Using CRISPR as a Gene Editing Tool for Validating Adaptive Gene Function in Tree Landscape Genomics
- 1Department of Environmental Science Policy and Management, University of California, Berkeley, Berkeley, CA, United States
- 2Innovative Genomics Institute (IGI), Berkeley, CA, United States
Anthropogenic activities have substantially modified natural forested ecosystems around the world through species exploitation, land-use changes, soil degradation, pollution and introduction of exotic species. The impacts of these activities are being exacerbated today by climate change that is expected to become more severe over the coming decades. Modern landscape genomics has made advances in identifying genes that are associated with phenotypic expression, but they have been unable to prove that the associations are more than correlative. The threats to biological diversity raised by climate change, underscore the need to have an improved understanding of the genetic basis of phenotypic traits. In sedentary, long-lived tree species this becomes of utmost importance, as the success of populations is likely to depend, in large part, on existing standing genetic variation. The most recent technologies of gene editing (CRISPR/Cas9) promise to be an elegant approach that will move forest tree genomics to the next level, by allowing the rigorous testing of gene function and its role in the adaptation of trees to their environment. This perspectives paper looks at how genome editing technologies can be used to advance our understanding of the role genes play in adaptation to climate change in woody plants. We discuss the different CRISPR modes than can be used in studies of adaptive traits in perennial species.
Introduction
Understanding the genetic basis of adaptation is a fundamental question in ecology and evolution that has taken on new significance with the threat of climate change. Climate change is recognized as one of the most serious challenges to the future health of ecosystems worldwide (Pereira et al., 2010). Increasing temperatures have already led to changes in species' distributions (Gonzalez et al., 2010), modified phenology of biological processes that can disrupt ecosystem functioning (Parmesan, 2007; Cook et al., 2012) and led to increased extinction risk through decreases in population size (Cahill et al., 2013). The recent drought-related mortality of mixed conifer forests in the southern Sierra Nevada of California underscores the seriousness of climatic stress at a regional level (Potter, 2016). If climate continues to change as predicted (Settele et al., 2014), many populations will fall outside of their habitat niche and, in the absence of migration or adaptation, these populations will be extirpated. For long-lived trees, the potential to escape unfavorable climate through migration is limited and unlikely to lead to successful establishment of populations that track climatic changes (Aitken et al., 2008). Adaptation requiring new mutations will be unlikely for species with long generation times, so evolutionary success will depend on standing genetic variation. Earlier common garden studies have indicated that many tree species harbor substantial adaptive potential that is commonly spatially organized along climatic clines (Chiune et al., 2006). However, little is known of the underlying molecular regulation of phenotypic adaptation to climate variables.
The study of adaptive variation in forest tree populations has a long history, with formal provenance studies dating to the Eighteenth century. Modern landscape genomics has brought this rich history to the modern era of molecular biology, with a view to understanding the evolution and interaction of natural selection and past demographic processes that have led to the genetic architecture among populations today. Advances offered by next generation sequencing (NGS) have allowed an unprecedented opportunity to gain whole genome data that should provide new tools to overcome many of the problems faced in understanding the genetic basis of adaptation to environmental variation in forest trees. Recent publications of whole genome sequences of an important number of forest tree species (Tuskan et al., 2006; Neale et al., 2014; Plomion et al., 2015) represent a major advance in providing new tools for identifying the genes and/or genomic intervals controlling adaptive traits for ecology and evolution in forest tree species. This has led to the adoption of two broad approaches to identifying the molecular basis of adaptation in non-domesticated plants: (a) genome wide association studies (GWAS) and, (b) sequence variation in candidate genes associated with climate adaptation. By exploring correlations between molecular markers such as single nucleotide polymorphisms (SNPs) and climatic gradients, the GWAS approach aims to detect genomic regions affected by spatially divergent selection (Neale and Kremer, 2011). However, massive resequencing needed for population studies requires genome reduction methods, with the result that some genomic regions that harbor adaptive variation escape documentation. The candidate gene approach offers the advantage that targeted sequencing can permit large scale multiplexing of samples to detect sequence variations associated with climatic gradients. However, results from both GWAS and the candidate gene approach are only correlative with environmental stress. Today, the challenge for ecological geneticists is to prove the role of allelic variants on fitness (Barret and Hoekstra, 2011) through testing and validation of candidate genes with their hypothesized phenotype. To address these new challenges, gene editing, such as genetic knockouts and gene transformation, promises to become a very attractive alternative to association studies in demonstrating direct links between candidate gene function and phenotypic traits for plant adaptation to natural environments (Busov et al., 2003). Although there are significant obstacles to working with woody plants, such as the long generation time, we believe that gene editing will become a vital tool for understanding and managing the genetic variants that play a role in adaptation to climatic stress and the challenges that trees will face by climate change (Nellemann et al., 2009).
