Evolution of Regeneration in Animals: A Tangled Story
- 1Laboratory of Regenerative Medicine, FSBI National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of the Russian Federation, Moscow, Russia
- 2Histology Department, Pirogov Russian National Research Medical University, Ministry of Healthcare of the Russian Federation, Moscow, Russia
- 3Laboratory of Growth and Development, FSBSI Scientific Research Institute of Human Morphology, Moscow, Russia
- 4Histology Department, Medical Institute, Peoples’ Friendship University of Russia (RUDN University), Moscow, Russia
The evolution of regenerative capacity in multicellular animals represents one of the most complex and intriguing problems in biology. How could such a seemingly advantageous trait as self-repair become consistently attenuated by the evolution? This review article examines the concept of the origin and nature of regeneration, its connection with the processes of embryonic development and asexual reproduction, as well as with the mechanisms of tissue homeostasis. The article presents a variety of classical and modern hypotheses explaining different trends in the evolution of regenerative capacity which is not always beneficial for the individual and notably for the species. Mechanistically, these trends are driven by the evolution of signaling pathways and progressive restriction of differentiation plasticity with concomitant advances in adaptive immunity. Examples of phylogenetically enhanced regenerative capacity are considered as well, with appropriate evolutionary reasoning for the enhancement and discussion of its molecular mechanisms.
Nothing in biology makes sense except in the light of evolution
T.G. Dobzhansky
If there were no regeneration there could be no life
R.J. Goss
Introduction
Animal regeneration is a subject of continuous scientific interest. The first experimental studies on regeneration were carried out in the 18th century (Reaumur, 1712; Tremblay, 1744). Despite the remarkable progress in the field (Bely and Nyberg, 2010; Zattara et al., 2019), we have to face the fact that regenerative capacity varies colossally among the animal taxa. Despite the enormous amount of experimental data on regeneration, the mechanisms of its evolution remain largely uncertain.
The first attempts to understand the laws that drive the evolution of regenerative capacity in animals date to the 19th century. Since then, the so-called first rule of regeneration (“the regenerative capacity of animals decreases with an increase in anatomical complexity”) was re-formulated by many authors independently (Vorontsova and Liosner, 1960). The first counter-examples of phylogenetically enhanced regenerative capacity in animals date back to the 19th century as well.
August Weismann (1834–1914) was the first to propose comprehensive evolutionary reasoning for the diverse regeneration potential in animals. He postulated that regenerative capacity is an adaptive trait that is subject to phylogenetic alterations and therefore may vary considerably among the taxa. According to Weismann, the regenerative capacity of a particular organ depends on three factors: anatomical and physiological complexity, the frequency of damage to the organ, and its significance for survival (Weismann, 1893, 1899). In the 20th century, similar views were expressed by Arthur Edwin Needham, who also emphasized the relevance of environmental conditions (for instance, he believed that aquatic environments are highly favorable for regeneration) (Needham, 1952). Needham’s remarks on the adaptive value of high regenerative capacity, particularly on its ambiguous evolutionary feasibility and controversial impact on survival, represent an important addition to Weismann’s concept. According to Needham, the routes of adaptation to the damaging factors are multiple. Even under conditions of frequent damage to an organ, its regeneration would not necessarily be the unique or least expensive adaptive mechanism; the compensations for the loss may include the enhanced breeding capacity, as well as the effective avoidance of the damage through enhanced mobility (Needham, 1952).
Despite the long history of the subject, the evolution of regenerative capacity in animals is far from being fully understood (Bely and Nyberg, 2010). In a broad sense, the problematics of contemporary experimental studies and theoretical investigations in the field have been set up by Weismann (1893, 1899) and Needham (1952). It includes the questions like whether regeneration is a primitive or adaptive trait, what is the role of damage frequency in the evolution of regenerative capacity, what is the role of the environment, what are the reasons for the dynamic evolutionary alterations in regenerative capacity, is it appropriate to consider regeneration as a direct correlate of asexual reproduction, etc. The answers to these and other old questions in their contemporary perspective are the subject of this review.
Contribution of Russian Scientists to the Theory of Regeneration
The first comprehensive Russian studies in the field of regeneration date back to the early 20th century. We should mention the research by K. N. Davydov, performed on acorn worms Ptychodera minuta and Ptychodera clavigera. Davydov was one of the first to express the idea of the similarity between regeneration and embryonic development; his conclusions were based on the comparison of the process of anterior regeneration in P. minuta and P. clavigera with embryonic development (Davydov, 1903).
By the 1930s, several large scientific centers for the study of regeneration were formed in Russia. One of those was headed by academician A. A. Zavarzin. Scientific activities of his team had a pronounced evolutionary dimension; their principal findings include the archetypal similarity of skeletal muscle regeneration (with the involvement of myoblasts) in representatives of different taxa (Zavarzin, 1938).
Another famous team focused on studying regeneration in invertebrates (predominantly Porifera) was headed by B. P. Tokin (Tokin, 1969; Korotkova, 1988). B. P. Tokin reckoned that the term «regeneration» was historically coined as a generic notion encompassing multiple different phenomena. He believed that restoration of lost parts (extremities or organs) proceeds by a different scenario and obeys other laws than the so-called «somatic embryogenesis» —formation of a whole organism from a limited number of preserved cells or small tissue fragments. In this regard, B. P. Tokin and colleagues proposed a broader concept of «regulation» which was a unifying term for regeneration per se and «somatic embryogenesis» (Tokin, 1969). This idea was subsequently criticized by Liosner, who questioned the criteria for the distinction between the regeneration of body parts and «somatic embryogenesis». L. D. Liosner justly pointed that in many cases the distinction is vague, e.g., the restoration of body terminus in many invertebrates (cnidarians, planarians, annelids, etc.) satisfies the definitions of both regeneration and somatic embryogenesis (Liozner, 1975).
Another key term that B. P. Tokin was operating with was «integration»—a universal measure of adaptive fitness showing a tendency to a continuous increase in the course of phylogenesis. B. P. Tokin believed that the ability to regenerate body parts increases evolutionary along with “integration” (as indicated by the high regeneration rates characteristic of the integument and internal organs in vertebrates), while the capacity of asexual reproduction and somatic embryogenesis decreases (Tokin, 1969). Tokin’s views on the origin of regenerative capacity should be mentioned as well: he believed that physiological regeneration arose very early based on the properties and metabolic needs of primitive living systems, while reparative regeneration evolved later, based on the principles of physiological regeneration and subsequent evolution of metabolic pathways and defense mechanisms of the body (Tokin, 1969).
Another influential Russian team working on fundamental problems of regeneration was the laboratory headed by M. A. Vorontsova and L. D. Liosner (the Laboratory of Growth and Development at the Institute of Human Morphology Russian Academy of Medical Sciences, Moscow). The scope of their scientific interest within the field of animal regeneration was extremely diverse. Initially, the model choice was confined to limb regeneration in amphibians, with the main focus on the balance of destruction and proliferation and the role of mitogenic radiation in these processes; a series of such studies was published in the Wilhelm Roux’ Archiv für Entwicklungsmechanik der Organismen (Blacher et al., 1933; Liosner et al., 1936). Later on, the focus of scientific interest eventually shifted toward the regeneration of internal organs, notably parenchymal organs, in amphibians and ultimately in mammals. The vast experimental data on the regeneration of different organs (kidneys, liver, lungs, testes, ovaries, etc.) allowed a number of important fundamental generalizations (Vorontsova and Liosner, 1960; Liozner, 1974). In particular, Vorontsova found out that all parenchymal organs regenerate in a similar way; to describe this; the term «regenerative hypertrophy» was introduced. Regenerative hypertrophy—compensation of the loss by, respectively, cell proliferation or the increase in the size of individual cells without restoration of the initial morphogenetic complexity (Vorontsova and Liosner, 1960; Liozner, 1974). V. F. Sidorova showed that cellular mechanisms of postnatal regeneration of parenchymal organs correspond to postnatal growth rather than embryonic development, as no additional structural units (lobules, acini, and nephrons) are formed after the resection (Sidorova, 1964, 1978). A. G. Babaeva demonstrated the key role of the immune system in regeneration, notably the ability of lymphocytes to stimulate or suppress the repair processes in mammals (Babaeva, 1989, 1990). In the works of G. B. Bolshakova and her co-workers began a new research area—the study of the regeneration of the internal organs of mammalian fetuses; it was shown that in the prenatal period, myocardial injury in 16-day-old rat fetuses causes an increase in the proliferation of cardiomyocytes away from the injury zone, while the formation of connective tissue in the damaged zone is slow, which turns out to be unfavorable on the survival of such animals in the postnatal period (Bolshakova, 2008). A. V. Elchaninov showed that after resection of the liver of rat fetuses, the proliferation of hepatocytes is also activated and the liver mass is restored, while, in contrast to the postnatal period, without an increase in the ploidy and size of hepatocytes (Elchaninov and Bolshakova, 2011a, b, 2012).
Findings of other Russian research teams that worked successfully on specific fundamental issues of animal regeneration should be mentioned as well. These include the influence of pigment epithelium of the retina in its regeneration in tailed amphibians studied by Mitashov (1996) and the role of polyploidy in liver regeneration/myocardium repair in mammals demonstrated by Brodsky and Uryvaeva (1977).
The Origins of Regeneration
From the very beginning of regeneration studies, two opposing opinions have been expressed about its origin. Some experts qualified regeneration as a primary property of living systems (A. E. Needham and T. H. Morgan adhered to this point) (Needham, 1952), while others believed that it had emerged as a trait in some primitive organisms along with multicellularity (Weismann, 1893, 1899; Bely and Nyberg, 2010). The second opinion implies the understanding of regeneration as an epiphenomenon—inducible re-play of a program, which underlies a particular morphogenetic process (asexual reproduction, growth, and embryogenesis) and is used repeatedly in the case of damage (Garza-Garcia et al., 2010; Tiozzo and Copley, 2015).
Except for the radically different interpretation of very early events, these two theories are mutually consistent, as both allow viewing regeneration in terms of fundamental homology and account for the employment of recognizable, highly conservative patterns (which can be loosely defined as intensive physiological maintenance of the remnant complemented by active reconstruction of the missing part). Repair processes in different organisms have much in common, for example, rapid re-epithelialization of the damaged site, activation of cell proliferation, activation of matrix metalloproteinases, scavenging and regulatory activities of macrophages and other cells of the immune system (Elchaninov et al., 2018, 2019), the impact of the nervous system, etc. Repair processes may involve dedifferentiation and transdifferentiation of cells and, notably, activation of a stereotype genetic program (Fumagalli et al., 2018; Darnet et al., 2019; Mehta and Singh, 2019).
