Diversification of Aeonium Species Across Macaronesian Archipelagos: Correlations Between Genome-Size Variation and Their Conservation Status
- 1Linking Landscape, Environment, Agriculture and Food (LEAF), Instituto Superior de Agronomia (ISA), Universidade de Lisboa, Lisbon, Portugal
- 2Faculdade de Ciências, Centre for Ecology, Evolution and Environmental Changes (cE3c), Universidade de Lisboa, Lisbon, Portugal
- 3Forest Research Center (CEF, Instituto Superior de Agronomia (ISA), Universidade de Lisboa, Lisbon, Portugal
- 4Jardín de Aclimatación de La Orotava, Instituto Canario de Investigaciones Agrarias (ICIA), Puerto de La Cruz, Spain
- 5Jardín Botánico Canario “Viera y Clavijo”-Unidad Asociada CSIC (Cabildo de Gran Canaria), Las Palmas de Gran Canaria, Spain
- 6Faculty of Life Sciences, Madeira Botanical Group (GBM), University of Madeira, Funchal, Portugal
- 7InBio, Research Network in Biodiversity and Evolutionary Biology, CIBIO-Azores, Pólo dos Açores, Ponta Delgada, Portugal
The rich endemic flora of the Macaronesian Islands places these oceanic archipelagos among the top biodiversity hotspots worldwide. The radiations that have determined the evolution of many of these insular lineages resulted in a wealth of endemic species, many of which occur in a wide range of ecological niches, but show small distribution areas in each of them. Aeonium (Crassulaceae) is the most speciose lineage in the Canary Islands (ca. 40 taxa), and as such can be considered a good model system to understand the diversification dynamics of oceanic endemic floras. The present study aims to assess the genome size variation within Aeonium distribution, i.e., the Macaronesian archipelagos of Madeira, Canaries and Cabo Verde, and analyse it together with information on distribution (i.e., geography and conservation status), taxonomy (i.e., sections), morphological traits (i.e., growth-form), geological data (i.e., island's geological age), and environmental variables (i.e., altitude, annual mean temperature, and precipitation). Based on extensive fieldwork, a cytogeographic screening of 24 Aeonium species was performed. The conservation status of these species was assessed based on IUCN criteria. 61% of the taxa were found to be threatened (4% Endangered and 57% Vulnerable). For the first time, the genome size of a comprehensive sample of Aeonium across the Macaronesian archipelagos was estimated, and considerable differences in Cx-values were found, ranging from 0.984 pg (A. dodrantale) to 2.768 pg (A. gorgoneum). An overall positive correlation between genome size and conservation status was found, with the more endangered species having the larger genomes on average. However, only slight relationships were found between genome size, morphological traits, and environmental variables. These results underscore the importance of characterizing the cytogenomic diversity and conservation status of endemic plants found in Macaronesian Islands, providing, therefore, new data to establish conservation priorities.
Introduction
The Macaronesian Region belongs to the Mediterranean basin hotspot, which comprises one of the 36 worldwide biodiversity hotspots (Myers et al., 2000 and updates at http://www.biodiversityhotspots.org). Macaronesia includes five volcanic archipelagos (i.e., Azores, Madeira, Selvages, Canaries, and Cabo Verde) located in the North-eastern Atlantic Ocean on both sides of the Tropic of Cancer. These oceanic islands comprise a great number of habitat types with contrasting ecological conditions due to their biogeographical context (Azores is quite isolated, ca. 1500 km west of Portugal, to ca. 100 km from Fuerteventura to Morocco), a broad range of altitudes (3718 m in Tenerife), and their wide range of geological ages (from ~0.27 My in Pico to 29.5 My in Selvages) (Juan et al., 2000; Carracedo et al., 2002; Ávila et al., 2016). They were never connected to the mainland and were colonized by long-distance dispersal (García-Verdugo et al., 2019). This contributes to an enormous floristic diversity with a high number of plant endemics (ca. 900 taxa), and a great prevalence of secondary woodiness (Carlquist, 1974), highlighting several large plant lineages [e.g., Aeonium alliance (four genera, ca. 61 species), Echium L. (27 species), Sideritis L. (subgenus Marrubiastrum Tourn. ex Moench, 23 species), Crambe L. (section Dendrocrambe DC., 14 species), and Sonchus alliance (six genera, ca. 31 species)] (Caujapé-Castells et al., 2017).
The biodiversity of the Macaronesian Islands has become one of the most threatened worldwide (Médail and Quézel, 1997) due to the generally restricted geographical distribution of the taxa (Romeiras et al., 2016a) and the threats that they are subjected to (e.g., habitat loss or alterations caused by human impact, invasive species, overexploitation of resources or climate change) (Caujapé-Castells et al., 2010). The combination of the above-mentioned features with the high endemicity index makes this region very important in terms of conservation (Whittaker and Fernández-Palacios, 2007).
Aeonium Webb & Berthel. (Crassulaceae, Sempervivoideae) is the most diverse and the largest plant lineage endemic to the Macaronesian Islands, with 44 insular taxa, of which 41 on the Canary Islands, two on Madeira, one on Cabo Verde, and the remaining species occur in Morocco (one species) and East Africa (two species) (Supplementary Table 1). Aeonium comprises a perennial succulent and rosette-forming genus endowed with a huge morphological, ecological, and physiological variation among the several species (Liu, 1989; Jorgensen and Olesen, 2001).
According to Bañares (2015), seven sections can be recognized (Figure 1): Aeonium sect. Aeonium comprises mainly woody branched subshrubs provided by leaves with glands in adaxial surface and ciliate margin, and ovoid inflorescences of yellow flowers; Aeonium sect. Leuconium includes woody subshrubs, with dense to sparse branches or single monocarpic rosetted plants, with variously reddish margins on glaucous and ciliate leaves, conical to semiglobose inflorescences, and whitish to reddish petals; Aeonium sect. Goochiae englobes herbaceous perennial plants with yellow or pink petals without nectaries; Aeonium sect. Chrysocome includes small densely branched subshrubs with conspicuous longitudinal brownish stripes on the leaf abaxial face, lax inflorescences with few yellow flowers without nectaries; Aeonium sect. Canariensia includes large branching rosettes with pubescent leaves at margin, lax multi-flowered inflorescences of pale-yellow flowers with nectaries; Aeonium sect. Petrothamnium includes herbaceous perennials with flowers of yellow petals, without nectaries; and Aeonium sect. Greenovia corresponding to branching-rosette or single-rosette monocarpic plants with leaves of hyaline margin, densely leafy, and compact umbels of yellow 18- to 32- merous flowers.

Figure 1. Morphological diversity in Aeonium s.l. sections. (A,B): Section Aeonium (A. arboreum subsp. holochrysum, and A. gorgoneum). (C,D): Section Canariensia (A. canariense subsp. canariense; and A. glandulosum). (E): Section Chrysocome (A. spathulatum). (F): Section Goochia (A. lindleyi subsp. lindleyi). (G,H): Section Greenovia (A. aureum; and A. diplocyclum). (I–L): Section Leuconium (A. davidbramwelii; A. gomerense; A. urbicum; and A. nobile). (M–O): Section Petrothamnium (A. glutinosum; A. saundersii; and A. sedifolium). All species pictured have been sampled in the current study and by some of the authors (MB; GR; MR).
Three species i.e., A. balsamiferum, A. gomerense, and A. saundersii, are protected by the Bern Convention (Appendix I) and two, i.e., A. gomerense and A. saundersii are listed as priority species by the “Habitat Directive” (92/43/EEC, Annex II). Only four species appear in the IUCN Red List: A. gorgoneum and A. gomerense as EN and A. saundersii and A. balsamiferum as VU (IUCN, 2020) whilst Moreno-Saiz and coord (2008), in the Spanish Red List using the IUCN criteria, included A. mascaense as EW (Extinct in Wild), eight species as VU and seven species near threatened (NT) (Supplementary Table 1).
Nevertheless, appropriate conservation actions should be taken to improve the status of the remaining taxa, which were not yet evaluated (Moreno-Saiz and coord, 2008; IUCN, 2020) (Supplementary Table 1). In addition, none of the Aeonium species is listed in the Appendices of CITES (the Convention on International Trade in Endangered Species of Wild Fauna and Flora; https://www.cites.org/eng) (Supplementary Table 1).
Knowing the distribution of genetic diversity is of chief importance for the conservation of native populations (Francisco-Ortega et al., 2000). Nevertheless, investigations of infraspecific variation and population structure, and therefore of recent eco-evolutionary processes and potential drivers for species divergence in Aeonium, have not taken place yet (Harter et al., 2015). Furthermore, the molecular phylogenies published by Mort et al. (2002) and by Thiv et al. (2010) are largely unresolved, and several clades show very weak support. There is thus a glaring need for additional studies using large-scale sampling and wide sets of molecular markers to understand the evolutionary patterns within the Macaronesian Region (Romeiras et al., 2019).
The use of genome size data is an approach in active growth for several research areas, like taxonomy, systematics, ecology, and evolutionary biology (Kron et al., 2007). In recent years, cytogenomic studies comprise estimates for 7058 angiosperms (Leitch et al., 2019). Across the group of angiosperms, the size of the nuclear genome varies greatly (e.g., Pellicer et al., 2018), ranging from 1C = 0.065 pg in Genlisea aurea A.St.-Hil. (Lentibulariaceae; Greilhuber et al., 2006) to 1C = 152.23 pg in Paris japonica Franch (Melanthiaceae; Pellicer et al., 2010). This variation is reported to be caused predominantly through polyploidy and amplification of non-coding repetitive DNA, especially retrotransposons (Schubert and Vu, 2016). These expansion processes are compensated by mechanisms that result in a decrease in genome size; however, this aspect is poorly understood, and it is prospectively hypothesized to be based in recombinational phenomena, such as homologous or illegitimate recombination (Grover and Wendel, 2010; Pellicer et al., 2018; Qiu et al., 2019). Moreover, several studies revealed relationships between genome size and seed weight (Chung et al., 1998), plant phenology (Grime and Mowforth, 1982), life history (Bennett, 1972), sensitivity to frost (MacGillivray and Grime, 1995), and mean temperature (Suda et al., 2003, 2005).
