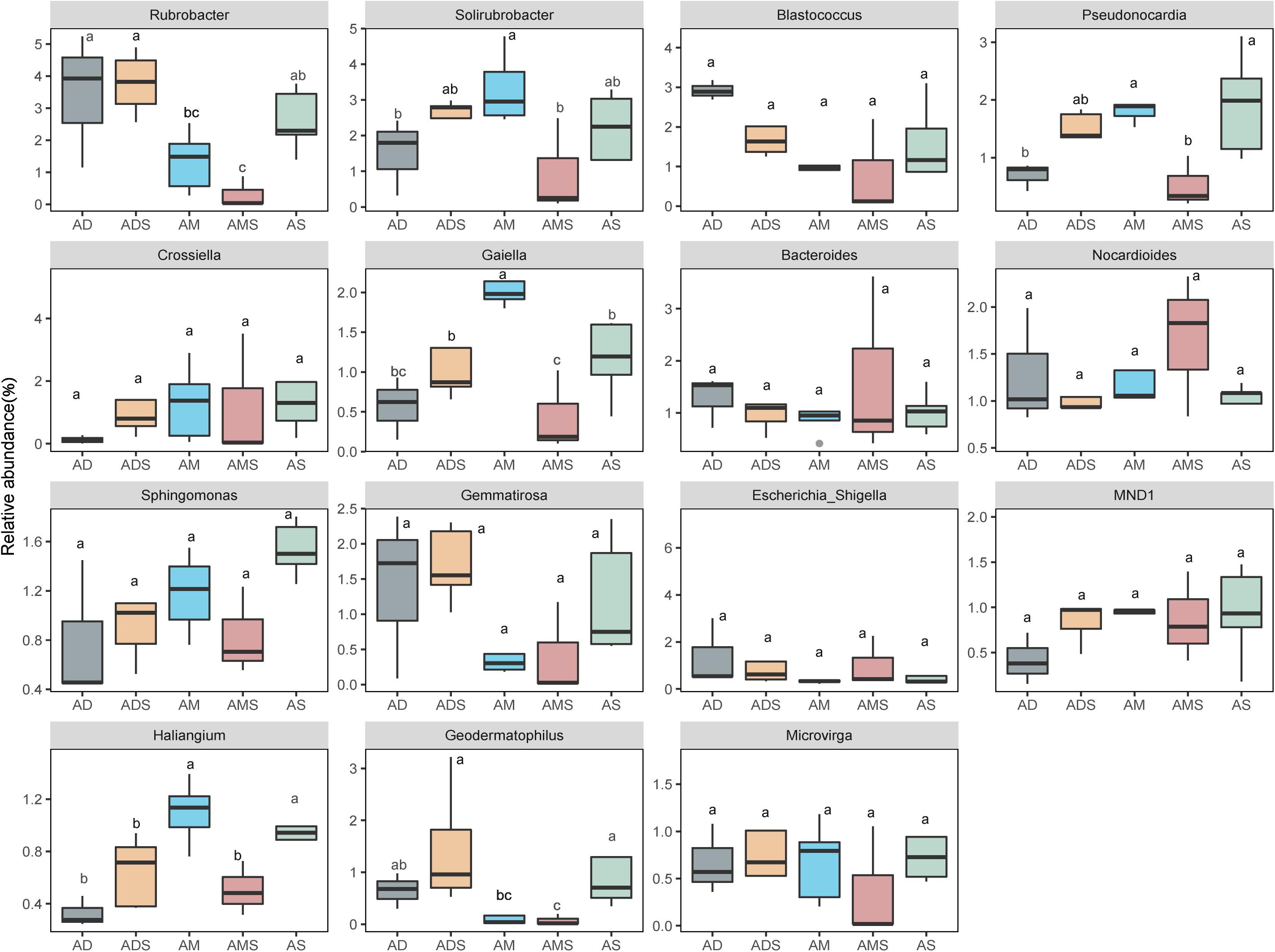
Figure 3. Differences in soil bacteria among alpine grasslands at the genus level on the Tibetan Plateau. AD, alpine desert; ADS, alpine desert steppe; AM, alpine meadow; AMS, alpine meadow steppe; AS, alpine steppe. Different lowercase letters marking the same genus represent significant differences between two alpine grasslands (P < 0.05).
The random forest model revealed the indicator bacteria at the genus level, the genus with high essential values was also the genus with high relative abundance. According to the ordering of essential values (Figure 4), Rubrobacter and Solirubrobacter were extremely sensitive indicator bacteria in the alpine grasslands (importance > 1); Blastococcus, Pseudonocardia, Crossiella, and Gaiella also had high values of importance.
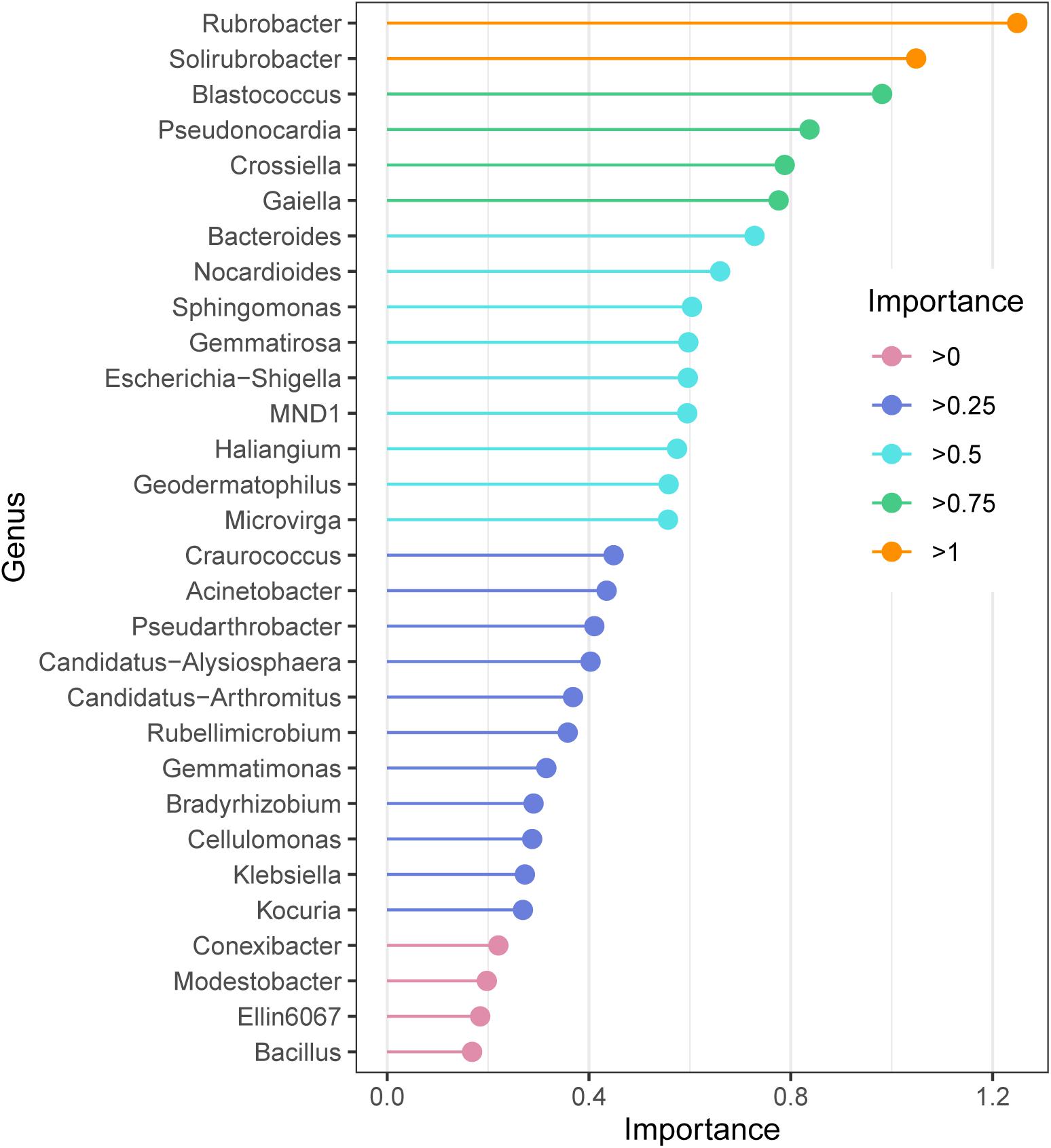
Figure 4. Species importance point map according to the importance of the top 30 soil bacteria at the genus level of alpine grasslands on the Tibetan Plateau.