Moreover, the diversity of views on the origin of regeneration is more of historical interest, as early studies considered this process only at the level of tissues and organs while understandably neglecting the corresponding phenomena at subcellular levels. With the current state of knowledge, it is difficult to ignore the events and processes of restoration and maintenance of intracellular integrity, including the continuous renewal of organelles, turnover of the membranes, duplication of centrioles, division of mitochondria, disassembly and reassembly of the nuclear envelope during mitosis, etc. A unicellular organism devoid of any ability to regenerate would be maladaptive if viable at all; therefore, the direct association of regenerative capacity with multicellularity is hardly reasonable.
Vorontsova and Liosner (1960) distinguished several types of regeneration which had evolved separately; this point has been reflected in recent studies (Bely and Nyberg, 2010). For example, the regeneration of various components of organs, the regeneration of whole organs, and the regeneration of the entire body from a fragment represent different types of regeneration. Some of these types are continuously preserved by evolution, while others become eliminated (for example, the regeneration of the entire body from a fragment).
Despite the distinct common features, repair processes in different animal taxa may take dramatically different ways (Alvarado and Tsonis, 2006). These ways are most commonly distinguished by the scale of damage-induced cell proliferation and its contribution to the morphogenesis, with the extremes called morphallaxis and epimorphosis (the terms were introduced by T. H. Morgan) (Figures 1A–C). Morphallaxis proceeds by a spatial reorganization of the remnant at the initial stages of repair; for example, Hydra regenerates by morphallaxis (Figure 1A) (Alvarado and Tsonis, 2006). The opposite way, epimorphosis, proceeds through the formation of regeneration blastema composed of low-differentiated cells with high proliferative capacity (Figure 1C). Epimorphosis is characteristic of limb regeneration in tailed amphibians (Caudata) and to a certain extent also of planarian regeneration (Figure 1B) (Gurley et al., 2008). Currently, most experts agree that the clear distinction between epimorphosis and morphallaxis hardly makes sense, as any real regeneration is usually a combination of both (Agata et al., 2007; Bely and Nyberg, 2010). For instance, the oral pole regeneration in Hydra is distinctly epimorphic (Chera et al., 2009; Galliot and Ghila, 2010).
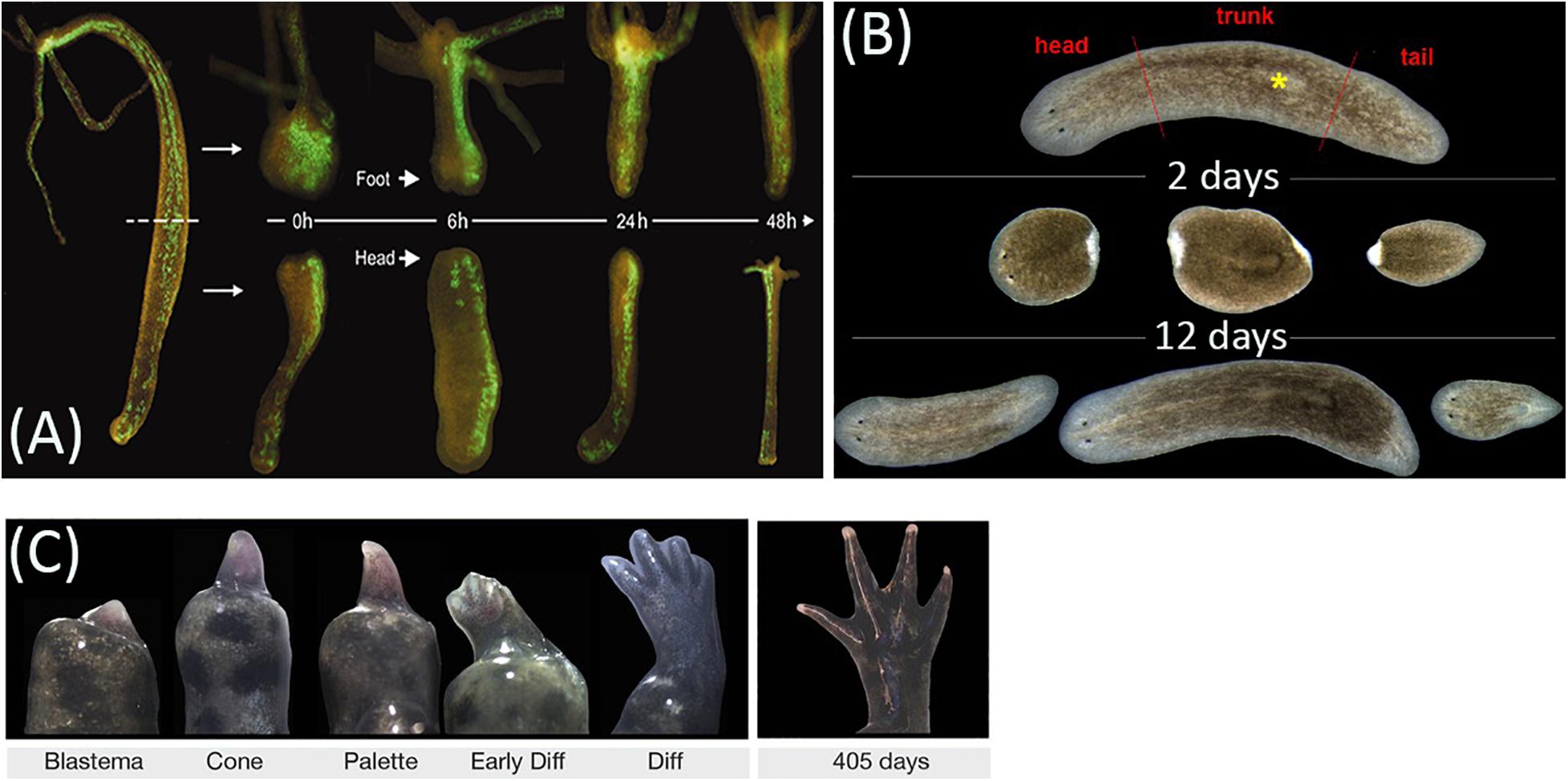
Figure 1. Animal regeneration models. (A) Regeneration of head and foot in transgenic Hydra vulgaris by morphallaxis. (B) Regeneration timing in planarian Schmidtea mediterranea. (C) Epimorphic limb regeneration in axolotl Ambystoma mexicanum. Adapted from, respectively, Wittlieb et al. (2006); Adell et al. (2014), and Monaghan et al. (2014), under CC-BY. The asterisk labels the pharynx.
The apparent phylogenetic primacy of morphallaxis is indirectly indicated by its broad representation in both bilaterians and non-bilaterians, whereas epimorphosis is specific for bilaterians (Bely and Nyberg, 2010). Considering their similarity, it can be assumed that epimorphosis evolved on the basis of morphallaxis (Agata et al., 2007; Ben Khadra et al., 2018; Ferrario et al., 2018).
It should be noted that the overall homology of regeneration mechanisms in animals is not that obvious. The mechanisms of regeneration in distant taxa can differ beyond recognition, as can be illustrated by the diverse genesis of regeneration blastema in invertebrates (Das, 2015; Bertemes et al., 2020) and vertebrates (Seifert and Muneoka, 2018; Muneoka and Dawson, 2020).
In planarians, the formation of blastema results from the proliferation of neoblasts in response to amputation (Bertemes et al., 2020); in crustaceans and insects, wound blastema is formed from the migrating epidermal cells that undergo dedifferentiation (Mito et al., 2002; Das, 2015; Bando et al., 2018).
Phylogenetic plasticity of regeneration mechanisms in Caudata, with optional stem cell involvement and varying contributions of dedifferentiation and transdifferentiation, should be noted (Seifert and Muneoka, 2018; Muneoka and Dawson, 2020). For instance, in newts, myoblasts are formed by fragmentation of muscle fibers, whereas in axolotls, they form by differentiation of myosatellite cells found within the blastema (Seifert and Muneoka, 2018; Muneoka and Dawson, 2020).
Based on these findings, K. Muneoka et al. reckon that regenerative capacity in vertebrates evolved independently in different taxa originating from a hypothetical common tetrapod ancestor incapable of limb regeneration. The authors use this concept to describe the evolution of epimorphic limb regeneration in amphibians (Seifert and Muneoka, 2018; Muneoka and Dawson, 2020) and suggest a similar scenario for the evolution of regenerative capacity in mammals, with their ability to partially restore the terminal phalanx of a finger by forming a blastema-like structure through remodeling and growth of bone tissue, which is different from the mechanisms of blastema formation in amphibians (Seifert and Muneoka, 2018; Muneoka and Dawson, 2020).
It should also be noted that, in mammals, cellular sources of the wound blastema of the terminal phalanx differ in an age-dependent manner. In mouse embryos at advanced developmental stages, wound blastema is a derivative of chondrogenic cells of the terminal phalanx, which express Msx1, Msx2, Dlx5, and Bmp4 markers. A similar amputation performed in the neonatal period promotes the formation of the wound blastema as a derivative of mesenchymal cells located predominantly beneath the nail organ and expressing Msx1, while the blastema cells express Bmp2 and Bmp7 (Seifert and Muneoka, 2018; Muneoka and Dawson, 2020).
The diversity of cellular mechanisms of blastema formation has been emphasized by Brockes et al. whose theory of regeneration origin and evolution is based on two assumptions: (1) regeneration employs the highly conservative principal mechanisms of growth, development, and maintenance of tissue homeostasis universally found in animals, and ensuring the capability of self-repair in certain species/taxa and (2) these highly conservative cellular mechanisms are governed and regulated by a relatively small number of taxon-specific genes responsible for the pronounced regenerative capacity (Garza-Garcia et al., 2010).
The first of these points is consistent with the evidence on the molecular invariance of morphogenetic processes (i.e., various types of morphogenesis involve similar regulatory cascades) (Cary et al., 2019; Mehta and Singh, 2019). The second point (existence of “principal regulator” genes) is less evident; notable examples include fgf20 proposed as a primary regulator of fin regeneration in Danio rerio (Whitehead et al., 2005; Poss, 2010). A taxon-specific protein Prod1 (Geng et al., 2015), found in newts and salamanders but missing in D. rerio, Xenopus, and mammals, participates in the neural control over regeneration and patterning (Garza-Garcia et al., 2010; Geng et al., 2015; Muneoka and Dawson, 2020). The presence of Prod1 orthologs in Ambystoma mexicanum and Ambystoma maculatum places its origin before the divergence of Salamandridae and Ambystomidae (Garza-Garcia et al., 2010). In a planarian Schmidtea mediterranea, 15% of 1065 genes associated with homeostasis and regeneration have no homologs in other organisms and are considered taxon-specific (Reddien et al., 2005). According to Brockes et al., this group of genes is likely to comprise principal regulators that determine the ability to regenerate (Garza-Garcia et al., 2010).
The concept of principal regulators has also been indirectly supported by a comparative genomic study encompassing 132 species of multicellular animals with different regeneration capacities. A group of 118 highly conservative genes, 96% of which encoded Jumonji C (JmjC) domain-containing proteins, have been found specific for the «highly regenerative» species. The evolutionary loss of such genes has been associated with a dramatic decrease in regenerative capacity (Cao et al., 2019).
