- 1Guangdong Province Famous Fish Reproduction Regulation and Breeding Engineering Technology Research Center, Fisheries College of Guangdong Ocean University, Zhanjiang, China
- 2Key Laboratory of Freshwater Fish Reproduction and Development (Ministry of Education), Key Laboratory of Aquatic Science of Chongqing, School of Life Sciences, Southwest University, Chongqing, China
Gonadal somatic cell-derived factor (Gsdf) is a member of the TGF-β superfamily, which exists mainly in fishes. Homozygous gsdf mutations in Japanese medaka and zebrafish resulted in infertile females, and the reasons for their infertility remain unknown. This study presents functional studies of Gsdf in ovary development using CRISPR/Cas9 in Nile tilapia (Oreochromis niloticus). The XX wild type (WT) female fish regularly reproduced from 12 months after hatching (mah), while the XX gsdf -/- female fish never reproduced and were infertile. Histological observation showed that at 24 mah, number of phase IV oocyte in the XX gsdf -/- female fish was significantly lower than that of the WT fish, although their gonadosomatic index (GSI) was similar. However, the GSI of the XX gsdf -/- female at 6 mah was higher than that of the WT. The mutated ovaries were hyperplastic with more phase I oocytes. Transcriptome analysis identified 344 and 51 up- and down-regulated genes in mutants compared with the WT ovaries at 6 mah. Some TGF-β signaling genes that are critical for ovary development in fish were differentially expressed. Genes such as amh and amhr2 were up-regulated, while inhbb and acvr2a were down-regulated in mutant ovaries. The cyp19a1a, the key gene for estrogen synthesis, was not differentially expressed. Moreover, the serum 17β-estradiol (E2) concentrations between XX gsdf -/- and WT were similar at 6 and 24 mah. Results from real-time PCR and immunofluorescence experiments were similar and validated the transcriptome data. Furthermore, Yeast-two-hybrid assays showed that Gsdf interacts with TGF-β type II receptors (Amhr2 and Bmpr2a). Altogether, these results suggest that Gsdf functions together with TGF-β signaling pathway to control ovary development and fertility. This study contributes to knowledge on the function of Gsdf in fish oogenesis.
1 Introduction
Gonadal somatic cell-derived factor (Gsdf) is a secretory protein of the transforming growth factor β (TGF-β) superfamily, which exists in most vertebrates but is lost in some tetropods including mammals (1). First cloned in the rainbow trout (Oncorhynchus mykiss) 15 years ago, Gsdf has since been widely studied, especially in teleosts (2). Gsdf is predominantly expressed in the somatic cells of ovaries and testes, but its expression is male-dominant in most fish species (1).
In Philippine medaka (Oryzias luzonensis) and sablefish (Anoplopoma fimbria), the gsdfY that evolved from autosomal gsdf via allelic diversification is the male sex determination gene (3, 4). Autosomal gsdf is required for testicular differentiation in Japanese medaka (O. latipes) and Nile tilapia (Oreochromis niloticus), as XY gsdf -/- individuals undergo sex reversal from male to female (5–7). However, the gsdf homozygous mutation does not cause the male to female sex reversal in zebrafish (Danio rerio), indicating that the Gsdf function in male sex differentiation is not conserved among fish species (8).
In Japanese medaka, gsdf is located within an evolutionarily conserved gene cluster containing several genes preferentially expressed in previtellogenic oocytes, indicating the possible Gsdf role in ovary development (9). Most gsdf -/- female Japanese medaka are infertile, but few females are sub-fertile, suggesting that Gsdf is also important for oogenesis (10), yet gsdf -/- female zebrafish are infertile (8). The homozygous mutation of gsdf causes ovarian hyperplasia and early-stage oocyte-arrest in zebrafish and Japanese medaka (8, 10). However, the detailed mechanisms of infertility in gsdf -/- female fish and the role of Gsdf in ovary development remain unknown.
Oogenesis involves ovary formation, development and maturation of female gametes, and finally ovulation (11). The hormones secreted by the hypothalamic-pituitary-gonadal (HPG) axis regulate oogenesis in vertebrates, including fish (12–15). Moreover, many genes of the TGF-β signaling pathway expressed in the HPG axis are critical in oogenesis and hormone regulation. The TGF-β signaling pathway consists of several extracellular ligands (phylogenetically divided into TGF-β and BMP groups), cell membrane receptors (types I and II), intracellular Smad proteins, and other functionally related components (16–18). For example, in zebrafish, mutation of three genes from the TGF-β signaling pathway (amh, bmpr1bb (alk6b), and bmpr2a) causes ovary hyperplasia and accumulation of immature oocytes (19–21).
In Japanese medaka, the hotei mutation of amhr2 dysregulates germ cells and arrests follicular development at an early stage (22, 23). Similarly, amh and amhr2 mutations in Nile tilapia cause hypertrophic ovaries with increased primary follicles (24). Serum estradiol (E2) levels decrease in the amh and amhr2 female mutants, and E2 administration fails to rescue ovary development (24). In mice, Amhr2 is expressed in the brain and pituitary (25). Additionally, in mice and humans, Amh directly regulates the secretion of GnRH neurons in the hypothalamus, hence increasing Lh pulsatility and secretion (25). In Nile tilapia, both amh and amhr2 are expressed in the brain and pituitary besides the gonads (24). Decrease of serum E2 and down-regulation of the expression of brain gnrh3, pituitary fsh and lh, and ovary fshr and lhr were observed in amh and amhr2 null Nile tilapia, which was attributed to the down-regulation of the Fsh/Lh signaling (24). Similarly, the expression of cyp19a1a in the ovary and E2 in the body significantly decreased in the female gsdf -/- zebrafish (8). While the relationship between Gsdf, Fsh/Lh and E2 in fish remains unclear, these reproduction-related hormones and the TGF-β signaling genes are attractive enigmas.
Candidate Gsdf receptors are unknown, but Gsdf probably shares known TGF-β receptors with other ligands since TGF-β family members share the limited type I and II receptors. Synteny analysis of zebrafish, spotted gar (Lepisosteus oculatus), and humans indicate that gsdf, bmp15, and gdf9 were originally paralogs (8). The Gsdf protein might also bind to similar Bmp15 and Gdf9 receptors. As previously mentioned, amh, amhr2, bmpr1bb, and bmpr2a mutants cause ovary hyperplasia with immature oocytes, which phenocopy the gsdf female mutants in fish (19–24). The phenotype similarity of these TGF-β signaling mutants indicates that these genes might influence each other via similar signaling pathways. Moreover, Gsdf might share similar receptors with other TGF-β ligands, but evidence to identify the Gsdf receptors is lacking.
The Nile tilapia is an important farmed fish with an XX-XY sex determination system (26). Gene editing technology has been established in Nile tilapia, and the availability of high-quality genome sequences makes this species an excellent model for studying gene function (27–30). Previously, we established a gsdf knockout line in Nile tilapia and illustrated its role in male sex differentiation, while its function in ovary development is still unclear (7). Thus, this study analyzed the XX gsdf homozygous mutant fish to determine the gonadal histology, gonadal gene expression and serum steroid hormone levels. In addition, yeast-two-hybrid assays were performed to identify the Gsdf receptors. This study sought to decipher the plausible reasons for the infertility of the female gsdf mutants. This study contributes to knowledge on the function of gsdf in regulating fish oogenesis.
2 Materials and Methods
2.1 Animals and Screen of XX gsdf Mutants
In the previous study, the Nile tilapia XY gsdf F0 mutants were obtained using CRISPR/Cas9 at the Southwest University, China (7). Male and female siblings of heterozygous F1 mutants (gsdf +/-) carrying the same mutation (4 bp deletion) were transferred and reared in Guangdong Ocean University, China. The 4 bp deletion occurs at the first gsdf exon, which causes a frame-shift mutation, making Gsdf dysfunctional (7).
The Nile tilapia were reared in aerated recirculating freshwater tanks at 26~28°C. XX gsdf -/- and gsdf +/+ progenies were obtained by crossing an XX gsdf +/- female with XY gsdf +/- male fish. Genotyping using a sex-linked marker (Marker-5) determined the genetic sex of each fish (31, 32). The gsdf mutant fish were detected using the BstNI enzyme, polyacrylamide gels electrophoresis (PAGE), or Sanger sequencing (Figure S1). Primer sequences are listed in Table S1. The study did not involve endangered or protected species. All animal experiments involving tilapia were performed according to the protocol of the Animal Research and Ethics Committee of Guangdong Ocean University.
2.2 Sampling and Histological Analysis
The morphology and histology of XX gsdf +/+ and gsdf -/- fish ovaries were analyzed at 6 and 24 months after hatching (mah) to study the role of gsdf in ovary development. The body and gonad weight were measured to determine the gonadosomatic index (GSI, gonad weight/body weight). The ovaries of the XX gsdf +/+ and gsdf -/- fish were dissected, and one part of each ovary was snap-frozen in liquid nitrogen and stored at −80°C for RNA extraction. The other part was fixed with Bouin’s solution for 24 h at room temperature. The fixed gonads were embedded in paraffin and used for histological analysis. Tissue blocks were sectioned at 5 µm thickness, and the sections were stained with hematoxylin and eosin (HE).
2.3 Oocyte Count at Different Phases
The number of oocytes at different development phases in the ovaries of XX gsdf +/+ and gsdf -/- fish was compared at 6 and 24 mah. The histological classification of the oocyte phase was performed as previously described (33). Briefly, the oocyte numbers at different phases were counted from photos of the HE-stained sections of the XX gsdf +/+ and gsdf -/- ovaries.