Alpha Diversity of Soil Bacterial Community in Alpine Grasslands
Six alpha diversity indices, the PD whole tree index, Chao1 index, Good’s coverage index, observed species index, Shannon index, and Simpson index, were calculated to compare soil microbial community differences among the five different alpine grassland types (Table 1). The Good’s coverage indices in all alpine grasslands were higher than 0.97, and the Good’s coverage index of the AD was significantly higher than that of other alpine grasslands (P < 0.05), indicating that that our sequenced sample was sufficient to reveal the true diversity. Significance in the other five alpha diversity indices among alpine grassland types demonstrated a similarity in alpha diversity among the five investigated alpine grasslands.
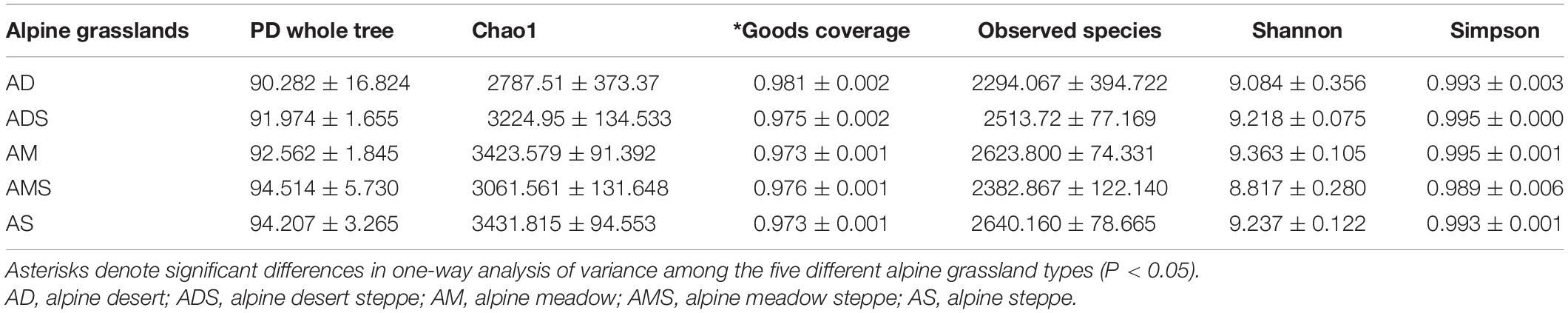
Table 1. Alpha diversity indices of soil bacterial communities (mean ± SE) in alpine grasslands on the Tibetan Plateau.
Beta Diversity of Soil Bacterial Community in Alpine Grasslands
The NMDS based on Bray–Curtis dissimilarities was conducted to reflect the soil bacterial beta diversity of the alpine grasslands on the Tibetan Plateau. The results showed that OTUs were clustered within alpine grasslands (Figure 5), indicating a clear separation of soil bacterial communities among five alpine grassland ecosystems (stress = 0.0284). For instance, the Bray–Curtis dissimilarity from the AM to the AD was relatively far, highlighting the significant difference of soil bacterial communities between them. In addition, the Bray–Curtis dissimilarity of AS, ADS, and AMS was close, implying the similar soil bacteria community structures, especially the relative abundance of genus. Furthermore, the ANOSIM highlighted the differences in soil bacterial communities in alpine grasslands (Table 2). The soil bacteria community of AM was substantially different from those of AMS, ADS, and AD (P < 0.05). Moreover, the soil bacterial community at AS was different from that at AD, and there were also even differences between the two transition states alpine grasslands (AMS and ADS).
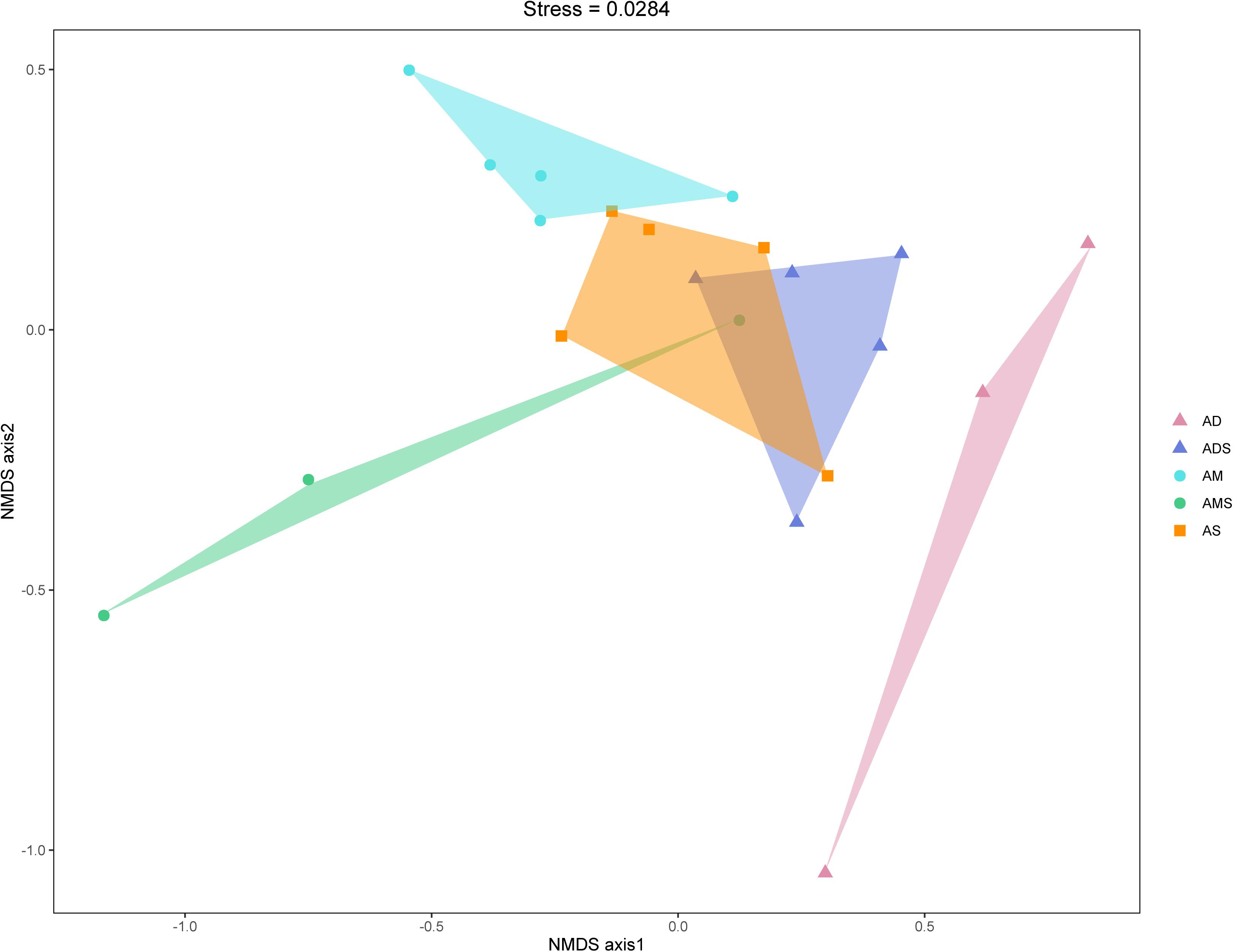
Figure 5. Non-metric multidimensional scaling (NMDS) analysis of the soil bacteria in alpine grasslands on the Tibetan Plateau. Different shaped dots represent the samples from different alpine grassland types, and the bumps of the samples bound the faces of different colors. AD, alpine desert; ADS, alpine desert steppe; AM, alpine meadow; AMS, alpine meadow steppe; AS, alpine steppe.
