- Jiangsu Key Laboratory for Biodiversity and Biotechnology, College of Life Sciences, Nanjing Normal University, Nanjing, China
G protein-coupled receptors (GPCRs) are the largest and most versatile family of transmembrane receptors in the cell and they play a vital role in the regulation of multiple physiological processes. The family Miridae (Hemiptera: Heteroptera) is one of the most diverse families of insects. Until now, information on GPCRs has been lacking in Miridae. Apolygus lucorum, a representative species of the Miridae, is an omnivorous pest that occurs worldwide and is notorious for causing serious damage to various crops and substantial economic losses. By searching the genome, 133 GPCRs were identified in A. lucorum. Compared with other model insects, we have observed GPCR genes to be remarkably expanded in A. lucorum, especially focusing on biogenic amine receptors and neuropeptide receptors. Among these, there is a novel large clade duplicated from known FMRFamide receptors (FMRFaRs). Moreover, the temporal and spatial expression profiles of the 133 genes across developmental stages were determined by transcriptome analysis. Most GPCR genes showed a low expression level in the whole organism of A. lucorum. However, there were a few highly expressed GPCR genes. The highly expressed LW opsins in the head probably relate to nocturning of A. lucorum, and the expression of Cirl at different times and in different tissues indicated it may be involved in growth and development of A. lucorum. We also found C2 leucine-rich repeat-containing GPCRs (LGRs) were mainly distributed in Hemiptera and Phthiraptera among insects. Our study was the first investigation on GPCRs in A. lucorum and it provided a molecular target for the regulation and control of Miridae pests.
1 Introduction
G protein-coupled receptors (GPCRs) are in a large family of protein cell surface receptors that detect molecules outside the cell and activate cellular responses (1, 2). GPCRs are found only in eukaryotes, namely, yeast, choanoflagellates, and animals (3). Based on sequence homology and functional similarity, GPCRs can be grouped into six families (4, 5): Family-A (rhodopsin-like); Family-B (secretin receptor family); Family-C (metabotropic glutamate/pheromone); Family-D (fungal mating pheromone receptors); Family-E (cyclic AMP receptors); and Family-F (frizzled/smoothened). These receptors are involved in a wide variety of physiological processes (6), namely, visual sensation (7), taste (8), smell sensation (9), behavioral and mood regulation (10), regulation of immune system activity and inflammation (11, 12), and autonomic nervous system transmission (10, 13). Because of their crucial roles in the regulation of multiple physiological processes, GPCRs are an important drug target (14) and approximately 34% (15) of all Food and Drug Administration (FDA) approved drugs target 108 members of this family.
With the continuous innovation of next-generation sequencing technology and bioinformatics, systematic identification research about GPCRs has been reported in several insects (16–21). Among Hemiptera, it has been reported in Acyrthosiphon pisum (22), Aphis craccivora (23), Cimex lectularius (24), Diaphorina citri (25), Nilaparvata lugens (26), and Rhodnius prolixus (27). The family Miridae (Hemiptera: Heteroptera), which includes plant bugs, leaf bugs, or grass bugs, is one of the most diverse families of insects, including over 11,000 species in more than 1,300 genera (4, 28). It is the largest family of true bugs belonging to Hemiptera and new members of Miridae are being described constantly. Mirids exhibit a wide range of food preferences and behaviors, including phytophagy, carnivory, and omnivory. Some mirids exhibit significant economic impacts and some are pests of food and fiber crops, whereas others are beneficial species used as biological control agents (29). Although Miridae is the largest family of Hemiptera and exhibits a complex habit, there has been little information reported on GPCRs.
Apolygus lucorum (Miridae) is an omnivorous pest that occurs worldwide and is notorious for the serious damage it causes in various crops and its substantial economic losses (30, 31). Recently, the genome of A. lucorum had been reported, which provided convenient in-depth studies of this pest (32). In the present research, using bioinformatics analysis, we screened the genes encoding GPCRs from the genome of A. lucorum. The expression profiles of all GPCRs were also determined by using public transcriptome data. These results allowed us to make comparisons of GPCR systems in different insect species and to provide relevant information for further functional studies in A. lucorum. Our study was the first investigation of GPCRs in A. lucorum, which may become the basis for further investigation of the function of miridae GPCRs.
2 Materials and Methods
2.1 Identification of A. lucorum GPCRs
A. lucorum protein sequences were retrieved from the NCBI Genome database (https://www.ncbi.nlm.nih.gov/assembly/GCA_009739505.2/) (32). Based on previous studies and records in Flybase (http://www.flybase.org/) (33), the GPCRs of Drosophila melanogaster (34), A. pisum (22), Bombyx mori (17), Tribolium castaneum (16), and Pediculus humanus humanus (18) were collected. By using D. melanogaster GPCRs as references and A. lucorum protein sequences as queries, BLASTP searches (35) were performed with a cut-off e-value of 1e−5 to look for all GPCR candidates. Then, seven-transmembrane (7TM) domain and annotation information was adopted as the basic criteria for all GPCR candidates. The GPCR candidates in which the number of 7TM domains was more than four or the annotation information indicated it was a GPCR were retained. The remaining GPCR candidates were also confirmed by means of BLASTX analysis in the UniProtKB/Swiss-Prot database. Using all GPCRs that we collected, pre-phylogenetic analysis with the maximum likelihood method was the final criteria to remove non-GPCRs from candidate pools. If a candidate showed fewer genetic relationships with known GPCRs by phylogenetic analysis and the hit sequences in BLASTX analysis indicated they were not GPCRs, they were classified as a non-GPCR and removed from our analysis.
2.2 Structural Analyses, Annotation Information, and Gene Locations of GPCRs
The 7TM domains for all GPCR candidates were predicted with the server TMHMM (v2.0) (36) from the Centre for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/TMHMM/). Functional annotations of the target proteins were done using InterProScan (37). In addition, the chromosomal location of each GPCR candidate was extracted from the genome annotation file of A. lucorum.
2.3 Phylogenetic Analysis
Partial GPCRs of R. prolixus and C. lectularius that also belonged to Heteroptera were also obtain based on previous study (24, 27). GPCRs from D. melanogaster, A. pisum, C. lectularius, and R. prolixus were assigned to a family/subfamily according to previous results (22, 24, 27, 34). Putative A. lucorum GPCRs were classified into different families/subfamilies according to the families to which their orthologous proteins were assigned. Amino acid sequences of the putative A. lucorum GPCRs in each family/subfamily were aligned with receptors of the same family/subfamily in D. melanogaster, A. pisum, C. lectularius, and R. prolixus using MAFFT v7 (38). Phylogeny tests were accomplished using the bootstrap method with 1,000 replications to reconstruct maximum likelihood (ML) trees using IQ-TREE (39) and the best-fit tree model was determined with ModelFinder (40). It should be noted that the GPCRs of R. prolixus and C. lectularius were uncompleted, which were composed of opsins, biogenic amine receptors, and neuropeptide GPCRs. For the Drosophila sequences, the name of the GPCRs were used, while for A. pisum and R. prolixus, the protein names were same as in previous work (22, 27), and for C. lectularius, the accession numbers in NCBI were used. The GPCRs of A. lucorum identified in this work were numbered according to their families.
2.4 Expression Analysis
To study the expression profiles of the GPCRs, a total of 39 transcriptome data of A. lucorum were downloaded from the genome project of A. lucorum (Accession: PRJNA526332) in the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/) (30, 41), which included egg and different tissues (leg, head, body, mouthpart, wing, and gut) of nymphs and adults. Each tissue contained three biological replicates. In detail, we downloaded the SRA data first and then we used an SRA-Toolkit to split the paired-end reads. Clean reads were obtained from the raw data using Trimmomatic (42) to remove reads with quality scores lower than 10 and adapter sequences. To analyze gene expression profiles, clean reads of each sample were mapped to A. lucorum gene sets using hisat2 (43), and then the TPM value (44) of each putative GPCR gene was calculated with featureCounts (45). These TPM expression values were scaled and served to generate a cross-sample normalized trimmed mean of the M-values (TMM) gene expression matrix (46). Finally, the heatmap was drawn in ITOL (https://itol.embl.de) (47) using the normalized matrix. The value used for each sample was the mean of three independent biological replicates.