Gene Editing Technologies
The new and promising genome editing (GE) systems developed in the last few years represent a great opportunity for molecular ecologists to achieve target-specific manipulation of genes of interest in the genomes of non-model plants. Genome editing technologies using ZFN (zinc finger nuclease) and TALEN (transcription activator-like effector nuclease) can generate genome modifications. However, these technologies are, either labor intensive, or expensive as the targeting mechanisms are all based on protein-nucleic acid interactions, thereby requiring a custom-designed protein for each gene locus of interest. Recent advances in the study of prokaryotic adaptive immune systems provide an alternative genome editing strategy (Mussolino and Cathomen, 2013).
In the late 1980s, a group of researchers described a strange genomic topology consisting of 32 nucleotides of unique sequence. This was the first known description of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR; Ishino et al., 1987). It was 17 years before additional work proposed that CRISPR loci act together with Cas genes to provide adaptive immunity (Mojica et al., 2009). In 2012, the molecular mechanism by which Cas proteins and CRISPR arrays worked, was elucidated by the Doudna group (Jinek et al., 2012) and the potential of CRISPR-Cas systems was realized. The first demonstration of RNA-guided mutation in eukaryotic cells occurred in 2013 (Cong et al., 2013). The CRISPR/Cas9 system is recognized now as an extremely potent editing technology to induce targeted DBS in eukaryotes, which previously could only be accomplished with considerable difficulty. In addition, this approach has led numerous research groups to recognize the robustness, affordability and ease of engineering with this RNAi-based technique. Recently, CRISPR/Cas9 has emerged as the most important tool for genome engineering due to its simple structure, design flexibility, high efficiency, and its applicability to a wide of range organisms. This technology plays an important role in genetic engineering, which is the reason why the CRISPR/Cas9 system has screen platforms that can be operated in various formats, such as the knockout, knockdown and activation screens. With this approach it is possible to edit single to multiple genes by knocking-in or knocking-out genes of a host genome. Thus, introgression of multiple traits or modification of metabolic pathways can be achieved, where it is possible to edit several genes simultaneously by the introduction of DSBs (double stranded breaks) at several sites (Mao et al., 2013). Since 2012, when it was recognized that CRISPR/Cas9 could be used for targeted genome editing and provide a means to introduce precise modifications at a desired locus in the genome of any living organism (Jinek et al., 2012), the number of publications has risen sharply. More than 2,300 publications pertaining to CRISPR were published within the period 2012–2016.
CRISPR/Cas9 in Perennial Species
The application of CRISPR/Cas9 genome editing in plants has lagged behind that of other organismal groups, although it offers great promise in the study of plant functional genomics and in the development of novel phenotypes in economically important crop plants. To date, the technology has been successfully applied to a number of model herbaceous systems including Arabidopsis thaliana and Nicotiana benthamiana (Li et al., 2013), Oryza sativa (Noman et al., 2016), tomato (Ueta et al., 2017), potato (Wang et al., 2015), soybean (Jacobs et al., 2014), and Zea mays (Svitashev et al., 2015). Its application in woody species are few and limited to those that are well characterized by having whole genome sequences and efficient transformation systems, such as Populus (Fan et al., 2015), Citrus sinensis (Jia and Wang, 2014), Duncan grapefruit (Jia et al., 2016) and Malus domestica (Nishitani et al., 2016). See Table 1 for list of tree species successfully transformed) Although the application of CRISPR/Cas9 is likely to be focused initially on commercially important crop plants, the increasing number of undomesticated species for which whole genomes are being sequenced will allow the technology to be used more broadly across the plant kingdom (see Figure 1 for pipeline adapted to evolutionary and ecological studies).