When examining the genome size variation across Macaronesian endemic plants, the limited range of 1C-values, from 0.19 pg to 9.52 pg (Suda et al., 2003, 2005; Leitch et al., 2019) emerges as a major signature. Since most of these taxa have a relatively small amount of DNA, DNA shrinkage might be evolutionarily advantageous in an insular context, considering the strong selective pressures acting in the Macaronesian archipelagos (Suda et al., 2005). Several studies point out that larger genomes are maladaptive (e.g., Lynch and Conery, 2003; Knight et al., 2005; Leitch et al., 2019;). This can be explained by the fact that DNA replication is slow and, consequently, also the cell cycle and individual development (Bennett, 1972). Larger genomes are also associated with populations of small effective sizes, where the effect of natural selection is not effective (Lynch et al., 1993). Vinogradov (2003) showed that threatened plant species, with declining populations, are characterized by average larger genomes and slower rates of diversification than their congeners, stating that the surplus of non-coding DNA seems to be correlated with the likelihood of extinction.
Cytogenomic studies by Suda et al. (2005) have estimated the genome size for 12 species of Aeonium, which ranges from 0.93 pg in A. dodrantale to 1.36 pg in A. lindleyi. These previous estimates furnish invaluable insight, yet many questions surrounding the evolution of genome size variation in this genus have not been satisfactorily resolved.
The present study aims to assess the genome size variation within the distribution of 24 Aeonium species native to the Macaronesian archipelagos of Madeira, Canaries and Cabo Verde, and analyse this along with information on the distribution (i.e., geography and conservation status), taxonomy (i.e., sections), morphological traits (i.e., growth-form), geological data (i.e., island's geological age), and environmental variables (i.e., altitude, annual mean temperature, and precipitation). We hypothesize that larger genome size (C-content) is positively correlated with the degree of threat, as it underlies a much lower resilience in the species and, therefore, it makes conservation actions mandatory.
Methods
Study Area
This study encompasses the full distribution area of Aeonium, i.e., the Macaronesian archipelagos of the Canaries, Madeira, and Cabo Verde (Figure 2). The Canary Islands is the largest archipelago of Macaronesia and includes seven main islands. It is characterized by a semi-arid climate, with Lanzarote and Fuerteventura being the driest islands (García-Herrera et al., 2003). Madeira archipelago consists of two inhabited islands, the island of Madeira and the island of Porto Santo. Madeira is characterized by a great variety of microclimates, including Mediterranean and temperate climates, while Porto Santo is Mediterranean, more homogeneous and predominantly arid (Menezes de Sequeira et al., 2013). Cabo Verde archipelago includes 10 islands and is the southernmost archipelago of Macaronesia, located c. 1350 km south-west of the Canary Islands (Figure 2). The climate of this archipelago is tropical dry, and the altitude and the north-east trade winds are key factors in shaping species distribution (Neto et al., 2020). The knowledge of the Cabo Verde flora lagged far behind the central Macaronesian archipelagos of the Canaries and Madeira, and there is still incomplete knowledge about the taxonomy and geographic distribution of most species (Romeiras et al., 2020).
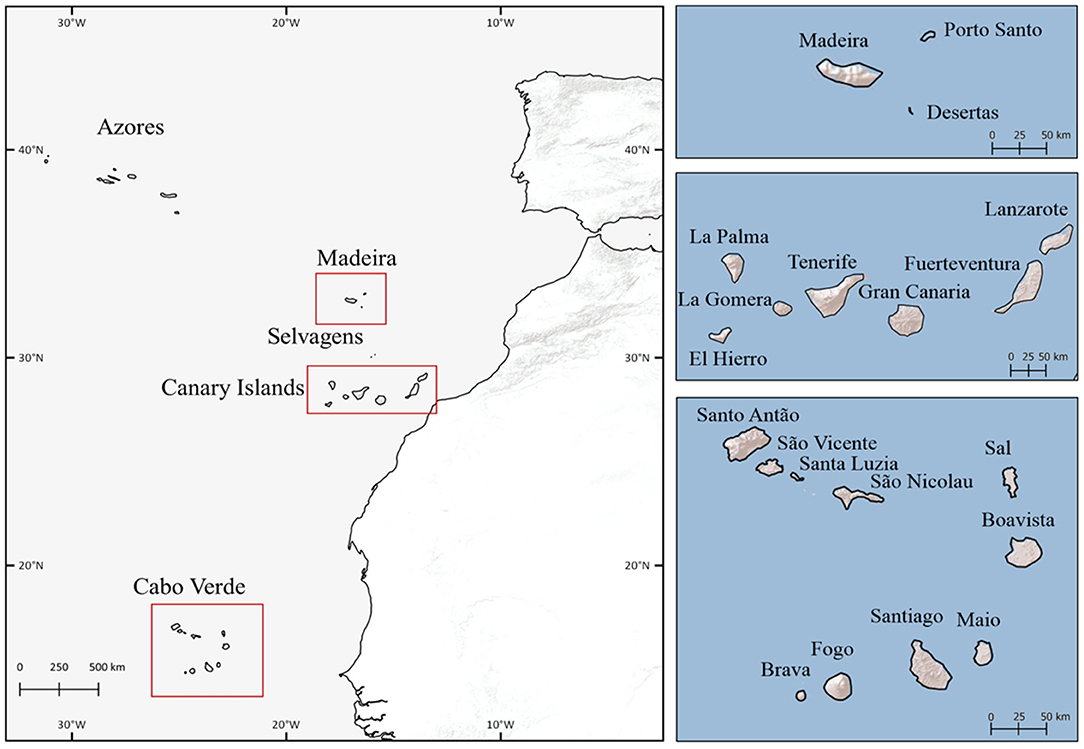
Figure 2. The five archipelagos of Macaronesia Region. The archipelagos included in study area are marked with a red line and represented in further detail in the right side.
Sampling
Sampling on the Canaries, Madeira and Cabo Verde took place from February to June 2019, involving the collection of 24 Aeonium species for cytogenomic analysis. For each species, three fresh leaves were collected from different individuals (n = 36) of each sampled location in the Canary, Madeira, and Cabo Verde Islands (Supplementary Table 2).
The distributional data were obtained from the data available on the Global Biodiversity Information Facility (GBIF; https://www.gbif.org/) and BIOTA (Gobierno de Canarias, 2020), i.e., 2772 records for 23 Aeonium species. The records without geographical coordinates, inaccurate taxonomic data, and duplicated records were discarded to avoid inaccurate predictions. The remaining records were projected and validated using Google Earth Pro 7.3.2.5491 and fieldwork screening. The final dataset, including the 2410 occurrence records belonging to 23 species were used in the analysis. Data concerning species ecology and threats was retrieved from bibliographic sources (Moreno-Saiz and coord, 2008; Bañares, 2015) and online databases (IUCN, 2020).
Herbarium vouchers are deposited in the Herbarium of Instituto Superior de Agronomia of the University of Lisbon, João de Carvalho e Vasconcellos (LISI), Botanical Museum of the University of Mainz—Herbarium (MJG), and Herbarium of the University of Basel (BAS).
Climatic and Geological Variables
The climatic variables [annual mean temperature (°C) and precipitation (mm/year)] used were extracted from CHELSA dataset version 1.2 (Climatologies at High Resolution for the Earth's Land Surface Areas, available at https://chelsa-climate.org/) (Karger et al., 2017). The island ages were obtained from Ávila et al. (2016).
Red List Assessments
The conservation status of the Aeonium species (at the specific level) from Canary and Madeira Islands were evaluated according to the IUCN Red List Categories and Criteria (IUCN Standards Petitions Committee, 2019), for further details see: http://www.iucnredlist.org/documents/RedListGuidelines.pdf. The native species of Cabo Verde (A. gorgoneum) was already assessed and classified as Endangered (Catarino et al., 2017) in the IUCN Red List. The assessments are based on five criteria to evaluate each taxon (A–E). According to our data, criterion B (i.e., restricted distribution and decline, fluctuations and/or fragmentation) and criterion D (i.e., very small or restricted populations) were the most used. Data/Information on population decline, fragmentation and decline in area, extent and quality of habitat were based on the authors' field experience and available data (e.g., Bañares, 2015; IUCN, 2020).
For each Aeonium species, we considered the number of subpopulations (quantified by the number of islands of occurrence of a taxon); the number of locations (estimated from the number of geographically or ecologically distinct areas of occurrence); the area of occupancy (AOO); and the extent of occurrence (EOO). For the last two parameters, we used the GeoCAT software (Bachman et al., 2011).
The conservation status assessment was performed in RAMAs Red List software v.2.0 (Akçakaya et al., 2001) according to IUCN Red List Criteria, ascribing the Red List category to each taxon considering the uncertainty that arises when several parameters are unknown (Akçakaya et al., 2000).
To keep consistency across evaluations, different attitudes toward risk and uncertainty can be taken by considering a precautionary or evidentiary attitude to risk based on risk tolerance (RT) parameter. RT ranges from 0 for risk-averse (precautionary attitude) to one for risk-prone (evidentiary attitude) (Akçakaya et al., 2000). As suggested to Romeiras et al. (2016a) for islands, a RT = 0.6, revealing an evidentiary attitude, was adopted.