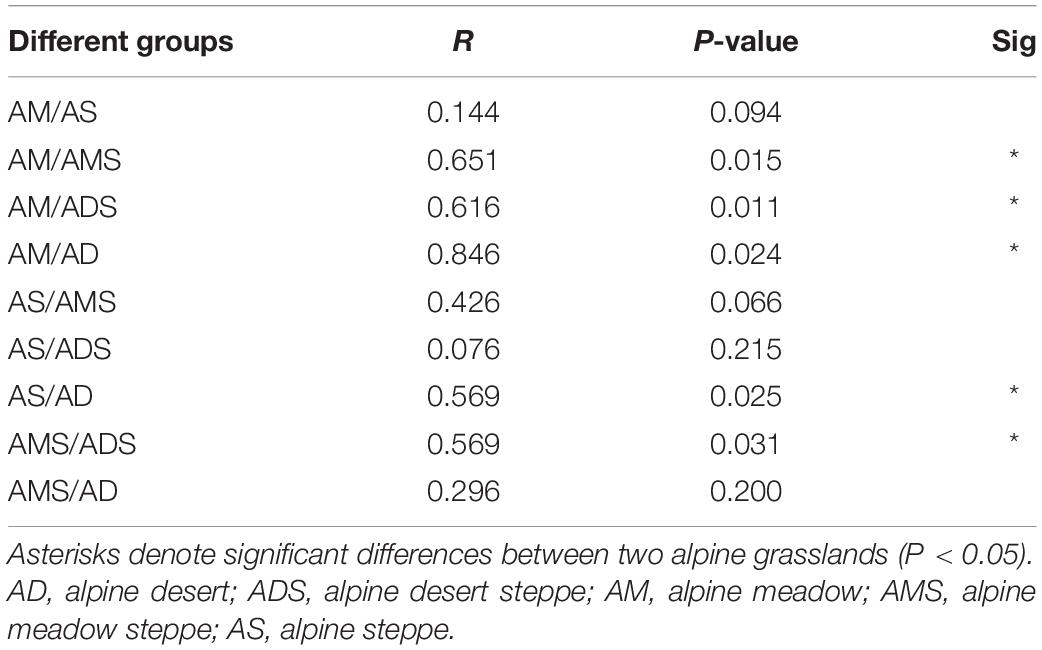
Table 2. Analysis of similarities (ANOSIM) of soil bacteria in alpine grasslands on the Tibetan Plateau.
Relationships Between Soil Bacterial Community and the Environmental Factors
The RDA showed that the variations in the soil bacterial community were determined by the environmental factors (Figure 6). The permutation test indicated significance in the ordination diagram (P = 0.003). Environmental variables had an explanation rate of 41.69% for the soil bacterial community, with the first and second axes accounting for 24.49 and 17.2% of the variation, respectively. The results also revealed correlations between each genus and environmental variations; the dominant factors for different soil bacteria genera were different. For example, soil pH was positively correlated to Nocardioides, Escherichia-Shigella, MND1, and Haliangium, but negatively correlated to Rubrobacter, Solirubrobacter, Gemmatirosa, and Geodermatophilus. In addition, the differences in sampling points can also be reflected by environmental variables, and the soil bacterial abundance at different points also showed variability. For instance, the projection point of the corresponding sample points in AM on the corresponding vector of Pseudonocardia was in front of that of AD, implying that the Pseudonocardia has greater abundance potential in the AM than in AD. Environmental factors, especially SOM, TN, TP, pH, and SWC, were closely related to the soil bacterial community (P < 0.05) (Table 3). The difference in environmental factors in alpine grasslands can explain the differences in soil bacterial communities among different alpine grassland types.