The evolutionary relationship between morphallaxis and epimorphosis is disputable. The assumption on their intrinsic homology was expressed by Bely and Nyberg (2010). This point of view is supported by the non-random incidence of both regeneration modes among animal taxa, as well as the fundamental similarity of the cellular processes underlying them. However, the depth of this similarity varies, and the mechanisms can be fundamentally different. Moreover, the terms «morphallaxis» and «epimorphosis», in the sense that Morgan (who coined them) put into them, do not take into account the overall mechanistic diversity of regenerative processes in the animal kingdom; as a result, phenomena of different nature are combined under one term. In this regard, some authors propose to abandon the use of terms «morphallaxis» and «epimorphosis». For instance, K. Agata suggested new terms «distalization» and «intercalation» (Agata et al., 2007; Tiozzo and Copley, 2015). Recent findings indicate striking diversity of regulation and implementation of regenerative processes at molecular and cellular levels; even within a taxonomic group, the mechanisms of regenerative response may vary significantly. In this regard, the concept of homology as related to regeneration becomes a distinct complex problem (Tiozzo and Copley, 2015).
The question of the origin of reparative regeneration is closely related to the problem of how physiological regeneration (i.e., the non-injury-induced restorative processes) and reparative regeneration relate to each other. In general, physiological regeneration is defined as the restoration of organs, tissues, cells, and subcellular structures lost during their normal life cycle or when performing their functions (Vorontsova and Liosner, 1960). In modern understanding, physiological regeneration is inherent in all tissues and cells; however, it proceeds in different forms. The phenomena of physiological regeneration include desquamation of epidermal cells, renewal of the intestinal epithelium, restoration of the uterine mucosa during the menstrual cycle, etc. (Carlson, 2007). B. P. Tokin viewed physiological regeneration as a mechanistic basis and direct evolutionary precursor to reparative processes. In an extreme interpretation (currently only of historical interest), reparative regeneration is an enhanced version of physiological regeneration. This simplification is due to the fact that cell proliferation, observed in some tissues under normal conditions and activated after injury, was the only measurable sign of regeneration. Currently, it is obvious that reparative regeneration differs in mechanisms from physiological regeneration and according to some views evolves as epiphenomenon which partially employs both the principles of physiological regeneration and the highly conserved molecular and cellular mechanisms of embryonic development and growth (Goss, 1992; Tiozzo and Copley, 2015).
Anyway, there is no doubt that regeneration as a process arose very early in the evolution and therefore involves highly conserved cellular mechanisms of morphogenesis. The intrinsic similarity of regeneration processes with asexual reproduction (Vorontsova and Liosner, 1960; Martinez et al., 2005; Kawamura et al., 2008; Burton and Finnerty, 2009; Zattara and Bely, 2016), growth (Bely and Wray, 2001; Gurley et al., 2008), and embryonic development (Martin and Parkhurst, 2004; Ghosh et al., 2008; Vogg et al., 2019) has been repeatedly noted.
Regeneration and Asexual Reproduction
Indeed, it is quite difficult not to link regeneration with asexual reproduction (Vorontsova and Liosner, 1960; Martinez et al., 2005; Brockes and Kumar, 2008; Kawamura et al., 2008; Burton and Finnerty, 2009; Zattara and Bely, 2016). In many organisms, regeneration can be morphologically indistinguishable from asexual reproduction by budding or fission. The mechanisms of asexual reproduction could be “easily” adapted for regeneration; the key difference is the stimuli that trigger these processes. Such concept has been supported by molecular studies of regeneration and asexual reproduction in hydras, planarians, annelids, and other invertebrates (Martinez et al., 2005; Mehta and Singh, 2019; Reddy et al., 2019a, b) revealing specific involvement of stem cells and generically similar roles of Wnt-signaling in these two processes (Mehta and Singh, 2019).
Ultimately, the phenomenon of restoration of the entire body from a fragment can be considered as asexual reproduction (Tokin, 1969). B. P. Tokin viewed the decreasing capacity for asexual reproduction as a direct correlate (and reflection) of the loss in regenerative capacity.
The resemblance of asexual reproduction with regeneration in invertebrates is remarkable. However, despite the rich recent history of comparative studies on the histological level, only a limited number of specific molecular findings support the intrinsic similarity of the two processes. The positive examples include similar expression of Pl-en in the nervous system, as well as Pl-Otx1 and Pl-Otx2 in the anterior body wall, foregut, and nervous system, of the annelid worm Pristina leidyi during regeneration and asexual reproduction (Bely and Wray, 2001). Also, Hydra shows a similar expression of HyBMP5-8b, a BMP5-8 ortholog involved in axial patterning and formation of tentacles, in budding and regeneration (Reinhardt et al., 2004). However, despite the outward similarity of asexual reproduction with regeneration, these two processes evolved separately. For instance, the closest common ancestor of Annelida was probably capable of regenerating the anterior and posterior ends of the body but was devoid of the ability to reproduce itself asexually (Zattara and Bely, 2011, 2016). In Nematostella vectensis, molecular markers expressed during asexual reproduction and regeneration significantly overlap; however, no expression of regeneration markers Nv-otxC and anthox1 is observed during asexual reproduction (Burton and Finnerty, 2009).
Regeneration and Embryogenesis
K. N. Davydov was one of the first to express the idea of the similarity between regeneration and embryonic development; his conclusions were based on the comparison of the process of anterior regeneration in P. minuta and P. clavigera with embryonic development (Davydov, 1903). The relationship between regeneration and embryogenesis is of particular importance for evolutionary biology, as it allows experimental investigation of the emergence of new structures. Sánchez Alvarado and coauthors developed an original view of this problem (Sánchez Alvarado, 2000; Elliott and Sánchez Alvarado, 2013). According to his opinion, the limb development in arthropods and vertebrates is governed by similar molecular cascades. However, the closest common ancestors of arthropods and vertebrates had no limbs at all. What factors, then, predetermined the homology? (Sánchez Alvarado, 2000; Elliott and Sánchez Alvarado, 2013).
The answer to this question can be obtained by studying regeneration. The similarity of embryonic limb buds with regeneration blastema is evident both histologically and at the level of molecular signaling cascades (Galis et al., 2003). In planarians, the blastema contains key components of molecular pathways regulating the establishment of anterior–posterior (Wnt-signaling), dorsal–ventral (BMP-pathway), and medial–lateral polarities (Sánchez Alvarado, 2000; Elliott and Sánchez Alvarado, 2013; Karami et al., 2015). According to Sánchez Alvarado and coauthors opinion, «the molecular processes underlying blastema formation and regeneration have been co-opted by sexually reproducing animals for the production of new structures such as limbs during the evolution of their developmental processes» (Sánchez Alvarado, 2000; Elliott and Sánchez Alvarado, 2013).
In molecular terms, embryonic development and regeneration are very different. N. vectensis shows no asymmetric expression of Hox-like genes (characteristic of embryogenesis) during asexual reproduction or regeneration (Burton and Finnerty, 2009). In zebrafish, the epimorphic regeneration of fins requires fgf20a expression, which is not required for fin development (Whitehead et al., 2005). In Xenopus, three Abdominal B-type Hox genes XHoxc10, XHoxa13, and XHoxd13 show different expression patterns in regenerating and developing limbs (Christen et al., 2003). The similarities and differences of embryonic development, asexual reproduction, and regeneration are consistent with the idea that the capacities of asexual reproduction and regeneration evolved on the basis of signaling pathways of growth and development; however, the “borrowing” was selective and proceeded in a variety of ways.
Apparently, signaling pathways governing regeneration and asexual reproduction in primitive animals were eventually redirected for the performance of other tasks, e.g., limb development (Sánchez Alvarado, 2000; Elliott and Sánchez Alvarado, 2013).
Evolutionary Maintenance of Regenerative Capacity
Regardless of the character of regeneration origins at the most ancient stages of evolution (whether it was a primary or secondary property of animals), this property was propagated in diverse forms throughout the animal kingdom.
The problem of maintaining regenerative capacity during evolution is one of the key ones. However, there are very few specific experimental studies. Initially, the very idea of maintaining the ability to regenerate, the role of the frequency of damage in this process was developed by Weismann (1893, 1899), and further tested in the works of Morgan T.H. (1901). Further insight into the role of injury and the value of regeneration in the fitness of a species was developed by Needham (1952) and Goss (1969).
According to the classical reasoning, frequent damage to an organ is favorable for the maintenance of its regenerative capacity (Weismann, 1893, 1899), given that its loss will significantly reduce the individual’s fitness and the overall costs are not detrimental for the species (Needham, 1952; Goss, 1969).
At the initial stages of evolution, aggressive environmental conditions apparently played a principal role in maintaining the regenerative capacity (Wulff, 2006). Indeed, a high frequency of damage is typical for some groups of highly regenerative organisms in natural environments, to the extent that the majority of individuals in wild populations show distinct signs of damage and repair (Clark et al., 2007; Bely and Nyberg, 2010). However, the high regenerative capacity may be preserved even at low frequencies of damage. T. H. Morgan, in his classical studies on hermit crabs, showed that the rudimentary hind limbs, hidden in the shell and rarely damaged unless the shell is broken (in which case the animal would likely perish), regenerate in the same way as front limbs (exposed to the environment and frequently damaged or autotomized) (Morgan T., 1901; Morgan T.H., 1901; Sunderland, 2010). Noteworthy, hydras, and planarians, with their remarkable regenerative capacities, show no signs of active repair in the wild (Bely and Nyberg, 2010). As emphasized by Needham, regeneration would never be the only adaptive response to frequent damage. Instead, the species may enhance its reproductive potential; the animals may also develop mobility, protective coloration, exoskeleton, etc. (Needham, 1952).
Theoretically, as already noted, the severity of damage must be balanced by the cost of the regenerative process. Excessive severity of damage will kill the animal, whereas its insignificance for the normal functioning (due to dispensability or redundancy of the damaged structure) will eliminate the need for regeneration. However, in practice, it is rather difficult to determine the cost of damage, as well as the cost of regeneration for a particular organism (Tiozzo and Copley, 2015). Several studies indicate that regeneration is indeed associated with significant energy expenditures (Naya et al., 2007) and functional opportunity costs that affect the survival and reproductive capacity of the organism (Bernardo and Agosta, 2005; Maginnis, 2006; Suzuki et al., 2019). Complex adaptive reactions (e.g., autotomy, which helps to minimize the loss of biological fluids and tissues when attacked by predators) can reduce the cost of damage thus increasing the feasibility of regeneration (Maginnis, 2006; Mcgaw, 2006; Bateman et al., 2008). In the general case, the regeneration is feasible when its benefits and rates override the possible negative effects from the existence of functionally immature and burdensome intermediate structures (Ramos et al., 2004; Dupont and Thorndyke, 2006; Barr et al., 2019) or incomplete/deviant recovery in cases of atypical regeneration (Lailvaux et al., 2009; Bely and Nyberg, 2010).