Nuclear DNA Content Estimation
Nuclear DNA content was estimated performing a flow cytometry (FCM) analysis of propidium iodide-stained nuclei using Solanum lycopersicum L. (2C = 1.96 pg; DoleŽel et al., 1992) as standard. The fresh young leaves were chopped with a razor blade in a Petri dish containing 1 ml of Woody Plant Buffer (WPB 0,2 M Tris-HCl, 4 mM MgCl2, 1% Triton X-100, Na2EDTA 2 mM, NaCl 86 mM, sodium metabisulfite 20 mM, 1% PVP-10, pH 7.5; Loureiro et al., 2007). The nuclear suspension obtained was filtered with a 30 μm nylon filter and a 0.25 μg ml−1 solution of propidium iodide (PI; Sigma-Aldrich, USA) was added for DNA staining. The resulting mixture was analyzed in a flow cytometer CyFlow Space (Sysmex, Görlitz, Germany). The acquisition of numeric data and fluorescence graphs was made by Partec FloMax software v2.4d (Sysmex, Görlitz, Germany), as described by Guilengue et al. (2020). From the analyzed sample, the diploid quantity of DNA (in pg, per nucleus) was estimated using the formula:
Statistical Analysis
Statistical analyses were performed in R (R Core Team, 2019). Descriptive statistics were calculated for each sampled species, namely mean and standard deviation (SD) of the nuclear DNA amount (2C-value, pg). For the data on 2C-value, descriptive analysis was performed using the boxplot statistical algorithm performed by “ggplot2” package. Comparisons of genome size average values were subjected to analysis of variance (ANOVA) and the means were compared using the Tukey test (“TukeyHSD” function; “agricolae” package; de Mendiburu and de Mendiburu, 2019). The significance level for all statistical tests were set to the 95% confidence interval. Comparisons of genome size average values with growth-form, section, archipelago, island, and conservation status were performed using descriptive statistics, due to the highly unbalanced data distribution. Relationships between 2C-value and climate variables and islands' age were performed by Spearman-rank correlation coefficient.
Results
Conservation Status of Aeonium Species in Canary and Madeira Islands
The conservation status of Aeonium taxa in Canary and Madeira Islands are summarized in Table 1. Nine (39%) of the 23 Aeonium species assessed were classified as of Least Concern, and 14 (61%) were assigned in threat categories: one (4%) as Endangered and 13 (57%) as Vulnerable. Criterion D was applied, defining as Vulnerable a species where the number of mature individuals is <1000 and/or AOO <20 km2 or the number of known locations is <5 (Table 1). On the other hand, Criterion B attributed the category “Endangered” to one species (A. gomerense), those having a limited geographical range with AOO <20 km2 and/or EOO <200 km2, being single-island endemics (SIEs). The largest AOO and/or EOO values (Table 1) are displayed by A. arboreum and A. canariense; these species occur on five islands and were evaluated as LC. The smallest AOO and EOO values were for A. aizoon and A. volkerii, both included in the “Vulnerable” category due to the stability of the number of mature individuals within their populations. Nearly 61% of the species assessed (14) are single-island endemics (SIEs); within these, 50% (7 species) have AOO <20 km2 and EOO <200 km2.
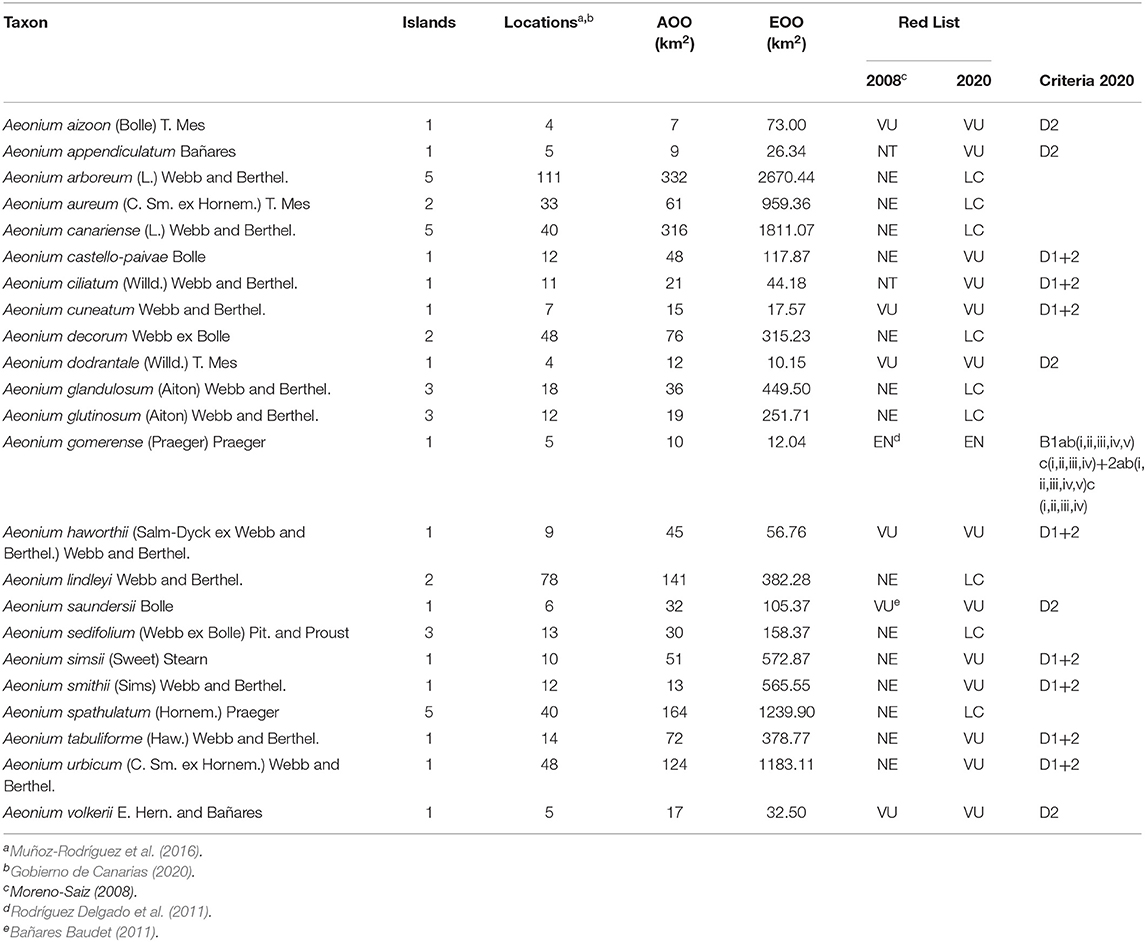
Table 1. Assessment of conservation status of the 23 Aeonium species from Canary and Madeira Islands and Red List categories, criteria, and the main parameters used.
The AOO value for the most threatened species in this assessment (A. gomerense, the only EN) is 10 km2, and for VU species it ranged from 7 to 124 km2. The EOO value is 12.04 km2 for the EN species and it ranged between 10.15 and 1183.11 km2 for the VU species (Table 1; Figure 3).
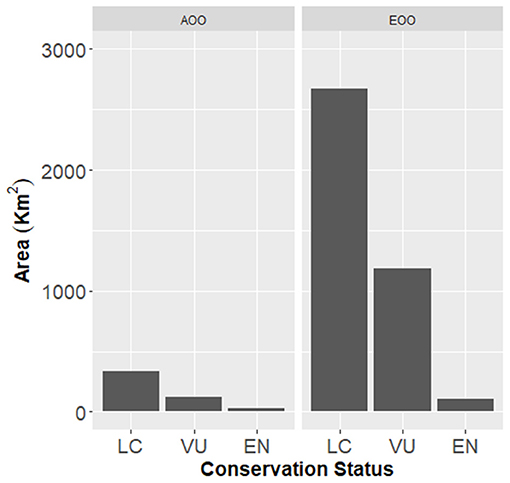
Figure 3. Species geographical range (AOO, area of occupancy; EOO, extent of occurrence) for the 23 Aeonium species classified under IUCN categories (LC, Least Concern; VU, Vulnerable; EN, Endangered).
The distribution of the 23 species evaluated shows that the two from the Madeira archipelago which occur in several islands are associated with the LC category. The remaining 21 species belong to the Canary archipelago, home to the largest number of threatened species, most of which are SIEs, which in turn have restricted AOO and EOO and are associated with the EN and VU threat categories, qualified by Criteria B and D, respectively. All species categorized as non-threatened (LC) have relatively high EOO values once they are well-distributed among several islands of the archipelago (Figure 3).
Compared to an assessment carried out previously with 39% (9) of the 23 species assessed here (Moreno-Saiz and coord, 2008; Rodríguez Delgado et al., 2011) (see Table 1), the conservation statuses emerging from our data are generally worsened, given the increase in the number of threatened species: the six evaluated as VU maintained their status; the two evaluated as NT passed to VU; the only one evaluated with EN, which underwent a re-evaluation in 2011, A. gomerense, maintained this category. Of the 14 species not evaluated previously, five are evaluated as VU and nine as LC (Figure 4). It should be noted that none of the species decreased its threat status, and the risk of extinction increased in the time interval between the two assessments.