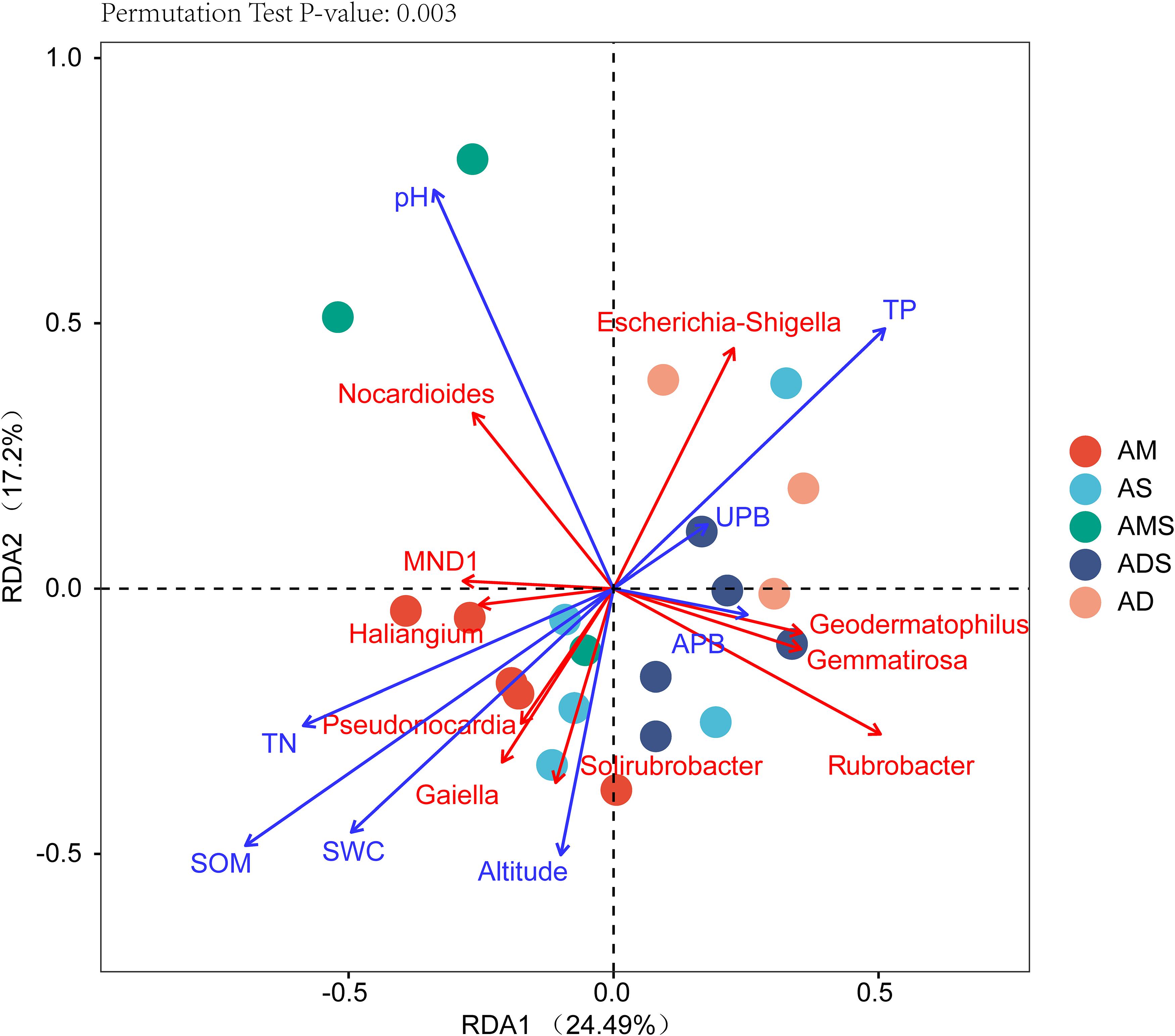
Figure 6. Redundancy analysis (RDA) of soil bacterial communities and environmental factors in alpine grasslands on the Tibetan Plateau. Points of the same color with a symbol denote sampling sites of an alpine grassland type. The blue arrows represent environmental factors, the red arrows represent soil bacteria (genus), and the cosine of the angle within two vector arrows represents the correlation between vectors. AD, alpine desert; ADS, alpine desert steppe; AM, alpine meadow; AMS, alpine meadow steppe; AS, alpine steppe; APB, aboveground plant biomass; UPB, underground plant biomass; SOM, soil organic matter; TN, total nitrogen; TP, total phosphorus; SWC, soil water content.
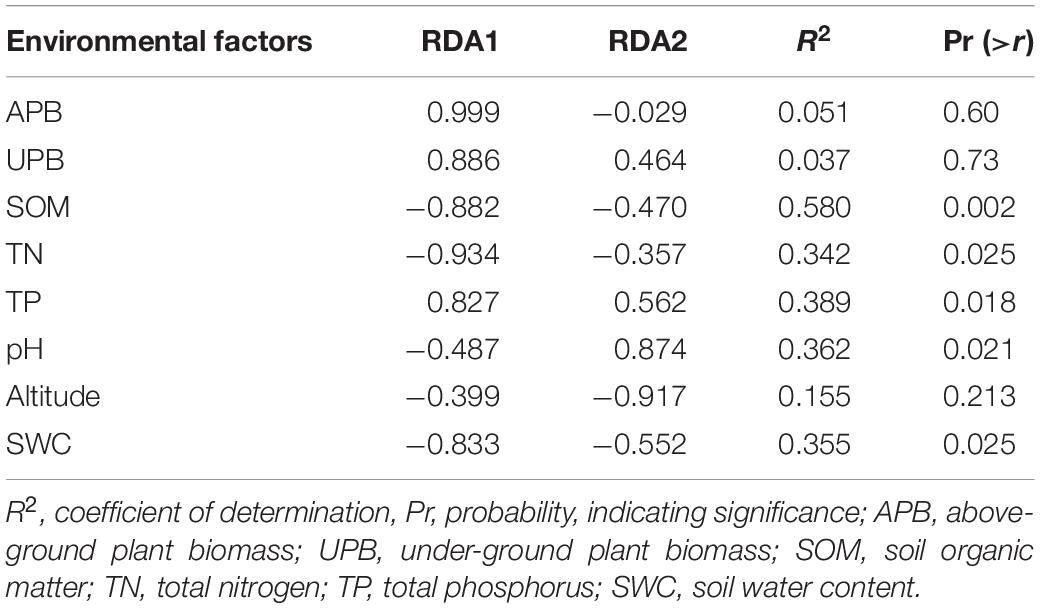
Table 3. Significance of explanatory variables and the variance values in redundancy analysis (RDA).
Discussion
Soil Microbial Community Structures in Alpine Grasslands
Alpine ecosystems are susceptible to climate change and human disturbance, causing variability in soil bacterial composition and diversity (Donhauser and Frey, 2018). Soil bacteria play an essential role in the cycling organic matter in nature, soil formation, and soil fertility. Simultaneously, various secondary metabolites produced by soil bacteria during their life processes are also crucial to the soil ecological environment (Chen et al., 2020). In our study, the dominant bacteria at the phylum level were Actinobacteria, Proteobacteria, Gemmatimonadetes, Bacteroidetes, Firmicutes, and Acidobacteria, accounting for approximately 94% of the total abundance of the bacterial community. Despite differences in relative abundance, Actinobacteria, Proteobacterial, Acidobacteria, and Chloroflexi were also the dominant bacteria found by Chu et al. (2016) and Zhou et al. (2019).
Actinobacteria constitute one of the largest bacterial phyla in many types of soil ecosystems (Barka et al., 2015). They often live as plant commensals (Fitzpatrick et al., 2018), nitrogen-fixing symbionts in soil, involved in the SOM formation and biogeochemical cycles (Yu et al., 2020), and are also a significant source of extracellular enzymes and secondary metabolites (Alvarez et al., 2017). In the present study, the relative abundance of Acidobacteria was higher in all five alpine grasslands. Proteobacteria, one of the main phyla of ammonifying microorganisms, were also abundant in the five alpine grasslands, especially in AM, AMS, and AS, which might be due to the different vegetation types and their litters, resulting in the structural difference of the dominant species of Proteobacteria (Chen et al., 2020). Gemmatimonadetes, Entotheonellaeota, and Fusobacteria were also found in alpine grassland soils. Gemmatimonadetes and Entotheonellaeota are also adapted to low soil moisture or arid environments (DeBruyn et al., 2011; Wilson et al., 2014; Zethof et al., 2020) that are similar to our sampling area conditions. Fusobacteria are often found in fecal microbiota, which were probably introduced by ruminants (Wu et al., 2016).