Due to the difficulties and contradictions of adaptationism (when applied on its own), alternative hypotheses were proposed to explain the evolutionary maintenance of regenerative capacity. In this regard, pleiotropic effects and phylogenetic inertia represent particularly important factors that should be discussed separately.
In an evolutionary context, the term «pleiotropy» refers to the maintenance of regenerative capacity of an organ in close association with some other important morphogenetic process, for example, asexual reproduction, growth, embryogenesis, or regeneration of another organ (possibly regulated by the same genetic frameworks). Pleiotropy implies default activation of related morphogenetic processes; for instance, in cnidarians and flatworms, the mechanisms of regeneration and normal growth are intrinsically similar (Alvarado and Tsonis, 2006; Bosch, 2007).
The concept of phylogenetic inertia refers to cases when regenerative capacity confers no distinct selective advantages to the species, nor shows distinct associations with any other morphogenetic process. In such cases, regeneration is preserved for the reason of insufficient selection pressure (or time) for its elimination. This concept provides a valuable description for the evolution of regenerative capacity in annelids, some of which retained the capacity while others lost it (Bely and Wray, 2001; Bely, 2006).
Evolutionary Enhancement of Regenerative Capacity
It should be noted that evolutionary enhancement of regenerative capacity is rare. Nevertheless, the distinct minor trends can be illustrated by the enhanced regenerative capacity of muscle liver tissues and in mammals and birds compared with amphibians (Liozner, 1974; Carlson, 2005) and the enhanced regeneration of extremities in arthropods compared with other ecdysozoans (Maruzzo and Bortolin, 2013). Another famous example is the regeneration of the tail in lizards (Garza-Garcia et al., 2010) and high skin regeneration in the spiny mouse, Acomys (Brant et al., 2016). Despite these impressive examples of the enhanced regenerative capacity, their mutual relationship is too distant to allow comprehensive investigation of common evolutionary patterns.
One of the most productive strategies in tracing the evolutionary dynamics of regenerative capacity is to compare closely related species with different regenerative capacities (Bely and Sikes, 2010; Zattara et al., 2019). Phylum Nemertea is one of the most promising in this aspect, as all of its studied species are capable of regenerating the posterior portion of the body, while only some of them can regenerate the anterior terminus (Bely et al., 2014; Zattara and Bely, 2016). The findings indicate that the common ancestor of Nemertea was capable of regenerating the posterior portion, but not the anterior terminus. In the evolution of Nemertea, this capacity was reinforced in at least four instances, as revealed by facile regeneration of the anterior terminus in corresponding species (one among Palaeonemertea and three among Pilidiophora; Zattara et al., 2019). The repeated events of enhancement were apparently promoted by repeated emergence of certain traits which allowed the transition (probably, the long-term survival of decapitated individuals) (Zattara et al., 2019). Mechanistically, the enhancement may result from the activation of some embryonic developmental programs in adults. Such assumption is consistent with the experiments on the embryos of Nemertopsis bivittata, which, after being cut into two parts, develop into two individuals (whereas the adults of this species are non-regenerative) (Martindale and Henry, 1995). Such mechanisms can be highly conserved; cf. the organizing roles of Wnt/β-catenin signaling during apical regeneration in Hydra and early development in vertebrates (Guder et al., 2006; Reddy et al., 2019a; Vogg et al., 2019).
An Evolutionary Decline in Regenerative Capacity
The decline in regenerative capacity is a very strong phylogenetic trend, the examples of which can be found in any phylum (Bely and Nyberg, 2010; Lai and Aboobaker, 2018). However, its accurate comparative assessment in different groups of animals is complicated (Bely, 2010; Bely and Sikes, 2010).
Meanwhile, mechanistic reasons for the decline, though much discussed, remain understudied. In the view of adaptationists, regenerative capacity may be alleviated as a direct consequence of low damage frequency (Baumiller and Gahn, 2004). However, this view has not been supported by experimental findings, efficient regeneration of rudimentary limbs in hermit crabs reported by T. Morgan. The same applies to the regeneration of internal organs, which, according to A. Weismann, should regenerate poorly (Weismann, 1893, 1899). In the 20th century, this concept was criticized by M. A. Vorontsova, L. D. Liosner, and their followers (Vorontsova and Liosner, 1960; Liozner, 1974).
In addition, a decline in regenerative capacity may occur as a result of a significant change in the adaptive value of the organ. In case of dramatic gain in adaptive value, damage to the organ may kill the individual without giving regeneration a chance. However, a decrease in the adaptive value of an organ may also promote a decline in its regenerative capacity, as it happens with a multiplication of identical or similar structures, e.g., the alleviated capacity of limb regeneration in certain arachnids (Brautigam and Persons, 2003).
Regenerative capacity may also decrease in a pleiotropic manner. Galis et al. (2003) suggest that the regenerative capacity of vertebrate limbs evolves in connection with their embryonic development. In the case of the early onset of limb development, its formation coincides with basic morphogenetic events involving complex interactions of multiple embryonic structures. As a consequence, the limb develops under powerful inducing effects of somites, lateral plate mesoderm, etc., but not as a self-organizing structure. Accordingly, the regenerative capacity of the definitive limb is reduced (Galis et al., 2003).
When the onset of limb development is delayed until the completion of fundamental inductive interactions between the primary germ layer derivatives (somites, neural tube, etc.), the autonomously developing limbs will be regenerative. This concept can be illustrated by the delayed limb development in Caudata (whose capacity for limb regeneration is renowned). Opposite examples include the fins of sharks and lungfish, as well as the limbs of birds and mammals, which develop from early anlagen and regenerate poorly. At the same time, the concept does not account for the poor limb regeneration in Anura, whose limbs develop fairly late, but regenerate well in larvae only (Galis et al., 2003). However, adult Anura are not completely devoid of the ability to regenerate limbs: in Rana temporaria and Rana clamitans, limb regeneration can be obtained after additional damaging effects on the wound surface (Polezhaev, 1946), while in Xenopus laevis, the same effect can be achieved by blocking proton channels and limiting the duration of local immune responses (Adams et al., 2007; Fukazawa et al., 2009).
Close to the concept under consideration is the concept of modules, a network of genes that control the behavior of cells taken from evo-devo. Defining the concept of modularity is not a trivial task. In developmental biology, the hypothesis of modules assumes the division of a developing organism into functional or organizational subunits that have pronounced morphological isolation, for example, somites, or correspond to a certain part of the body of an adult, such as a limb kidney (Bolker, 2000). Raff (1996) listed the following module characteristics: it should have a discrete genetic specification, hierarchical organization, interactions with other modules, a particular physical location within a developing organism, and the ability to undergo transformations on both developmental and evolutionary time scales (Raff, 1996).
In connection with the problem of the evolution of regeneration, this concept implies the idea of developmental constraint, i.e., restraints on phenotype production due to limited interaction among modules. For example, an increase in the complexity of the structure at the histological level can prevent the propagation of gradients of morphogens or bioelectric signals, which can lead to a decrease in the regenerative capacity (Tiozzo and Copley, 2015).
The interplay of regeneration and immunity represents a special issue (Mescher et al., 2017). The advent of adaptive immunity apparently collided with the pronounced regenerative capacity. In the highly regenerative Caudata, many components of adaptive immunity are underdeveloped; for example, compared with tailless amphibians, they lack antiviral immunity (Cotter et al., 2008; Murawala et al., 2012). Significant upgrade of the adaptive immune system during metamorphosis in Anura is consistent with the observed decline in the regenerative capacity of the adult individuals compared with the larvae (Robert and Ohta, 2009; Godwin and Rosenthal, 2014). In Anura, the immune system undergoes significant developmental changes. Prior to metamorphosis, it is functionally immature, as indicated by larval repertoires of T cell and B cell receptors, low expression of MHC I, low levels of B cell-mediated responses and antibody production, the negligible activity of natural killer cells, and low activity of helper and killer T cells. Metamorphosis is associated with a significant upgrade of these indicators; in addition, it brings the capacity of MHC II-dependent activation of helper T cells (Robert and Ohta, 2009). The increase in activity of natural killer cells and T cells in tailless amphibians leads to enhanced antitumor and antiviral immunity, which apparently costs them their regenerative potential.
Similar patterns are observed in mammals, with the pronounced regenerative capacity (manifested in scarless wound healing and myocardial regeneration) confined to certain stages of fetal development (Porrello et al., 2011; Vivien et al., 2016). The pronounced regenerative capacity of fetal skin and myocardium can be associated with certain functional properties of the developing immune system. It has been demonstrated that during this period the body more readily develops a Th2-mediated anti-inflammatory response than pro-inflammatory reactions (Sattler and Rosenthal, 2016). The shifted balance apparently favors a full-value compensation of the defect in line with its immediate tissue environment rather than its replacement with fibrous tissue. Apart from the plausible role of T cell-mediated responses, the influence of innate immunity should be considered as well. The development of organs is accompanied by their colonization with macrophages of bone marrow origin as opposed to primary populations of embryonic macrophages, which may also affect the regenerative capacity (Epelman et al., 2014; Elchaninov et al., 2019, 2020). Apparently similar reasons explain the high skin regeneration in the spiny mouse, Acomys. So they have an almost complete absence of macrophages and a low level of pro-inflammatory cytokines in their skin wounds (Brant et al., 2016).
Thus, it can be noted that evolutionary maturation of the immune system leads to a decrease in the regenerative potential, as illustrated by the inability of frogs to regenerate limbs after metamorphosis, as well as the extinction of scarless healing of skin wounds in mammals.
The reverse correlation between adaptive immunity and regenerative capacity (Godwin et al., 2017) may reflect the important role of under-, trans-, or dedifferentiated cells in regeneration (considered in the next section). It has been suggested that the advanced adaptive immunity (characteristic of Anura, birds, and mammals) is poorly compatible with the presence of non-differentiated cells, which are considered compromised and become eliminated along with foreign cells. The constant immune pressure on the populations of cells with high differentiation potential negatively affects the regenerative capacity (Godwin et al., 2017).
Another reason for the decline in regenerative capacity may be the high energy cost of this process. In animals with a short lifespan, individuals invest more resources in reproduction, which leads to a decrease in regenerative potential; this apparently has happened to certain species of lizards (Fox and McCoy, 2000; Bernardo and Agosta, 2005). A similar relationship between reproduction and regeneration can be observed in species with asexual reproduction, e.g., annelids who have lost the capacity of anterior regeneration (Bely and Wray, 2001; Bely, 2010; Zattara and Bely, 2013). Regeneration may affect the development; for instance, it significantly delays the metamorphosis in fruit flies, cockroaches, butterflies, and crabs, which can also adversely affect survival (Suzuki et al., 2019). Another possible cause for the decline in regenerative capacity is warm-bloodedness (Goss, 1969), which is closely related to the evolution of adaptive immunity, hard skeleton (Wulff, 2006), and finite growth (Bely and Wray, 2001; Bely, 2010).