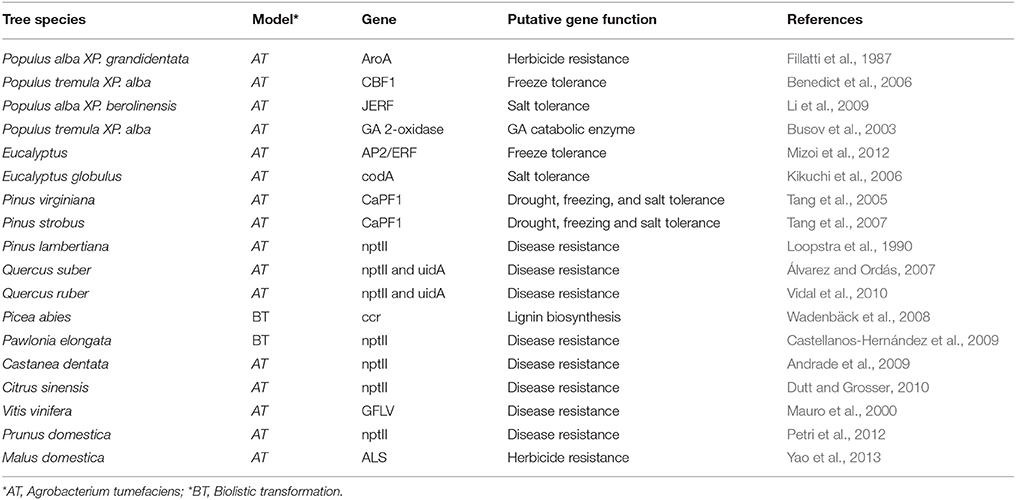
Table 1. List of tree species successfully transformed genetically and potential candidates to be genome edited with CRISPR/Cas9.
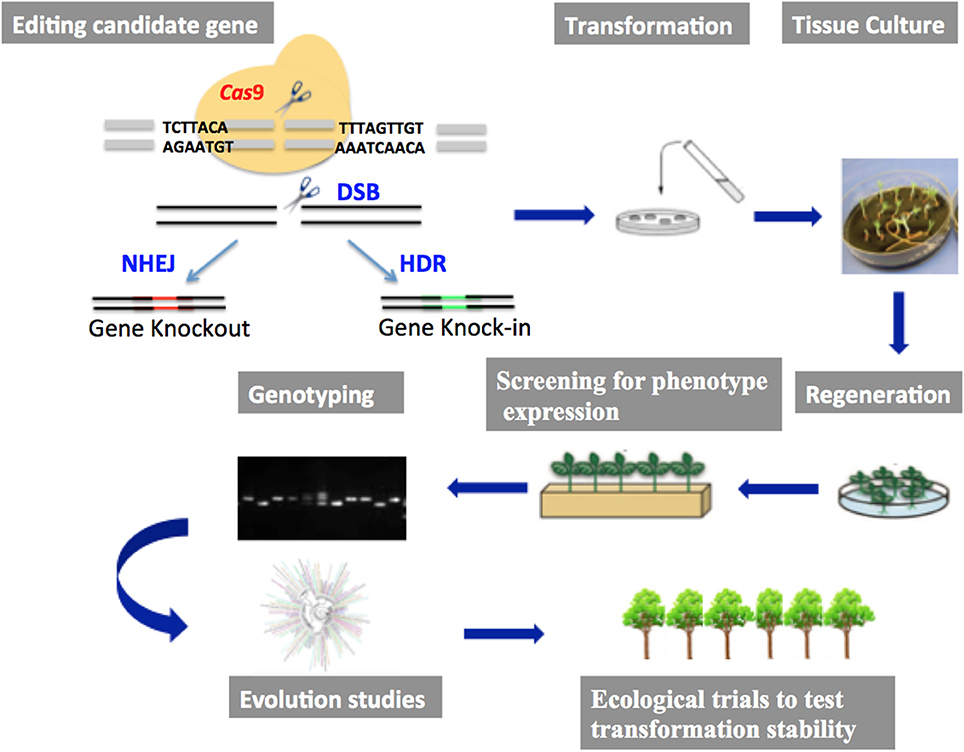
Figure 1. Pipeline for gene editing using CRISPR/Cas9, showing the essential steps in the gene editing approach to landscape genomics and evolutionary studies.