At the genus level, the average relative abundance of the dominant species was all lower than 2.5%, and the diverse community composition might be related specific functions of soil bacteria. For example, Rubrobacter was abundant in AD, ADS, and AS; a previous study showed that Rubrobacter contains many highly specific proteins that play a vital role in the evolution of bacteriochlorophyll-based photosynthesis (Gupta and Khadka, 2016). Geodermatophilus is abundant in ADS and AS, as it often thrived in nutrient-poor habitats, such as the surface of calcarenite stones or sandy desert soils (Sghaier et al., 2016). Pseudonocardia has received extensive attention for degrading recalcitrant cyclic ether pollutants, mutualism with soil organisms, as well as producing antibiotics (Tanvir et al., 2016; Li et al., 2018; Ma et al., 2018); it is abundant in AM and AS. The Gaiella, as a good indicator of C:N in the soil (Hermans et al., 2017), and the Haliangium, involving in soil denitrification and playing an essential role in the biochemical cycling of N and fermentation of C sources (Lévesque et al., 2020), were both found with the higher relative abundance in AM.
Based on the soil bacterial communities in alpine grasslands on the Tibetan Plateau, soil bacterial taxonomic compositions and alpha diversity were similar among the five different alpine grassland types. There were significant differences in the relative abundance at the genus level among the alpine grassland types, especially for the Rubrobacter, Solirubrobacter, Pseudonocardia, Gaiella, Haliangium, and Geodermatophilus. The parallel composition of soil bacteria in different alpine grasslands reflected their strong adaptability to the environment. In contrast, the relative abundance of some soil bacteria varied among the five alpine grasslands at the genus levels, suggesting a filtering effect of environmental factors. The relative abundance of soil bacteria changed significantly in other alpine soils (Rime et al., 2016; Malard and Pearce, 2018). For example, the relative abundance of soil bacteria in different alpine edaphic conditions showed significant differences, especially for Chloroflexi, whose relative abundance significantly increased with elevation and indicated a high colonization potential in the Swiss Alps (Adamczyk et al., 2019).
Soil Bacterial Diversity in Alpine Grasslands
The vegetation and soil nutrients were heterogeneous among the five alpine grasslands on the Tibetan Plateau, which may lead to variations in soil bacterial diversity. However, we did not detect significant differences in the alpha diversity of soil bacteria among the alpine grassland types, although supported by other studies on the Tibetan Plateau and other regions (French et al., 2017; Zhou et al., 2019; Su et al., 2020). For instance, there were no differences in the alpha diversity of microbes in five degraded AS in Qinghai Province, China (Zhou et al., 2019). Furthermore, a study in southeast England found that bacterial diversity and specific beneficial taxa did not change when the land-use changed (French et al., 2017). Ramirez et al. (2014) reported that soil bacterial diversity in Central Park in New York City was very similar to the multiple locations globally.
It seems that two possible reasons can account for the similar alpha diversity of soil bacteria in alpine grasslands. First, the alpha diversity may be influenced by soil bacterial colonization strategies, and some bacteria have large population sizes with greater dispersal ability. For instance, Actinobacteria OTUs were found to contribute up to 52% of the soil bacterial communities in Australian and northern Antarctica soils, indicating the remarkable dispersion ability of Actinobacteria (Araujo et al., 2020). Second, nutrient restriction may also affect soil bacterial alpha diversity in alpine grasslands, with C, N, phosphorus (P), and especially SOC, acting as limiting factors. Ma et al. (2019) reported that total bacteria slightly increased with P addition only when plants remained N limited, and most bacterial taxa were limited by resources other than N and P. Hence, similar species richness and evenness usually occurred due to constraints of the same resource (Ho et al., 2017).
Bray–Curtis dissimilarities represent beta diversity, and the results showed the differentiation of soil microbial communities among the five types of alpine grasslands. The significant difference in soil bacterial communities between AM and AD was demonstrated because the Bray–Curtis dissimilarity from AM to AD was relatively far. Furthermore, similar results have also been found in New Zealand, where the bacteria communities showed that heterogeneity in more than 3,000 soil samples of different land-use types and soil bacterial communities could be a good indicator of different land-use types with up to 83% accuracy (Hermans et al., 2020). Environmental factors drive soil bacterial community beta diversity; other studies also found that soil pH (Shen et al., 2020), nutrients such as N (Luo et al., 2020), and climate change (Shi et al., 2020) were important determinants. In a certain sense, the range of environmental conditions and some soil characteristics such as pH were closely correlated with the geographical range of the samples (Chu et al., 2010; Barberán et al., 2014), indicating that our classification of alpine grasslands may need to be reconsidered, to provide more accurate predictions on the differentiation of soil microbial communities between different types of alpine grasslands.
Response of Soil Bacteria to Environmental Factors
Soil bacterial community structures differed among the five different alpine grassland types, despite the similarities in the soil species composition. High consistency of soil bacterial alpha diversity and differentiation of beta diversity were found in the alpine grasslands. These results may indirectly reveal the extensive adaptability of soil bacteria to the environment. In order to explore the reasons for the differences in soil bacterial communities, correlations between the soil bacterial compositions and environmental factors were determined using RDA. We found that environmental factors, including SOM, TN, TP, SWC, and soil pH, were closely related to the soil bacterial community. Environmental factors were also found to have both positive and negative effects on different alpine soil bacteria. For instance, the result reflected that the soil pH was positively correlated with Nocardioides, while it was negatively correlated with Rubrobacter.
Soil organic matter provides important sustenance to soil bacteria; the vegetation compositions in alpine grasslands implies multiple litters, which might contribute to the different structures of soil bacterial communities (Leff et al., 2018; Hicks et al., 2019; Yang et al., 2019). Meanwhile, soil N content can affect the microbial mineralization of SOM (Carrillo et al., 2017; Hicks et al., 2019). In the present study, TN, TP, and SWC had a high explanatory power for soil bacteria. Similar results were also found in an AM and a poplar plantation, where RDA revealed that soil pH, extractable organic carbon, and extractable organic nitrogen accounted for most of the variability in the bacterial communities (Zhang et al., 2017; Sun et al., 2020). A study on Mt. Kilimanjaro in East Africa found that bacterial diversity had a U-shaped pattern across the mountain gradient, and pH could explain about 12% of the soil bacteria (Shen et al., 2020). Soil pH may affect soil microorganisms by indirectly influencing the form of available nutrients, and is closely correlated with temperature and vegetation changes along the elevational gradient (Xu et al., 2014). Finally, in alpine grasslands, SWC may affect nutrient solubility and indirectly affect soil microbial communities.
Conclusion
Based on 16S rRNA gene sequencing, we compared soil bacterial composition and diversity among the five types of alpine grasslands on the Tibetan Plateau. We found that the compositions of soil bacteria in different alpine grasslands were similar; however, the community structure varied. The alpha diversity index showed no significant differences in the alpha diversity of soil bacterial communities, implying that there were similarities in the bacterial richness and evenness among different alpine grasslands. The results from NMDS and ANOSIM assessing the beta diversity among alpine grasslands revealed that the soil bacterial communities in AM diverged significantly from the rest of the alpine grassland types, except AS. Moreover, the soil bacterial communities in AS and AD were dissimilar, and soil bacterial communities varied in two alpine grassland transition types, AMS and ADS. The soil environmental factors were examined to explain the soil bacterial community variations by RDA, highlighting that the distribution pattern of soil bacteria was influenced by some environmental factors, including SOM, TN, TP, pH, and SWC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR13370619.