Elucidation of mechanisms that determine the decline of regenerative capacity is challenging, especially given the varying degree of such effects in the evolution. It was noted that in certain groups of animals, e.g., annelids, regeneration is reduced to wound healing, amphibians and fish tend to exhibit hypomorphic regeneration, whereas reptiles may show either decreased rates of recovery or confinement of repair to certain stages of ontogeny (Vorontsova and Liosner, 1960; Han et al., 2003, 2008; Seifert and Muneoka, 2018). In planarian Dendrocoelum lacteum, cross-cut at a certain level, tail fragments are incapable of regenerating the head. It has been found that the restriction is due to the uninhibited Wnt/b-catenin signaling in such fragments and that ectopic suppression of Wnt/b-catenin signaling makes them capable of anterior regeneration (Liu et al., 2013; Maden, 2018). Similarly, the lack of anterior regeneration observed in certain annelids has been associated with low expression of nanos (Bely and Sikes, 2010).
Differentiation Status as a Correlate of Regenerative Capacity
According to Weismann’s theory, the regenerative capacity decreases as the structural and functional organization becomes more complex. In other words, Weismann believed that complex structural patterns are poorly compatible with regeneration, which requires pronounced tissue plasticity and a sufficient degree of freedom for the reconstruction.
Despite the vagueness and controversy of the term “organization complexity” as applied to animals, differentiation plasticity of cells is certainly connected with regeneration capacity.
The terms «transdifferentiation», «dedifferentiation», and «redifferentiation» have a rich history of scientific usage. The issue of their exact meanings and, in general, whether their use makes sense, is still open. Despite the long controversy, the definitions vary. Literally, dedifferentiation is the loss of structural and functional specialization; accordingly, redifferentiation may be understood as reacquisition of its previous differentiated phenotype by a particular cell (Odelberg, 2004, 2005; Grigoryan, 2016). «Transdifferentiation» is a particularly controversial term. Some experts use it loosely, even to describe a transition between derivatives of the same germ layer, for example, the transition between cholangiocyte and hepatocyte (Michalopoulos, 2011). Others use it in a narrower sense, to describe a transition between germ layers; the examples include the transition of the coelomic epithelium into gut epithelium during gut regeneration in holothurians (Dolmatov et al., 2019) and the transition of pigment cells of the iris into epithelial cells of the lens (Grigoryan, 2016). «Dedifferentiation» implies explicit transition to a low-differentiated state with high proliferative activity. A classic example of dedifferentiation is observed during regeneration of the retina from the pigment epithelium in newts, during which the epithelial cells lose melanin granules, enter proliferation, and differentiate into neurons (Mitashov, 1996); the whole sequence, however, can be justly classified as redifferentiation or even transdifferentiation. Formation of the wound blastema during regeneration of newt limbs also involves dedifferentiation, with muscle fibers losing their striation and undergoing fragmentation to become myoblasts (Odelberg, 2005).
Differentiation plasticity of cells at the site of injury (or directed to it) is closely related to the extent of remodeling in response to damage, with the extremes termed morphallaxis («blastema-less» regeneration) and epimorphosis (which involves the formation of blastema). For instance, the diploblastic Hydra can be considered as an organism that is constantly in a state of regeneration (Sánchez Alvarado, 2000; Martínez and Bridge, 2012). In Hydra, non-differentiated pluripotent cells of the gastric column are constantly proliferating and changing their location within the body (Sánchez Alvarado, 2000; Bosch, 2007; Vogg et al., 2019). According to some expert opinions, these cells may be considered as a hidden permanent analog of the blastema. The constant «circulation» of such cells in Hydra’s body provides a reasonable alternative to their emergency accumulation at the site of damage (which would be an epimorphic feature). Moreover, the constant presence of non-differentiated progenitors enables the triggering of determination and differentiation processes immediately after damage, which is typical for morphallaxis (Sánchez Alvarado, 2000).
In triploblastic animals, the evolution of an expanded system of cell differentiation checkpoints posed critical restrictions on the pluripotency. In planarians (considered as the most primitive triploblastic animals), the only pluripotent cells are neoblasts. In the case of damage to the planarian body, neoblasts actively proliferate and form blastema. It is believed that the cells involved in the restoration of the entire body from a fragment have similar properties in different groups of animals (endowed with such capacity). These cells are marked with RNA/protein-rich structures referred to as nuage, germ plasm, or chromatoid bodies (nuGPCB) which typically contain the expression products of germline-associated genes of Vasa, Nanos, Piwi, Tudor, Pumilio, and Bruno families. In invertebrates, non-differentiated cells are also typically marked by high expression of PIWI/piRNA genes, which ensures genome stability (Tiozzo and Copley, 2015; Lai and Aboobaker, 2018).
In more complex triploblastic animals, e.g., tailed amphibians, the pluripotency is restricted even further. These animals lack a reserve of pluripotent cells, which emerge during regeneration as a result of dedifferentiation and transdifferentiation of the pre-existing differentiated cells (Alvarado and Tsonis, 2006; Brockes and Kumar, 2008; Li et al., 2015). In tailless amphibians and salamanders, the potency of accumulating non-differentiated cells in response to injury is dramatically reduced or restricted to the larval stages (Agata and Inoue, 2012). Relative contributions of dedifferentiation and transdifferentiation to regeneration remain disputable, partly due to the pluralism of definitions for these processes in different settings (Galliot and Ghila, 2010). The majority of experts agree that dedifferentiation and transdifferentiation characteristically occur during regeneration in Hydra, as well as during Wolffian regeneration of the lens in Caudata (Galliot and Ghila, 2010; Henry and Hamilton, 2018). Transdifferentiation of coelomic epithelial cells into enterocytes can be observed during regeneration in sea cucumbers (Dolmatov et al., 2019; Boyko et al., 2020). At the same time, the cells of regenerating limbs in tailed amphibians have been shown to retain their key differentiation determinants (Kragl et al., 2009; Slack, 2017).
According to a number of authors, the ability of cells to return to the cell cycle is closely related to the concept of cell plasticity (Galliot and Ghila, 2010). In the course of evolution in some animals, the regulation of the cell cycle became more complicated, the appearance of additional checkpoints, which in turn could cause a decrease in the regenerative capacity.
In the course of a comparative study of the mechanisms of regulation of the cell cycle, it was found that 23 cyclins are encoded in the Saccharomyces cerevisiae genome, which regulates six proline-directed serine/threonine protein kinases. Cdc28 is required for driving the cell cycle. The multifunctional kinase Pho85 regulates G1 progression and other intracellular processes. In humans, 13 members of the CDK-family (cyclin-dependent kinase) have been found to interact with 29 cyclins and cyclin-related proteins (Malumbres and Barbacid, 2005). A family of five proteins (known as Ringo or Speedy) has been found in vertebrates but not in S. cerevisiae, Caenorhabditis elegans, or Drosophila melanogaster (Nebreda, 2006).
It has been found that CDK7, CDK8, and CDK9 are not very different from their yeast orthologs. CDK4 and CDK6 first appeared in multicellular organisms. The increased number of cyclins in the mammalian genome has resulted in a large variety of CDK–cyclin complexes. However, only 10 cyclins (three D-type, two E-type, two A-type, and three B-type cyclins) are known to be directly involved in driving the mammalian cell cycle (Malumbres and Barbacid, 2009).
The control of the mitotic cycle in the nuclei of muscle fibers in Anamnia and mammals is carried out with the involvement of different amounts of regulatory proteins. It was found that in non-amniotic vertebrates, one INK4 gene functions, which is responsible for the synthesis of cyclin-dependent kinase inhibitor 2 (p16Ink4). At the same time, mammals have two Ink4 genes (Ink4a which produces p16INK4a, and ARF, and Ink4b which produces p15INK4b). p16INK4a and p15INK4b block cyclin-dependent kinases 4 and 6 (CDK4,6) activity under normal conditions. In mammals, there is an additional mechanism of inhibition of the cell cycle re-entry by alternate open reading frame (ARF) through tumor protein p53. Under normal conditions, maintenance of chromosomes 2 (MCM2) ubiquitinates p53 and targets it for destruction (Seifert et al., 2012).
Despite the limitations in proliferative potential and phenotypic plasticity, mammalian tissues present with certain examples of dedifferentiation. However, these examples are most often associated with pathological processes, to leave alone tumorigenesis. For example, under conditions of severe viral or toxic liver damage, cholangiocytes are prone to dedifferentiation, with subsequent redifferentiation to cholangiocytes or transdifferentiation to hepatocytes (Michalopoulos, 2011). Another effect of viral or toxic liver damage on cell differentiation status is the loss of lipid droplets by Ito cells and their transition to myofibroblasts (Unanue, 2007).
Conclusion
In the course of the evolution of certain animal taxa, more and more checkpoints were added to the regulation of the cell cycle and exit from it. These checkpoints are maintained by the expanded system of cyclins and cyclin-dependent kinases with associated gene-and-protein networks and circuits (Malumbres and Barbacid, 2009; Seifert et al., 2012). The establishment of complex multilevel control of the mitotic cycle was inevitably coupled to enhanced control of the differentiation status; this association represents a major cause for the decline in regenerative capacity in vertebrates. An eventual increase in the activity of metabolic processes in warm-blooded animals allowed neither the preservation of non-differentiated cells in sufficiently high numbers nor the massive waves of dedifferentiation fraught with tumorigenesis (Sánchez Alvarado, 2000; Li et al., 2015).
Regeneration is a complex and diversified process inherent to the life at different levels of its organization. For obvious reasons, morphologically advanced cases of regeneration (such as restoration of the entire body from a fragment or regeneration of amputated limbs) draw more attention than others. As a consequence, a limited number of regeneration model organisms are used for research: zebrafish, newts, hydra, and planaria. In this case, the same type of damage is very often used—amputation, which narrows our understanding of regeneration and its evolution. Almost nothing is known about the mechanisms of regeneration in such animals after toxic damage, viral or bacterial. This is often considered in the relevant sections of microbiology, toxicology, and is not taken into account by regeneration researchers.
The evolution of regeneration can be studied by various approaches (Vorontsova and Liosner, 1960; Bely and Nyberg, 2010). The methodology involves a reduction of the phenomenon to particular events assigned to different levels of the organization and classified accordingly, with appropriate accounting for their relative contributions in a single model. Moreover, it is obvious that the evolution of regeneration is not a unidirectional process. Despite a major trend of the decline in regenerative capacity with the increasing organizational complexity, the phenomenon is modified in a variety of ways and never completely eliminated. For instance, mammals, who have suffered a pronounced phylogenetic decline in regenerative capacity, are capable of restoring neither amputated limbs nor other external appendages (the repair is limited to wound healing). At the same time, regeneration of certain organs and structures in mammals is morphologically consistent and results in complete functional recovery; characteristic examples include the restoration of the auricle tissue after a perforating wound (Williams-Boyce and Daniel, 1986) and restoration of the liver mass after massive resections (Bangru and Kalsotra, 2020).