2.5 Classification of Gene Duplication Types
MCScanX (48) was used to classified the duplication types of different duplicate GPCR genes. First, the homology with different genes in the genome of A. lucorum was determined by a whole-genome BLASTP analysis with a max target seqs of 5 and a cut-off e-value of 1e−5. Then, the homology with different genes and the chromosomal location were combined and all genes were classified into various types, including the segmental duplication, and tandem duplication. Finally, the duplication types of GPCRs were extracted based on these results. All visualized works were accomplished in TBtools (49).
3 Results
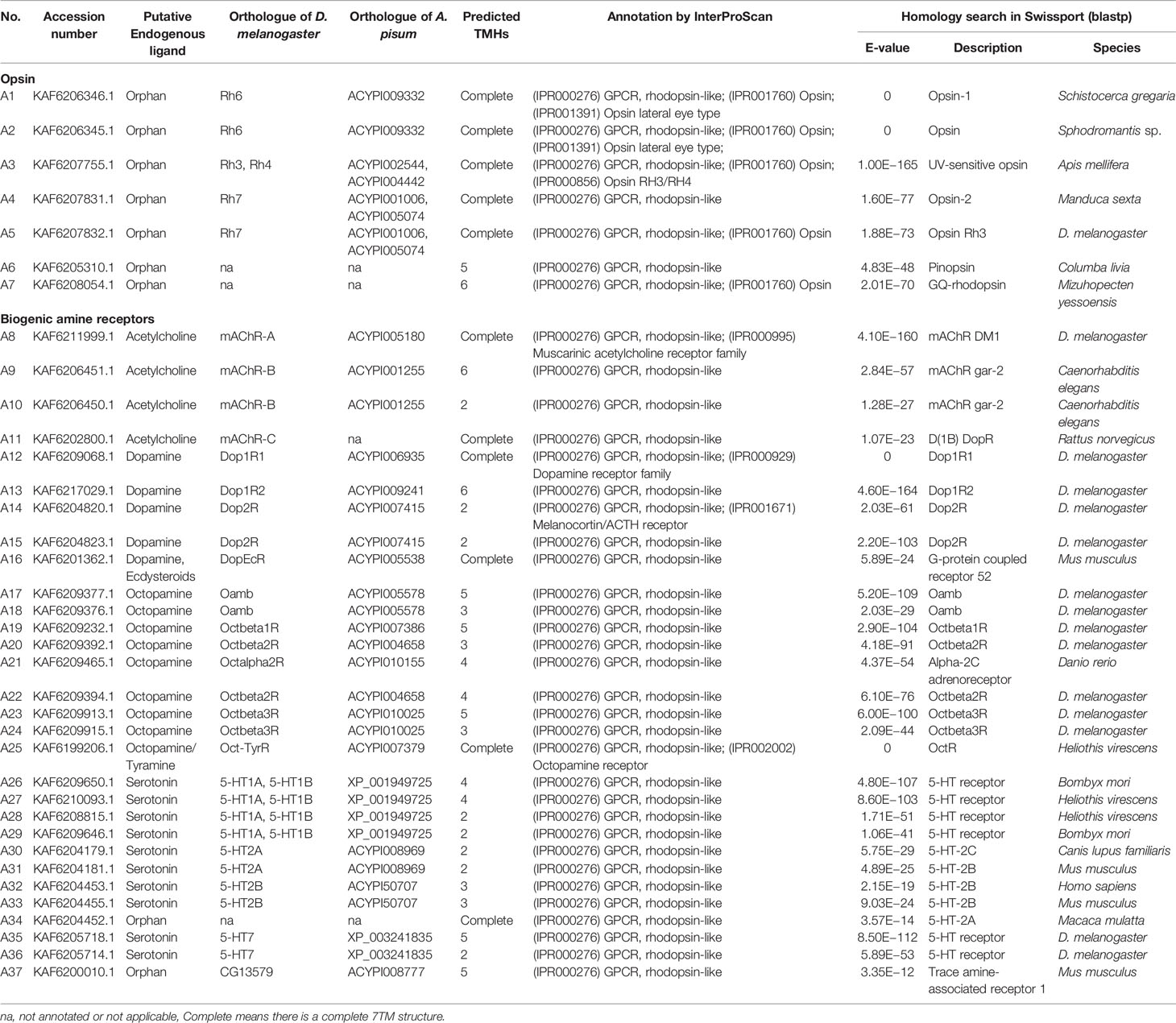
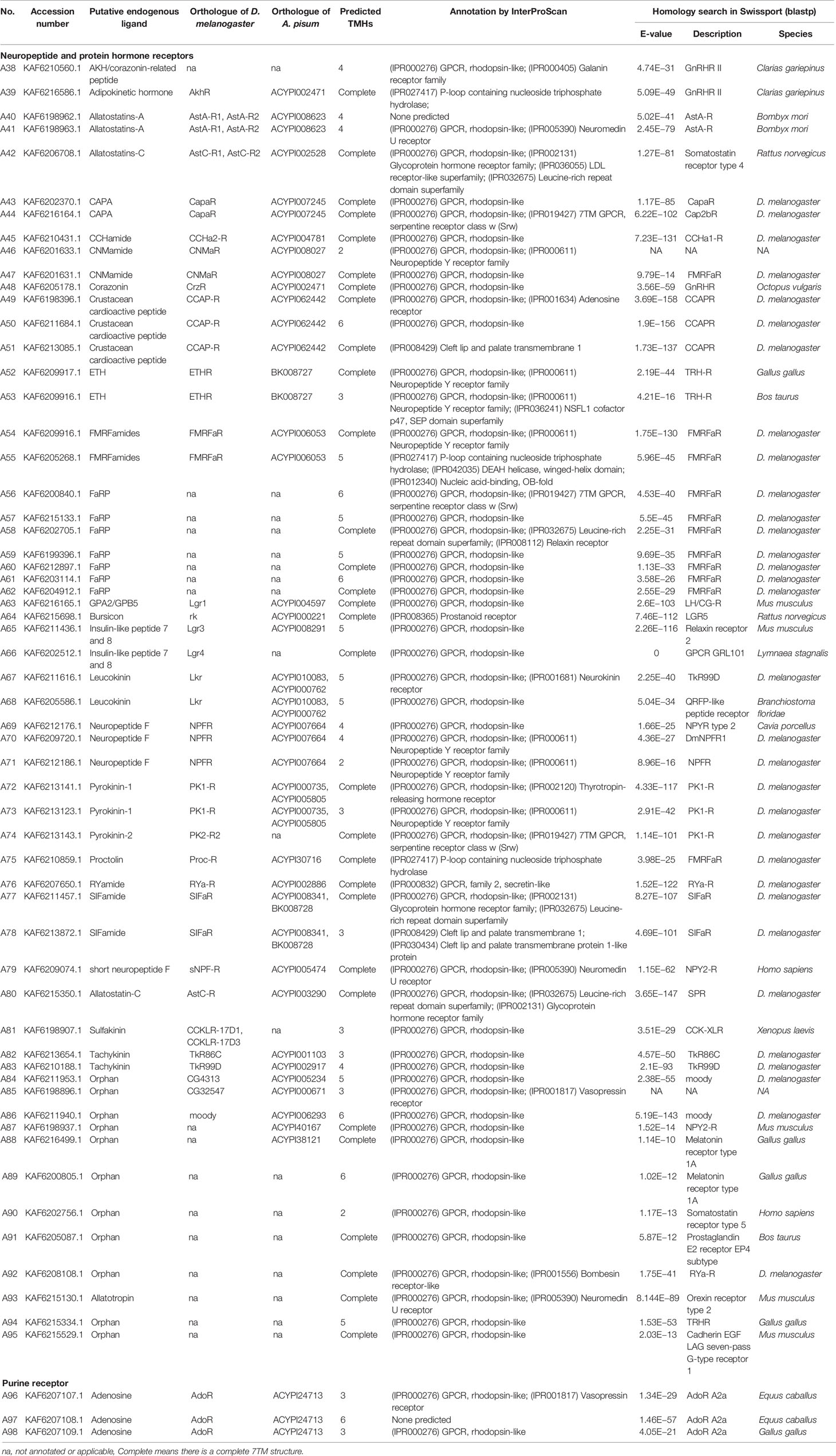
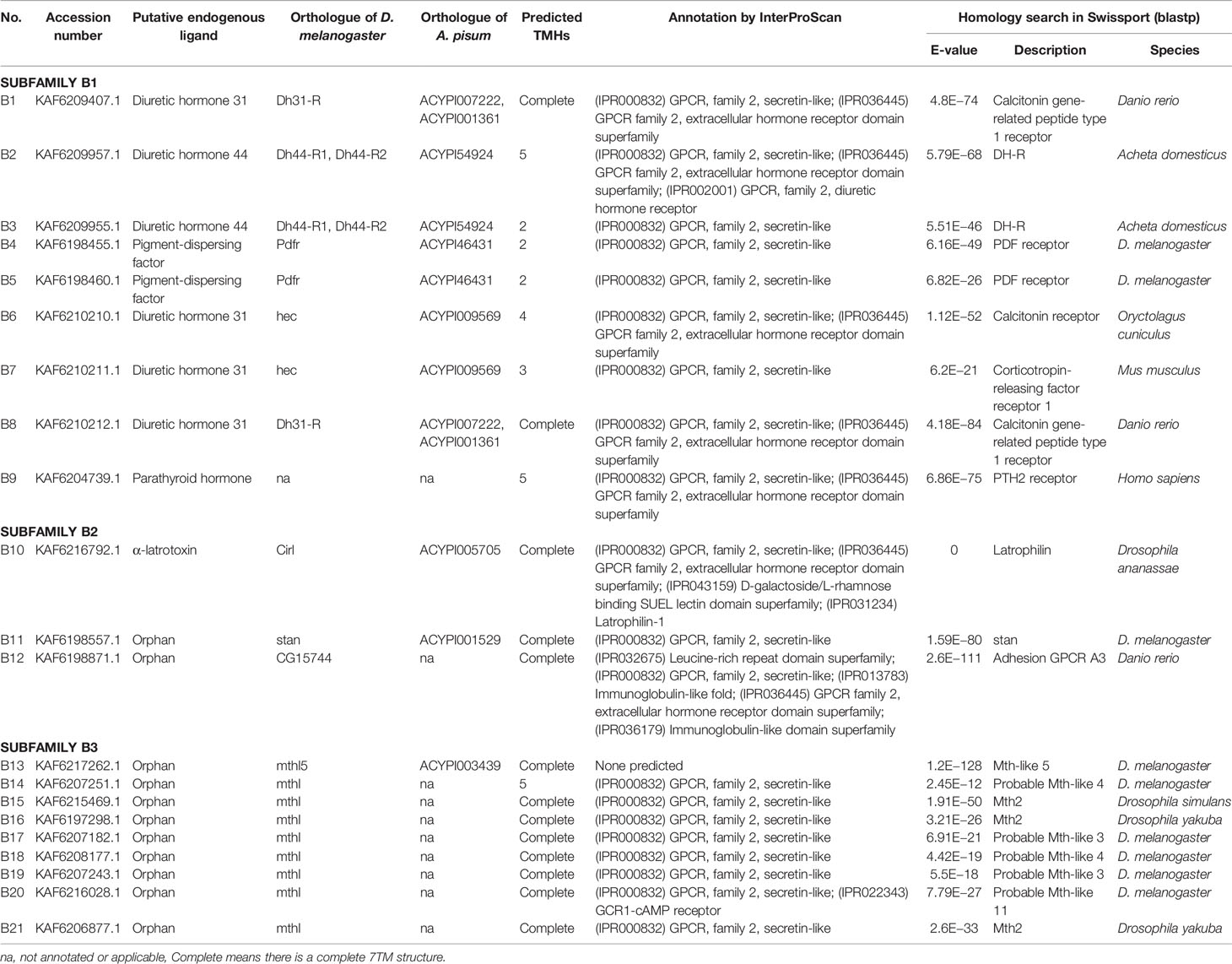