Author Contributions
XL planned and designed the study. HJ, YH, and ZW performed the experiments and conducted fieldwork. HM and YC analyzed the data and wrote the manuscript. All authors contributed critically to the drafts and gave the final approval for publication.
Funding
This study was supported by the National Natural Science Foundation of China (41877338 and 31901198).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.630722/full#supplementary-material
Footnotes
References
Adamczyk, M., Hagedorn, F., Wipf, S., Donhauser, J., Vittoz, P., Rixen, C., et al. (2019). The soil microbiome of GLORIA mountain summits in the swiss Alps. Front. Microbiol. 10:1080. doi: 10.3389/fmicb.2019.01080
Alvarez, A., Saez, J. M., Davila Costa, J. S., Colin, V. L., Fuentes, M. S., Cuozzo, S. A., et al. (2017). Actinobacteria: current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 166, 41–62. doi: 10.1016/j.chemosphere.2016.09.070
Araujo, R., Gupta, V. V. S. R., Reith, F., Bissett, A., Mele, P., and Franco, C. M. M. (2020). Biogeography and emerging significance of Actinobacteria in Australia and Northern Antarctica soils. Soil Biol. Biochem. 146:107805. doi: 10.1016/j.soilbio.2020.107805
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Barberán, A., Ramirez, K. S., Leff, J. W., Bradford, M. A., Wall, D. H., and Fierer, N. (2014). Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 17, 794–802. doi: 10.1111/ele.12282
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2015). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford England) 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carrillo, Y., Bell, C., Koyama, A., Canarini, A., Boot, C. M., Wallenstein, M., et al. (2017). Plant traits, stoichiometry and microbes as drivers of decomposition in the rhizosphere in a temperate grassland. J. Ecol. 105, 1750–1765. doi: 10.1111/1365-2745.12772
Chao, A., and Bunge, J. (2002). Estimating the number of species in a stochastic abundance model. Biometrics 58, 531–539. doi: 10.1111/j.0006-341X.2002.00531.x
Chao, A., Chiu, C.-H., and Jost, L. (2016). “Phylogenetic diversity measures and their decomposition: a framework based on hill numbers,” in Biodiversity Conservation and Phylogenetic Systematics: Preserving our evolutionary heritage in an extinction crisis, eds R. Pellens, and P. Grandcolas, (Cham: Springer International Publishing), 141–172.
Che, R., Qin, J., Tahmasbian, I., Wang, F., Zhou, S., Xu, Z., et al. (2018). Litter amendment rather than phosphorus can dramatically change inorganic nitrogen pools in a degraded grassland soil by affecting nitrogen-cycling microbes. Soil Biol. Biochem. 120, 145–152. doi: 10.1016/j.soilbio.2018.02.006
Chen, H., Zhu, Q., Peng, C., Wu, N., Wang, Y., Fang, X., et al. (2013). The impacts of climate change and human activities on biogeochemical cycles on the Qinghai-Tibetan Plateau. Glob. Change Biol. 19, 2940–2955. doi: 10.1111/gcb.12277
Chen, L., Xiang, W., Wu, H., Ouyang, S., Zhou, B., Zeng, Y., et al. (2019). Tree species identity surpasses richness in affecting soil microbial richness and community composition in subtropical forests. Soil Biol. Biochem. 130, 113–121. doi: 10.1016/j.soilbio.2018.12.008
Chen, Q., Niu, B., Hu, Y., Luo, T., and Zhang, G. (2020). Warming and increased precipitation indirectly affect the composition and turnover of labile-fraction soil organic matter by directly affecting vegetation and microorganisms. Sci. Total Environ. 714:136787. doi: 10.1016/j.scitotenv.2020.136787
Chen, Y. L., Ding, J. Z., Peng, Y. F., Li, F., Yang, G. B., Liu, L., et al. (2016). Patterns and drivers of soil microbial communities in Tibetan alpine and global terrestrial ecosystems. J. Biogeogr. 43, 2027–2039.
Chu, H., Fierer, N., Lauber, C. L., Caporaso, J. G., Knight, R., and Grogan, P. (2010). Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x
Chu, H., Sun, H., Tripathi, B. M., Adams, J. M., Huang, R., Zhang, Y., et al. (2016). Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ. Microbiol. 18, 1523–1533. doi: 10.1111/1462-2920.13236
Ciccazzo, S., Esposito, A., Borruso, L., and Brusetti, L. (2016). Microbial communities and primary succession in high altitude mountain environments. Ann. Microbiol. 66, 43–60. doi: 10.1007/s13213-015-1130-1
Contreras-Cornejo, H. A., Macías-Rodríguez, L., del-Val, E., and Larsen, J. (2016). Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: interactions with plants. FEMS Microbiol. Ecol. 92:fiw036. doi: 10.1093/femsec/fiw036
Cui, Y., Bing, H., Fang, L., Wu, Y., Yu, J., Shen, G., et al. (2019). Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 338, 118–127. doi: 10.1016/j.geoderma.2018.11.047
DeBruyn, J. M., Nixon, L. T., Fawaz, M. N., Johnson, A. M., and Radosevich, M. (2011). Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl. Environ. Microbiol. 77, 6295–6300. doi: 10.1128/AEM.05005-11
Diamond, S., Andeer, P. F., Li, Z., Crits-Christoph, A., Burstein, D., Anantharaman, K., et al. (2019). Mediterranean grassland soil C–N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms. Nat. Microbiol. 4, 1356–1367. doi: 10.1038/s41564-019-0449-y
Donhauser, J., and Frey, B. (2018). Alpine soil microbial ecology in a changing world. FEMS Microbiol. Ecol. 94:fiy099. doi: 10.1093/femsec/fiy099
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Evans, S. E., Dueker, M. E., Logan, J. R., and Weathers, K. C. (2019). The biology of fog: results from coastal maine and namib desert reveal common drivers of fog microbial composition. Sci. Total Environ. 647, 1547–1556. doi: 10.1016/j.scitotenv.2018.08.045
Felipe, M. (2020). agricolae: Statistical Procedures for Agricultural Research. Available: https://CRAN.R-project.org/package=agricolae (accessed June 6, 2020).
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115:E1157. doi: 10.1073/pnas.1717617115
French, K. E., Tkacz, A., and Turnbull, L. A. (2017). Conversion of grassland to arable decreases microbial diversity and alters community composition. Appl. Soil Ecol. 110, 43–52. doi: 10.1016/j.apsoil.2016.10.015
Frindte, K., Pape, R., Werner, K., Löffler, J., and Knief, C. (2019). Temperature and soil moisture control microbial community composition in an arctic-alpine ecosystem along elevational and micro-topographic gradients. ISME J. 13, 2031–2043. doi: 10.1038/s41396-019-0409-9
Gupta, R. S., and Khadka, B. (2016). Evidence for the presence of key chlorophyll-biosynthesis-related proteins in the genus Rubrobacter (Phylum Actinobacteria) and its implications for the evolution and origin of photosynthesis. Photosynth. Res. 127, 201–218.