2.4 Fertility Analysis
XX gsdf -/- and gsdf +/+ individuals over 10 mah were cultured in tanks with circulating water at 26°C and fed twice a day (08:00 am and 16:00 pm). The tanks had a layer of crude river sand laid at the bottom for the fish to nest and spawn. Every day from 12:00 to 18:00 hours, when the fish started nesting on the sand, evidenced by their protruding genital papilla, the fish were taken out and squeezed slightly on the abdomen to strip their eggs into a clean bowl. The collected eggs were artificially fertilized using sperms from wild-type XY male fish, and the embryos were hatched at 26~28°C.
2.5 Transcriptome Analysis
A transcriptome analysis was performed on the XX gsdf -/- ovaries from fish at 6 mah to understand the causes of delayed oocyte development, infertility of the XX gsdf -/- fish, and the gsdf function in ovary development. Eight ovaries were sampled from XX gsdf +/+ (n=4) and gsdf -/- (n=4) fish. Total RNA was extracted from the ovaries using the RNeasy Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The mRNA was enriched with a poly-A tail using oligo (dT) coupled to magnetic beads, and the mRNA was randomly interrupted to about 200 bp short fragments.
The first-strand cDNA was synthesized in the M-MuLV reverse transcriptase system (M0253S, NEB, US), and the second-strand was synthesized from dNTPs in the DNA polymerase I system (E7530, NEB, US). These short fragments were ligated to sequencing adapters and sequenced on Illumina Hiseq 2500 platform following the manufacturer’s protocol (Illumina, CA, USA). The raw reads were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database with BioProject number PRJNA703049 (SRR13752974-SRR13752981).
Adapter sequences were removed. The reads containing poly-N (unable to determine base information) and low-quality reads were also removed using fastp (version 0.18.0). Finally, the adapters read with more than 10% unknown nucleotides and over 50% low-quality nucleotides (Q-value ≤ 20) were also removed. Clean reads from each library were aligned to the reference genome using the HISAT2 program (34). The Oreochromis niloticus (Orenil1.0) reference genome was downloaded from Ensembl (http://www.ensembl.org/Oreochromis_niloticus/Info/Index). Gene expression levels were estimated using the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) method. The Stringtie assembler (35) reconstructed the transcripts from HISAT2, which were quantified using RSEM (36).
Differential expression analysis was performed using the DESeq2 package (37). Significant differentially expressed genes (DEGs) were identified at |log2(gsdf -/- FPKM/gsdf +/+ FPKM)| > 1.0 with < 0.05 false discovery rate (FDR). Gene Ontology (GO)(http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the significant DEGs. The GO and KEGG enriched terms with P <0.05 were considered significantly enriched.
2.6 Real-Time PCR (qPCR)
Ovaries of the XX gsdf +/+ and gsdf -/- fish were dissected at 6 mah for qPCR gene expression assays to validate the transcriptome results. Total RNA (1.0 μg) was extracted using the Rneasy Mini Kit (Qiagen, Hilden, Germany) and reverse transcribed to cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (RRO41A, Takara Bio, Kusatsu-Shiga, Japan). Real-time PCR was performed using the SYBR Green qPCR Mix (Dongsheng Biotech, Guangdong, China) following the manufacturer’s instructions on a LightCycler real-time quantitative PCR system (Roche, Basel, Switzerland). The Nile tilapia β-actin gene was used to normalize the gene expressions. The relative abundance of the genes was evaluated using the formula R = 2−ΔΔCt (38). Primer sequences used for real-time PCR are listed in Table S1.
2.7 Immunofluorescence (IF) Analysis
At 6 mah, the ovaries of gsdf +/+ and gsdf -/- XX fish were dissected, and the gonads were fixed in Bouin’s solution for 24 hours at room temperature, dehydrated, and embedded in paraffin. Tissue blocks were sectioned at 5 μm thickness, and immunohistochemistry was performed using the Gsdf, Amh and Cyp19a1a antibodies diluted at 1:1000, 1:500 and 1:2000, respectively. Gsdf and Amh antibodies were obtained from previous studies (7, 26), where their specificities were checked. The Cyp19a1a antibody was provided by Professor Yoshitaka Nagahama, National Institute for Basic Biology Okazaki, Japan (39).
For IF, Alexa Fluor 586-conjugated secondary antibodies (Invitrogen, MA, USA) were diluted at 1:500 in blocking solution and incubated with tissue to detect primary antibodies. The nuclei were stained by 4’,6’-diamidine-2-phenylindole-dihydrochloride (DAPI) (Invitrogen, MA, USA). Immunohistochemical analysis was performed as described previously. Photographs were taken under an FV3000 Laser confocal microscope (Olympus, Tokyo, Japan). Finally, the positive signals were quantified using Image J software (National Institute of Health, MD, USA) following the instructions provided.
2.8 Measurement of Serum Steroid Hormones
The influence of gsdf mutation on the serum steroid hormone levels was analyzed. Blood samples were collected from the caudal vein of XX gsdf +/+ and gsdf -/- fish using syringes at 6 and 24 mah. The blood samples were stored at 4°C overnight and centrifuged at 5,000 g for 5 min at 4°C. The upper layer serum was collected and stored at -80°C until use. The serum 17β-estradiol (E2), 11-keto Testosterone (11-KT), Testosterone (T), and Pregnenolone (P5) levels were measured using Enzyme Immunoassay (EIA) Kits (E2, 11-KT and T kits from Cayman Chemical Company (MI, USA), and the P5 kit from Abnova (Taipei, China) following the manufacturer’s instructions.
2.9 Yeast-Two-Hybrid Assays
Yeast-two-hybrid assays were performed to identify the possible Gsdf receptor. Firstly, the gsdf was constructed into the pBT3-SUC bait vector. Concurrently, the amhr2 and bmpr2a were constructed into the pPR3 prey vector. Next, the bait plasmid + pPR3-N, bait and prey plasmid, bait plasmid + POST-Nubal, pTSU2-APP+pNubG-Fe65 (positive control), and pTSU2-APP+pPR3-N (negative control) were co-transformed into NMY51 yeast strain. The bait plasmids + pPR3-N functioned as autoactivation assays, and bait plasmids + POST-Nubal functionally validated the ubiquitin system. After that, yeasts were cultured in DDO (Lack of LEU and TRP) liquid medium to the optical density (OD600) of ~0.4-0.8. Cultures were titrated 1:10, 1:100, 1:1,000, and 1:10,000 in saline solution.
Then, 10 μl of each dilution was seeded on DDO, TDO (Lack of LEU, TRP and HIS) and QDO (Lack of LEU, TRP, HIS and ADE) plates. The seeded plates were incubated at 30°C for 72 h to observe the yeast growth. Yeast growth assays estimated the protein-protein interaction strength on plates lacking HIS (suitable for weak protein-protein interactions) or ADE (suitable for strong protein-protein interactions).
2.10 Statistical Analysis
All data are presented as mean ± SD from at least three independent samples collected from different fish, and Student’s t-test analyzed the statistical significance. The significance level was P < 0.05, and statistical tests were performed with the Statistical Package for Social Sciences (SPSS) 19.0 (IL, USA).
3 Results
3.1 gsdf Homozygous Mutation Causes Infertility in Female Nile Tilapia
At 6 and 24 mah, there was no observable morphological difference between XX gsdf +/+ and gsdf -/- fish (Figure S2). The GSI of XX gsdf -/- was significantly higher than that of XX gsdf +/+ fish at 6 mah (Figures 1A, a–c, Table S2). The number of phase I oocytes in the XX gsdf +/+ ovaries was significantly lower than that of XX gsdf -/- ovaries at 6 mah. Nevertheless, the number of phase II oocytes in the XX gsdf +/+ fish was substantially higher than that of the XX gsdf -/- fish (Figures 1A, a’, b’, d). These results indicate that the ovaries of XX gsdf -/- fish were hyperplastic, and the oocytes were arrested at the primary growth stage.