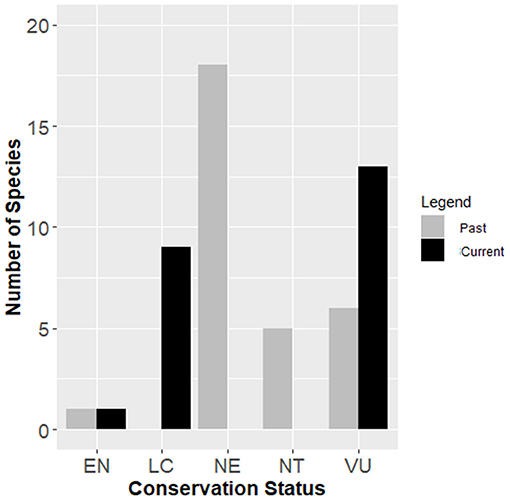
Figure 4. Status change of the Aeonium genus in the IUCN Red List from the previous inventories to the present assessment. IUCN Red List categories: NE, Not Evaluated; LC, Least Concern; NT, Near Threatened; VU, Vulnerable; EN, Endangered. Number of species assessed in each category (total assessments: nine in previous and 23 in the current). All species assessed in assessment are illustrated (see Table 1).
Genome Size Variation Across Aeonium Species
In the present study, we present the genome size estimations for 24 Aeonium species, of which ten had been previously evaluated by Suda et al. (2005): A. aizoon; A. arboreum; A. canariense; A. ciliatum; A. dodrantale; A. lindleyi; A. sedifolium; A. smithii; A. spathulatum, and A. tabuliforme. Notably, their results are consistent with our estimations. Table 2 shows the results on genome size, alongside growth-form, section and conservation status of these 24 Aeonium species endemic to Macaronesia.
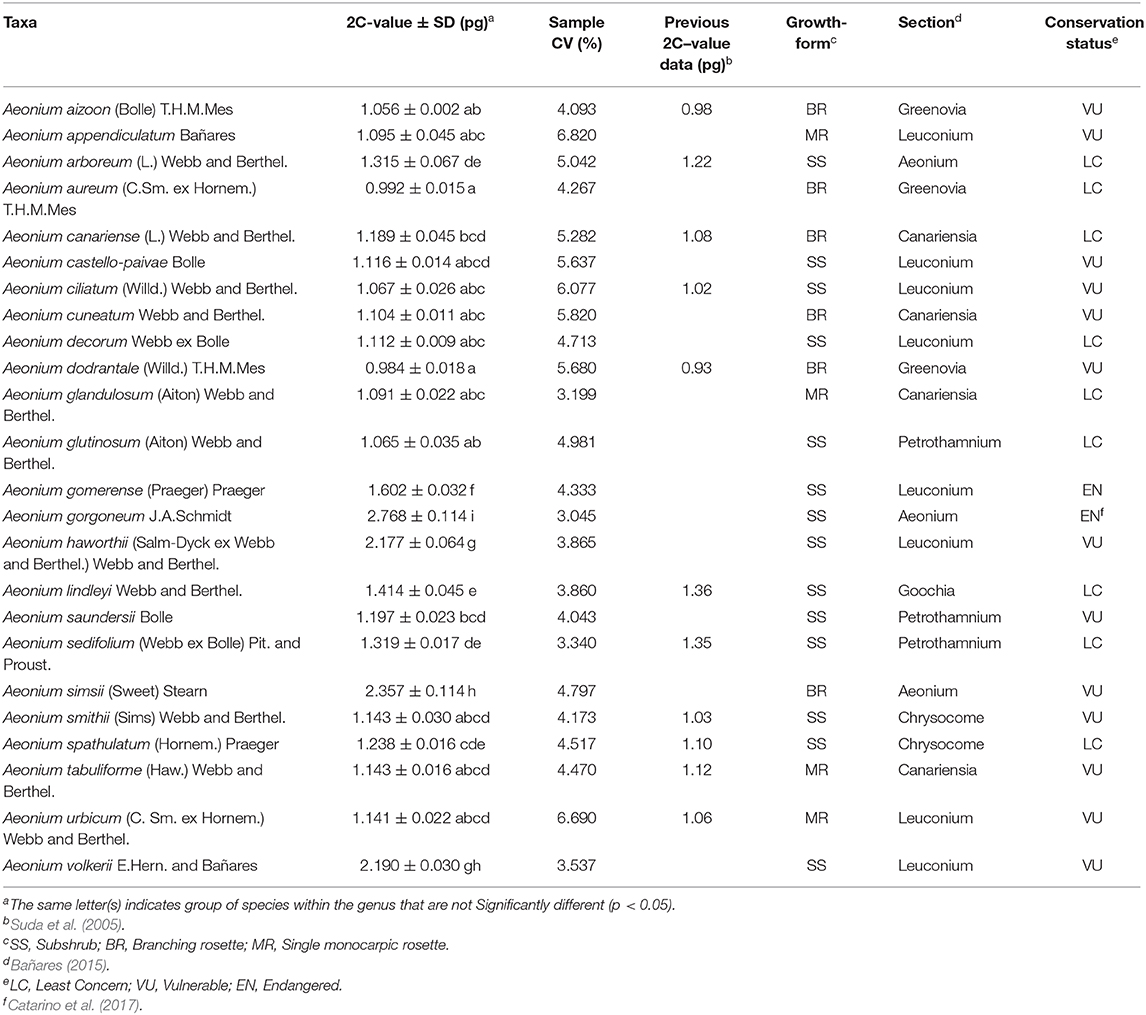
Table 2. Average 2C-value in picograms with standard deviation (SD), sample coefficient of variation, previous 2C-value estimates in picograms, growth-form, section, and conservation status of 24 Aeonium (Crassulaceae) taxa from Macaronesia.
Genome size determinations based on flow-cytometric techniques produced histograms of fluorescence of G0/G1 peaks with coefficients of variation (CV) ranging from 1.54% to 10.52% (mean 4.359%) for the taxa analyzed, and 0.76–7.18% (mean 3.367%) for internal standards. Figure 5 illustrates flow histograms presenting the final genome size estimation. According to Tavares et al. (2014), in order to eliminate partial nuclei and other types of debris, polygonal regions in dot-plots of SSC (side light scatter) vs. FL (fluorescence pulse integral in linear scale) was done to present more consistent nuclei in terms of size and shape. This approach improved CV values of DNA fluorescence peaks and, consequently, the quality of histograms yielded for the analyses of genome sizes.
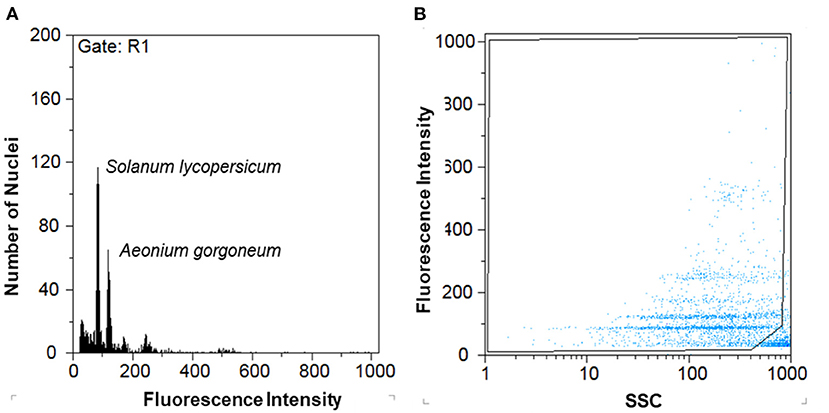
Figure 5. Flow cytometry outputs: (A) Flow cytometric histogram of relative fluorescence intensities from propidium iodide-stained Aeonium gorgoneum nuclei using Solanum lycopersicum (2C-value = 1.96 pg DNA) as an internal reference standard. (B) Dot-plot of SSC vs. FL presenting the gating made to exclude as much as possible partial nuclei and other types of debris.
The mean 2C nuclear DNA content for the sampled Aeonium species set range from 0.984 pg in A. dodrantale (Canary Islands) to 2.768 pg in A. gorgoneum (Cabo Verde archipelago). The genome size of Aeonium species is quite uniform (mean 1.596 ± 0.677 pg, n = 36), being A. gorgoneum, A. haworthii, A. simsii, A. volkerii, and A. gomerense the exceptions with the largest 2C-values (Table 2).
Figure 6A shows the genome size variation along the growth-forms: subshrubs (1.684 ± 0.673 pg, n = 23), branching-rosette (1.267 ± 0.488 pg, n = 7), and single-rosette monocarpic plants (1.111 ± 0.029 pg, n = 5). On average, the subshrubs are endowed with large genomes, followed by branching-rosette and single-rosette monocarpic plants.
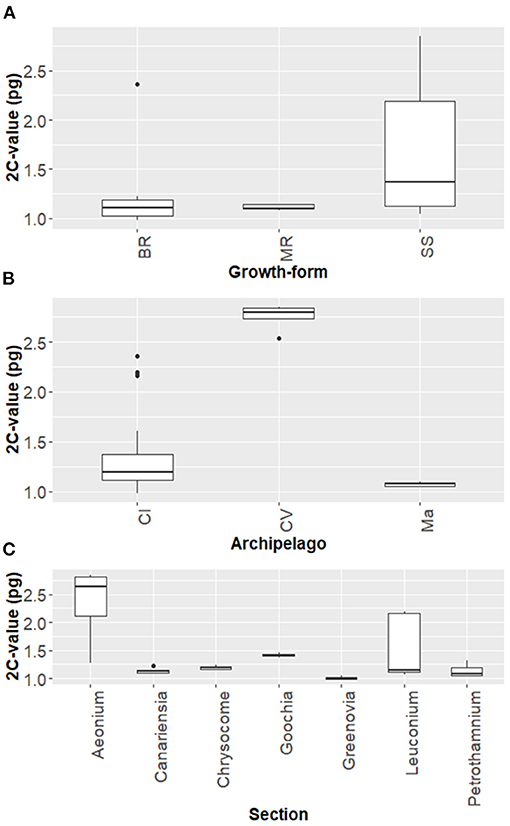
Figure 6. Boxplot diagram showing the genome size variation across 24 Aeonium species according to: (A) Growth-form (BR, Branching rosette; MR, Monocarpic rosette; SS, Subshrub); (B) Archipelago (CI, Canary Islands; CV, Cabo Verde; Ma, Madeira); and (C) Section; The gray box represents the 25th, 50th (median), and 75th percentiles, while whiskers represent the 10th and 90th percentiles with minimum and maximum observations. The black dots represent the outliers. Boxplots show 25th and 75th percentile, while error bars 1st and 99th percentile.