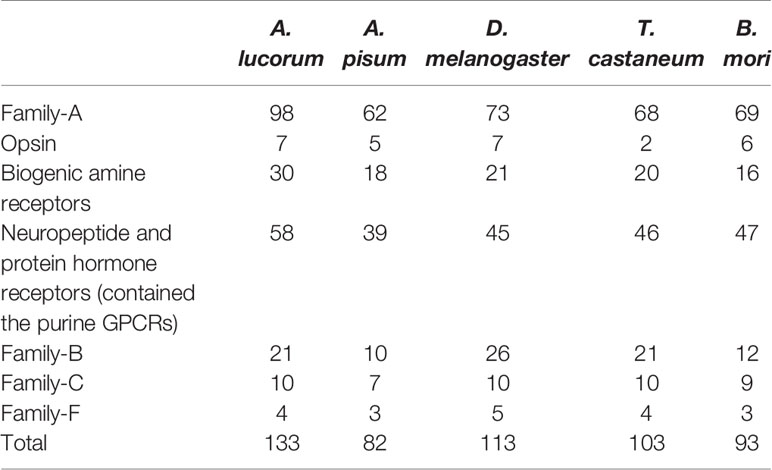
A total of 133 putative GPCRs were identified in A. lucorum. These GPCRs were classified into four families and included 98 family-A members, 21 family-B members, 10 family-C members, and four family-F members (Tables 1–4 and Table S1). Based on the protein sequences, phylogenetic trees were reconstructed for each GPCR family/subfamily of A. lucorum, R. prolixus, A. pisum, and D. melanogaster. All GPCRs were quantified with the TPM values obtained from transcriptomic data. The expression profile of each GPCR across developmental stages was also present in the phylogenetic trees of each GPCR family/subfamily (Figures 1–3, and Figures S1–S3). The chromosomal locations of all GPCRs are shown in Figure 4.