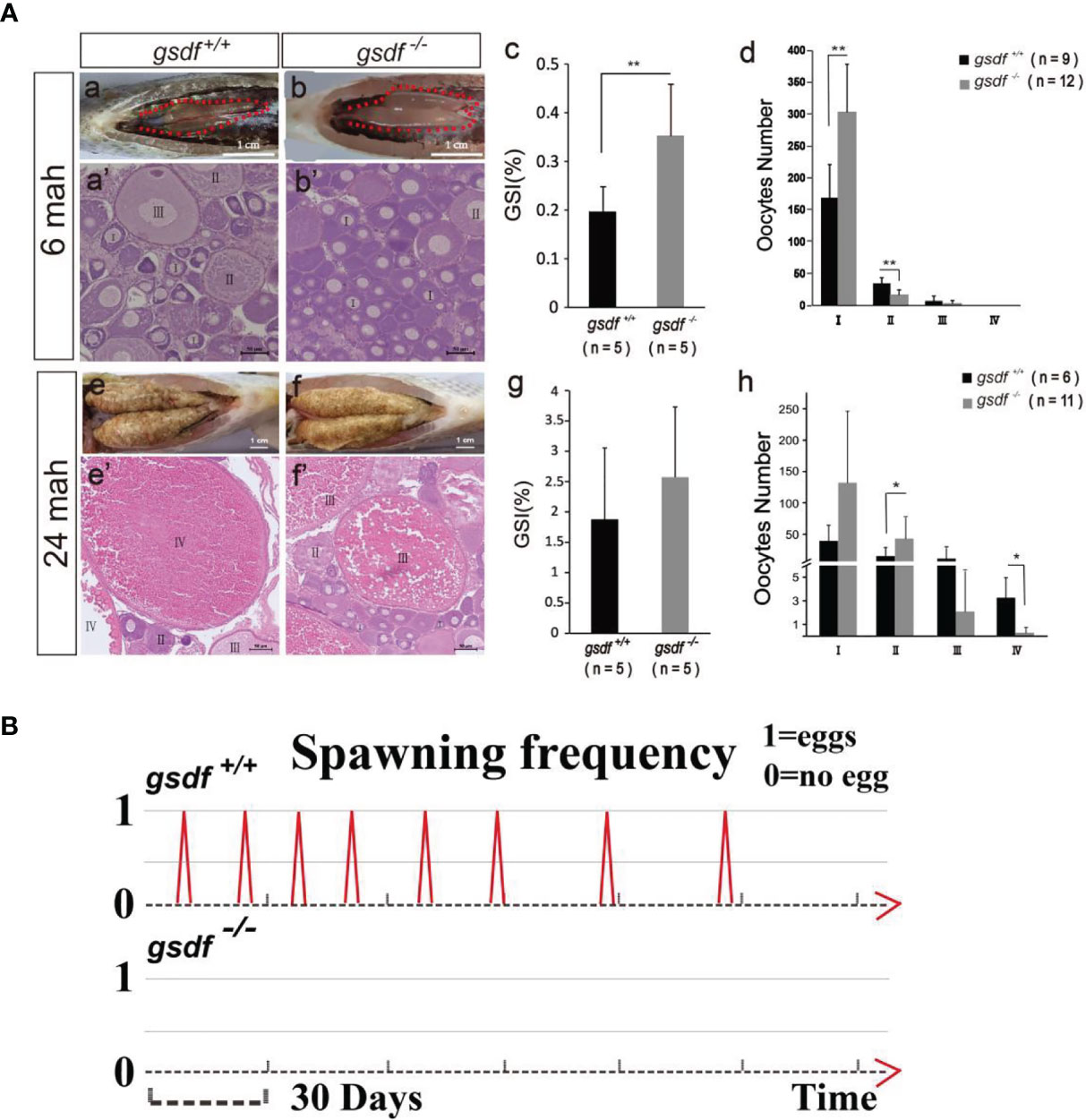
Figure 1 Morphological and histological analyses of ovaries (A) and spawning frequency (B) from XX gsdf +/+ and gsdf -/- tilapia. The fish were dissected at 6 and 24 mah. Results are presented as mean ± SD. * and ** indicate significant differences at P < 0.05 and P < 0.01, respectively. (B) The spawning frequency of 12-months old XX gsdf +/+ and gsdf -/- fish were observed for half a year. More than ten XX gsdf -/- fish were observed for spawning, while none reproduced.
There were many maturing oocytes in XX gsdf +/+ fish ovaries at 24 mah, while the ovaries of XX gsdf -/- fish had very few mature and many white dead oocytes (Figures 1A, e-f). Mutant gonads contained less-maturing oocytes, but the GSI between XX gsdf +/+ and gsdf -/- fish at 24 mah were similar, indicating that the mutant gonads were still hyperplastic with more immature oocytes (Figure 1A, g). The HE staining showed that XX gsdf -/- ovaries had more phase II oocytes than XX gsdf +/+ ovaries at 24 mah. However, the number of phase IV oocytes in the XX gsdf -/- ovaries was significantly lower than that of the XX gsdf +/+ ovaries (Figures 1A, e’-f’, h). These results indicate that the development of oocytes in XX gsdf -/- fish ovaries was delayed and immature at 24 mah, corresponding to the previtellogenic phase.
The XX gsdf +/+ fish became sexually mature at approximately one year and spawned eggs continuously, with the shortest interval at 14 days. Additionally, the spawning, fertilization, and hatching are normal, with normally-developed fry. However, the XX gsdf -/- fish (n=10) showed no spawning, indicating their sterility (Figure 1B).
3.2 Loss of gsdf Alters Gene Expression in the Ovaries of XX gsdf-/- Fish
The histology difference in the ovaries of XX gsdf +/+ and gsdf -/- fish are observably different at 24 mah. Thus, the ovaries of the 6-months old fish were chosen for gene expression analysis to identify the effect of gsdf mutation on gene expressions.
A quality control analysis of raw sequences using the fastp software filtered out low quality reads, resulting in 22.96 and 22.38 Gb of cleaned transcriptome data from the ovaries of XX gsdf +/+ and gsdf -/- fish, respectively. The average Q20 and Q30 of the clean data were 97.80% and 93.65%, respectively. An average of 95.6% and 94.7% clean reads from XX gsdf +/+ and gsdf -/- fish were mapped to the reference genome. The sequencing and mapping statistics for the eight transcriptomes are listed in Table S3. There were 13,133, and 13,722 genes (FPKM > 1) expressed in the ovaries of the XX gsdf +/+ and gsdf -/- fish, receptively. A total of 395 DEGs were identified in XX gsdf +/+ and gsdf -/- ovaries at 6 mah (FDR <0.05). The ovaries of XX gsdf -/- fish had 344 and 51 up-regulated and down-regulated genes, respectively (Table S4).
The DEGs enriched GO terms at the three main functional categories, including biological process (24 terms), molecular function (11 terms), and cellular component (16 terms) (Figure 2A). Under biological process, the DEGs significantly enriched cellular process (GO:0009987, 209 genes) (187 genes up-regulated and 22 genes down-regulated), single-organism process (GO:0044699, 204 genes) (182 genes up-regulated and 22 genes down-regulated), and biological regulation (GO:0065007, 163 genes) (146 genes up-regulated and 17 genes down-regulated) terms. Within the molecular function, the DEGs enriched binding (GO:0005488, 181 genes) (161 genes up-regulated and 20 genes down-regulated) and catalytic activity (65 genes up-regulated and eight genes down-regulated) (GO:0003824, 73 genes). In the cellular component, the DEGs were significantly enriched in cell (GO:0005623, 170 genes) (152 genes up-regulated and 18 genes down-regulated), cell part (GO:0044464, 170 genes) (152 genes up-regulated and 18 genes down-regulated) and organelle (GO:0043226, 146 genes) (127 genes up-regulated and 19 genes down-regulated) GO term (Figure 2A). The DEGs that enriched these GO terms are listed in Table S5.
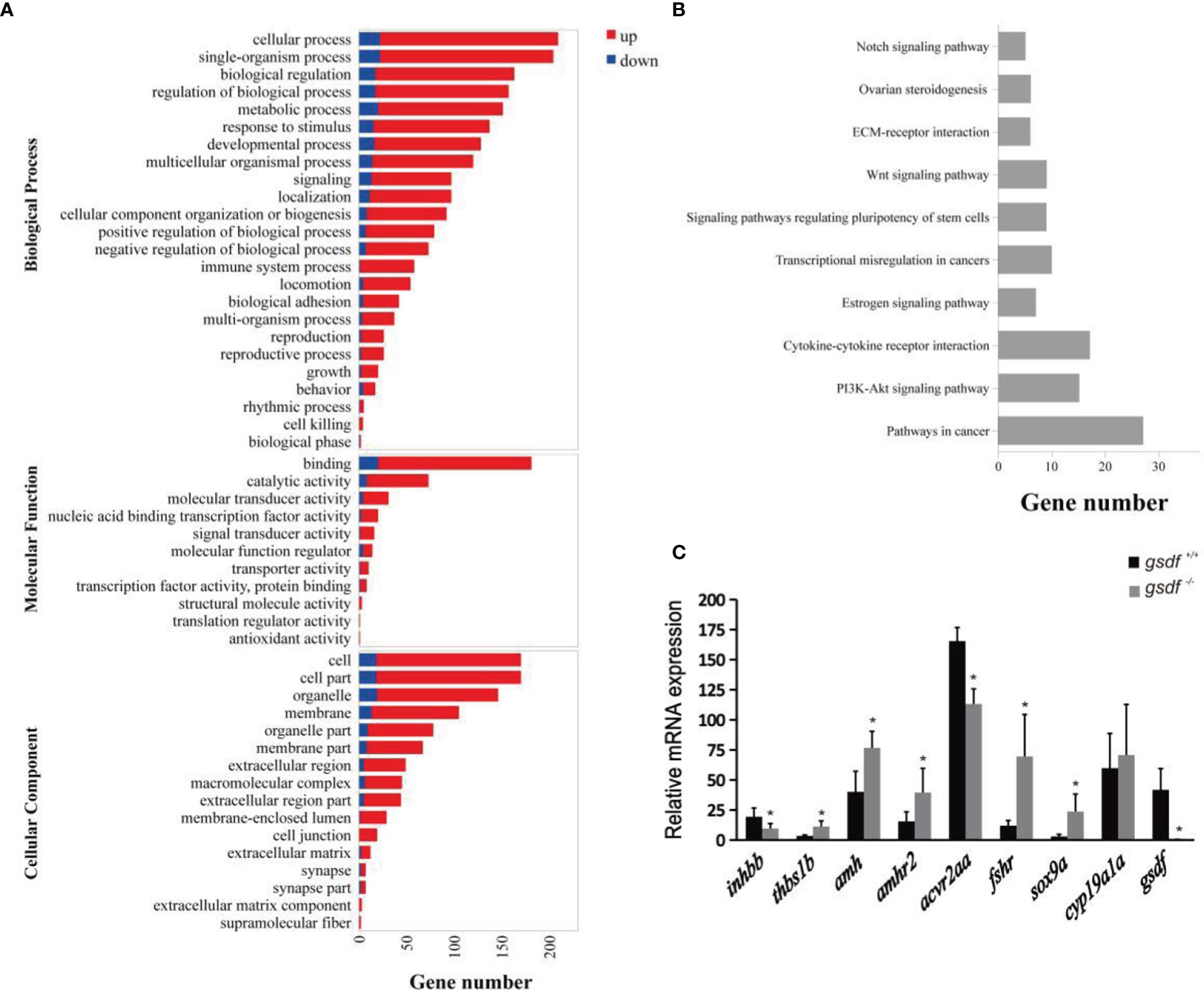
Figure 2 Transcriptome analysis of gene expression in ovaries of XX gsdf -/- and gsdf +/+ tilapia at 6 mah. (A) GO enrichment analysis of DEGs. Red and blue indicate up-regulated and down-regulated DEGs, respectively. (B) The top ten enriched KEGG pathways of the DEGs. (C) qPCR validation of the transcriptome data. All the examined genes showed similar expression patterns with the transcriptome data. Results are presented as mean ± SD. * indicates significant difference at P < 0.05.