Comparison of mean diploid nuclear DNA content across the several studied archipelagos (Figure 6B) stand out differences between Cabo Verde and the other two studied archipelagos, with larger genomes found in Cabo Verde (2.750 ± 0.128 pg, n = 5), followed by Canary Islands (1.362 ± 0.411 pg, n = 25) and Madeira archipelago (1.072 ± 0.023 pg, n = 5). The Canary Islands harbor the greatest number of Aeonium species and the greatest range of mean 2C-values (0.984 pg in A. dodrantale to 2.357 pg in A. simsii). Moreover, a negative correlation between 2C-values and island geological age was found (r = −0.59; p < 0.001; n = 35).
Average 2C-values ± SD for the sectional division of Aeonium genus (Figure 6C): sect. Aeonium−2.342 ± 0.656 pg, sect. Canariensia−1.134 ± 0.053 pg, sect. Chrysocome−1.191 ± 0.067 pg, sect. Goochia−1.414 ± 0.058 pg, sect. Greenovia−1.011 ± 0.040 pg, sect. Leuconium−1.520 ± 0.522 pg, and sect. Petrothamnium−1.140 ± 0.118 pg. As shown, the sections Aeonium and Leuconium have a broad genome size variation, whilst Greenovia section have the lowest and less variable nuclear DNA amount.
Correlations between 2C-value and environmental variables (annual mean temperature and precipitation) and altitude were performed for the species from Tenerife Island, where the number of Aeonium species is higher. We found a negative tendency between 2C-value and annual mean precipitation (r = −0.73; p = 0.005; n = 13). On the other no significant tendencies respecting altitude (r = −0.14; p = 0.66; n = 13) and annual mean temperature (r = 0.025; p = 0.94; n = 13).
From the 24 Aeonium species analyzed, 15 were found to be threatened, being two species EN (A. gomerense and A. gorgoneum) and 13 species VU (See Table 2). The remaining eight species are categorized as LC. Figure 7 presents the differences of 2C – values within IUCN categories, highlighting that EN species (2.557 ± 0.482) have, on average, larger genome sizes, followed by VU (1.425 ± 0.530) and LC (1.190 ± 0.141) species with lower genome sizes.
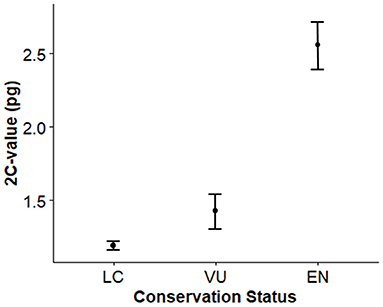
Figure 7. Differences in genome size among Aeonium species of different conservation status. LC, Least Concern; VU, Vulnerable; EN, Endangered.
Discussion
Conservation Status of Aeonium Species in the Macaronesian Islands
The Macaronesian flora has become one of the most threatened worldwide (e.g., 41% of Canaries' flora, 27% of Madeira's flora, and 78% of Cabo Verde endemic flora are endangered; Médail and Quézel, 1997; Romeiras et al., 2016b). Our study revealed that most assessed Aeonium species endemic to Macaronesia are single island endemics, with very restricted AOO and EOO, being A. gomerense classified as EN, and 13 taxa as VU. According to Lagabrielle et al. (2009), the small population sizes and limited geographical distribution ranges are the reasons for island endemic species to be among the most threatened in the world. These features have often been stimulated by limited habitat availability and by unique intrinsic biological traits resulting from prolonged evolutionary isolation, sometimes followed by secondary contact and hybridization (Caujapé-Castells et al., 2017).
Other threats affecting the Macaronesian floras are the gathering of plants for ornamental purposes, invasive alien species, grazing, and climate change (Moreno-Saiz et al., 2015; Romeiras et al., 2016b). For instance, A. gomerense is affected by the intensive grazing, competition with alien species [e.g., Opuntia ficus-indica (L.) Mill.], landslides, gathering for ornamental or selling purposes, and by hybridization with A. castello-paivae (Rodríguez Delgado et al., 2011). Also, A. saundersii populations are strongly affected by grazing (Bañares Baudet, 2011) while A. gorgoneum is threatened by gathering for medicinal purposes (Catarino et al., 2017).
According to the authors' field surveys experience, some species without previous conservation status assessment result to be threatened (i.e., more restricted in distribution and with smaller population effective sizes). For instance, the distribution of A. appendiculatum is limited by fire with A. decorum; A. smithii, and A. volkerii are threatened by harvesting for ornamental purposes; finally, A. glutinosum and A. glandulosum are threatened by harvesting for ornamental purposes (including the typical Madeira's flower festival), by introduction of Crassulaceae invasive species and possible hybridization or by competition in the same habitat, concreting and cleaning slopes, and the release of rabbits.
It should be noted that, although many of the species of Aeonium have a great ornamental interest, none of them is included in the Appendices of CITES (the Convention on International Trade in Endangered Species of Wild Fauna and Flora; https://www.cites.org/eng). The information on species conservation status is critical to prioritize conservation efforts, guiding the strategic expansion of the network of protected areas and the effective allocation of conservation resources to maximize the protection of Macaronesian biodiversity (Romeiras et al., 2016a). Several conservation actions have been already undertaken by managers to prevent species extinction, namely the establishment of Protected Areas (PA): within the Macaronesian islands. The Madeira archipelago has the largest proportion of protected area (67%), followed by the Canary archipelago with 40% and lastly Cabo Verde with only 0.16% of protected area (Caujapé-Castells et al., 2010; Romeiras et al., 2016b).
Regarding the Canary archipelago, all threatened Aeonium species analyzed are included in protected areas; however, a major effort is still needed in order to protect them effectively. Currently, the regional PA network encompasses 146 different areas (see https://www.gobiernodecanarias.org/planificacionterritorial/temas/informacion-territorial/enp/). Specifically, Anaga and Teno Rural Parks of Tenerife Island host the greatest endemic species richness in the Canary archipelago (Reyes-Betancort et al., 2008) and belong to the Important Plant Areas (IPA) as defined by del-Valle et al. (2004). However, this kind of protected area is managed mainly for sustainable uses of natural ecosystems, and it is not easy to reconcile human progress with conservation priorities. Furthermore, these areas also hold the greatest diversity of endemic and threatened Aeonium species, whose protection is critical for the conservation of range-restricted species from this genus. All things considered, there is an urgent need for creation and implementation of conservation measures in these regions with a special focus in Teno and Anaga that need a re-assessment of protected area status at least in some parts to reach effective protection of these species.
Genome Size Variation Across Aeonium Species
Aeonium is the most diverse and the largest endemic plant lineage in the Canary archipelago with 41 endemic taxa. Thus far, there is a low number of studies focusing on the cytogenomic variation in the island context of Macaronesia (but see Suda et al., 2003, 2005). Comparatively to Suda et al. (2005), the current study summarizes new data for 13 Aeonium species endemic to Canary Islands, Madeira and Cabo Verde, including: A. appendiculatum; A. aureum, A. castello-paivae, A. cuneatum, A. decorum, A. glandulosum, A. glutinosum, A. gomerense, A. gorgoneum, A. haworthii, A. saundersii, A. simsii, and A. volkerii (Table 2). At the species level, our estimates and those by Suda et al. (2005) make available 26 C-values for the genus, just missing nine of the currently recognized Macaronesian species (A. balsamiferum, A. davidbramwellii, A. diplocyclum, A. hierrense, A. lancerottense, A. percarneum, A. pseudourbicum, A. undulatum, and A. valverdense). Thus, this work substantially improves the global knowledge of nuclear DNA content in Aeonium, doubling the number of previously available C-value estimates for the biggest plant lineage of Macaronesia.
We found a range of mean 1C–values from 0.492 pg (A. dodrantale) to 1.384 pg (A. gorgoneum). According to genome size classes created by Leitch et al. (1998), all the present estimation of the haploid nuclear genome size (1C-value) is included in the ‘very small' class (<1.40 pg). As previously evidenced by Suda et al. (2005) for Canary Islands' endemic flora and considering the strong selective pressures of oceanic islands (see García-Verdugo et al., 2015), small genome sizes confer selective advantages because they reduce genetic instability. Thus, our results are cogent with the existing knowledge in bolstering the predominance of small genome sizes on islands.
According to the literature on Aeonium (e.g., Mes and Hart, 1996; Jorgensen and Olesen, 2001; Mort et al., 2002, 2007) and our field observations, all the sampled species show a great variation in growth-form. Our data do not reveal significant differences for genome size among the several perennial growth-forms. This finding is in line with Suda et al. (2005) observations for Aeonium that detect no differences in genome size among the perennial species. Our results follow the pattern described for angiosperms by Beaulieu et al. (2010), i.e., that woody species have small and less variable genome sizes, taking into account that distinct degrees of woodiness evolved in almost all the Aeonium genus (e.g., Mort et al., 2002).