How Different CRISPR Modes Can be Used in Studies of Adaptive Traits in Woody Plants
Testing Candidate Gene Function: Knock-Outs
Removal of gene function provides an important tool for testing the role of candidate genes in the expression of phenotypic traits that, in turn, can be assessed for fitness. The knock-out function in gene editing is achieved when double-strand breaks (DSB) are repaired by non-homologous end-joining (NHEJ) that commonly results in INDELS. A mutant form lacking the targeted gene is obtained because the resulting frameshift in the open reading frame interferes with reading of the gene sequence. The CRISPR/Cas9 system has been used successfully to knock-out genes in model plants and some well-documented crop plants. One problem with the knock-out process is that mosaicism may result if only some of the cells developing from the tissue being transformed are mutated. This has been successfully overcome in Arabidopsis thaliana by targeting early developmental stages using egg-cell promoters (Wang et al., 2015). Recently, Tsutsui and Higashiyama (2017) obtained high-efficiency transformation of the phytoene desaturase 3 (PDS3) gene in A. thaliana by targeting early embryonic meristematic tissue, producing albino phenotypes in both foliage and flowers after gene knock-out. The precision of gene editing using the CRISPR/Cas9 system has been demonstrated in a study of flower color in Japanese morning glory (Ipomoea nil) (Watanabe et al., 2017). The gene dihydroflavonol-4-reductase-B (DFR-B) codes for an anthocyanin biosynthesis enzyme that is responsible for flower color in morning glory. DFR is a tandemly arrayed gene family (DFR-A, DFR-B and DFR-C), so by targeting DFR-B, the risk of off-target modifications can be tested in the adjacent orthologous genes. Mutations in the targeted DFR-B gene included single base insertions, or deletions of more than two bases giving frameshifts that knocked out the DFR-B gene and produced white flowers lacking anthocyanin. The DFR-A and DFR-C genes were left intact, confirming the accuracy of the CRISPR/Cas9 system and the importance of design of the sgRNA and the PAMnGG motif that binds CRISPR/Cas9 to the target DNA.
Early focus of gene editing technologies will focus on genes of major effect, but these may be less important in plant adaptation to climate stress, where many genes of small effect appear to be the rule. One of the great advantages of the CRISPR/Cas9 system is multiplexing single guide RNAs (sgRNA) that simultaneously target different gene loci (Cong et al., 2013). This approach, not only allows testing of multiple genes simultaneously, but offers the possibility to study gene interactions in a way that has not been possible until now. The risk of off-target mutations becomes greater in multiplexed systems, but a recent study targeting 14 genomic loci of the GOLVEN (GLV/RGF/CLEL) gene family in Arabidopsis thaliana found no significant off-site effects, suggesting that in Arabidopsis, off-target effects were no more likely in multiplexed systems than in single gene knock-outs (Peterson et al., 2016).
Much of the focus of CRISPR/Cas9 technology has been on transcribed genes, however, promoter sequences that regulate gene expression may contribute significantly to adaptive potential of species under continuous environmental change through their effects on quantitative traits. Rodríguez-Leal et al. (2017) used CRISPR/Cas9 genome editing of promoters to generate a diverse array of cis-regulatory alleles that provided quantitative variation in a number of crop yield traits in tomato.
Testing the Role of Allelic Variants: Knock-ins
In landscape genomics, candidate gene loci are identified by allelic variants correlated with environmental variables and by outlier measures of population divergence (e.g., FST). However, these approaches identify potential loci and alleles important in adaptation, but are not a proof of function. Ideally, allelic variants should be tested under controlled conditions to quantify their effect. The knock-in mode of CRISPR/Cas9 could provide a tool that would allow reciprocal transfer of alleles, while leaving the rest of the genome unaltered. Gene knock-outs are technically easier to perform than knock-ins that are best achieved with the alternative homology-directed repair (HDR) system. HDR requires a repair template that has the desired sequence that could be a single nucleotide, or a longer sequence.
Compared to NHEJ, HDR has received much less attention, as it remains challenging to obtain very high efficiency. Providing sufficient donor DNA is one constraint. Wang et al. (2016) report achieving 19.4% efficiency by using geminiviral vectors to deliver the donor DNA in rice.