Hermans, S. M., Buckley, H. L., Case, B. S., Curran-Cournane, F., Taylor, M., and Lear, G. (2017). Bacteria as emerging indicators of soil condition. Appl. Environ. Microbiol. 83:e02826-16. doi: 10.1128/aem.02826-16
Hermans, S. M., Buckley, H. L., Case, B. S., Curran-Cournane, F., Taylor, M., and Lear, G. (2020). Using soil bacterial communities to predict physico-chemical variables and soil quality. Microbiome 8:79. doi: 10.1186/s40168-020-00858-1
Hicks, L. C., Meir, P., Nottingham, A. T., Reay, D. S., Stott, A. W., Salinas, N., et al. (2019). Carbon and nitrogen inputs differentially affect priming of soil organic matter in tropical lowland and montane soils. Soil Biol. Biochem. 129, 212–222. doi: 10.1016/j.soilbio.2018.10.015
Hill, T. C. J., Walsh, K. A., Harris, J. A., and Moffett, B. F. (2003). Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43, 1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x
Ho, A., Di Lonardo, D. P., and Bodelier, P. L. E. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93:fix006. doi: 10.1093/femsec/fix006
Jari Oksanen, F., Blanchet, G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. Available: https://CRAN.R-project.org/package=vegan (accessed July 5, 2019).
Jiao, S., Yang, Y., Xu, Y., Zhang, J., and Lu, Y. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Karlowsky, S., Augusti, A., Ingrisch, J., Hasibeder, R., Lange, M., Lavorel, S., et al. (2018). Land use in mountain grasslands alters drought response and recovery of carbon allocation and plant-microbial interactions. J. Ecol. 106, 1230–1243. doi: 10.1111/1365-2745.12910
Leff, J. W., Bardgett, R. D., Wilkinson, A., Jackson, B. G., Pritchard, W. J., De Long, J. R., et al. (2018). Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. Isme J. 12, 1794–1805. doi: 10.1038/s41396-018-0089-x
Lévesque, V., Rochette, P., Hogue, R., Jeanne, T., Ziadi, N., Chantigny, M. H., et al. (2020). Greenhouse gas emissions and soil bacterial community as affected by biochar amendments after periodic mineral fertilizer applications. Biol. Fertil. Soils 56, 907–925. doi: 10.1007/s00374-020-01470-z
Li, M., Yang, Y., He, Y., Mathieu, J., Yu, C., Li, Q., et al. (2018). Detection and cell sorting of Pseudonocardia species by fluorescence in situ hybridization and flow cytometry using 16S rRNA-targeted oligonucleotide probes. Appl. Microbiol. Biotechnol. 102, 3375–3386. doi: 10.1007/s00253-018-8801-3
Lu, X., Ma, S., Chen, Y., Yangzom, D., and Jiang, H. (2018). Squalene found in alpine grassland soils under a harsh environment in the Tibetan plateau, China. Biomolecules 8:154.
Lu, X., Yan, Y., Sun, J., Zhang, X., Chen, Y., Wang, X., et al. (2015). Carbon, nitrogen, and phosphorus storage in alpine grassland ecosystems of Tibet: effects of grazing exclusion. Ecol. Evol. 5, 4492–4504. doi: 10.1002/ece3.1732
Luo, Z., Liu, J., Jia, T., Chai, B., and Wu, T. (2020). Soil bacterial community response and nitrogen cycling variations associated with subalpine meadow degradation on the loess plateau, China. Appl. Environ. Microbiol. 86, e180–e120. doi: 10.1128/aem.00180-20
Ma, B., Cai, Y., Bork, E. W., and Chang, S. X. (2018). Defoliation intensity and elevated precipitation effects on microbiome and interactome depend on site type in northern mixed-grass prairie. Soil Biol. Biochem. 122, 163–172. doi: 10.1016/j.soilbio.2018.04.015
Ma, B., Zhou, X., Zhang, Q., Qin, M., Hu, L., Yang, K., et al. (2019). How do soil micro−organisms respond to N, P and NP additions? Application of the ecological framework of (co−)limitation by multiple resources. J. Ecol. 107, 2329–2345.
Malard, L. A., and Pearce, D. A. (2018). Microbial diversity and biogeography in Arctic soils. Environ. Microbiol. Rep. 10, 611–625. doi: 10.1111/1758-2229.12680
Peters, M. K., Hemp, A., Appelhans, T., Becker, J. N., Behler, C., Classen, A., et al. (2019). Climate–land-use interactions shape tropical mountain biodiversity and ecosystem functions. Nature 568, 88–92. doi: 10.1038/s41586-019-1048-z
Ramirez, K. S., Leff, J. W., Barberán, A., Bates, S. T., Betley, J., Crowther, T. W., et al. (2014). Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proceedings. Biol. Sci. 281:20141988. doi: 10.1098/rspb.2014.1988
Ren, C., Zhang, W., Zhong, Z., Han, X., Yang, G., Feng, Y., et al. (2018). Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 61, 750–758. doi: 10.1016/j.scitotenv.2017.08.110
Reyon, D., Tsai, S. Q., Khayter, C., Foden, J. A., Sander, J. D., and Joung, J. K. (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465. doi: 10.1038/nbt.2170
Rime, T., Hartmann, M., and Frey, B. (2016). Potential sources of microbial colonizers in an initial soil ecosystem after retreat of an alpine glacier. ISME J. 10, 1625–1641. doi: 10.1038/ismej.2015.238
Scarlett, K., Denman, S., Clark, D. R., Forster, J., Vanguelova, E., Brown, N., et al. (2020). Relationships between nitrogen cycling microbial community abundance and composition reveal the indirect effect of soil pH on oak decline. ISME J. [Epub ahead of print]. doi: 10.1038/s41396-020-00801-0
Selmants, P. C., Adair, K. L., Litton, C. M., Giardina, C. P., and Schwartz, E. (2016). Increases in mean annual temperature do not alter soil bacterial community structure in tropical montane wet forests. Ecosphere 7: e01296.
Sghaier, H., Hezbri, K., Ghodhbanegtari, F., Pujic, P., Sen, A., Daffonchio, D., et al. (2016). Stone-dwelling actinobacteria Blastococcus saxobsidens, Modestobacter marinus and Geodermatophilus obscurus proteogenomes. ISME J. 10, 21–29.