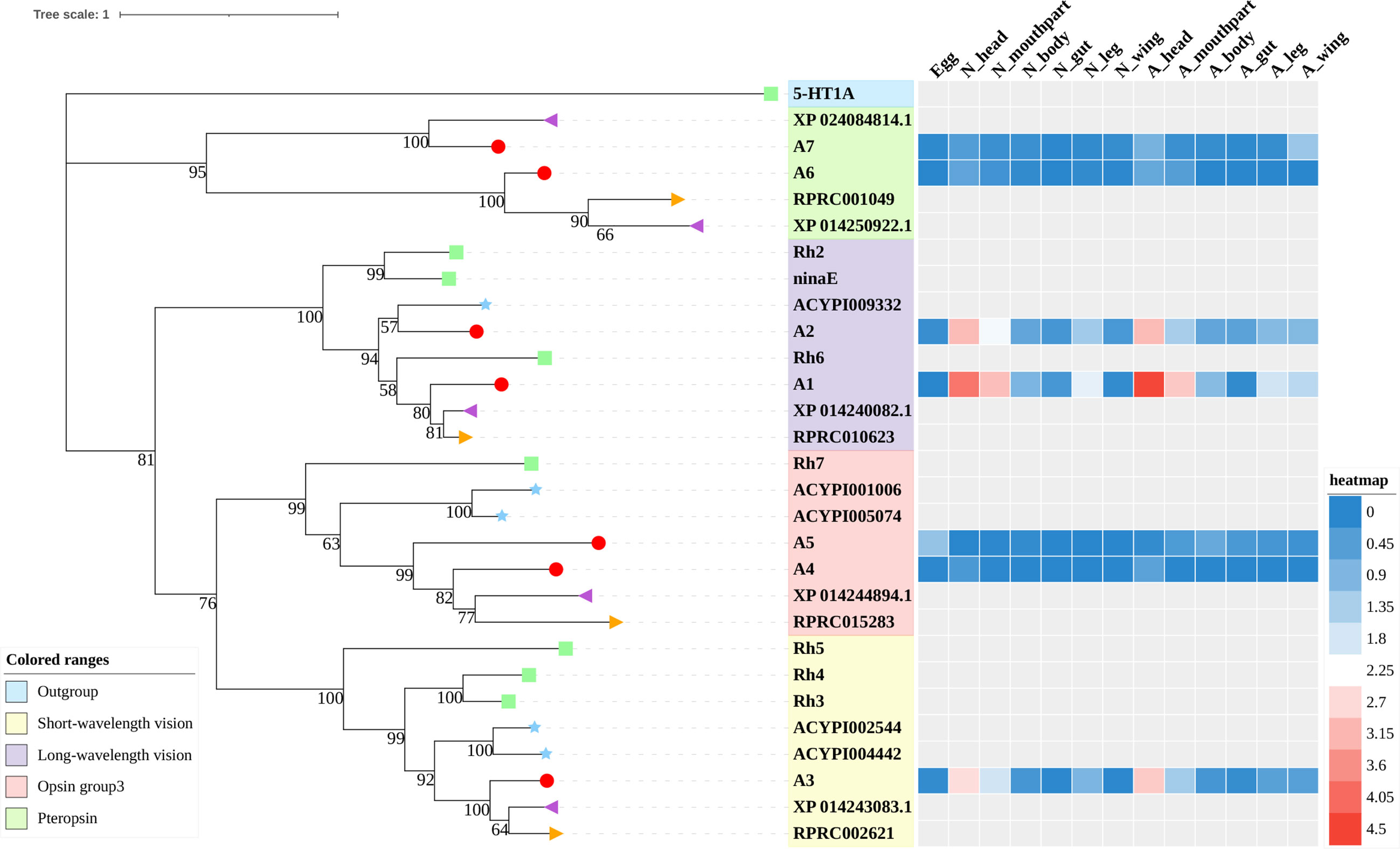
Figure 1 Phylogenetic tree reconstruction of opsin GPCRs from D. melanogaster (green square), A. pisum (blue star), R. prolixus (orange right triangle), C. lectularius (purple left triangle), and A. lucorum (red circle) inferred from maximum likelihood (ML) analysis. Numbers at nodes on the tree were the bootstrap values (below 50 are not shown). The tree was rooted by the D. melanogaster biogenic amine receptor 5-HT1A. Expression profiles of A. lucorum GPCR genes from different tissues are shown in the corresponding branch side. The transcription level of each gene is represented by a square with a color that codes for the values of Lg (TPM+1). N, nymph; A, adult.
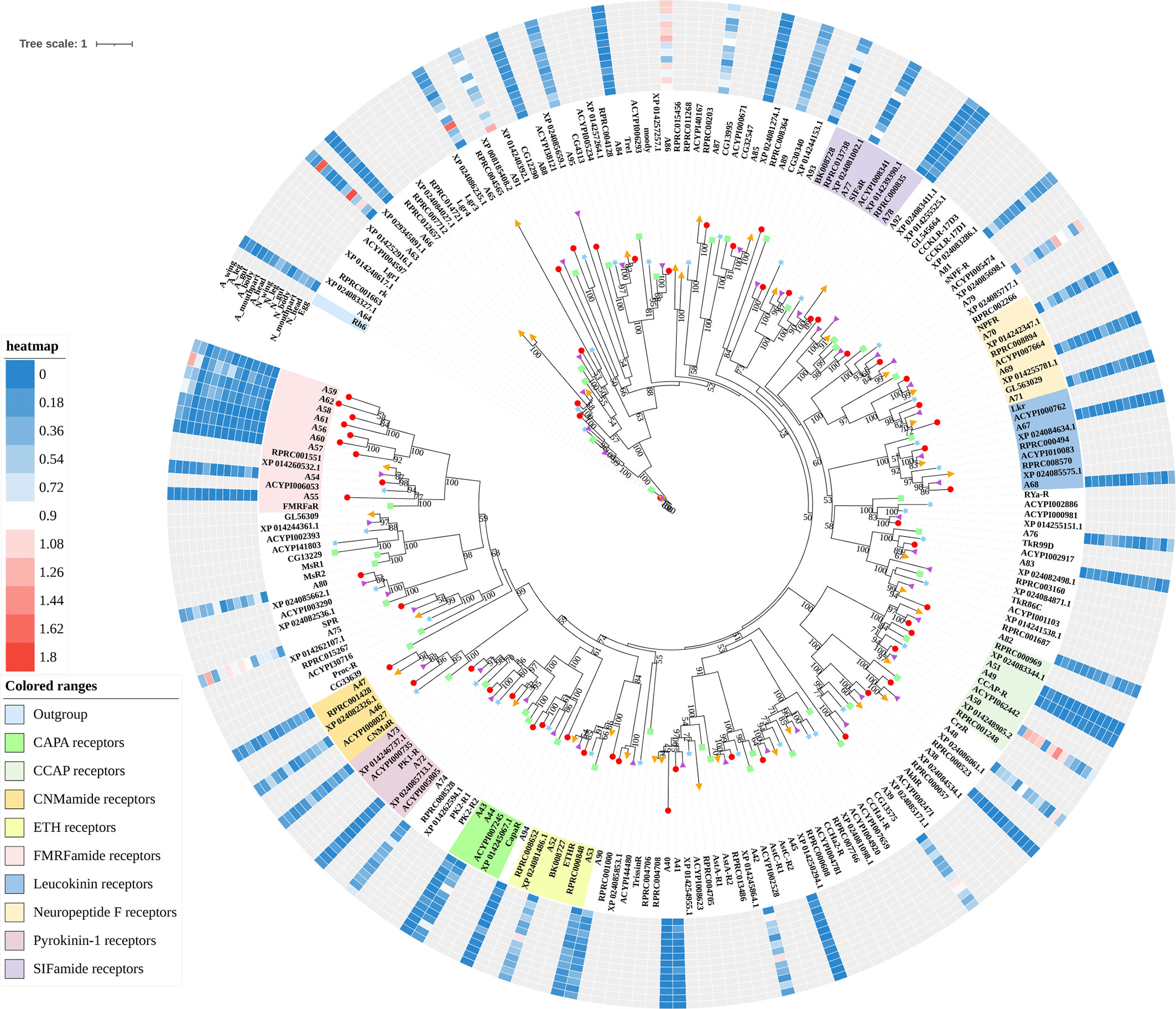
Figure 2 Phylogenetic tree reconstruction of neuropeptide and protein hormone receptors from D. melanogaster (green square), A. pisum (blue star), R. prolixus (orange right triangle), C. lectularius (purple left triangle), and A. lucorum (red circle) inferred from maximum likelihood (ML) analysis. Numbers at nodes on the tree were the bootstrap values (below 50 are not shown). The tree was rooted by the D. melanogaster opsin GPCR Rh6. Expression profiles of A. lucorum GPCR genes from different tissues are shown in the corresponding branch side. The transcription level of each gene is represented by a square with a color that codes for the values of Lg (TPM+1). N, nymph; A, adult.