Genetic Transformation Technologies in Undomesticated Species
Genetic transformation is an indispensable tool in plant molecular breeding and functional genomics research and is a critical step in genome editing. Genes of economical and agronomical importance that might be difficult to integrate into elite genotypes via natural processes because of mating barriers or, low heritability, have been successfully incorporated through genetic transformation.
The first successful attempt to genetically transform a woody plant was the introduction of the AroA gene coding 5-enol-puryvate 3-phosphate via Agrobacterium tumefaciens mediated gene transfer in the Populus hybrid (Populus alba x grandidentata) (Fillatti et al., 1987). Genetic transformation systems have now been successfully applied to a number of forest tree species to improve traits such as tree architecture (Busov et al., 2003), salt tolerance (Li et al., 2009), lignin content (Stout et al., 2014) and biotic and abiotic stress response (Tang et al., 2005, 2007). However, despite the major benefits that transgenic approaches can bring to the improved understanding of adaptive traits, the incorporation and adoption of transformation methods for ecological research has been slow. There are several reasons why this has been particularly slow for woody plants. First, the time needed to genetically transform tissue in woody plants is greater than in many other organismal groups and protocols need to be established for the regeneration of whole plants. Commonly, cells that are readily transformed cannot be regenerated, and vice versa. Second, the high cost of transformation protocols, requires substantial investment in infrastructure that must be adapted to the regulatory legislation of each country. Third, the social obstacles facing genetically modified organisms (GMO) have discouraged many researchers from pursuing this avenue for fundamental and applied research, in favor of other directions with fewer impediments.
However, despite all the promising results offered by CRISPR/Cas9, the scientific community should not forget that these experiments have been carried out and tested at a small scale. It will be very important to plan and set up long-term field trials under natural conditions to avoid unintended results, such as happened in the past, when some annual plants manipulated, for a targeted intended change in a given gene had unexpected effects. However, the historic accumulation of phenotypic data on woody trees by many research groups and the results of almost 25 years existence of transgenic forest trees, have proved that, after initial greenhouse phenotypic characterization, the inserted gene/s showed stable expression under natural conditions causing no/minor unexpected effects to the environment (Häggman et al., 2013). Thus, we assume that the new genome editing approach will have the potential of success, in particular in regard to the accelerated climate change, and to identify important economically and ecological novel traits more rapidly than ever.
Finally, there is a hope that genome edited events will not be classified as GMO (Hartung and Schiemann, 2014) and can have a different regulatory policy as the GMO. A legal guidance on how to define plants produced by exploring novel genome editing techniques is urgently needed. In the meantime, the debate concerning ethical issues and the consideration if the new genome editing techniques should come, or not, under the GMO legislation is still underway. In Europe, the case is in the hands of the EU Court of Justice (ECJ), which will decide whether plants that are gene-edited should be regulated as if they were genetically modified. Although this decision is not expected before 2018, some EU countries, such as Sweden, allowed its researchers to go ahead and move forward with their research, others, like Germany, are still waiting for the ECJ decision. On the other hand, Canada and US did not code genome-edited plants as GMO. In fact, several products such as non-browning mushroom or waxy corn entered the market in the USA with no resistance from anti-GMO protestor (Waltz, 2016) and have given the green light by the US regulatory system for commercialization since no genetic material from a different organism was inserted in their genomes (Ossola, 2016). We expect that, as plants generated by GE technologies do not leave signs of transgenesis, they should not be considered as GMO, and as a consequence, do not provoke an irresolvable social and political debate as it happened with GMO plants.
Conclusion and Future Perspectives
In the future, the application of CRISPR-Cas9 in genome editing may prove to be very important in ecological and evolutionary genetics to confirm candidate gene function and to test gene effects in nature. Ecologists and plant scientists can benefit from using this technique as it has the advantage that it can instantaneously generate alteration to genes without creating t-DNA insertion. With the recent availability of whole genome sequences and efficient transformation protocols for many woody plants, we anticipate remarkable advances will be feasible in terms of genome engineering and landscape genomics in plant ecology. Furthermore, the advance of bioinformatics tools aiding the design of trait specific guided RNA would undoubtedly lead the GE system to broaden its application to more plant species. However, GE technologies will need to be integrated with other disciplines in order to understand the molecular regulation of phenotypic adaptation to climate variables. It is becoming clear that the benefit of CRISPR-Cas depends on a range of ecological variables, but much more work needs to be done if we are to understand and manipulate the evolution of CRISPR-Cas immunity in the lab, let alone in nature.