The KEGG enrichment analysis showed that 178 DEGs enriched 244 signaling pathways. The top four enriched KEGG pathways include cancer, cytokine-cytokine receptor interaction, ovarian steroidogenesis, and estrogen signaling pathway (Figure 2B and Table S6). In the steroidogenesis pathway, two genes, fshr and hsd17b1, were significantly up-regulated in mutant fish. Several DEGs were enriched in the TGF-β signaling pathway, including inhbb, acvr2aa, thbs1b, amh and amhr2. In addition, qPCR confirmed that gsdf, inhbb and acvr2aa were significantly down-regulated, while thbs1b, amh, amhr2, fshr, and sox9a were up-regulated considerably in gsdf -/- ovaries, verifying the gonadal transcriptomic data (Figure 2C).
3.3 Immunofluorescence of Gsdf, Cyp19a1a and Amh Expression in the Ovaries of XX gsdf-/- Fish at 6 mah
Firstly, the expression of Gsdf positive signal was detected in the ovaries of XX gsdf +/+ fish. While the Gsdf signal was absent in the XX gsdf -/- fish ovaries at 6 mah, confirming that the Gsdf protein was wholly removed from the XX gsdf knockout fish ovary (Figures 3A–C). Estradiol (E2) is critical for fish ovary development, and cyp19a1a encodes aromatase, the rate-limited enzyme for E2 synthesis. The gonad transcriptome and qPCR analyses showed that the expression of cyp19a1a in gsdf -/- and wild-type fish are indistinguishable. Therefore, an IF analysis was performed to confirm the protein level expression of Cyp19a1a. The Cyp19a1a signal was present in the interstitial and follicle cells of the ovaries from both XX gsdf +/+ and gsdf -/- fish. The fluorescence signal intensity between the knockout and wild-type ovaries was similar (Figures 3D–F). Amh positive signals were also observed in the follicle cells of the ovaries of both XX gsdf +/+ and gsdf -/- fish at 6 mah (Figures 3G, H). Moreover, the quantitative Amh positive signal in the XX gsdf -/- ovary was significantly higher than the XX gsdf +/+ ovary at 6 mah, consistent with the qPCR results (Figure 3I).
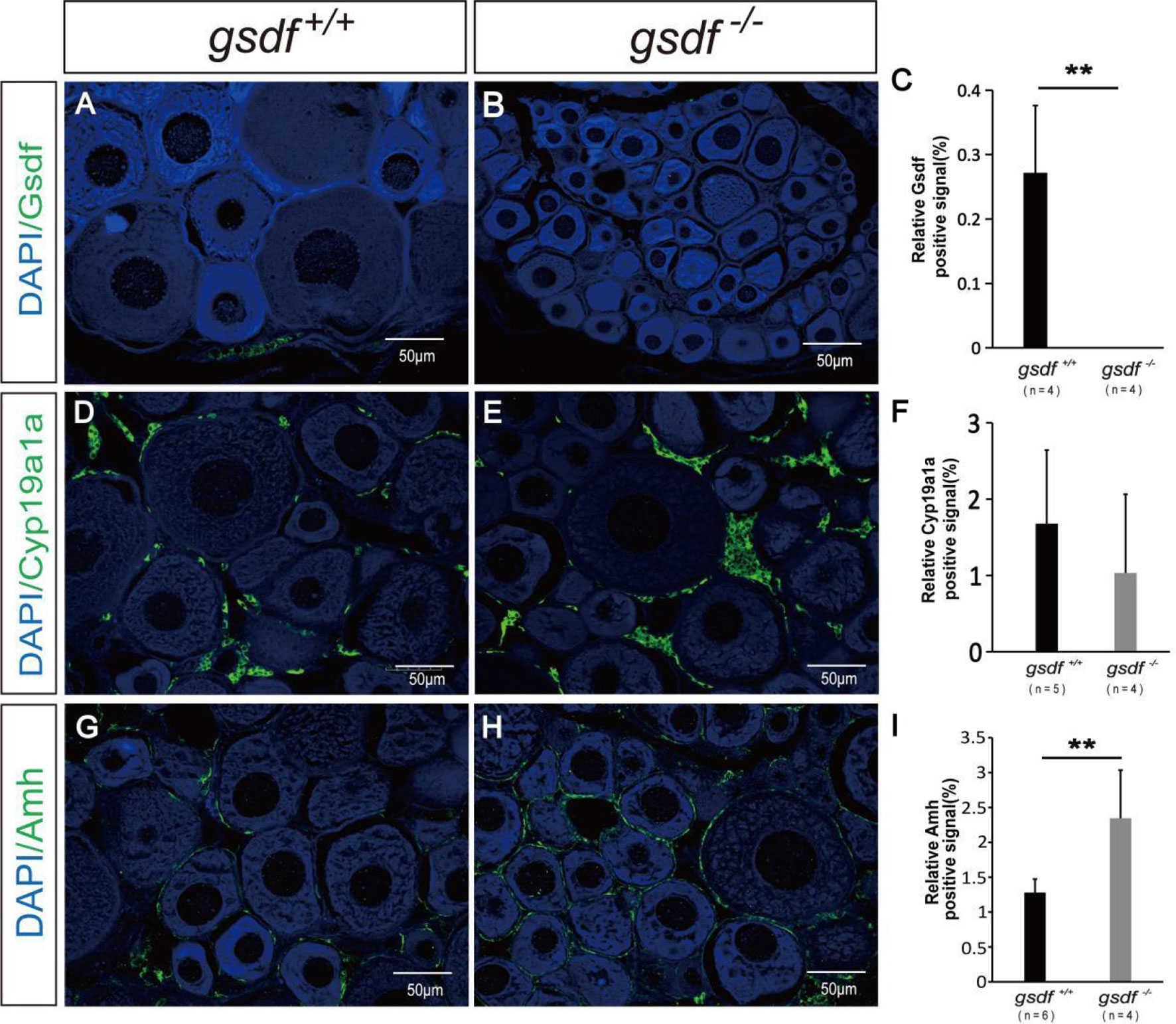
Figure 3 Immunofluorescence analysis of Gsdf, Cyp19a1a and Amh in ovaries of XX gsdf -/- and gsdf +/+ tilapia at 6 mah. Gsdf (A, B), Cyp19a1a (D, E) and Amh (G, H). Green fluorescence represents positive signal. Blue fluorescence represents the DAPI signal. The statistical analysis of the positive signal intensity (C, F, I). Results are presented as mean ± SD. ** indicates significant difference at P < 0.01.
3.4 The Serum Steroid Hormones Level in XX gsdf+/+ and gsdf -/- Fish
The serum steroid hormone concentration was examined in the XX gsdf +/+ and gsdf -/- fish at 6 and 24 mah to detect whether steroid hormones affect female sterility. There was no significant difference in E2, 11-KT and T levels in sera from XX gsdf +/+ and gsdf -/- fish at both 6 and 24 mah (Figures 4A, B). However, the P level in XX gsdf +/+ was significantly higher than gsdf -/- fish at both 6 and 24 mah.