Evolutionary studies on regeneration involve overcoming certain biases. Regrettably, the studies on regenerative capacity are still linked to a limited number of animal models and species. Importantly, in natural habitats, the organs may be damaged by disease rather than mechanically, which dramatically affects the course of regeneration. Regeneration of pathologically altered organs has been experimentally studied in mammals; for other animal taxa, the corresponding data are fragmentary or missing.
Author Contributions
AE, GS, and TF contributed the text. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the Russian Science Foundation (Grant No. 17-15-01419).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, D. S., Masi, A., and Levin, M. (2007). H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development 134, 1323–1335. doi: 10.1242/dev.02812
Adell, T., Saló, E., Van Loon, J. J. W. A., and Auletta, G. (2014). Planarians sense simulated microgravity and hypergravity. Biomed Res. Int. 2014:679672. doi: 10.1155/2014/679672
Agata, K., and Inoue, T. (2012). Survey of the differences between regenerative and non-regenerative animals. Dev. Growth Differ. 54, 143–152. doi: 10.1111/j.1440-169X.2011.01323.x
Agata, K., Saito, Y., and Nakajima, E. (2007). Unifying principles of regeneration I: epimorphosis versus morphallaxis. Dev. Growth Differ. 49, 73–78. doi: 10.1111/j.1440-169X.2007.00919.x
Alvarado, A. S., and Tsonis, P. A. (2006). Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 7, 873–884. doi: 10.1038/nrg1923
Babaeva, A. G. (1989). Cellular and humoral immunity factors as regulators of regenerative morphogenesis. Ontogenez 20, 453–460.
Babaeva, A. G. (1990). Limfotsity kak reguliatory proliferatsii i differentsirovki kletok nelimfoidnykh organov. Vestn. Akad. Med. Nauk. SSSR 2, 43–45.
Bando, T., Mito, T., Hamada, Y., Ishimaru, Y., Noji, S., and Ohuchi, H. (2018). Molecular mechanisms of limb regeneration: insights from regenerating legs of the cricket gryllus bimaculatus. Int. J. Dev. Biol. 62, 559–569. doi: 10.1387/ijdb.180048ho
Bangru, S., and Kalsotra, A. (2020). Cellular and molecular basis of liver regeneration. Semin. Cell Dev. Biol. 100, 74–87. doi: 10.1016/j.semcdb.2019.12.004
Barr, J. I., Boisvert, C. A., Somaweera, R., Trinajstic, K., and Bateman, P. W. (2019). Re-regeneration to reduce negative effects associated with tail loss in lizards. Sci. Rep. 9:18717. doi: 10.1038/s41598-019-55231-6
Bateman, P. W., Fleming, P. A., Bateman, P. W., and Bennett, N. (2008). To cut a long tail short: a review of lizard caudal autotomy studies carried out over the last 20 years. J. Zool. 277, 1–14. doi: 10.1111/j.1469-7998.2008.00484.x
Baumiller, T. K., and Gahn, F. J. (2004). Testing predator-driven evolution with Paleozoic crinoid arm regeneration. Science 305, 1453–1455. doi: 10.1126/science.1101009
Bely, A. E. (2006). Distribution of segment regeneration ability in the Annelida. Integr. Comp. Biol. 46, 508–518. doi: 10.1093/icb/icj051
Bely, A. E. (2010). Evolutionary loss of animal regeneration: pattern and process. Integr. Comp. Biol. 50, 515–527. doi: 10.1093/icb/icq118
Bely, A. E., and Nyberg, K. G. (2010). Evolution of animal regeneration: re-emergence of a field. Trends Ecol. Evol. 25, 161–170. doi: 10.1016/j.tree.2009.08.005
Bely, A. E., and Sikes, J. M. (2010). Latent regeneration abilities persist following recent evolutionary loss in asexual annelids. Proc. Natl. Acad. Sci. U.S.A. 107, 1464–1469. doi: 10.1073/pnas.0907931107
Bely, A. E., and Wray, G. A. (2001). Evolution of regeneration and fission in annelids: insights from engrailed- and orthodenticle-class gene expression. Development 128, 2781–2791.
Bely, A. E., Zattara, E. E., and Sikes, J. M. (2014). Regeneration in spiralians: evolutionary patterns and developmental processes. Int. J. Dev. Biol. 58, 623–634. doi: 10.1387/ijdb.140142ab
Ben Khadra, Y., Sugni, M., Ferrario, C., Bonasoro, F., Oliveri, P., Martinez, P., et al. (2018). “Regeneration in stellate echinoderms: Crinoidea, Asteroidea and Ophiuroidea,” in Results and Problems in Cell Differentiation, eds M. Kloc and J. Kubiak (Cham: Springer), 285–320. doi: 10.1007/978-3-319-92486-1_14
Bernardo, J., and Agosta, S. J. (2005). Evolutionary implications of hierarchical impacts of nonlethal injury on reproduction, including maternal effects. Biol. J. Linn. Soc. 86, 309–331. doi: 10.1111/j.1095-8312.2005.00532.x
Bertemes, P., Grosbusch, A. L., and Egger, B. (2020). No head regeneration here: regeneration capacity and stem cell dynamics of Theama mediterranea (Polycladida, Platyhelminthes). Cell Tissue Res. 379, 301–321. doi: 10.1007/s00441-019-03094-8
Blacher, L. J., Irichimowitsch, A. I., Liosner, L. D., and Woronzowa, M. A. (1933). Resorptionsprozesse als quelle der formbildung - IX. Einfluss der mitogenetischen Strahlen auf die Geschwindigkeit der regeneration. Wilhelm Roux. Arch. Entwickl. Mech. Org. 127, 339–352. doi: 10.1007/BF01390722
Bolker, J. A. (2000). Modularity in development and why it matters to evo-devo. Am. Zool. 40, 770–776. doi: 10.1093/icb/40.5.770
Bolshakova, G. B. (2008). Cardiomyocyte proliferation in rat fetuses under normal conditions and after heart injury. Bull. Exp. Biol. Med. 145, 487–489. doi: 10.1007/s10517-008-0125-3
Bosch, T. C. G. (2007). Why polyps regenerate and we don’t: towards a cellular and molecular framework for Hydra regeneration. Dev. Biol. 303, 421–433. doi: 10.1016/j.ydbio.2006.12.012
Boyko, A. V., Girich, A. S., Tkacheva, E. S., and Dolmatov, I. Y. (2020). The Eupentacta fraudatrix transcriptome provides insights into regulation of cell transdifferentiation. Sci. Rep. 10:1522. doi: 10.1038/s41598-020-58470-0
Brant, J. O., Yoon, J. H., Polvadore, T., Barbazuk, W. B., and Maden, M. (2016). Cellular events during scar-free skin regeneration in the spiny mouse, Acomys. Wound Repair Regen. 24, 75–88. doi: 10.1111/wrr.12385
Brautigam, S. E., and Persons, M. H. (2003). The effect of limb loss on the courtship and mating behavior of the wolf spider Pardosa milvina (Araneae: Lycosidae). J. Insect Behav. 16, 571–587. doi: 10.1023/A:1027311625059
Brockes, J. P., and Kumar, A. (2008). Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 24, 525–549. doi: 10.1146/annurev.cellbio.24.110707.175336
Brodsky, W. Y., and Uryvaeva, I. V. (1977). Cell polyploidy: its relation to tissue growth and function. Int. Rev. Cytol. 50, 275–332. doi: 10.1016/S0074-7696(08)60100-X
Burton, P. M., and Finnerty, J. R. (2009). Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Dev. Genes Evol. 219, 79–87. doi: 10.1007/s00427-009-0271-2
Cao, P.-L., Kumagai, N., Inoue, T., Agata, K., and Makino, T. (2019). JmjC domain-encoding genes are conserved in highly regenerative metazoans and are associated with planarian whole-body regeneration. Genome Biol. Evol. 11, 552–564. doi: 10.1093/gbe/evz021
Carlson, B. M. (2005). Some principles of regeneration in mammalian systems. Anat. Rec. B. New Anat. 287, 4–13. doi: 10.1002/ar.b.20079
Cary, G. A., Wolff, A., Zueva, O., Pattinato, J., and Hinman, V. F. (2019). Analysis of sea star larval regeneration reveals conserved processes of whole-body regeneration across the metazoa. BMC Biol. 17:16. doi: 10.1186/s12915-019-0633-9
Chera, S., Ghila, L., Dobretz, K., Wenger, Y., Bauer, C., Buzgariu, W., et al. (2009). Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell 17, 279–289. doi: 10.1016/j.devcel.2009.07.014
Christen, B., Beck, C. W., Lombardo, A., and Slack, J. M. W. (2003). Regeneration-specific expression pattern of three posterior Hox genes. Dev. Dyn. 226, 349–355. doi: 10.1002/dvdy.10231
Clark, M. S., Dupont, S., Rossetti, H., Burns, G., Thorndyke, M. C., and Peck, L. S. (2007). Delayed arm regeneration in the antarctic brittle star Ophionotus victoriae. Aquat. Biol. 1, 45–53. doi: 10.3354/ab00004
Cotter, J. D., Storfer, A., Page, R. B., Beachy, C. K., and Voss, S. R. (2008). Transcriptional response of Mexican axolotls to Ambystoma tigrinum virus (ATV) infection. BMC Genom. 9:493. doi: 10.1186/1471-2164-9-493
Darnet, S., Dragalzew, A. C., Amaral, D. B., Sousa, J. F., Thompson, A. W., Cass, A. N., et al. (2019). Deep evolutionary origin of limb and fin regeneration. Proc. Natl. Acad. Sci. U.S.A. 116, 15106–15115. doi: 10.1073/pnas.1900475116
Das, S. (2015). Morphological, molecular, and hormonal basis of limb regeneration across pancrustacea. Integr. Compar. Biol. 55, 869–877. doi: 10.1093/icb/icv101
Davydov, K. N. (1903). Observations of the regeneration processes at Enteropneumata. Notes Acad. Sci. 22, 1–10.