Author Contributions
AF and RD contributed equally to the ideas expressed here and to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Innovative Genomics Institute (IGI) grants.
References
Aitken, S. N., Yeaman, S., Holliday, J. A., Wang, T., and Curtis-McLane, S. (2008). Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95–111. doi: 10.1111/j.1752-4571.2007.00013.x
Álvarez, R., and Ordás, R. J. (2007). Improved genetic transformation protocol for cork oak (Quercus suber L). Plant Cell Tissue Organ Culture 91, 45–52. doi: 10.1007/s11240-007-9276-6
Andrade, G. M., Nairn, C. J., Le, H. T., and Merckle, S. A. (2009). Sexually mature transgenic American chestnut trees via embryogenic suspension-based transformation. Plant Cell Rep. 28, 1385–1397. doi: 10.1007/s00299-009-0738-7
Barret, R. D., and Hoekstra, H. E. (2011). Molecular spandrels: tests of adaptation at the genetic level. Nat. Rev. Genet. 12, 767–780. doi: 10.1038/nrg3015
Benedict, C., Skinner, J. S., Meng, R., Chang, Y., Bhalerao, R., Huner, N. P., et al. (2006). The CBF1-dependent low temperature signalling pathway regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29, 1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x
Busov, V. B., Meilan, R., Pearce, D. W., Ma, C., Rood, S. B., and Strauss, S. H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 132, 1283–1291. doi: 10.1104/pp.103.020354
Cahill, A. E., Aiello-Lammens, M. E., Fisher-Reid, M. C., Hua, X., Karanewsky, C. J., Ryu, H. Y., et al. (2013). How does climate change cause extinction? Proce. R. Soc. B. 280:20121890. doi: 10.1098/rspb.2012.1890
Castellanos-Hernández, O. A., Rodríguez-Sahagún, A., Acevedo-Hernández, G. J., Rodríguez-Garay, B., Cabrera-Ponce, J., et al. (2009). Transgenic Paulownia elongata S Y Hu plants using biolistic-mediated transformation. Plant Cell Tissue Organ Culture 99, 175–181. doi: 10.1007/s11240-009-9590-2
Chiune, I., Rehfeldt, G. E., and Aitken, S. N. (2006). Height growth determinants and adaptation to temperature in pines: a case study of Pinus contorta and Pinus monticola. Can. J. For. Res. 36, 1059–1066. doi: 10.1139/x06-005
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cook, B. I., Wolkovich, E. M., and Parmesan, C. (2012). Divergent responses to spring and winter warming drive community level flowering trends. Proc. Natl. Acad. Sci. U.S.A. 109, 9000–9005. doi: 10.1073/pnas.1118364109
Dutt, M., and Grosser, J. W. (2010). An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of Citrus. Plant Cell Reports 29, 1251–1260. doi: 10.1007/s00299-010-0910-0
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Fillatti, J., Sellmer, J., McCown, B., Haissig, B., and Comai, L. (1987). Agrobacterium mediated transformation and regeneration of Populus. Mol. General Genet. 206, 192–199.
Gonzalez, P., Neilson, R. P., Lenihan, J. M., and Drapek, R. J. (2010). Global patterns in the vulnerability of ecosystems to vegetation shifts due to climate change. Global Ecol. Biogeogr. 19, 755–768. doi: 10.1111/j.1466-8238.2010.00558.x
Häggman, H., Raybould, A., Borem, A., Fox, T., Handley, L., Hertzberg, M., et al. (2013). Genetically engineered trees for plantation forests: key considerations for environmental risk assessment. Plant Biotechnol. J. 11, 785–798. doi: 10.1111/pbi.12100
Hartung, F., and Schiemann, J. (2014). Precise plant breeding using new genome editing techniques: opportunities, safety and regulations in the EU. Plant J. 78, 742–752. doi: 10.1111/tpj.12413
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433.