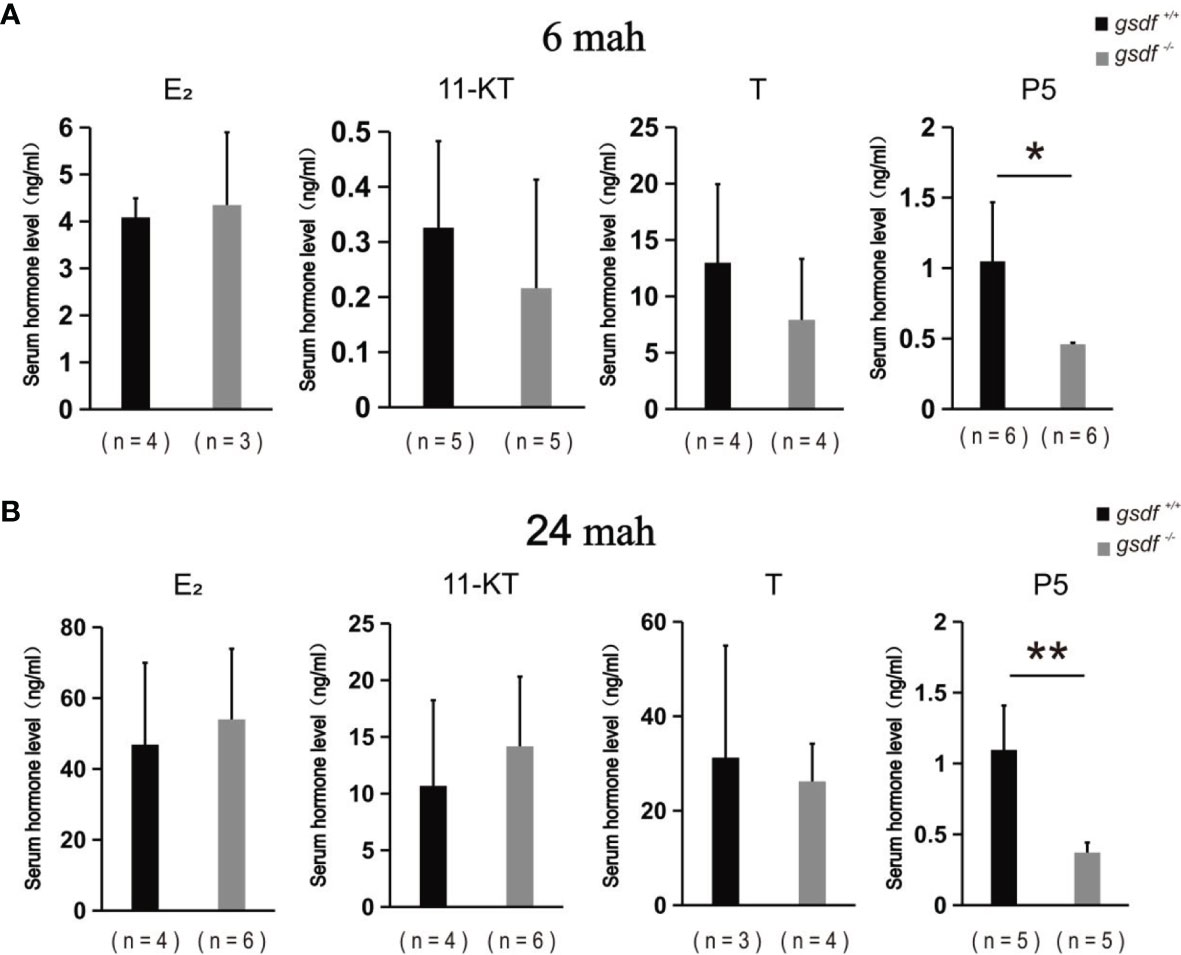
Figure 4 Serum E2, 11-KT, T and P5 levels of XX gsdf +/+ and gsdf -/- tilapia at 6 (A) and 24 (B) mah. Results are presented as mean ± SD. * and ** above the error bar indicate significant difference at P < 0.05 and P < 0.01, respectively.
3.5 Characterization of Gsdf Receptors
Yeast-two-hybrid assays verified the interaction between Gsdf and Amhr2, and Gsdf and Bmpr2a. NMY51 yeast co-transformed with the selected plasmids grew on the DDO plate, indicating successful transformation of the plasmids into yeast. The yeast co-transformed with the negative control plasmids did not grow on TDO and QDO plates, indicating that they did not self-activate the expression of HIS3 and ADE2 reporter genes, respectively (Figure 5). Whereas the yeast co-transformed with the positive control plasmids grew on the TDO and QDO plates. In addition, the yeast co-transformed with pBT3-SUC-gsdf + Post-Nubal plasmids grew on TDO and QDO plates, indicating the functional role of the ubiquitin system. The yeast co-transformed with pBT3-SUC-gsdf + pPR3-N did not grow on TDO plates, while the yeast co-transformed with pBT3-SUC-gsdf + pPR3-N-amhr2, and pBT3-SUC-gsdf + pPR3-N-bmpr2a plasmids grew on TDO plates, indicating the successful co-transformation and activation of the HIS3 reporter gene. However, the yeast co-transformed with pBT3-SUC-gsdf + pPR3-N-amhr2, and pBT3-SUC-gsdf + pPR3-N-bmpr2a plasmids did not grow on QDO plates, implying that the ADE2 reporter gene was not activated. These results showed a weak interaction between Gsdf and Amhr2, and Gsdf and Bmpr2a.
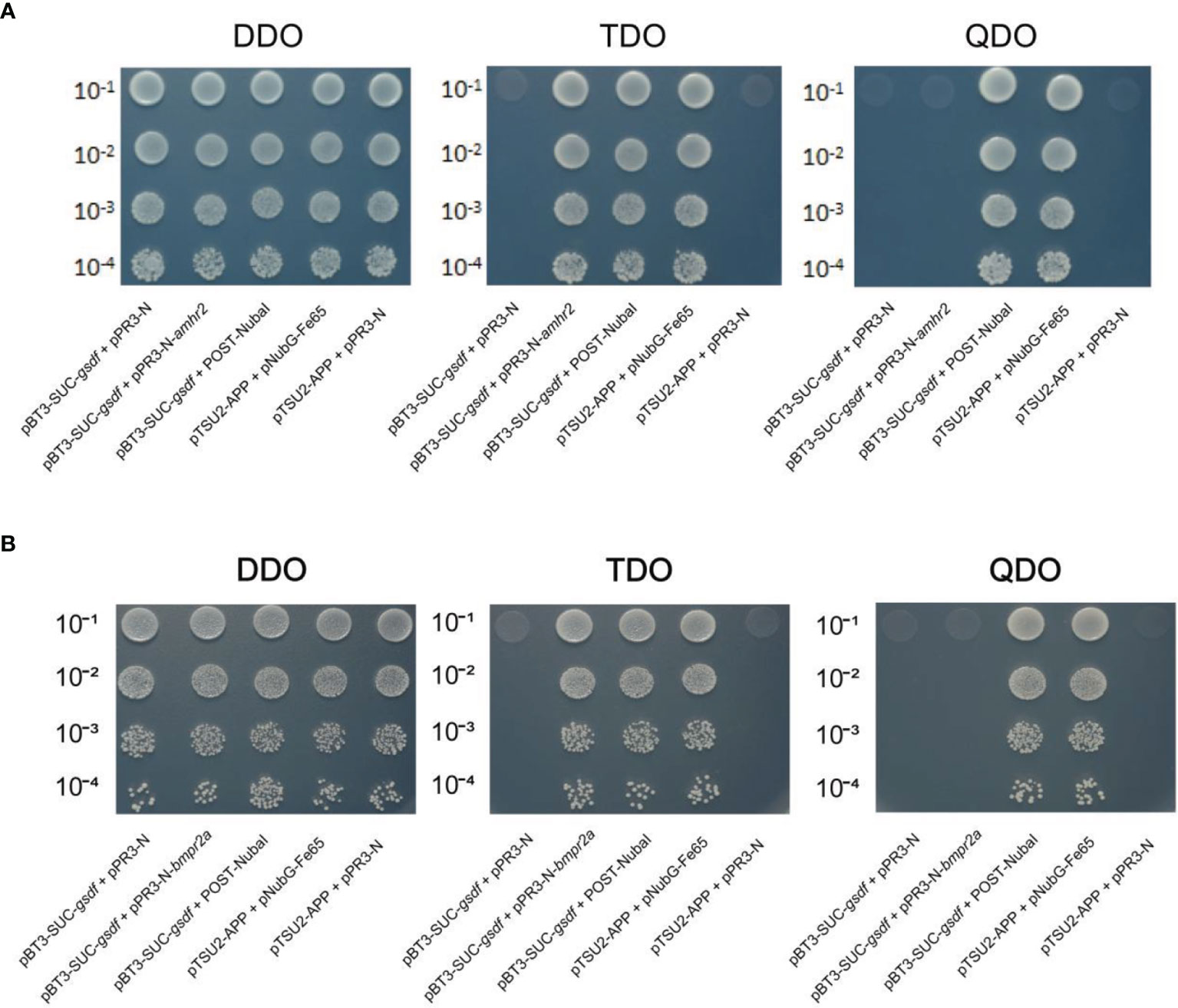
Figure 5 Yeast-two-hybrid assays for Gsdf (bait) interaction with the prey [Amhr2 (A) and Bmpr2a (B)] proteins. The hybridized yeast cell that survived on TDO and QDO plates indicate weak and strong positive interactions, respectively. The pBT3-SUC-gsdf + pPR3-N was set for detection of self-activation, while pBT3-SUC-gsdf + POST-Nubal was performed for functional verification of the ubiquitin system. The pTSU2-APP + pNubG-Fe65 and pTSU2-APP + pPR3-N were used as the positive and negative control, respectively.
4 Discussion
The Gsdf protein has remained a research focus since its first discovery in rainbow trout 15 years ago (1, 2). The Gsdf protein is mainly expressed in somatic cells of gonads, and gsdf functions in testicular differentiation in Japanese medaka and Nile tilapia (5–7). Loss of gsdf caused infertility in female zebrafish and Japanese medaka, indicating that Gsdf is also essential for ovary development in fish. This study analyzed the phenotype of gsdf mutant and the possible role of gsdf in ovary development in female Nile tilapia.
4.1 Phenotypes of gsdf Mutants in Fish
In Japanese medaka, the ovaries of the gsdf female mutant accumulate numerous small and previtellogenic oocytes while lacking the mature vitellogenic oocytes resulting in low fertility or infertility (10). In zebrafish, most gsdf mutant females lay a few eggs at 4.5 mpf (month post-fertilization) but become sterile at 18 mpf (8). In zebrafish, mutant ovaries have more younger oocytes than the wild type. Mutant ovaries at 18 mpf lack mature oocytes, indicating that gsdf is required for oocyte maturation and inhibits the accumulation of young ovarian follicles in zebrafish. Similarly, the gsdf homozygous mutated ovaries of Nile tilapia had an increased number of primary oocytes at 24 mah, lacked mature oocytes and finally became infertile. Gene knockout study in two Perciformes (Japanese medaka and Nile tilapia) and one Cypriniforme fish (zebrafish) species concluded that gsdf is functionally conserved for regulating young oocyte maturity and is critical for fertility in female fish (8, 10). In Nile tilapia, Gsdf is mainly expressed in the somatic cell around the oogonia (7, 40). Gsdf may act as a paracrine factor and directly control the proper proliferation of the oogonia, or control the transition of the oogonia into the oocyte. Gsdf may also act as an autocrine factor and could indirectly influence the oogonia and oocyte via somatic cell and germ cell interaction. It is interesting to reveal how Gsdf is involved in oocyte development. In doing so, it is necessary to identify its receptors and where they are localized.