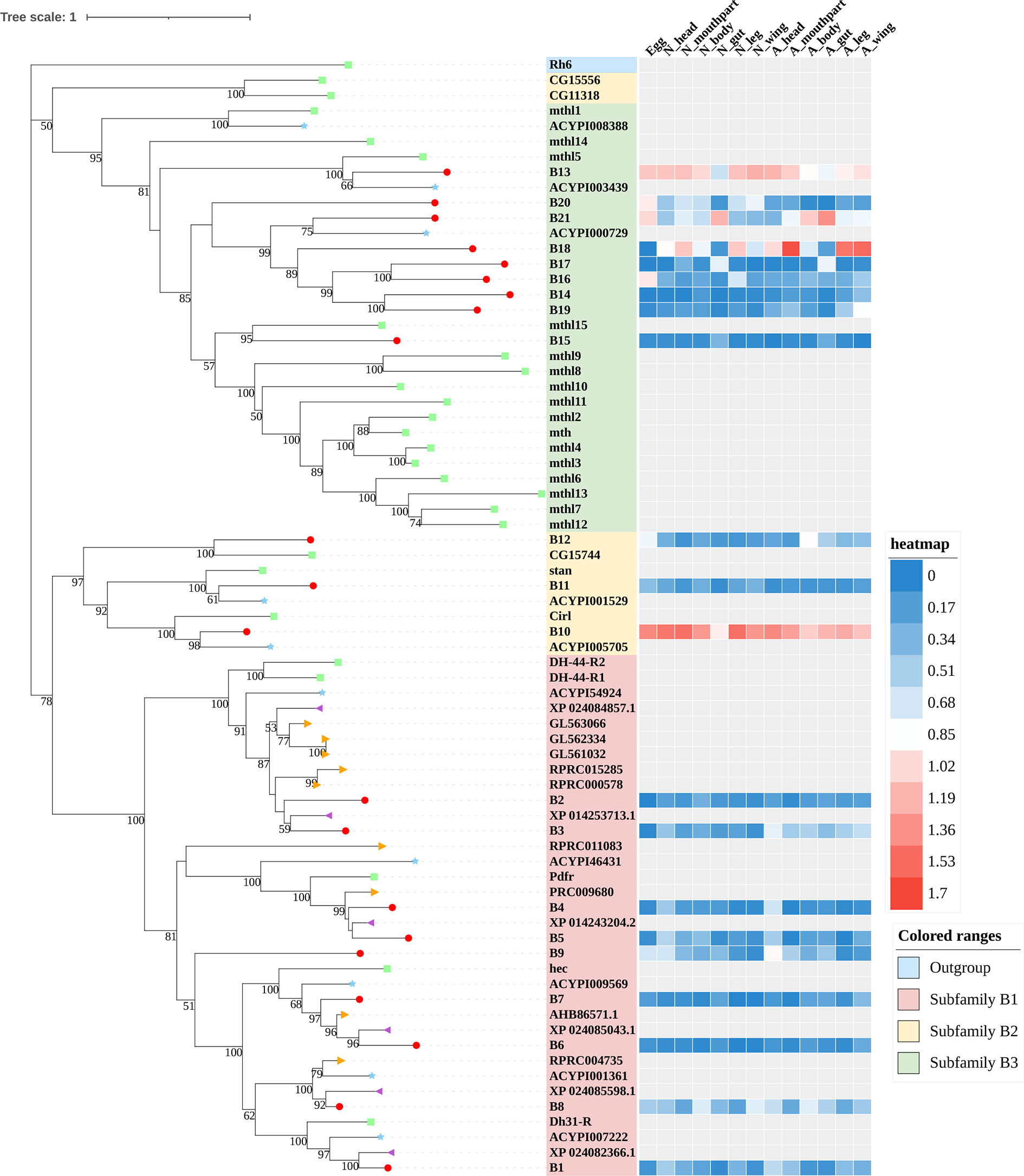
Figure 3 Phylogenetic tree reconstruction of Family-B GPCRs from D. melanogaster (green square), A. pisum (blue star), R. prolixus (orange right triangle), C. lectularius (purple left triangle), and A. lucorum (red circle) inferred from maximum likelihood (ML) analysis. Numbers at nodes on the tree were the bootstrap values (below 50 are not shown). The tree was rooted by the D. melanogaster opsin GPCR Rh6. Expression profiles of A. lucorum GPCR genes from different tissues are shown in the corresponding branch side. The transcription level of each gene is represented by a square with a color that codes for the values of Lg (TPM+1). N, nymph; A, adult.
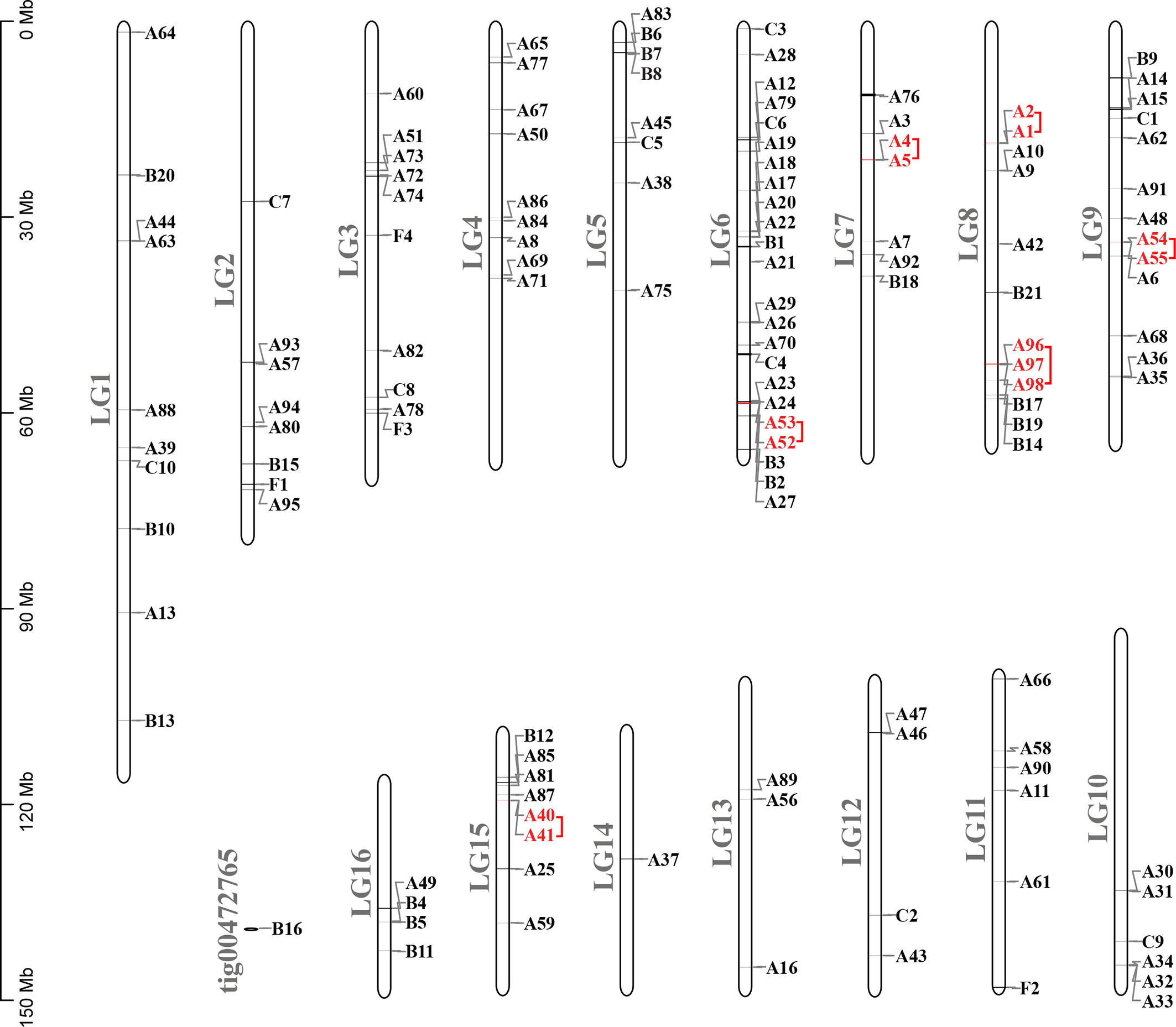
Figure 4 Chromosomal locations and tandem duplicated gene pairs of the 133 putative GPCR genes. Each was mapped to the chromosome based on its physical location. The chromosome number (LG1–LG16, tig00472765) is indicated at the left. The tandem duplicated genes were outlined with red color.