The correlation of environmental factors and genome size has been debated for years and is of great interest to the scientific community (e.g., Suda et al., 2005). Macaronesian Islands, more specifically the Canary Islands, represent an ideal study case to understand the relationships between environmental factors and genome size. The great diversity of habitat types (e.g., from xerophytic to wet laurel forests and alpine zone) with particular climatic conditions justify the above-mentioned (e.g., Juan et al., 2000). Suda et al. (2005) results reveal that Aeonium genus showed relationships between genome size and environmental variables, specifically positive with altitude and annual mean temperature, and negative with annual mean precipitation. Furthermore, for Argyranthemum Webb (Asteraceae; Francisco-Ortega et al., 1995), Suda et al. (2003) also found significant positive and negative relationships between the size of the genome and the mean annual temperature and mean annual precipitation, respectively. The latter is in line with our result of a decrease of 2C-value with annual mean precipitation. On the contrary, we found no significant relationships with respect to altitude and annual mean temperature. However, further studies should be done with a more comprehensive sampling, that is, with a large number of replicates in several different climatic strata of the islands where the genus occurs.
By comparing the average nuclear DNA content for the several Macaronesian archipelagos, two groups can be distinguished corresponding to Cabo Verde (where the species with the largest genome occurs) and Canary/Madeira Islands. Our results are in agreement with the hypothesis that Aeonium has its center of origin in the Canary Islands, which host species with a much broader range of genome sizes that possibly colonized the other archipelagos and back-colonized African mainland (Mort et al., 2002; Thiv et al., 2010). A first possibility to explain this dichotomy is that the species with the largest genome size colonized the Cabo Verde archipelago, where the annual mean temperature is higher and, contrarily, the annual mean precipitation is lower than at the other archipelagos (Cropper and Hanna, 2014), thus agreeing with Suda et al. (2005) finding of a negative correlation between genome size and annual mean precipitation for Aeonium. Hence, we could assume that a larger genome is advantageous under warmer temperatures (Grime et al., 1985) and lower precipitation (e.g., Suda et al., 2003). Agreeing with our results, large-genome Berberis species prefer habitats providing higher temperatures and less rainfall (Bottini et al., 2000). Similar findings have been observed in the southernmost Atriplex halimus L. populations (Walker et al., 2005).
Another possible explanation for the larger genomes in Cabo Verde archipelago can be that it represents the most recent colonization of the genus (Kim et al., 2008) and, as such, it is bound to have undergone more demographic bottlenecks, and a faster accumulation of transposable elements (e.g., Brookfield and Badge, 1997). This possibility is also in accordance with the pattern of DNA content variation found in Hawaiian Schiedea Cham. & Schltdl., where the younger islands hosted the most recent colonization processes and have the largest genome sizes (Kapralov et al., 2009). Notably, our data reveal that the species that occur in the youngest islands of Cabo Verde (Santo Antão and São Vicente) also have the highest genome sizes.
Our genome size results seem to be compatible with the sectional segregation of Aeonium proposed by Bañares (2015). Thus, on average, the 2C values obtained appear to be distinct between sections, being Aeonium sect. Greenovia the most conserved section in terms of small genome size and the Aeonium sect. Aeonium the one with the largest and most variable genomes sizes. Furthermore, our results are also generally in line with the phylogeny of Mort et al. (2002), as it can be discerned that species that diverged recently tend to have a larger genome size, e.g.: A. haworthii (2.18 pg); A. gorgoneum (2.74 pg); A. volkeri (2.19 pg). Therefore, hybridization followed by reproductive isolation, might have contributed to the diversification of the Aeonium genus, as has been hypothesized by Fjellheim et al. (2009) for the evolution of the genus Argyranthemum. Larger genomes incur on added energetic replication costs, but the higher proportion of transposable elements that is associated with such larger genomes results in additional genetic diversity creation factors, which may help circumventing the genetic bottlenecks associated to recent speciation events, as denoted by diverse examples from the genera Beta (Monteiro et al., 2018) or Colletotrichum (Silva et al., 2012; Pires et al., 2016).
Our results support a direct relationship between degree of threat and genome sizes, whereby species with larger genomes are more vulnerable to extinction as pointed by Vinogradov (2003). Larger nuclear DNA contents are associated with larger cell size, implying slower replication of DNA and, consequently, a slower rate of cell division (e.g., Knight and Beaulieu, 2008; Pandit et al., 2014). These effects negatively influence plant development, and are associated with populations of small effective size (Lynch and Conery, 2003). Conversely, small genomes are associated with an increased colonization potential (Lavergne et al., 2010) and are usually found in widely variable (Knight et al., 2005) and/or ephemeral environments (Rejmánek, 1996). Furthermore, Aeonium species with larger amounts of nuclear DNA tend to have small fragmented populations of reduced effective size and therefore short geographic range size (e.g., A. haworthii, A. simsii, and A. volkerii). Given that island floras are extremely vulnerable to threats such as human interference and invasive species (Bramwell, 1990), species with smaller genomes would tend to be more resilient, although species with larger genomes may contain additional genetic diversity creation factors associated to transposable elements.
Our study emphasizes genome size as a potential tool for conservation, since species with a larger genome size appear to be less resilient and, as such, more prone to risk of extinction. Further efforts to both increase the sampling to perform other cytogenomic analyses, and more robust phylogenies are still necessary for a deeper and wider comprehension of species diversity, and for answering broader questions on evolutionary, ecological and conservation aspects of the largest plant lineage endemic of the Macaronesian Islands, particularly in the Canary Islands, where the genus has its center of origin, but also its most threatened and range-restricted taxa.
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
MR and MB: conceptualization. MB, PdS, and JR-B: field surveys (Canaries). MR: field surveys (Cabo Verde). GR and MS: field surveys (Madeira). MB, SC, JR-B, JC-C, and MR: conservation assessments. PT, GR, and MB: cytogenomic analysis. MB: results analysis and writing—original draft preparation. MB, JR-B, MS, JC-C, PT, and MR: writing—review and editing. All authors have reviewed and approved the submitted version of this manuscript.
Funding
This research was funded by Fundação para a Ciência e a Tecnologia (FCT) and Aga Khan Development Network (AKDN) under the project CVAgrobiodiversity/333111699. Fellowship to SC (SFRH/BD/120054/2016), PdS (PD/BD/128367/2017), and research units: UID/AGR/04129/2021 (LEAF) and UID/BIA/00329/2021 (cE3c) were funded by Portuguese National Funds through FCT, Portugal.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the supported provided by Fundação para a Ciência e a Tecnologia (FCT, Portugal) and Aga Khan Development Network (AKDN). Also our gratitude to the different regional and insular authorities to provide us with collection permits.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.607338/full#supplementary-material
References
Akçakaya, H. R., Ferson, S., Burgman, M. A., Keith, D. A., Mace, G. M., and Todd, C. R. (2000). Making consistent IUCN classifications under uncertainty. Conserv. Biol. 14, 1001–1013. doi: 10.1046/j.1523-1739.2000.99125.x
Akçakaya, H. R., Root, W. T., and Ferson, S. (2001). RAMAS® Red List: Threatened Species Classifications Under Uncertainty. Version 2.0. Setauket, NY: Applied Biomathematics.
Ávila, S. P., Melo, C., Berning, B., Cordeiro, R., Landau, B., and da Silva, C. M. (2016). Persististrombus coronatus (Mollusca: Strombidae) in the lower Pliocene of Santa Maria Island (Azores, NE Atlantic): paleoecology, paleoclimatology, and paleobiogeographic implications. Palaeogeogr. Palaeoclimatol. Palaeoecol. 441, 912–923. doi: 10.1016/j.palaeo.2015.10.043
Bachman, S., Moat, J., Hill, A. W., de la Torre, J., and Scott, B. (2011). Supporting Red List threat assessments with GeoCAT: geospatial conservation assessment tool. Zookeys 150, 117–126. doi: 10.3897/zookeys.150.2109
Bañares Baudet, A. (2011). Aeonium saundersii. The IUCN Red List of Threatened Species 2011: e.T162264A5566168. doi: 10.2305/IUCN.UK.2011-1.RLTS.T162264A5566168.en
Bañares, Á. (2015). Las plantas suculentas (Crassulaceae) endémicas de las Islas Canarias. Santa Cruz de Tenerife: Editorial Turquesa.
Beaulieu, J. M., Smith, S. A., and Leitch, I. J. (2010). On the tempo of genome size evolution in angiosperms. J. Bot. 2010:989152. doi: 10.1155/2010/989152
Bennett, M. D. (1972). Nuclear DNA content and minimum generation time in herbaceous plants. Proc. R. Soc. Lond. B. Biol. Sci. 181, 109–135. doi: 10.1098/rspb.1972.0042
Bottini, M. C. J., Greizerstein, E. J., Aulicino, M. B., and Poggio, L. (2000). Relationships among genome size, environmental conditions, and geographical distribution in natural populations of NW Patagonian species of Berberis L. (Berberidaceae). Ann. Bot. 86, 565–573. doi: 10.1006/anbo.2000.1218
Bramwell, D. (1990). Conserving biodiversity in the Canary Islands. Ann. Missouri Bot. Gard. 77, 28–37. doi: 10.2307/2399622
Brookfield, J. F., and Badge, R. M. (1997). Population genetics models of transposable elements. Genetica 100, 281–294. doi: 10.1023/A:1018310418744
Carlquist, S. (1974). Island Biology. New York, NY: Columbia University Press. doi: 10.5962/bhl.title.63768
Carracedo, J. C., Pérez Torrado, F. J., Ancochea, E., Meco, J., Hernán, F., Cubas, C. R., et al. (2002). “Cenozoic volcanism II: the Canary Islands,” in The Geology of Spain, eds W. Gibbons and T. Moreno (London: Geological Society Publishing House), 439–472.