4.2 Receptors of Gsdf in Nile Tilapia
Genomics synteny analysis showed that gsdf is a paralog of the BMP members (8). Consistently, the yeast-two-hybrid results from this study showed that the Nile tilapia Gsdf binds to type II receptor in BMP signal pathway in vitro. Our experimental results confirmed that Gsdf belongs to the BMP group, and it shares the type II receptors with other BMP ligands, which was further proved by gene mutation analysis of TGF-β ligands and receptors.
Similar phenotypes of gsdf mutation were observed in other TGF-β ligand/receptor mutants in fish species. In Japanese medaka, the homozygous hotei mutation of amhr2 causes early-stage follicle arrest, the mutant ovaries become hypertrophic, and most mutant female fish become sterile (22). In zebrafish, young amh mutants lay very few eggs, while older mutants become infertile with accumulated non-vitellogenic follicles in exceedingly large ovaries (19). Amhr2 was evolutionarily lost in zebrafish; thus, Amh binds with another type II receptor of the TGF-β signaling pathway (41). Knockouts of bmpr2a phenocopied the phenotypes of amh mutants in male and female zebrafish, including gonadal hypertrophy, hyperproliferation of germ cells, and retarded gametogenesis. Thus, Bmpr2a is considered a possible type II receptor for Amh in zebrafish (21). Besides, the XX Nile tilapia with amh or amhr2 deficiency displays hypertrophic ovaries with many primary oocytes but fails to reproduce (24).
Both gsdf and amh are expressed in the granulosa cells of the fish ovary, and both act as paracrine factors which might influence the neighboring germ cells (7, 24). In Nile tilapia, the amhr2 is expressed in granulosa cells, oogonia and phase I oocytes, indicating that amh regulates germ cells directly via its type II receptor, Amhr2. This is also supported by the similar phenotypes observed in amh and amhr2 null female Nile tilapia. However, there is still no direct evidence showing that fish Amh binds with Amhr2, as it is with mammals (24). Herein, yeast-two-hybrid experiments showed that Gsdf binds with Amhr2. Therefore, Gsdf might control germ cell development via interactions with Amhr2 in Nile tilapia. The oocytes are unable to grow into stage IV either in the amh or amhr2 null ovaries at the adult stage (24), while stage IV oocyte was found in gsdf null ovaries in the present study. The similar amh and amhr2 null female phenotype suggests that Amh, instead of Gsdf, probably is the main ligand of Amhr2 in Nile tilapia. Considering that gsdf, amh, amhr2, and bmpr2a showed similar phenotypes of ovary development in different fish species, we deduced that gsdf and amh share similar receptors and control similar downstream genes in different fish species. Their downstream genes control germ cell proliferation and differentiation in the ovary.
4.3 Possible Mechanisms of Gsdf Signal in Regulation of Ovary Development
4.3.1 The Influence of Gsdf on Steroid Hormone Synthesis in the Ovary
Estrogens synthesized in somatic and germ cells are critical for normal folliculogenesis of fish ovaries (42, 43). In Nile tilapia, mutations of amh, amhr2, foxh1, and tsp1a down-regulate cyp19a1a expression in the ovary and serum E2 levels (24, 44, 45). The decreased E2 levels caused abnormal oocyte development in the mutants. However, exogenous E2 supplementation in amh and amhr2 female mutants failed to rescue the folliculogenesis arrest caused by apoptosis of follicle cells (24).
E2 levels in the whole body of gsdf -/- female zebrafish were lower than those of the wild-type fish at 11 and 20 mpf (19). The ovary cyp19a1a expression also significantly decreased in the gsdf -/- female zebrafish. In the current study, both ovary cyp19a1a and serum E2 levels of gsdf mutants were similar to the wild-type fish at 6 and 24 mah, indicating that Gsdf does not directly control estrogen synthesis in the ovary. The early-stage arrest of oocyte development and infertility of female gsdf -/- Nile tilapia may not be due to either deficiency or excessive estrogen. The estrogens interact with three receptors (Esr1, Esr2a, Esr2b) in Nile tilapia. Functional studies demonstrated that esr2a mutants show delayed ovary development (13). In contrast, the transcriptome data showed that none of these esrs`s expressions were altered in the gsdf mutants. However, we could not exclude the possibility that other molecular components in the estrogen signal pathway in the female gsdf -/- Nile tilapia were affected. Further studies should elucidate the underlying mechanism of gsdf mutation on the estrogen signaling pathway, which is critical for the regulation of oogenesis in different fish species.
Moreover, the serum pregnenolone levels significantly decreased in female gsdf -/- than the wild-type fish. The initial step of pregnenolone synthesis involves cholesterol transport to the inner mitochondrial membrane by steroidogenic acute regulatory protein (StAR). StAR is encoded by two paralogous genes, StAR1 and StAR2, of teleosts fish, including Nile tilapia (46). The cholesterol side-chain cleavage enzyme, coded by P450scc (Cyp11a1), converts cholesterol to pregnenolone (47). The present transcriptome results showed similar expression levels of StAR1, StAR2 and cyp11a1 between gsdf -/- and wild-type ovaries. Therefore, the decreasing serum pregnenolone levels of gsdf -/- fish are not attributable to these genes. However, protein-level expression of these genes is required to confirm these speculations. Additionally, pregnenolone is the intermediate product for most of steroids, including estrogen. Its decrease might change the activities of downstream enzymes. The decreased E2 level in gsdf -/- zebrafish and pregnenolone level in gsdf -/- Nile tilapia indicate that Gsdf controls ovary development by regulating certain steroid hormones in fish. Nevertheless, future studies should address the Gsdf regulatory mechanisms on steroid hormone synthesis in the different fish.
4.3.2 The Influence of Gsdf on Gonadotropin Synthesis
Sex steroid hormone synthesis, gonadal growth, and differentiation are regulated by gonadotropin signals through gonadal G-protein-coupled receptors (48). The gonadotropins include Fsh and Lh, and their mammalian receptors are Fshr and Lhcgr, respectively. Unlike in mammals, both Fsh and Lh interact with Fshr in zebrafish (49). Moreover, disrupting the fshr gene causes the arrest of ovarian follicles at the PG-PV transition. Later on, fshr is indispensable for folliculogenesis in zebrafish, while lhcgr may not be essential for female zebrafish reproduction, indicating that Fshr is more important for normal ovary development than Lhcgr in fish (49).
Besides, Fsh-deficient (fshb-/-) female zebrafish show delayed puberty onset but fertile, while Lh-deficient (lhb-/-) females show normal gonadal growth but are infertile from the failure to spawn (50). Pituitary lhb, ovary lhcgr and fshr decrease significantly in female gsdf -/- zebrafish (8). Similarly, pituitary lhb and fsh, and ovary lhr and fshr reduced considerably in female amh -/- and amhr2 -/- Nile tilapia (24). The decreased gonadotropin signals might be caused by early-stage follicle arrest in the amh or amhr2 mutant fish. The pituitary fshb was significantly down-regulated, and ovary fshr was significantly up-regulated in female gsdf -/- Nile tilapia (Figure S3). Gonadotropin signaling probably decreased in female gsdf -/- Nile tilapia, causing follicle arrest in the mutants. Altogether, gsdf possibly regulates ovary development via the gonadotropin signaling pathway in fish. However, besides gonads, amh and amhr2 are also expressed in the brain and pituitary of Nile tilapia, while gsdf is only expressed in the gonads (24, 40). Consequently, Gsdf might influence the pituitary gonadotropin signaling directly via its putative receptors (Amhr2 and Bmpr2a) if it could be transported to the pituitary or brain regions via the blood. Further studies are needed to support this speculation.
More evidence proves that the TGF-β signaling genes, besides Amh, gsdf and Amhr2 discussed above, are critical for regulating gonadotropin signaling pathways in vertebrates. The other two TGF-β subfamily members, Activin and Inhibin, antagonize each other to regulate pituitary fshb expression and Fsh secretion in vertebrates (51). Inhibin contains a unique inhibin-specific α subunit (Inha) and shares a common β subunit (Inhba and Inhbb) with Activin to form Inhibin A, or Inhibin B. Activins are homodimers of either Inhba or Inhbb or their heterodimers. In mammals, Activin stimulates, and Inhibin inhibits the Fsh section (51, 52).
Pituitary expression of fshb significantly increases in inha-/- female zebrafish compared with wild-type fish (52). However, the expression of ovarian inha, fshr, and lhr decreases significantly in female gsdf -/- Japanese medaka (10). Previously, IHC analysis showed positive Fshb and Lhb signals in cells of XY gsdf-null pituitaries compared with the wild-type males or females, indicating an increased Fsh and Lh secretion in medaka (53). In this study, ovary inhbb and pituitary fshb genes were significantly down-regulated in female gsdf -/- Nile tilapia. Altogether, gsdf mutations possibly influence pituitary fsh gene expression in fish via the Activin and Inhibin signaling pathway.