3.1 Family-A GPCRs
Insect family-A GPCRs include opsins, biogenic amine receptors, neuropeptide and protein hormone receptors, and purine GPCRs (17, 18, 22). In this study, 98 family-A GPCRs were identified in the genome of A. lucorum, and these receptors were composed of seven opsins, 30 biogenic amine receptors, 58 neuropeptide and protein hormone receptors, and three purine GPCRs (Tables 1, 2).
3.1.1 Opsins
Color vision in insects is based on the expression of different opsins in photoreceptive cells. Opsins are members of the family-A GPCRs and are coupled to light-sensitive chromophores in animal photoreceptors (50). Three groups of opsins have been reported in D. melanogaster: one related to long-wavelength (LW) vision (including Rh1, Rh2, and Rh6), another group related to short-wavelength (SW) vision (Rh3, Rh4, and Rh5), and a third group including only Rh7 (34, 51). A fourth group of invertebrate opsins, named pteropsins, has been found in Apis mellifera (50) and R. prolixus (27), which was missing from the genome of D. melanogaster and A. pisum.
In this study, seven putative opsins were identified in A. lucorum. The phylogenetic analysis suggested that A1 and A2 are related to the LW opsin, A3 is related to the SW opsin, A4 and A5 belong to a third group, and A6 and A7 are close to pteropsins (Figure 1). Four groups of invertebrate opsins were also identified in the A. lucorum genome. According to the expression profile, opsins were expressed at the highest levels in the head and mouthpart tissue, which is corresponding to their biological function. Among the four types of opsins detected in A. lucorum, the A1 showed the highest expression in the head tissue of adults with a transcripts per kilobase of exon model per million mapped reads (TPM) of 28,787 (Figure 5) followed by A3 with a TPM of 770.
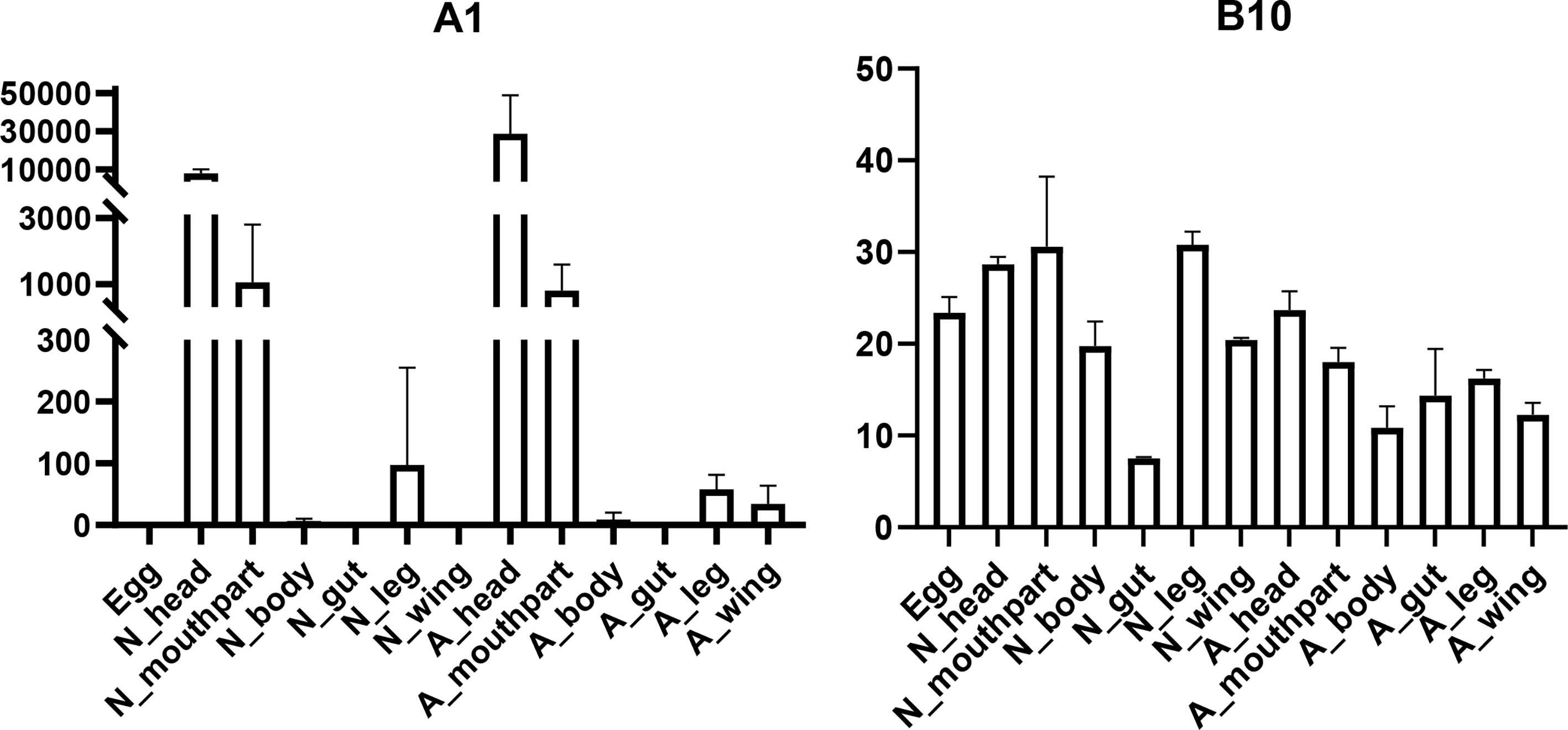
Figure 5 Expression patterns of A1 and B10 in different tissues by transcriptome analysis. The vertical bars indicate standard errors of the mean (n = 3).
3.1.2 Biogenic Amine Receptors
The known biogenic amines that act as ligands for GPCRs in insects contain acetylcholine, dopamine, serotonin, octopamine, and tyramine (27). Here, we identified 30 biogenic amine receptors in A. lucorum. Based on phylogenetic analysis and sequence similarity, A8–11 are receptors for acetylcholine; A12–16 are dopamine-like receptors; A17–24 are orthologs of the octopamine receptors; A26–33 and A35–36 were identified as the serotonin-like receptors; and A25 is the GPCR that could be stimulated by two structurally related endogenous ligands, octopamine and tyramine (Figure S1). Additionally, A34 and A37 are orphan receptors of this subfamily in A. lucorum, and are orthologs of RPRC011175 and CG13579, respectively. However, two tyramine receptors (TyrR and TyrRII) are likely to be missing in all three heteropteran insects. A25 is the only tyramine receptor in A. lucorum. Compared with opsins, the expression level of biogenic amine receptors is much lower. A36 showed the highest expression in gut tissues of adults with a TPM of 11. In FlyBase (33), we found 5-HT7, the ortholog gene of A36 in D. melanogaster, was also expressed in the digestive system.