Jacobs, J. Z., Ciccaglione, K. M., Tournier, V., and Zaratiegui, M (2014). Implementation of the CRISPR-Cas9 system in fission yeast. Nat. Commun. 5:5344. doi: 10.1038/ncomms6344
Jia, H., and Wang, N (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806. doi: 10.1371/journal.pone.0093806
Jia, H., Orbovic, V., Jones, J. B., and Wang, N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccDpthA4: dCsLOB1.3 infection. Plant Biotech. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kikuchi, A., Kawaoka, A., Shimazaki, T., Yu, X., Ebinuma, H., and Watanabe, K. N. (2006). Trait stability and environmental biosafety assessments on three transgenic Eucalyptus lines (Eucalyptus camaldulensis Dehnh codA 12-5B codA 12-5C codA 20-C) conferring salt tolerance. Breed Res. 8, 17–26.
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, Y., Su, X., Zhang, B., Huang, Q., Zhang, X., and Huang, R. (2009). Expression of jasmonic ethylene responsive factor gene in transgenic poplar tree leads to increased salt tolerance. Tree Physiol. 29, 273–279. doi: 10.1093/treephys/tpn025
Loopstra, C. A., Stomp, A. M., and Sederoff, R. R. (1990). Agrobacterium-mediated DNA transfer in sugar pine. Plant Mol. Biol. 15, 1–9.
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., Zhu, J. K., et al. (2013). Application of the CRISPR/Cas system for efficient genome engineering in plants. Mol. Plant. 6, 2008–2011. doi: 10.1093/mp/sst121
Mauro, M. C., Toutain, S., Walter, B., Pinck, L., Otten, L., Coutos-Thevenot, P., et al. (2000). High efficiency regeneration of grapevine plants transformed with the GFLV coat protein gene. Plant Sci. 112, 97–106. doi: 10.1016/0168-9452(95)04246-Q
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 1819: 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Mojica, F. J., Diez-Villaseñor, C., Garcia-Martinez, J., and Almendros, C. (2009). Short motif sequences determines the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Mussolino, C., and Cathomen, T. (2013). RNA guides genome engineering. Nat. Biotechnol. 31, 208–209. doi: 10.1038/nbt.2527
Neale, D. B., and Kremer, A. (2011). Forest tree genomics: growing resources and applications. Nat. Rev. Genet. 12, 111–122. doi: 10.1038/nrg2931
Neale, D. B., Wegrzyn, J. L., Stevens, K. A., Zimin, A. V., Puiu, D., Crepeau, M. W., et al. (2014). Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15:R59. doi: 10.1186/gb-2014-15-3-r59
Nellemann, C., MacDevette, M., Manders, T., Eickhout, B., Svihus, B., Prins, A., et al. (2009). The Environmental Food Crisis. The Environment's Role in Averting Future Food Crises. A UNEP Rapid Response Assessment. Arendal: UNDP.
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6:31481. doi: 10.1038/srep31481
Noman, A., Aqeel, M., and He, S. (2016). CRISPR-Cas9: Tool for qualitative and quantitative plant genome editing. Front. Plant Sci. 7:1740. doi: 10.3389/fpls.2016.01740
Parmesan, C. (2007). Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Glob. Chang. Biol. 13, 1860–1872. doi: 10.1111/j.1365-2486.2007.01404.x
Pereira, H. M., Leadley, P. W., Proenca, V., Alkemade, R., Scharlemann, J. P., Fernandez-Manjarrés, J. F., et al. (2010). Scenarios for global biodiversity in the 21st century. Science 330, 1496–1501. doi: 10.1126/science.1196624
Peterson, B. A., Haak, D. C., Nishimura, M. T., Teixeira, P. J., James, S. R., Dangl, J. L., et al. (2016). Genome-wide assessment of efficiency and Specificity in CRISPR/Cas9 mediated multiple site targeting in arabidopsis. PLoS ONE 11:e0162169. doi: 10.1371/journal.pone.0162169.