4.3.3 The Disorganized Gene Expression in gsdf Mutants
Gsdf mutation influences the same genes in different ways in different species, as mentioned above. We recognize that the TGF-β signaling pathway shares the same downstream proteins, such as Smad4, which combine with other Smads to form activated Smad complexes that move to the nucleus to stimulate or inhibit downstream genes. Therefore, these TGF-β ligands are tightly linked to complex gene regulatory networks. Nevertheless, absence of any TGF-β ligand negatively influences the whole TGF-β signaling pathway.
Furthermore, the numbers of TGF-β ligands vary in different species because of gene duplication or loss (16). Thus, deficient of a TGF-β ligand in one species substantially influences the species physiology and homeostasis. However, this phenomenon would improve our understanding of TGF-β ligand mutations in different species and their effects on gene expression. Although gsdf mutations exert differential influences on different genes in female fish from different species. The final, infertile phenotype of female mutants was consistent and conserved in three unrelated fish species (Japanese medaka, zebrafish, and Nile tilapia). The infertility of gsdf mutants is probably due to genetic toxicity caused by the disorganized TGF-β signaling in the ovary after mutation. The KEGG analysis enriched more than 20 genes in the cancer pathway and supported our speculation that the gsdf mutated fish is unhealthy and has a defect in the ovary. The gsdf mutated female fish could be used as an ovary development disorder model caused by TGF-β signaling deficiency in vertebrates.
5 Conclusion
This study demonstrated that female gsdf -/- Nile tilapia was infertile, and the development of oocytes was arrested at an early stage. There was no significant changes in serum estrogen level and ovarian cyp19a1a expression in gsdf knockout fish, indicating that infertility in gsdf mutants was not caused by estrogen deficiency. The expression of several TGF-β signaling genes changed significantly in the ovaries of gsdf knockout fish, which may be the reason for its infertility. In addition, two possible type II receptors of Gsdf were identified by yeast-two-hybrid assays for the first time. These results enrich our understanding of the role of Gsdf in the oogenesis of teleosts.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR13752974-SRR13752981.
Ethics Statement
The animal study was reviewed and approved by Animal Research and Ethics Committee of Guangdong Ocean University.
Author Contributions
D-NJ and Y-XP took part in the whole process of the experiment, and wrote the draft of this manuscript. X-YL, Y-QH, and UM participated in the experiments. M-HL, H-JS, and G-LL co-conceived the experiment. UM revised the draft. D-SW and D-NJ designed and supervised the experiments. All authors read and approved the final manuscript. All authors approved the final version to be published.
Funding
The authors thank the financial support provided by the Key Project of “Blue Granary Science and Technology Innovation” of the Ministry of Science and Technology (2018YFD0900202); the National Natural Science Foundation of China (Project Nos. 31702326, 32002367, 32072960 and 32172971).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Yoshitaka Nagahama (National Institute for Basic Biology Okazaki, Japan) for providing Cyp19a1a antibody. We thank the funding agency for supporting our study. We would also like to thank all lab members for helpful discussions and assistance in lab management.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.813320/full#supplementary-material
References
1. Hsu CW, Chung BC. Evolution, Expression, and Function of Gonadal Somatic Cell-Derived Factor. Front Cell Dev Biol (2021) 9:684352:684352. doi: 10.3389/fcell.2021.684352
2. Sawatari E, Shikina S, Takeuchi T, Yoshizaki G. A Novel Transforming Growth Factor-Beta Superfamily Member Expressed in Gonadal Somatic Cells Enhances Primordial Germ Cell and Spermatogonial Proliferation in Rainbow Trout (Oncorhynchus Mykiss). Dev Biol (2007) 301:266–75. doi: 10.1016/j.ydbio.2006.10.001
3. Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, et al. Tracing The Emergence of A Novel Sex-Determining Gene in Medaka, Oryzias Luzonensis. Genetics (2012) 191:163–70. doi: 10.1534/genetics.111.137497
4. Herpin A, Schartl M, Depincé A, Guiguen Y, Bobe J, Hua-Van A, et al. Allelic Diversification After Transposable Element Exaptation Promoted Gsdf as The Master Sex Determining Gene of Sablefish. Genome Res (2021) 31(8):1366–80. doi: 10.1101/gr.274266.120
5. Imai T, Saino K, Matsuda M. Mutation of Gonadal Soma-Derived Factor Induces Medaka XY Gonads to Undergo Ovarian Development. Biochem Bioph Res Co (2015) 467:109–14. doi: 10.1016/j.bbrc.2015.09.112
6. Zhang X, Guan G, Li M, Zhu F, Liu Q, Naruse K, et al. Autosomal Gsdf Acts as A Male Sex Initiator in The Fish Medaka. Sci Rep-UK (2016) 6:19738. doi: 10.1038/srep19738
7. Jiang DN, Yang HH, Li MH, Shi HJ, Zhang XB, Wang DS. Gsdf is A Downstream Gene of Dmrt1 That Functions in The Male Sex Determination Pathway of The Nile Tilapia. Mol Reprod Dev (2016) 83(6):497–508. doi: 10.1002/mrd.22642
8. Yan YL, Desvignes T, Bremiller R, Wilson C, Dillon D, High S, et al. The Gonadal Soma Controls Ovarian Follicle Proliferation Through Gsdf in Zebrafish. Dev Dynam (2017) 246:925–45. doi: 10.1002/dvdy.24579
9. Gautier A, Le Gac F, Lareyre JJ. The Gsdf Gene Locus Harbors Evolutionary Conserved and Clustered Genes Preferentially Expressed in Fish Previtellogenic Oocytes. Gene (2011) 472:7–17. doi: 10.1016/j.gene.2010.10.014
10. Guan G, Sun K, Zhang X, Zhao X, Li M, Yan Y. Developmental Tracing of Oocyte Development in Gonadal Soma-Derived Factor Deficiency Medaka (Oryzias Latipes) Using A Transgenic Approach. Mech Dev (2017) 143:53–61. doi: 10.1016/j.mod.2016.12.006
11. Lubzens E, Young G, Bobe J, Cerdà J. Oogenesis in Teleosts: How Fish Eggs Are Formed. Gen Comp Endocr (2010) 165(3):367–89. doi: 10.1016/j.ygcen.2009.05.022
12. Takahashi A, Kanda S, Abe T, Oka Y. Evolution of The Hypothalamic-Pituitary-Gonadal Axis Regulation in Vertebrates Revealed by Knockout Medaka. Endocrinology (2016) 157(10):3994–4002. doi: 10.1210/en.2016-1356
13. Yan LX, Feng HW, Wang FL, Lu BY, Liu XY, Sun LN, et al. Establishment of Three Estrogen Receptors (Esr1, Esr2a, Esr2b) Knockout Lines for Functional Study in Nile Tilapia. J Steroid Biochem (2019) 191:105379. doi: 10.1016/j.jsbmb.2019.105379
14. Kayo D, Zempo B, Tomihara S, Oka Y, Kanda S. Gene Knockout Analysis Reveals Essentiality of Estrogen Receptor β1 (Esr2a) for Female Reproduction in Medaka. Sci Rep-UK (2019) 9(1):8868. doi: 10.1038/s41598-019-45373-y
15. Zhu Y, Liu DT, Shaner ZC, Chen SX, Hong WS, Stellwag EJ, et al. Nuclear Progestin Receptor (Pgr) Knockouts in Zebrafish Demonstrate Role for Pgr in Ovulation But Not in Rapid Non-Genomic Steroid Mediated Meiosis Resumption. Front Endocrinol (2015) 6:37:37. doi: 10.3389/fendo.2015.00037
16. Zheng SQ, Long J, Liu ZL, Tao WJ, Wang DS. Identification and Evolution of TGF-β Signaling Pathway Members in Twenty-Four Animal Species and Expression in Tilapia. Int J Mol Sci (2018) 19(4):1154. doi: 10.3390/ijms19041154
17. Liu Z, Ning G, Xu R, Cao Y, Meng A, Wang Q. Fscn1 is Required for The Trafficking of TGF-β Family Type I Receptors During Endoderm Formation. Nat Commun (2016) 7:12603. doi: 10.1038/ncomms12603
18. Yan Y, Ning G, Li L, Liu J, Wang Q. The BMP Ligand Pinhead Together With Admp Supports The Robustness of Embryonic Patterning. Sci Adv (2019) 5(12):eaau6455. doi: 10.1126/sciadv.aau6455
19. Yan YL, Batzel P, Titus T, Sydes J, Desvignes T, Bremiller R, et al. A Hormone That Lost Its Receptor: Anti-Müllerian Hormone (Amh) in Zebrafish Gonad Development and Sex Determination. Genetics (2019) 213:529–53. doi: 10.1534/genetics.119.302365
20. Neumann JC, Chandler GL, Damoulis VA, Fustino NJ, Lillard K, Looijenga L, et al. Mutation in The Type IB Bone Morphogenetic Protein Receptor Alk6b Impairs Germ-Cell Differentiation and Causes Germ-Cell Tumors in Zebrafish. P Natl Acad Sci USA (2011) 108(32):13153–8. doi: 10.1073/pnas.1102311108