3.1.3 Neuropeptide and Protein Hormone Receptors
The rhodopsin-like neuropeptide and protein hormone receptors are the largest subfamily in the rhodopsin-like family (17, 22, 52). In this subfamily, 58 putative A. lucorum sequences were identified. Like other insects, A. lucorum rhodopsin-like neuropeptide and protein hormone receptors can be classified into 25 groups based on their ligands; i.e., adipokinetic hormone receptors (AKHR), AKH/corazonin-related peptide (ACP) receptors, allatotropin receptor (AT-R), allatostatin-A receptors (AstA-R), allatostatin-B receptors (AstB-R), allatostatin-C receptors (AstC-R), bursion receptor, corazonin receptors (CrzR), neuropeptide F receptors (NPFR), short neuropeptide F receptors (sNPFR), proctolin receptors (Proc-R), pyrokinin receptors (PK-R), leukokinin receptors (Lkr), cholecystokinin-like receptors (CCKLR), tachykinin receptors (TkR), CAPA receptors (CapaR), crustacean cardioactive peptide receptors (CCAP-R), CNMamide receptors (CNMaR), CCHamide receptors (CCHa-R), ecdysis triggering hormone receptors (ETHR), FMRFamide receptors (FMRFaR), GPA2/GPB5 receptors, SIFamide receptors (SIFaR), relaxin receptors, RYamide receptors (RYa-R), and several orphan GPCRs (Figure 2). Most of these neuropeptide receptors displayed one-to-one orthologous relationships between A. lucorum, R. prolixus, C. lectularius, A. pisum, and D. melanogaster, and all subtypes of leucine-rich repeat-containing GPCRs (LGR) were observed in A. lucorum (Figures 2 and 6). However, several duplications and losses of neuropeptide receptor genes were also observed in A. lucorum. It is worth mentioning that as many as nine A. lucorum GPCRs (A54–62) displayed strong evidence of an evolutionary kinship with the FMRFaRs of R. prolixus, C. lectularius, A. pisum, and D. melanogaster, indicating that a large clade may have duplicated from FMRFaRs in A. lucorum (Figures 2 and 7). Duplications of eight neuropeptide receptor genes (CapaR, CCAP-R, CNMaR, ETHR, Lkr, NPFR, PK1-R, and SIFaR) were identified in A. lucorum, and duplications of Lkr, PK1-R, ETHR, CCAP-R, NPFR, and SIFaR were also observed in R. prolixus or C. lectularius. The trapped in endoderm 1 (Tre1) receptors, trissin receptors (TrissinR), myosuppressin receptors (MsR), and other six orphan receptors were not found in the genome of A. lucorum. Instead, we found six orphan receptors (A89–92 and A94–95) that have not been reported in A. lucorum. The expression levels of neuropeptide and protein hormone receptors were higher than in biogenic amine receptors. The expression of A65 (LGR) in the bodies of nymphs was the highest in this subfamily with a TPM of 41. Moreover, A50 (CCAP-R) and A86 (moody) showed high expression levels in multiple tissues (TPM >10 at least in five tissues).
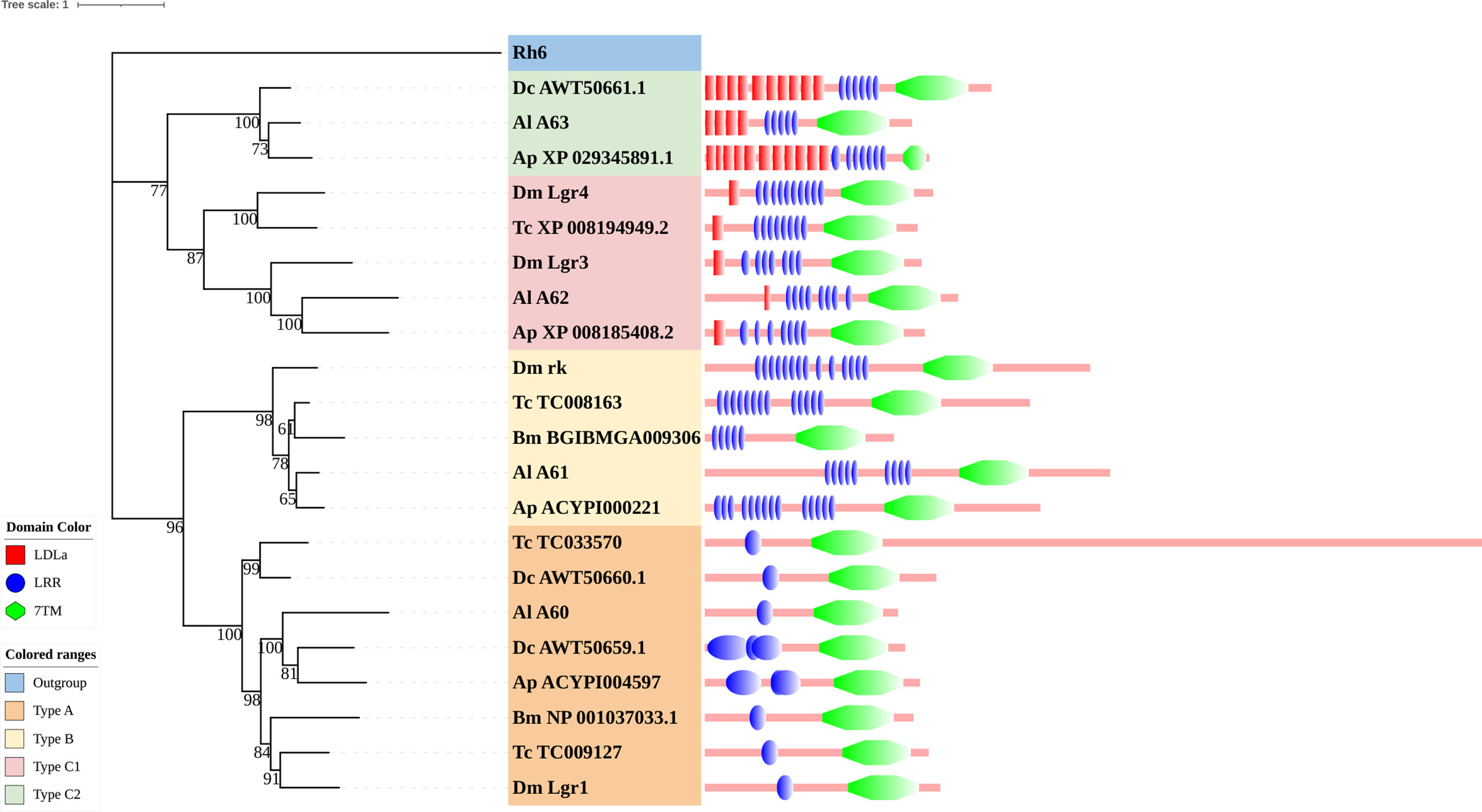
Figure 6 Phylogenetic tree and domains of different LGRs. Numbers at nodes on the tree were the bootstrap values (below 50 are not shown). The tree was rooted by the D. melanogaster opsin GPCR Rh6. Different LGR types were painted with different colors. Predicted domains of each sequence are shown in the corresponding branch side. Dc, D. citri; Al, A. lucorum; Ap, A. pisum; Dm, D. melanogaster; Tc, Tribolium castaneum; Bm, B. mori.