Petri, C., Scorza, R., and Srinivasan, C. (2012). Highly efficient transformation protocol for plum (Prunus domestica L). Methods Mol. Biol. 2012847:191–199. doi: 10.1007/978-1-61779-558-9_16
Plomion, C., Aury, J. M., Amselem, J., Alaeitabar, T., Barbe, V., Belser, C., et al. (2015). Decoding the oak genome: public release of sequence data, assembly, annotation and publication strategies. Mol. Ecol. Notes. 16, 254–265. doi: 10.1111/1755-0998.12425
Potter, C. S. (2016). Landsat image analysis of tree mortality in the southern Sierra Nevada region of California during the 2013-2015 drought. J. Earth Sci. Clim. Change 7:3. doi: 10.4172/2157-7617.1000342
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480. e8. doi: 10.1016/j.cell.2017.08.030
Settele, J., Scholes, R., Betts, R., Bunn, S., Leadley, P., Nepstad, D., et al. (2014). “Terrestrial and inland water systems,” in Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds C. B. Field, V. R. Barros, D. J. Dokken, K. J. Mach, M. D. Mastrandrea, T. E. Bilir, M. Chatterjee, K. L. Ebi, Y. O. Estrada, R. C. Genova, B. Girma, E. S. Kissel, A. N. Levy, S. MacCracken, P. R. Mastrandrea, and L. L. White (Cambridge, UK; New York, NY: Cambridge University Press), 271–359.
Stout, A., Davis, A., Domec, J. C., Yang, C., Shi, R., John, S., et al. (2014). Growth under field conditions affects lignin content and productivity in transgenic Populus trichocarpa with altered lignin biosynthesis. Biomass Bioenergy 68, 228–239. doi: 10.1016/j.biombioe.2014.06.008
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., Cigan, A. M., et al. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Tang, W., Charles, T. M., and Newton, R. J. (2005). Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 59, 603–617. doi: 10.1007/s11103-005-0451-z
Tang, W., Newton, R. J., Li, C., and Charles, T. M. (2007). Enhanced stress tolerance in transgenic pine expressing the pepper CaPF1 gene is associated with the polyamine biosynthesis. Plant Cell Rep. 26, 115–124. doi: 10.1007/s00299-006-0228-0
Tsutsui, H., and Higashiyama, T. (2017). pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56. doi: 10.1093/pcp/pcw191
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–604. doi: 10.1126/science.1128691
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7:507. doi: 10.1038/s41598-017-00501-4
Vidal, N., Mallón, R., Valladares, S., Meijomín, A. M., and Vieitez, A. M. (2010). Regeneration of transgenic plants by Agrobacterium-mediated transformation of somatic embryos of juvenile and mature Quercus robur. Plant Cell Rep. 29, 1411–1422. doi: 10.1007/s00299-010-0931-8
Wadenbäck, J., von Arnold, S., Egertsdotter, U., Walter, M. H., Grima-Pettenati, J., Goffner, D., et al. (2008). Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR). Transgenic Res. 17, 379–392. doi: 10.1007/s11248-007-9113-z
Waltz, E. (2016). CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 34:582. doi: 10.1038/nbt0616-582.
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, S., Zhang, S., Wang, W., Xiong, X., Meng, F., and Cui, X. (2015). Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 34, 1473–1476. doi: 10.1007/s00299-015-1816-7
Watanabe, K., Kobayashi, A., Endo, M., Sage-Ono, K., Toki, S., and Ono, M. (2017). CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B)locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 7:10028. doi: 10.1038/s41598-017-10715-1
Keywords: CRISPR/Cas9, ecological genomics, genome editing, forest trees, climate change, adaptation
Citation: Fernandez i Marti A and Dodd RS (2018) Using CRISPR as a Gene Editing Tool for Validating Adaptive Gene Function in Tree Landscape Genomics. Front. Ecol. Evol. 6:76. doi: 10.3389/fevo.2018.00076
Received: 25 January 2018; Accepted: 15 May 2018;
Published: 04 June 2018.
Edited by:
Rongling Wu, Pennsylvania State University, United StatesReviewed by:
Lifeng Zhu, Nanjing Normal University, ChinaLarissa Moriarty Williams, Bates College, United States
Copyright © 2018 Fernandez i Marti and Dodd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard S. Dodd, dodd@berkeley.edu
 Angel Fernandez i Marti
Angel Fernandez i Marti Richard S. Dodd
Richard S. Dodd