21. Zhang Z, Wu K, Ren Z, Ge W. Genetic Evidence for Amh Modulation of Gonadotropin Actions to Control Gonadal Homeostasis and Gametogenesis in Zebrafish and Its Noncanonical Signaling Through Bmpr2a Receptor. Development (2020) 147(22):dev.189811. doi: 10.1242/dev.189811
22. Morinaga C, Saito D, Nakamura S, Sasaki T, Asakawa S, Shimizu N, et al. The Hotei Mutation of Medaka in The Anti-Mullerian Hormone Receptor Causes the Dysregulation of Germ Cell and Sexual Development. P Natl Acad Sci USA (2007) 104(23):9691–6. doi: 10.1073/pnas.0611379104
23. Nakamura S, Watakabe I, Nishimura T, Picard JY, Toyoda A, Taniguchi Y, et al. Hyperproliferation of Mitotically Active Germ Cells Due to Defective Anti-Müllerian Hormone Signaling Mediates Sex Reversal in Medaka. Development (2012) 139(13):2283–7. doi: 10.1242/dev.076307
24. Liu XY, Xiao HS, Jie MM, Dai SF, Wang DS. Amh Regulate Female Folliculogenesis and Fertility in A Dose-Dependent Manner Through Amhr2 in Nile Tilapia. Mol Cell Endocrinol (2019) 499:110593. doi: 10.1016/j.mce.2019.110593
25. Cimino I, Casoni F, Liu X, Messina A, Giacobini P. Novel Role For Anti-Müllerian Hormone in The Regulation of GnRH Neuron Excitability and Hormone Secretion. Nat Commun (2016) 7:10055. doi: 10.1038/ncomms10055
26. Li MH, Sun YL, Zhao JE, Shi HJ, Zeng S, Ye K, et al. A Tandem Duplicate of Anti-Mullerian Hormone With A Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis Niloticus. PloS Genet (2015) 11(11):e1005678. doi: 10.1371/journal.pgen.1005678
27. Li MH, Yang HH, Zhao JU, Fang LL, Shi HJ, Li MR, et al. Efficient and Heritable Gene Targeting in Tilapia by CRISPR/Cas9. Genetics (2014) 197(2):591–3SI. doi: 10.1534/genetics.114.163667
28. Conte MA, Gammerdinger WJ, Bartie KL, Penman DJ. Kocher TD. A High Quality Assembly of The Nile Tilapia (Oreochromis Niloticus) Genome Reveals the Structure of Two Sex Determination Regions. BMC Genomics (2017) 18(1):341. doi: 10.1186/s12864-017-3723-5
29. Conte MA, Joshi R, Moore EC, Nandamuri SP, Gammerdinger WJ, Roberts RB, et al. Chromosome-Scale Assemblies Reveal the Structural Evolution of African Cichlid Genomes. GigaScience (2019) 8(4):4. doi: 10.1093/gigascience/giz030
30. Li MH, Dai SF, Liu XY, Xiao HS. Wang DS. A Detailed Procedure for CRISPR/Cas9-Mediated Gene Editing in Tilapia. Hydrobiologia (2020) 848(16):1–17. doi: 10.1007/s10750-020-04414-8
31. Sun YL, Jiang DN, Zeng S, Hu CJ, Ye K, Yang C, et al. Screening and Characterization of Sex-Linked DNA Markers and Marker-Assisted Selection in the Nile Tilapia (Oreochromis Niloticus). Aquaculture (2014) 433:19–27. doi: 10.1016/j.aquaculture.2014.05.035
32. Jiang DN, Kuang ZY, Yang KS, Huang YQ, Mustapha UF, Guo XZ, et al. Polymorphism in A Sex-Linked DNA Marker Located on LG23 in Hainan Strain of Nile Tilapia (Oreochromis Niloticus). J World Aquacult Soc (2022) 53:205–23. doi: 10.1111/jwas.12815
33. Coward K, Bromage NR. Histological Classification of Oocyte Growth and The Dynamics of Ovarian Recrudescence in Tilapia Zillii. J Fish Biol (2010) 53(2):285–302. doi: 10.1111/j.1095-8649.1998.tb00981.x
34. Kim D, Langmead B, Salzberg SL. HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12(4):357–60. doi: 10.1038/nmeth.3317
35. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie Enables Improved Reconstruction of A Transcriptome From RNA-Seq Reads. Nat Biotechnol (2015) 33(3):290–5. doi: 10.1038/nbt.3122
36. Li B, Dewey CN. RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without A Reference Genome. BMC Bioinf (2011) 12(1):323–3. doi: 10.1186/1471-2105-12-323
37. Love MI, Huber W, Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data With Deseq2. Genome Biol (2014) 15(12):550. doi: 10.1186/s13059-014-0550-8
38. Livak KJ, Schmittgen T. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔct Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
39. Kobayashi T, Kajiura-Kobayashi H, Nagahama Y. Induction of XY Sex Reversal by Estrogen Involves Altered Gene Expression in A Teleost, Tilapia. Cytogenet Genome Res (2003) 101(3-4):289–94. doi: 10.1159/000074351
40. Kaneko H, Ijiri S, Kobayashi T, Izumi H, Kuramochi Y, Wang DS, et al. Gonadal Soma-Derived Factor (Gsdf), A TGF-Beta Superfamily Gene, Induces Testis Differentiation in the Teleost Fish Oreochromis Niloticus. Mol Cell Endocrinol (2015) 415(C):87–99. doi: 10.1016/j.mce.2015.08.008
41. Rocha A, Zanuy S, Gomez A. Conserved Anti-Mullerian Hormone: Anti-Mullerian Hormone Type-2 Receptor Specific Interaction and Intracellular Signaling in Teleosts. Biol Reprod (2016) 94(6):141. doi: 10.1095/biolreprod.115.137547
42. Gohin M, Bodinier P, Fostier A, Chesnel F, Bobe J. Aromatase is Expressed and Active in The Rainbow Trout Oocyte During Final Oocyte Maturation. Mol Reprod Dev (2011) 78(7):510–8. doi: 10.1002/mrd.21335
43. Guiguen Y, Fostier A, Piferrer F, Chang CF. Ovarian Aromatase and Estrogens: A Pivotal Role for Gonadal Sex Differentiation and Sex Change in Fish. Gen Comp Endocr (2010) 165:352–66. doi: 10.1016/j.ygcen.2009.03.002
44. Jie M, Ma H, Zhou L, Wu J, Wang DS. Regulation of Female Folliculogenesis by Tsp1a in Nile Tilapia (Oreochromis Niloticus). Int J Mol Sci (2020) 21(16):5893. doi: 10.3390/ijms21165893
45. Tao WJ, Shi HJ, Yang J, Diakite HD, Kocher TD, Wang DS. Homozygous Mutation of Foxh1 Arrests Oogenesis Causing Infertility in Female Nile Tilapia. Biol Reprod (2020) 102(3):758–69. doi: 10.1093/biolre/ioz225
46. Yu XG, Wu LM, Xie L, Yang SJ, Charkraborty T, Shi HJ, et al. Characterization of Two Paralogous Star Genes in a Teleost, Nile Tilapia (Oreochromis Niloticus). Mol Cell Endocrinol (2014) 392(1-2):152–62. doi: 10.1016/j.mce.2014.05.013
47. Wang XJ, Wen HS, Li Y, Lyu LK, Song M, Zhang Y, et al. Characterization of cyp11a1 and Its Potential Role in Sex Asynchronous Gonadal Development of Viviparous Black Rockfish Sebastes Schlegelii (Sebastidae). Gen Comp Endocr (2020) 302:113689. doi: 10.1016/j.ygcen.2020.113689
48. Levavi-Sivan B, Bogerd J, Mananos EL, Gomez A, Lareyre JJ. Perspectives on Fish Gonadotropins and Their Receptors. Gen Comp Endocr (2010) 165(3):412–37. doi: 10.1016/j.ygcen.2009.07.019
49. Zhang ZW, Lau SW, Zhang LL, Ge W. Disruption of Zebrafish Follicle-Stimulating Hormone Receptor (Fshr) But Not Luteinizing Hormone Receptor (Lhcgr) Gene by TALEN Leads to Failed Follicle Activation in Females Followed by Sexual Reversal to Males. Endocrinology (2015) 156(10):3747–62. doi: 10.1210/en.2015-1039
50. Zhang ZW, Zhu B, Ge W. Genetic Analysis of Zebrafish Gonadotropin (FSH and LH) Functions by TALEN-Mediated Gene Disruption. Mol Endocrinol (2015) 29(1):76–98. doi: 10.1210/me.2014-1256
51. Lu HJ, Zhao C, Zhu B, Zhang ZW, Ge W. Loss of Inhibin Advances Follicle Activation and Female Puberty Onset But Blocks Oocyte Maturation in Zebrafish. Endocrinology (2020) 161(12):1–19. doi: 10.1210/endocr/bqaa184
52. Makanji Y, Zhu J, Mishra R, Holmquist C, Wong WPS, Schwartz NB, et al. Inhibin at 90: From Discovery to Clinical Application, a Historical Review. Endocr Rev (2014) 35(5):747–94. doi: 10.1210/er.2014-1003
Keywords: TGF-β signal pathway, estrogen, CRISPR/Cas9, oogenesis, infertility, Nile tilapia
Citation: Jiang D-N, Peng Y-X, Liu X-Y, Mustapha UF, Huang Y-Q, Shi H-J, Li M-H, Li G-L and Wang D-S (2022) Homozygous Mutation of gsdf Causes Infertility in Female Nile Tilapia (Oreochromis niloticus). Front. Endocrinol. 13:813320. doi: 10.3389/fendo.2022.813320
Received: 22 December 2021; Accepted: 20 January 2022;
Published: 15 February 2022.
Edited by:
So-Youn Kim, University of Nebraska Medical Center, United StatesCopyright © 2022 Jiang, Peng, Liu, Mustapha, Huang, Shi, Li, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: De-Shou Wang, wdeshou@swu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Dong-Neng Jiang
Dong-Neng Jiang You-Xing Peng1†
You-Xing Peng1† Umar Farouk Mustapha
Umar Farouk Mustapha Hong-Juan Shi
Hong-Juan Shi Guang-Li Li
Guang-Li Li De-Shou Wang
De-Shou Wang