Shen, C., Gunina, A., Luo, Y., Wang, J., He, J. Z., Kuzyakov, Y., et al. (2020). Contrasting patterns and drivers of soil bacterial and fungal diversity across a mountain gradient. Environ. Microbiol. 22, 3287–3301. doi: 10.1111/1462-2920.15090
Shen, M., Piao, S., Dorji, T., Liu, Q., Cong, N., Chen, X., et al. (2015). Plant phenological responses to climate change on the Tibetan Plateau: research status and challenges. Natl. Sci. Rev. 2, 454–467. doi: 10.1093/nsr/nwv058
Shi, Y., Zhang, K., Li, Q., Liu, X., He, J.-S., and Chu, H. (2020). Interannual climate variability and altered precipitation influence the soil microbial community structure in a Tibetan Plateau grassland. Sci. Total Environ. 714:136794. doi: 10.1016/j.scitotenv.2020.136794
Steidinger, B. S., Crowther, T. W., Liang, J., Van Nuland, M. E., Werner, G. D. A., Reich, P. B., et al. (2019). Climatic controls of decomposition drive the global biogeography of forest-tree symbioses. Nature 569, 404–408. doi: 10.1038/s41586-019-1128-0
Su, Y.-g, Liu, J., Zhang, B.-c, Zhao, H.-m, and Huang, G. (2020). Habitat-specific environmental factors regulate spatial variability of soil bacterial communities in biocrusts across northern China’s drylands. Sci. Total Environ. 719:137479. doi: 10.1016/j.scitotenv.2020.137479
Sun, Y., Chen, H. Y. H., Jin, L., Wang, C., Zhang, R., Ruan, H., et al. (2020). Drought stress induced increase of fungi:bacteria ratio in a poplar plantation. CATENA 193:104607. doi: 10.1016/j.catena.2020.104607
Tanvir, R., Sajid, I., Hasnain, S., Kulik, A., and Grond, S. (2016). Rare actinomycetes Nocardia caishijiensis and Pseudonocardia carboxydivorans as endophytes, their bioactivity and metabolites evaluation. Microbiol. Res. 185, 22–35. doi: 10.1016/j.micres.2016.01.003
Tian, L., Zhao, L., Wu, X., Fang, H., Zhao, Y., Yue, G., et al. (2017). Vertical patterns and controls of soil nutrients in alpine grassland: implications for nutrient uptake. Sci. Total Environ. 60, 855–864. doi: 10.1016/j.scitotenv.2017.07.080
Wang, J., Deng, H., Wu, S.-S., Deng, Y.-C., Liu, L., Han, C., et al. (2019). Assessment of abundance and diversity of exoelectrogenic bacteria in soil under different land use types. CATENA 172, 572–580. doi: 10.1016/j.catena.2018.09.028
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wei, H., Peng, C., Yang, B., Song, H., Li, Q., Jiang, L., et al. (2018). Contrasting soil bacterial community, diversity, and function in two forests in China. Front. Microbiol. 9:1693. doi: 10.3389/fmicb.2018.01693
White, H. J., León-Sánchez, L., Burton, V. J., Cameron, E. K., Caruso, T., Cunha, L., et al. (2020). Methods and approaches to advance soil macroecology. Glob. Ecol. Biogeogr. 29, 1674–1690. doi: 10.1111/geb.13156
Wilson, M. C., Mori, T., Rückert, C., Uria, A. R., Helf, M. J., Takada, K., et al. (2014). An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62. doi: 10.1038/nature12959
Wu, J., Song, M., Ma, W., Zhang, X., Shen, Z., Tarolli, P., et al. (2019). Plant and soil’s δ15N are regulated by climate, soil nutrients, and species diversity in alpine grasslands on the northern Tibetan Plateau. Agric. Ecosyst. Environ. 281, 111–123. doi: 10.1016/j.agee.2019.05.011
Wu, L., Wen, C., Qin, Y., Yin, H., Tu, Q., Van Nostrand, J. D., et al. (2015). Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 15:125. doi: 10.1186/s12866-015-0450-4
Wu, X., Zhang, H., Chen, J., Shang, S., Wei, Q., Yan, J., et al. (2016). Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3–V4 region of the 16S rRNA gene. Appl. Microbiol. Biotechnol. 100, 3577–3586. doi: 10.1007/s00253-015-7257-y
Xu, M., Li, X., Cai, X., Gai, J., Li, X., Christie, P., et al. (2014). Soil microbial community structure and activity along a montane elevational gradient on the Tibetan Plateau. Eur. J. Soil Biol. 64, 6–14. doi: 10.1016/j.ejsobi.2014.06.002
Yang, F., Niu, K., Collins, C. G., Yan, X., Ji, Y., Ling, N., et al. (2019). Grazing practices affect the soil microbial community composition in a Tibetan alpine meadow. Land Degrad. Dev. 30, 49–59.
Yu, J., Liu, F., Tripathi, B. M., and Steinberger, Y. (2020). Changes in the composition of soil bacterial and fungal communities after revegetation with Caragana microphylla in a desertified semiarid grassland. J. Arid Environ. 182:104262. doi: 10.1016/j.jaridenv.2020.104262
Yu, L., Chen, Y., Sun, W., and Huang, Y. (2019). Effects of grazing exclusion on soil carbon dynamics in alpine grasslands of the Tibetan Plateau. Geoderma 353, 133–143. doi: 10.1016/j.geoderma.2019.06.036
Yuan, X.-L., Cao, M., Liu, X.-M., Du, Y.-M., Shen, G.-M., Zhang, Z.-F., et al. (2018). Composition and genetic diversity of the nicotiana tabacum microbiome in different topographic areas and growth periods. Int. J. Mol. Sci. 19:3421. doi: 10.3390/ijms19113421
Zethof, J. H. T., Bettermann, A., Vogel, C., Babin, D., Cammeraat, E. L. H., Solé-Benet, A., et al. (2020). Prokaryotic community composition and extracellular polymeric substances affect soil microaggregation in carbonate containing semiarid Grasslands. Front. Environ. Sci. 8:51. doi: 10.3389/fenvs.2020.00051
Zhang, K., Shi, Y., Jing, X., He, J.-S., Sun, R., Yang, Y., et al. (2016). Effects of short-term warming and altered precipitation on soil microbial communities in alpine grassland of the Tibetan Plateau. Front. Microbiol. 7:1032. doi: 10.3389/fmicb.2016.01032
Zhang, Y., Dong, S., Gao, Q., Liu, S., Ganjurjav, H., Wang, X., et al. (2017). Soil bacterial and fungal diversity differently correlated with soil biochemistry in alpine grassland ecosystems in response to environmental changes. Sci. Rep. 7:43077. doi: 10.1038/srep43077
Zhang, Y., Dong, S., Gao, Q., Liu, S., Zhou, H., Ganjurjav, H., et al. (2016). Climate change and human activities altered the diversity and composition of soil microbial community in alpine grasslands of the Qinghai-Tibetan Plateau. Sci. Total Environ. 562, 353–363. doi: 10.1016/j.scitotenv.2016.03.221
Zhong, W., Gu, T., Wang, W., Zhang, B., Lin, X., Huang, Q., et al. (2010). The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil 326, 511–522. doi: 10.1007/s11104-009-9988-y
Keywords: soil bacteria, soil microorganism, alpine grassland, Tibetan Plateau, 16S rRNA
Citation: Jiang H, Chen Y, Hu Y, Wang Z and Lu X (2021) Soil Bacterial Communities and Diversity in Alpine Grasslands on the Tibetan Plateau Based on 16S rRNA Gene Sequencing. Front. Ecol. Evol. 9:630722. doi: 10.3389/fevo.2021.630722
Received: 24 November 2020; Accepted: 20 January 2021;
Published: 16 February 2021.
Edited by:
Mauro Fois, University of Cagliari, ItalyReviewed by:
Jianshuang Wu, Chinese Academy of Agricultural Sciences, ChinaMan Kit Cheung, The Chinese University of Hong Kong, China
Copyright © 2021 Jiang, Chen, Hu, Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuyang Lu, xylu@imde.ac.cn
 Hongmao Jiang
Hongmao Jiang Youchao Chen
Youchao Chen Yang Hu
Yang Hu Ziwei Wang1,2
Ziwei Wang1,2  Xuyang Lu
Xuyang Lu