Dolmatov, I. Y., Shulga, A. P., Ginanova, T. T., Eliseikina, M. G., and Lamash, N. E. (2019). Metalloproteinase inhibitor GM6001 delays regeneration in holothurians. Tissue Cell 59, 1–9. doi: 10.1016/j.tice.2019.05.006
Dupont, S., and Thorndyke, M. C. (2006). Growth or differentiation? Adaptive regeneration in the brittlestar Amphiura filiformis. J. Exp. Biol. 209, 3873–3881. doi: 10.1242/jeb.02445
Elchaninov, A., Lokhonina, A., Nikitina, M., Vishnyakova, P., Makarov, A., Arutyunyan, I., et al. (2020). Comparative analysis of the transcriptome, proteome, and miRNA profile of kupffer cells and monocytes. Biomedicines 8:627. doi: 10.3390/biomedicines8120627
Elchaninov, A. V., and Bolshakova, G. B. (2011a). Dynamics of hepatocyte proliferation in regenerating fetal rat liver. Bull. Exp. Biol. Med. 151, 374–377. doi: 10.1007/s10517-011-1334-8
Elchaninov, A. V., and Bolshakova, G. B. (2011b). Reparative regeneration of rat fetal liver after partial hepatectomy. Bull. Exp. Biol. Med. 150, 383–386. doi: 10.1007/s10517-011-1148-8
Elchaninov, A. V., and Bolshakova, G. B. (2012). Proliferation and cell death of hepatocytes in regenerating fetal rat liver. Cell Tissue Biol. 6, 485–489. doi: 10.1134/S1990519X12050070
Elchaninov, A. V., Fatkhudinov, T. K., Usman, N. Y., Kananykhina, E. Y., Arutyunyan, I. V., Makarov, A. V., et al. (2018). Dynamics of macrophage populations of the liver after subtotal hepatectomy in rats. BMC Immunol. 19:23. doi: 10.1186/s12865-018-0260-1
Elchaninov, A. V., Fatkhudinov, T. K., Vishnyakova, P. A., Lokhonina, A. V., and Sukhikh, G. T. (2019). Phenotypical and functional polymorphism of liver resident macrophages. Cells 8:32. doi: 10.3390/cells8091032
Elliott, S. A., and Sánchez Alvarado, A. (2013). The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2, 301–326. doi: 10.1002/wdev.82
Epelman, S., Lavine, K. J., and Randolph, G. J. (2014). Origin and functions of tissue macrophages. Immunity 41, 21–35. doi: 10.1016/j.immuni.2014.06.013
Ferrario, C., Ben Khadra, Y., Czarkwiani, A., Zakrzewski, A., Martinez, P., Colombo, G., et al. (2018). Fundamental aspects of arm repair phase in two echinoderm models. Dev. Biol. 433, 297–309. doi: 10.1016/j.ydbio.2017.09.035
Fox, S. F., and McCoy, J. K. (2000). The effects of tail loss on survival, growth, reproduction, and sex ratio of offspring in the lizard Uta stansburiana in the field. Oecologia 122, 327–334. doi: 10.1007/s004420050038
Fukazawa, T., Naora, Y., Kunieda, T., and Kubo, T. (2009). Suppression of the immune response potentiates tadpole tail regeneration during the refractory period. Development 136, 2323–2327. doi: 10.1242/dev.033985
Fumagalli, M. R., Zapperi, S., and La Porta, C. A. M. (2018). Regeneration in distantly related species: common strategies and pathways. NPJ Syst. Biol. Appl. 4:5. doi: 10.1038/s41540-017-0042-z
Galis, F., Wagner, G. P., and Jockusch, E. L. (2003). Why is limb regeneration possible in amphibians but not in reptiles, birds, and mammals? Evol. Dev. 5, 208–220. doi: 10.1046/j.1525-142X.2003.03028.x
Galliot, B., and Ghila, L. (2010). Cell plasticity in homeostasis and regeneration. Mol. Reprod. Dev. 77, 837–855. doi: 10.1002/mrd.21206
Garza-Garcia, A. A., Driscoll, P. C., and Brockes, J. P. (2010). Evidence for the local evolution of mechanisms underlying limb regeneration in salamanders. Integr. Comp. Biol. 50, 528–535. doi: 10.1093/icb/icq022
Geng, J., Gates, P. B., Kumar, A., Guenther, S., Garza-Garcia, A., Kuenne, C., et al. (2015). Identification of the orphan gene Prod 1 in basal and other salamander families. Evodevo 6:9. doi: 10.1186/s13227-015-0006-6
Ghosh, S., Roy, S., Séguin, C., Bryant, S. V., and Gardiner, D. M. (2008). Analysis of the expression and function of Wnt-5a and Wnt-5b in developing and regenerating axolotl (Ambystoma mexicanum) limbs. Dev. Growth Differ. 50, 289–297. doi: 10.1111/j.1440-169X.2008.01000.x
Godwin, J. W., Pinto, A. R., and Rosenthal, N. A. (2017). Chasing the recipe for a pro-regenerative immune system. Semin. Cell Dev. Biol. 61, 71–79. doi: 10.1016/j.semcdb.2016.08.008
Godwin, J. W., and Rosenthal, N. (2014). Scar-free wound healing and regeneration in amphibians: immunological influences on regenerative success. Differentiation 87, 66–75. doi: 10.1016/j.diff.2014.02.002
Goss, R. J. (1992). The evolution of regeneration: adaptive or inherent? J. Theor. Biol. 159, 241–260. doi: 10.1016/S0022-5193(05)80704-0
Grigoryan, E. N. (2016). High regenerative ability of tailed amphibians (Urodela) as a result of the expression of juvenile traits by mature animals. Russ. J. Dev. Biol. 47, 83–92. doi: 10.1134/S1062360416020041
Guder, C., Pinho, S., Nacak, T. G., Schmidt, H. A., Hobmayer, B., Niehrs, C., et al. (2006). An ancient Wnt-dickkopf antagonism in Hydra. Development 133, 901–911. doi: 10.1242/dev.02265
Gurley, K. A., Rink, J. C., and Sánchez Alvarado, A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323–327. doi: 10.1126/science.1150029
Han, M., Yang, X., Farrington, J. E., and Muneoka, K. (2003). Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development 130, 5123–5132. doi: 10.1242/dev.00710
Han, M., Yang, X., Lee, J., Allan, C. H., and Muneoka, K. (2008). Development and regeneration of the neonatal digit tip in mice. Dev. Biol. 315, 125–135. doi: 10.1016/j.ydbio.2007.12.025
Henry, J. J., and Hamilton, P. W. (2018). Diverse evolutionary origins and mechanisms of lens regeneration. Mol. Biol. Evol. 35, 1563–1575. doi: 10.1093/molbev/msy045
Karami, A., Tebyanian, H., Goodarzi, V., and Shiri, S. (2015). Planarians: an in vivo model for regenerative medicine. Int. J. Stem Cells 8, 128–133. doi: 10.15283/ijsc.2015.8.2.128
Kawamura, K., Sugino, Y., Sunanaga, T., and Fujiwara, S. (2008). Multipotent epithelial cells in the process of regeneration and asexual reproduction in colonial tunicates. Dev. Growth Differ. 50, 1–11. doi: 10.1111/j.1440-169X.2007.00972.x
Korotkova, G. P. (1988). Integratsionnye mekhanizmy i morfogenez (k probleme évoliutsii ontogeneza). Zh. Obshch. Biol. 49, 464–475.
Kragl, M., Knapp, D., Nacu, E., Khattak, S., Maden, M., Epperlein, H. H., et al. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. doi: 10.1038/nature08152
Lai, A. G., and Aboobaker, A. A. (2018). EvoRegen in animals: time to uncover deep conservation or convergence of adult stem cell evolution and regenerative processes. Dev. Biol. 433, 118–131. doi: 10.1016/j.ydbio.2017.10.010
Lailvaux, S. P., Reaney, L. T., and Backwell, P. R. Y. (2009). Dishonest signalling of fighting ability and multiple performance traits in the fiddler crab Uca mjoebergi. Funct. Ecol. 23, 359–366. doi: 10.1111/j.1365-2435.2008.01501.x
Li, Q., Yang, H., and Zhong, T. P. (2015). Regeneration across metazoan phylogeny: lessons from model organisms. J. Genet. Genom. 42, 57–70. doi: 10.1016/j.jgg.2014.12.002
Liosner, L. D., Woronzowa, M. A., and Kusmina, N. A. (1936). Regenerationspotenz der Knochenlosen Extremität. Wilhelm Roux. Arch. Entwickl. Mech. Org. 134, 738–750. doi: 10.1007/BF00576070
Liozner, L. D. (1975). Ob évoliutsii regeneratsionnoǐ sposobnosti zhivotnykh. Usp. Sovrem. Biol. 79, 459–467.
Liozner, L. D. (ed.) (1974). Organ Regeneration: A Study of Developmental Biology in Mammals (Studies in Soviet Science), 1st Edn, Berlin: Springer, doi: 10.1007/978-1-4684-8456-4
Liu, S.-Y., Selck, C., Friedrich, B., Lutz, R., Vila-Farré, M., Dahl, A., et al. (2013). Reactivating head regrowth in a regeneration-deficient planarian species. Nature 500, 81–84. doi: 10.1038/nature12414
Maden, M. (2018). The evolution of regeneration - Where does that leave mammals? Int. J. Dev. Biol. 62, 369–372. doi: 10.1387/ijdb.180031mm
Maginnis, T. L. (2006). The costs of autotomy and regeneration in animals: a review and framework for future research. Behav. Ecol. 17, 857–872. doi: 10.1093/beheco/arl010
Malumbres, M., and Barbacid, M. (2005). Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30, 630–641. doi: 10.1016/j.tibs.2005.09.005
Malumbres, M., and Barbacid, M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166. doi: 10.1038/nrc2602
Martin, P., and Parkhurst, S. M. (2004). Parallels between tissue repair and embryo morphogenesis. Development 131, 3021–3034. doi: 10.1242/dev.01253
Martindale, M. Q., and Henry, J. Q. (1995). Modifications of cell fate specification in equal-cleaving nemertean embryos: alternate patterns of Spiralian development. Development 121, 3175–3185.