Catarino, S., Duarte, M. C., and Romeiras, M. M. (2017). Aeonium gorgoneum. The IUCN Red List of Threatened Species 2017: e.T107356246A107468052. doi: 10.2305/IUCN.UK.2017-3.RLTS.T107356246A107468052.en
Caujapé-Castells, J., García-Verdugo, C., Marrero-Rodríguez, Á., Fernández-Palacios, J. M., Crawford, D. J., and Mort, M. E. (2017). Island ontogenies, syngameons, and the origins and evolution of genetic diversity in the Canarian endemic flora. Perspect. Plant Ecol. Evol. Syst. 27, 9–22. doi: 10.1016/j.ppees.2017.03.003
Caujapé-Castells, J., Tye, A., Crawford, D. J., Santos-Guerra, A., Sakai, A., Beaver, K., et al. (2010). Conservation of oceanic island floras: present and future global challenges. Perspect. Plant Ecol. Evol. Syst. 12, 107–129. doi: 10.1016/j.ppees.2009.10.001
Chung, J., Lee, J. H., Arumuganathan, K., Graef, G. L., and Specht, J. E. (1998). Relationships between nuclear DNA content and seed and leaf size in soybean. Theor. Appl. Genet. 96, 1064–1068. doi: 10.1007/s001220050840
Cropper, T. E., and Hanna, E. (2014). An analysis of the climate of Macaronesia, 1865–2012. Int. J. Climatol. 34, 604–622. doi: 10.1002/joc.3710
de Mendiburu, F., and de Mendiburu, M. F. (2019). Package ‘agricolae'. R package version, (Lima: Universidad Nacional Agraria La Molina), 1–2.
del-Valle, E., Maldonado, J., Sainz, H., and Sánchez de Dios, R. (2004). Áreas importantes para la flora amenazada española. Atlas y Libro Rojo de la Flora Vascular Amenazada de España. Madrid: Dirección General de Conservación de la Naturaleza.
DoleŽel, J., Sgorbati, S., and Lucretti, S. (1992). Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 85, 625–631. doi: 10.1111/j.1399-3054.1992.tb04764.x
Fjellheim, S., Jørgensen, M. H., Kjos, M., and Borgen, L. (2009). A molecular study of hybridization and homoploid hybrid speciation in Argyranthemum (Asteraceae) on Tenerife, the Canary Islands. Bot. J. Linn. Soc. 159, 19–31. doi: 10.1111/j.1095-8339.2008.00947.x
Francisco-Ortega, J., Jansen, R. K., Crawford, D. J., and Santos-Guerra, A. (1995). Chloroplast DNA evidence for intergeneric relationships of the Macaronesian endemic genus Argyranthemum (Asteraceae). Syst. Bot. 20, 413–422. doi: 10.2307/2419801
Francisco-Ortega, J., Santos-Guerra, A., Kim, S. C., and Crawford, D. J. (2000). Plant genetic diversity in the Canary Islands: a conservation perspective. Am. J. Bot. 87, 909–919. doi: 10.2307/2656988
García-Herrera, R., Gallego, D., Hernández, E., Gimeno, L., Ribera, P., and Calvo, N. (2003). Precipitation trends in the Canary Islands. Int. J. Climatol. 23, 235–241. doi: 10.1002/joc.870
García-Verdugo, C., Caujapé-Castells, J., and Monroy, P. (2019). How repeatable is microevolution on islands? patterns of dispersal and colonization-related plant traits in a phylogeographical context. Ann. Bot. 123, 557–568. doi: 10.1093/aob/mcy191
García-Verdugo, C., Sajeva, M., La Mantia, T., Harrouni, C., Msanda, F., and Caujapé-Castells, J. (2015). Do island plant populations really have lower genetic variation than mainland populations? effects of selection and distribution range on genetic diversity estimates. Mol. Ecol. 24, 726–741. doi: 10.1111/mec.13060
Gobierno de Canarias (2020). Biota. [online] Biodiversidadcanarias.es. Available online at: https://www.biodiversidadcanarias.es/biota/ (accessed May 19, 2020).
Greilhuber, J., Borsch, T., Müller, K., Worberg, A., Porembski, S., and Barthlott, W. (2006). Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biol. 8, 770–777. doi: 10.1055/s-2006-924101
Grime, J. P., and Mowforth, M. A. (1982). Variation in genome size - an ecological interpretation. Nature 299, 151–153. doi: 10.1038/299151a0
Grime, J. P., Shacklock, J. M. L., and Band, S. R. (1985). Nuclear DNA contents, shoot phenology, and species co-existence in a limestone grassland community. New Phytol. 100, 434–445. doi: 10.1111/j.1469-8137.1985.tb02792.x
Grover, C. E., and Wendel, J. F. (2010). Recent insights into mechanisms of genome size change in plants. J. Bot. 2010:382732. doi: 10.1155/2010/382732
Guilengue, N., Alves, S., Talhinhas, P., and Neves-Martins, J. (2020). Genetic and genomic diversity in a tarwi (Lupinus mutabilis Sweet) germplasm collection and adaptability to Mediterranean climate conditions. Agronomy 10:21. doi: 10.3390/agronomy10010021
Harter, D. E., Thiv, M., Weig, A., Jentsch, A., and Beierkuhnlein, C. (2015). Spatial and ecological population genetic structures within two island-endemic Aeonium species of different niche width. Ecol. Evol. 5, 4327–4344. doi: 10.1002/ece3.1682
IUCN (2020). The IUCN Red List of Threatened Species. Version 2020-1. Available online at: https://www.iucnredlist.org (accessed March, 19, 2020).
IUCN Standards Petitions Committee (2019). Guidelines for Using the IUCN Red List Categories and Criteria. Version 14. Prepared by the Standards and Petitions Committee. Available online at: http://www.iucnredlist.org/documents/RedListGuidelines.pdf (accessed March 11, 2020).
Jorgensen, T. H., and Olesen, J. M. (2001). Adaptive radiation of island plants: evidence from Aeonium (Crassulaceae) of the Canary Islands. Perspect. Plant Ecol. Evol. Syst. 4, 29–42. doi: 10.1078/1433-8319-00013
Juan, C., Emerson, B. C., Orom,i, P., and Hewitt, G. M. (2000). Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends Ecol. Evol. 15, 104–109. doi: 10.1016/S0169-5347(99)01776-0
Kapralov, M. V., Stift, M., and Filatov, D. A. (2009). Evolution of genome size in Hawaiian endemic genus Schiedea (Caryophyllaceae). Trop. Plant Biol. 2, 77–83. doi: 10.1007/s12042-009-9029-2
Karger, D. N., Conrad, O., Böhner, J., Kawohl, T., Kreft, H., Soria-Auza, R. W., et al. (2017). Climatologies at high resolution for the earth's land surface areas. Sci. Data 4:170122. doi: 10.1038/sdata.2017.122
Kim, S.-C., McGowen, M. R., Lubinsky, P., Barber, J. C., Mort, M. E., and Santos-Guerra, A. (2008). Timing and tempo of early and successive adaptive radiations in Macaronesia. PLoS ONE 3:e2139. doi: 10.1371/journal.pone.0002139
Knight, C. A., and Beaulieu, J. M. (2008). Genome size scaling through phenotype space. Ann. Bot. 101, 759–766. doi: 10.1093/aob/mcm321
Knight, C. A., Molinari, N. A., and Petrov, D. A. (2005). The large genome constraint hypothesis: evolution, ecology, and phenotype. Ann. Bot. 95, 177–190. doi: 10.1093/aob/mci011
Kron, P., Suda, J., and Husband, B. C. (2007). Applications of flow cytometry to evolutionary and population biology. Annu. Rev. Ecol. Evol. Syst. 38, 847–876. doi: 10.1146/annurev.ecolsys.38.091206.095504
Lagabrielle, E., Rouget, M., Payet, K., Wistebaar, N., Durieux, L., Baret, S., et al. (2009). Identifying and mapping biodiversity processes for conservation planning in islands: a case study in Réunion Island (Western Indian Ocean). Biol. Conserv. 142, 1523–1535. doi: 10.1016/j.biocon.2009.02.022
Lavergne, S., Muenke, N. J., and Molofsky, J. (2010). Genome size reduction can trigger rapid phenotypic evolution in invasive plants. Ann. Bot. 105, 109–116. doi: 10.1093/aob/mcp271
Leitch, I. J., Chase, M. W., and Bennett, M. D. (1998). Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann. Bot. 82, 85–94. doi: 10.1006/anbo.1998.0783
Leitch, I. J., Johnston, E., Pellicer, J., Hidalgo, O., and Bennett, M. D. (2019). Plant DNA C-values Database (release 7.1, April 2019). Available online at: http://data.kew.org/cvalues/ (accessed August 9, 2019).
Liu, H.-Y. (1989). Systematics of Aeonium (Crassulaceae) (No. 3). Taichung: National Museum of Natural Science Special publications.
Loureiro, J., Rodriguez, E., DoleŽel, J., and Santos, C. (2007). Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann. Bot. 100, 875–888. doi: 10.1093/aob/mcm152
Lynch, M., Bürger, R., Butcher, D., and Gabriel, W. (1993). The mutational meltdown in asexual populations. J. Hered. 84, 339–344. doi: 10.1093/oxfordjournals.jhered.a111354
Lynch, M., and Conery, J. S. (2003). The origins of genome complexity. Science 302, 1401–1404. doi: 10.1126/science.1089370
MacGillivray, C. W., and Grime, J. P. (1995). Genome size predicts frost-resistance in British herbaceous plants–Implications for rates of vegetation response to global warning. Funct. Ecol. 9, 320–325. doi: 10.2307/2390580
Médail, F., and Quézel, P. (1997). Hot-spots analysis for conservation of plant biodiversity in the Mediterranean basin. Ann. Missouri Bot. Gard. 84, 112–127. doi: 10.2307/2399957
Menezes de Sequeira, M., Capelo, J., Jardim, R., and Pupo, A. (2013). “Comments on the vascular flora of madeira: history, recent advances, and relations with other Macaronesian archipelagos,” in Proceedings of the Amurga International Conferences on Island Biodiversity, eds J. Caujapé-Castells, G. Nieto Feliner, and J. M. Fernández Palacios (Las Palmas de Gran Canaria: Fundación Canaria Amurga-Maspalomas), 228–236.
Mes, T. H. M., and Hart, H. T. (1996). The evolution of growth-forms in the Macaronesian genus Aeonium (Crassulaceae) inferred from chloroplast DNA RFLPs and morphology. Mol. Ecol. 5, 351–363. doi: 10.1046/j.1365-294X.1996.00090.x
Monteiro, F., Frese, L., Castro, S., Duarte, M. C., Paulo, O. S., Loureiro, J., et al. (2018). Genetic and genomic tools to assist sugar beet improvement: the value of the crop wild relatives. Front. Plant Sci. 9:74. doi: 10.3389/fpls.2018.00074
Moreno-Saiz, J. C., and coord. (2008). Lista Roja 2008 de la flora vascular española. Madrid: Dirección General de Medio Natural y Política Forestal (Ministerio de Medio Ambiente, y Medio Rural y Marino, y Sociedad Española de Biología de la Conservación de Plantas).
Moreno-Saiz, J. C. M., Lozano, F. D., Gómez, M. M., and Baudet, Á. B. (2015). Application of the red list index for conservation assessment of Spanish vascular plants. Conserv. Biol. 29, 910–919. doi: 10.1111/cobi.12437
Mort, M. E., Solits, D. E., Soltis, P. S., Santos-Guerra, A., and Francisco-Ortega, J. (2007). Physiological evolution and association between physiology and growth form in Aeonium (Crassulaceae). Taxon 56, 453–464. doi: 10.1002/tax.562016
Mort, M. E., Soltis, D. E., Soltis, P. S., Francisco-Ortega, J., and Santos-Guerra, A. (2002). Phylogenetics and evolution of the Macaronesian clade of Crassulaceae inferred from nuclear and chloroplast sequence data. Syst. Bot. 27, 271–288. doi: 10.1043/0363-6445-27.2.271
Muñoz-Rodríguez, P., Munt, D. D., and Saiz, J. C. M. (2016). Global strategy for plant conservation: inadequate in situ conservation of threatened flora in Spain. Israel Journal of Plant Sciences, 63, 297–308.
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Fonseca, G. A. B., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Neto, C., Costa, J. C., Figueiredo, A., Capelo, J., Gomes, I., Vitória, S., et al. (2020). The role of climate and topography in shaping the diversity of plant communities in Cabo Verde Islands. Diversity 12:80. doi: 10.3390/d12020080
Pandit, M. K., White, S. M., and Pocock, M. J. (2014). The contrasting effects of genome size, chromosome number, and ploidy level on plant invasiveness: a global analysis. New Phytol. 203, 697–703. doi: 10.1111/nph.12799
Pellicer, J., Fay, M. F., and Leitch, I. J. (2010). The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 164, 10–15. doi: 10.1111/j.1095-8339.2010.01072.x
Pellicer, J., Hidalgo, O., Dodsworth, S., and Leitch, I. (2018). Genome size diversity and its impact on the evolution of land plants. Genes 9:88. doi: 10.3390/genes9020088
Pires, A. S., Azinheira, H. G., Cabral, A., Tavares, S., Tavares, D., Castro, M., et al. (2016). Cytogenomic characterization of Colletotrichum kahawae, the causal agent of Coffee Berry Disease, reveals diversity in minichromosome profiles and genome size expansion. Plant Pathol. 65, 968–977. doi: 10.1111/ppa.12479
Qiu, F., Baack, E. J., Whitney, K. D., Bock, D. G., Tetreault, H. M., Rieseberg, L. H., et al. (2019). Phylogenetic trends and environmental correlates of nuclear genome size variation in Helianthus sunflowers. New Phytol. 221, 1609–1618. doi: 10.1111/nph.15465
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/ (accessed September 10, 2019).
Rejmánek, M. (1996). A theory of seed plant invasiveness: the first sketch. Biol. Conserv. 78, 171–181. doi: 10.1016/0006-3207(96)00026-2
Reyes-Betancort, J. A., Guerra, A. S., Guma, I. R., Humphries, C. J., and Carine, M. A. (2008). Diversity, rarity, and the evolution and conservation of the Canary Islands endemic flora. An. Jard. Bot. Madrid 65, 25–45. doi: 10.3989/ajbm.2008.v65.i1.244
Rodríguez Delgado, O., García Gallo, A., Cruz Trujillo, G. M., Carqué Álamo, E., and Marrero Gómez, M. V. (2011). Aeonium gomerense. The IUCN Red List of Threatened Species 2011: e.T162065A5545550. doi: 10.2305/IUCN.UK.2011-1.RLTS.T162065A5545550.en
Romeiras, M. M., Carine, M., Duarte, M. C., Catarino, S., Dias, F. S., and Borda-de-Água, L. (2020). Bayesian methods to analyze historical collections in time and space: a case study using Cabo Verde Endemic Flora. Front. Plant Sci. 11:278. doi: 10.3389/fpls.2020.00278
Romeiras, M. M., Catarino, S., Filipe, A. F., Magalhães, M. F., Duarte, M. C., and Beja, P. (2016a). Species conservation assessments in oceanic islands: the consequences of precautionary versus evidentiary attitudes. Conserv. Lett. 9, 275–280. doi: 10.1111/conl.12212
Romeiras, M. M., Catarino, S., Gomes, I., Fernandes, C., Costa, J. C., Caujapé-Castells, J., et al. (2016b). IUCN red list assessment of the Cape Verde endemic flora: towards a global strategy for plant conservation in Macaronesia. Bot. J. Linn. Soc. 180, 413–425. doi: 10.1111/boj.12370
Romeiras, M. M., Pena, A. R., Menezes, T., Vasconcelos, R., Monteiro, F., Paulo, O. S., et al. (2019). Shortcomings of phylogenetic studies on recent radiated insular groups: a meta-analysis using Cabo Verde biodiversity. Int. J. Mol. Sci. 20:2782. doi: 10.3390/ijms20112782
Schubert, I., and Vu, G. T. (2016). Genome stability and evolution: attempting a holistic view. Trends Plant Sci. 21, 749–757. doi: 10.1016/j.tplants.2016.06.003
Silva, D. N., Talhinhas, P., Cai, L., Manuel, L., Gichuru, E., Loureiro, A., et al. (2012). Host-jump drives rapid and recent ecological speciation of the emergent fungal pathogen Colletotrichum kahawae. Mol. Ecol. 21, 2655–2670. doi: 10.1111/j.1365-294X.2012.05557.x
Suda, J., Kyncl, T., and Freiová, R. (2003). Nuclear DNA amounts in Macaronesian angiosperms. Ann. Bot. 92, 153–164. doi: 10.1093/aob/mcg104
Suda, J., Kyncl, T., and Jarolímová, V. (2005). Genome size variation in Macaronesian angiosperms: forty percent of the Canarian endemic flora completed. Plant Syst. Evol. 252, 215–238. doi: 10.1007/s00606-004-0280-6
Tavares, S., Ramos, A. P., Pires, A. S., Azinheira, H. G., Caldeirinha, P., Link, T., et al. (2014). Genome size analyses of Pucciniales reveal the largest fungal genomes. Front. Plant Sci. 5:422. doi: 10.3389/fpls.2014.00422
Thiv, M., Esfeld, K., and Koch, M. (2010). “Studying adaptive radiation at the molecular level: a case study in the Macaronesian Crassulaceae-Sempervivoideae,” in Evolution in Action, ed M. Glaubrecht (Berlin: Springer), 35–59.
Vinogradov, A. E. (2003). Selfish DNA is maladaptive: evidence from the plant Red List. Trends Genet. 19, 609–614. doi: 10.1016/j.tig.2003.09.010
Walker, D. J., Monino, I., Gonzalez, E., Frayssinet, N., and Correal, E. (2005). Determination of ploidy and nuclear DNA content in populations of Atriplex halimus (Chenopodiaceae). Bot. J. Linn. Soc. 147, 441–448. doi: 10.1111/j.1095-8339.2004.00379.x
Keywords: conservation, cytogenomics, DNA flow cytometry, flora, endemic species, oceanic islands, RAMAS Red List, threatened species
Citation: Brilhante M, Roxo G, Catarino S, dos Santos P, Reyes-Betancort JA, Caujapé-Castells J, Sequeira MM, Talhinhas P and Romeiras MM (2021) Diversification of Aeonium Species Across Macaronesian Archipelagos: Correlations Between Genome-Size Variation and Their Conservation Status. Front. Ecol. Evol. 9:607338. doi: 10.3389/fevo.2021.607338
Received: 17 September 2020; Accepted: 12 January 2021;
Published: 04 February 2021.
Edited by:
Yongpeng Ma, Chinese Academy of Sciences, ChinaReviewed by:
Jing Yang, Chinese Academy of Sciences, ChinaPablo Vargas, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Copyright © 2021 Brilhante, Roxo, Catarino, dos Santos, Reyes-Betancort, Caujapé-Castells, Sequeira, Talhinhas and Romeiras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria M. Romeiras, mmromeiras@isa.ulisboa.pt
 Miguel Brilhante
Miguel Brilhante Guilherme Roxo
Guilherme Roxo Sílvia Catarino
Sílvia Catarino Patrícia dos Santos2
Patrícia dos Santos2  J. Alfredo Reyes-Betancort
J. Alfredo Reyes-Betancort Juli Caujapé-Castells
Juli Caujapé-Castells Miguel Menezes Sequeira
Miguel Menezes Sequeira Pedro Talhinhas
Pedro Talhinhas Maria M. Romeiras
Maria M. Romeiras