Martínez, D. E., and Bridge, D. (2012). Hydra, the everlasting embryo, confronts aging. Int. J. Dev. Biol. 56, 479–487. doi: 10.1387/ijdb.113461dm
Martinez, V. G., Menger, G. J., and Zoran, M. J. (2005). Regeneration and asexual reproduction share common molecular changes: upregulation of a neural glycoepitope during morphallaxis in Lumbriculus. Mech. Dev. 122, 721–732. doi: 10.1016/j.mod.2004.12.003
Maruzzo, D., and Bortolin, F. (2013). “Arthropod regeneration,” in Arthropod Biology and Evolution: Molecules, Development, Morphology, ed. A. Minelli (Berlin: Springer-Verlag), 149–169. doi: 10.1007/978-3-642-36160-9_7
Mcgaw, I. J. (2006). Cardiovascular and respiratory responses associated with limb autotomy in the blue crab, Callinectes sapidus. Mar. Freshw. Behav. Physiol. 39, 131–141. doi: 10.1080/10236240600563321
Mehta, A. S., and Singh, A. (2019). Insights into regeneration tool box: an animal model approach. Dev. Biol. 453, 111–129. doi: 10.1016/j.ydbio.2019.04.006
Mescher, A. L., Neff, A. W., and King, M. W. (2017). Inflammation and immunity in organ regeneration. Dev. Comp. Immunol. 66, 98–110. doi: 10.1016/j.dci.2016.02.015
Michalopoulos, G. K. (2011). Liver regeneration: alternative epithelial pathways. Int. J. Biochem. Cell Biol. 43, 173–179. doi: 10.1016/j.biocel.2009.09.014
Mitashov, V. I. (1996). Mechanisms of retina regeneration in Urodeles. Int. J. Dev. Biol. 40, 833–844. doi: 10.1387/ijdb.8877458
Mito, T., Inoue, Y., Kimura, S., Miyawaki, K., Niwa, N., Shinmyo, Y., et al. (2002). Involvement of hedgehog, wingless, and dpp in the initiation of proximodistal axis formation during the regeneration of insect legs, a verification of the modified boundary model. Mech. Dev. 114, 27–35. doi: 10.1016/S0925-4773(02)00052-7
Monaghan, J. R., Stier, A. C., Michonneau, F., Smith, M. D., Pasch, B., Maden, M., et al. (2014). Experimentally induced metamorphosis in axolotls reduces regenerative rate and fidelity. Regeneration 1, 2–14. doi: 10.1002/reg2.8
Morgan, T. H. (1901). Regeneration and liability to injury. Science 14, 235–248. doi: 10.1126/science.14.346.235
Muneoka, K., and Dawson, L. A. (2020). Evolution of epimorphosis in mammals. J. Exp. Zool. Part B Mol. Dev. Evol. 2020:jez.b.22925. doi: 10.1002/jez.b.22925
Murawala, P., Tanaka, E. M., and Currie, J. D. (2012). Regeneration: the ultimate example of wound healing. Semin. Cell Dev. Biol. 23, 954–962. doi: 10.1016/j.semcdb.2012.09.013
Naya, D. E., Veloso, C., Muñoz, J. L. P., and Bozinovic, F. (2007). Some vaguely explored (but not trivial) costs of tail autotomy in lizards. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 146, 189–193. doi: 10.1016/j.cbpa.2006.10.014
Nebreda, A. R. (2006). CDK activation by non-cyclin proteins. Curr. Opin. Cell Biol. 18, 192–198. doi: 10.1016/j.ceb.2006.01.001
Odelberg, S. J. (2004). Unraveling the molecular basis for regenerative cellular plasticity. PLoS Biol. 2:20232. doi: 10.1371/journal.pbio.0020232
Odelberg, S. J. (2005). Cellular plasticity in vertebrate regeneration. Anat. Rec. Part B New Anat. 287, 25–35. doi: 10.1002/ar.b.20080
Polezhaev, L. W. (1946). The loss and restoration of regenerative capacity in the limbs of tailless amphibia. Biol. Rev. 21, 141–147. doi: 10.1111/j.1469-185X.1946.tb00319.x
Porrello, E. R., Mahmoud, A. I., Simpson, E., Hill, J. A., Richardson, J. A., Olson, E. N., et al. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. doi: 10.1126/science.1200708
Poss, K. D. (2010). Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 11, 710–722. doi: 10.1038/nrg2879
Raff, R. (1996). The Shape of Life: Genes, Development, and the Evolution of Animal Form, 1st Edn, Chicago: University of Chicago Press.
Ramos, M., Irschick, D. J., and Christenson, T. E. (2004). Overcoming an evolutionary conflict: removal of a reproductive organ greatly increases locomotor performance. Proc. Natl. Acad. Sci. U.S.A. 101, 4883–4887. doi: 10.1073/pnas.0400324101
Reaumur, R.-A. F. D. (1712). Sur les diverses reproductions qui se fontdans les Ecrivisses, les Omars, les Crabes, etc. et entre Autressur celles de leurs jambes et de Leurs ecailles. Mem. R. Sci. 12, 226–245.
Reddien, P. W., Bermange, A. L., Murfitt, K. J., Jennings, J. R., and Sánchez Alvarado, A. (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in Planaria. Dev. Cell 8, 635–649. doi: 10.1016/j.devcel.2005.02.014
Reddy, P. C., Gungi, A., Ubhe, S., Pradhan, S. J., Kolte, A., and Galande, S. (2019a). Molecular signature of an ancient organizer regulated by Wnt/β-catenin signalling during primary body axis patterning in Hydra. Commun. Biol. 2:434. doi: 10.1038/s42003-019-0680-3
Reddy, P. C., Gungi, A., and Unni, M. (2019b). Cellular and molecular mechanisms of hydra regeneration. Results Prob. Cell Differ. 68, 259–290. doi: 10.1007/978-3-030-23459-1_12
Reinhardt, B., Broun, M., Blitz, I. L., and Bode, H. R. (2004). HyBMP5-8b, a BMP5-8 orthologue, acts during axial patterning and tentacle formation in hydra. Dev. Biol. 267, 43–59. doi: 10.1016/j.ydbio.2003.10.031
Robert, J., and Ohta, Y. (2009). Comparative and developmental study of the immune system in Xenopus. Dev. Dyn. 238, 1249–1270. doi: 10.1002/dvdy.21891
Sánchez Alvarado, A. (2000). Regeneration in the metazoans: why does it happen? Bioessays 22, 578–590.
Sattler, S., and Rosenthal, N. (2016). The neonate versus adult mammalian immune system in cardiac repair and regeneration. Biochim. Biophys. Acta Mol. Cell Res. 183, 1813–1821. doi: 10.1016/j.bbamcr.2016.01.011
Seifert, A. W., Monaghan, J. R., Smith, M. D., Pasch, B., Stier, A. C., Michonneau, F., et al. (2012). The influence of fundamental traits on mechanisms controlling appendage regeneration. Biol. Rev. 87, 330–345. doi: 10.1111/j.1469-185X.2011.00199.x
Seifert, A. W., and Muneoka, K. (2018). The blastema and epimorphic regeneration in mammals. Dev. Biol. 433, 190–199. doi: 10.1016/j.ydbio.2017.08.007
Sidorova, V. F. (1964). Ob Osobennostiakh Vosstanovleniia i postnatal’nogo rosta nekotroykh. Usp. Sovrem. Biol. 57, 283–299.
Sidorova, V. F. (1978). The Postnatal Growth and Restoration of Internal Organs in Vertebrates. Littleton, MA: PSG Pub. Co.
Slack, J. M. (2017). Animal regeneration: ancestral character or evolutionary novelty? EMBO Rep. 18, 1497–1508. doi: 10.15252/embr.201643795
Sunderland, M. E. (2010). Regeneration: Thomas Hunt Morgan’s window into development. J. Hist. Biol. 43, 325–361. doi: 10.1007/s10739-009-9203-2
Suzuki, Y., Chou, J., Garvey, S. L., Wang, V. R., and Yanes, K. O. (2019). Evolution and regulation of limb regeneration in arthropods. Results Probl. Cell Differ. 68, 419–454. doi: 10.1007/978-3-030-23459-1_17
Tiozzo, S., and Copley, R. R. (2015). Reconsidering regeneration in metazoans: an evo-devo approach. Front. Ecol. Evol. 3:67. doi: 10.3389/fevo.2015.00067
Tokin, B. P. (1969). Asexual reproduction, somatic embryogenesis and regeneration. Zh. Obs. Biol. 30, 18–32.
Tremblay, A. (1744). Mémoires Pour Servir à l’histoire d’un Genre de Polypes d’eau Douce à bras en Forme de Cornes, 1st Edn, eds C. Jean and H. Verbeek Leyde (London: Nabu Press).
Unanue, E. R. (2007). Ito cells, stellate cells, and myofibroblasts: new actors in antigen presentation. Immunity 26, 9–10. doi: 10.1016/j.immuni.2007.01.001
Vivien, C. J., Hudson, J. E., and Porrello, E. R. (2016). Evolution, comparative biology and ontogeny of vertebrate heart regeneration. NPJ Regen. Med. 1:12. doi: 10.1038/npjregenmed.2016.12
Vogg, M. C., Beccari, L., Iglesias Ollé, L., Rampon, C., Vriz, S., Perruchoud, C., et al. (2019). An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. Nat. Commun. 10:312. doi: 10.1038/s41467-018-08242-2
Vorontsova, M., and Liosner, L. (1960). Asexual Propagation and Regeneration. New York, NY: Pergamon Press.
Whitehead, G. G., Makino, S., Lien, C.-L., and Keating, M. T. (2005). Fgf20 Is essential for initiating Zebrafish fin regeneration supporting online material. Science 310, 1957–1960. doi: 10.1126/science.1117637
Williams-Boyce, P. K., and Daniel, J. C. (1986). Comparison of ear tissue regeneration in mammals. J. Anat. 149, 55–63.
Wittlieb, J., Khalturin, K., Lohmann, J. U., Anton-Erxleben, F., and Bosch, T. C. G. (2006). Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 6208–6211. doi: 10.1073/pnas.0510163103
Wulff, J. L. (2006). Resistance vs recovery: morphological strategies of coral reef sponges. Funct. Ecol. 20, 699–708. doi: 10.1111/j.1365-2435.2006.01143.x
Zattara, E. E., and Bely, A. E. (2011). Evolution of a novel developmental trajectory: fission is distinct from regeneration in the annelid Pristina leidyi. Evol. Dev. 13, 80–95. doi: 10.1111/j.1525-142X.2010.00458.x
Zattara, E. E., and Bely, A. E. (2013). Investment choices in post-embryonic development: quantifying interactions among growth, regeneration, and asexual reproduction in the annelid Pristina leidyi. J. Exp. Zool. Part B Mol. Dev. Evol. 320, 471–488. doi: 10.1002/jez.b.22523
Zattara, E. E., and Bely, A. E. (2016). Phylogenetic distribution of regeneration and asexual reproduction in Annelida: regeneration is ancestral and fission evolves in regenerative clades. Invertebr. Biol. 135, 400–414. doi: 10.1111/ivb.12151
Zattara, E. E., Fernandez-Alvarez, F. A., Hiebert, T. C., Bely, A. E., and Norenburg, J. L. (2019). A phylum-wide survey reveals multiple independent gains of head regeneration in nemertea. Proc. R. Soc. B Biol. Sci. 286:20182524. doi: 10.1098/rspb.2018.2524
Keywords: evolution, regeneration, morphallaxis, epimorphosis, blastema, dedifferentiation
Citation: Elchaninov A, Sukhikh G and Fatkhudinov T (2021) Evolution of Regeneration in Animals: A Tangled Story. Front. Ecol. Evol. 9:621686. doi: 10.3389/fevo.2021.621686
Received: 26 October 2020; Accepted: 15 February 2021;
Published: 05 March 2021.
Edited by:
Stefano Tiozzo, Université Paris-Sorbonne, FranceReviewed by:
Eduardo E. Zattara, Indiana University Bloomington, United StatesAlexandra Bely, University of Maryland, College Park, United States
Copyright © 2021 Elchaninov, Sukhikh and Fatkhudinov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrey Elchaninov, elchandrey@yandex.ru
 Andrey Elchaninov
Andrey Elchaninov Gennady Sukhikh1
Gennady Sukhikh1  Timur Fatkhudinov
Timur Fatkhudinov