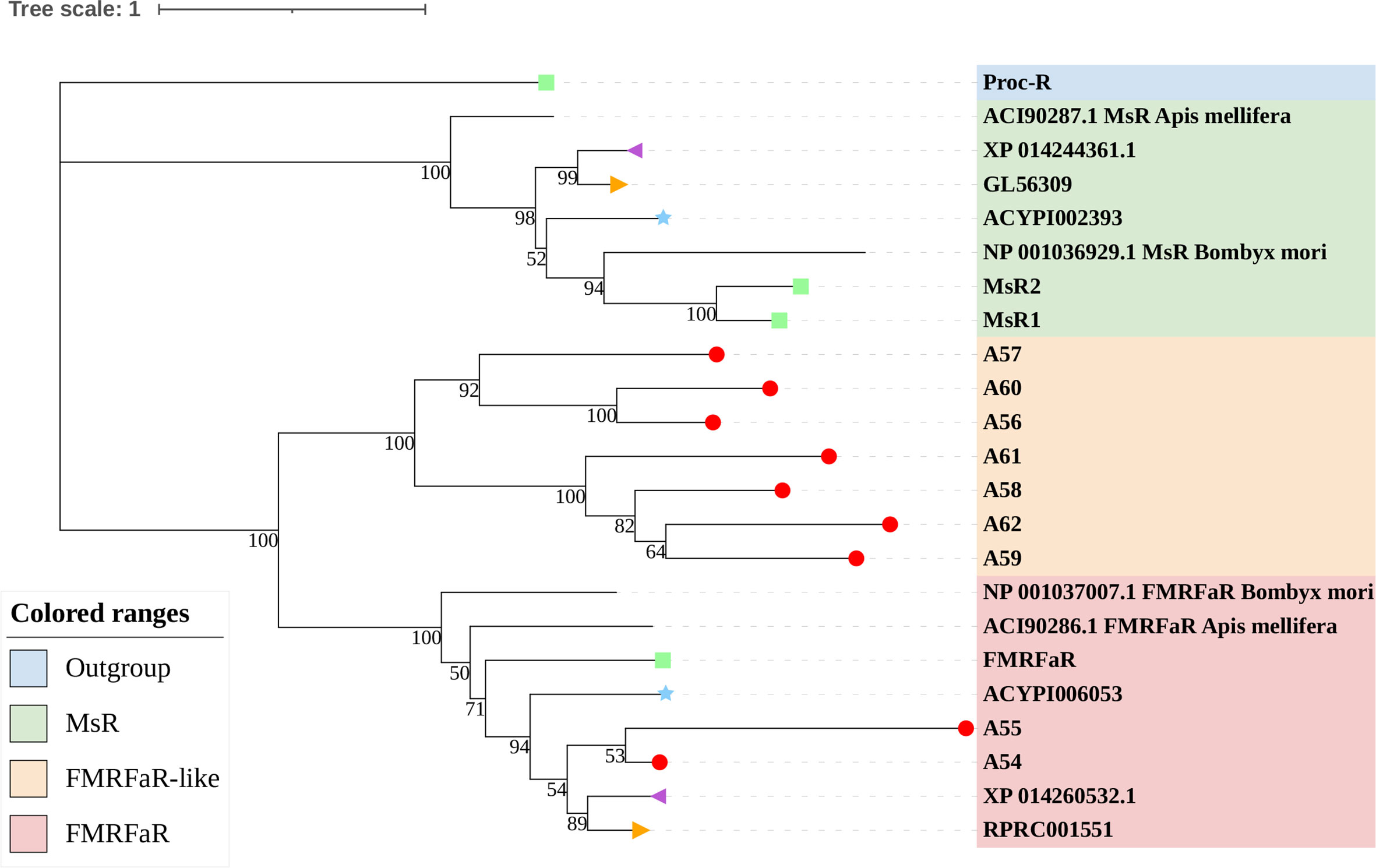
Figure 7 Phylogenetic tree reconstruction of FMRFaRs and MsRs from D. melanogaster (green square), A. pisum (blue star), R. prolixus (orange right triangle), C. lectularius (purple left triangle), A. lucorum (red circle), and other two model insects inferred from maximum likelihood (ML) analysis. Numbers at nodes on the tree were the bootstrap values. The tree was rooted by the D. melanogaster Proc-R.
3.1.4 Purine GPCRs
Only one receptor in this subfamily, adenosine receptor (AdoR), has been previously classified in this subfamily (17, 53). Here, three putative A. lucorum GPCRs (A96–98) were identified as AdoR, whereas there was only one member in D. melanogaster and A. pisum (Table 2). Purine GPCRs are activated by the binding of purine nucleotides or their derivatives (principally adenosine or ADP/ATP) (54, 55). Duplication of AdoR suggests that purinergic neural transmission may play a more important role in A. lucorum.
3.2 Family-B GPCRs
Family-B GPCRs play vital roles in many biological processes, including growth, development, and reproduction. They are characterized by long N-terminal domains, and they form a small group of receptors that are structurally and functionally divergent from other groups of GPCRs (56). Within this family, family-B GPCRs can be further subdivided into three subfamilies: B1–B3, which are greatly divergent in both function and structure. In total, 21 family-B GPCRs were identified from the genome of A. lucorum in this study, which consisted of nine B1 subfamily members, three B2 subfamily members, and nine B3 subfamily members (Table 3 and Figure 3).
The B1 subfamily is made of largely classical hormone receptors. It comprises three types of hormone receptors in D. melanogaster: diuretic hormone 31 receptor (DH31-R/hector), CRF-like diuretic hormone 44 (DH44-R), and pigment dispersing factor receptor (Pdfr) (57–60). In our study, all three types of hormone receptors were identified. The parathyroid hormone receptor (PTHR), which is involved in the calcium and phosphate homeostasis and bone growth in vertebrates, is also a subfamily-B1 GPCR (58). There are two PTHRs in mammals, which are involved in calcium and phosphate homeostasis and bone growth (61, 62). In insects, PTHR-like (PTHRL) have been identified from T. castaneum, A. mellifera, P. h. humanus, and N. lugens (18, 26, 57), but its counter-parts in D. melanogaster, B. mori, A. pisum, and A. gambiae are not found (17, 22, 34, 63). T. castaneum has two distinct PTHRLs (57), N. lugens possesses a pair of homologous PTHRLs (26), and A. mellifera only has one PTHRL (57). In our study, we also identified one PTHRL, B9, which shared a low e-value (1e−104 and 1e−111) with two PTHRLs in N. lugens. These results showed that genes coding for PTHR are divergent among insects.
The B2 subfamily is characterized by a long extracellular N-terminal domain and a GPCR proteolytic site (57, 58, 64). Based on phylogenetic analysis and sequence similarity, three receptors (B10–12) were classified in the B2 subfamily, which correspond to a calcium-independent receptor for α-latrotoxin (Cirl), starry night (stan), and CG15744, respectively. However, the orthologs for CG11318 and CG15556 were not identified in the genome of A. lucorum.
There is only one group of receptors in the B3 subfamily; i.e., Methuselah (mth)/Methuselah-like (mthl) (57, 58). This gene family is involved in the modulation of life span and stress responses. No counterpart for the mth gene family has been identified in vertebrates (57). In insects, the number in the B3 subfamily is highly variable (18, 57). In D. melanogaster, this subfamily can be divided into two groups based on their structure, 12 mth ectodomain‐positive members (mth, mthl2, mthl3, mthl4, mthl6, mthl7, mthl8, mthl9, mthl10, mthl11, mthl12, and mthl13) and four mth ectodomain‐negative members (mthl1, mthl5, mthl14, and mthl15) (65). In our study, nine receptors (B13–21) were identified in this family. Based on phylogenetic analysis, B13 and B20 may belong to the mth ectodomain‐positive group, and others may be members of the mth ectodomain‐negative group. At the mRNA level, B13, B18, and B21 showed a higher expression level than other mthl members, which indicated these three members of the B3 subfamily may play a more important role in A. lucorum.
3.3 Family-C GPCRs
Family-C GPCRs possess a large ligand-binding extracellular domain and form constitutive dimers (34, 66–68). There are three types of GPCRs in family-C, the glutamate and γ-amino butyric acid (GABA-B) receptors, the bride of sevenless (boss-type) receptors, and the metabotropic glutamate (mGlu) receptors. Until now, nine family-C GPCRs from D. melanogaster and seven from A. pisum have been reported. By using these reference sequences, 10 family-C members (Table S1 nad Figure S2) of A. lucorum were identified here.
Like A. pisum (22), there are two GABA-B receptors (C1 and C2) in A. lucorum. C1 shares a 78% sequence similarity with D. melanogaster GABA-B-R1, while C2 has a 44% sequence identity with D. melanogaster GABA-B-R2. The orthologous gene to D. melanogaster GABA-B-R3 has not been found in two Hemiptera insects. The boss-type receptor was first identified as a ligand for sevenless tyrosine kinase, which was involved in eye differentiation in D. melanogaster. Subsequently, boss has been implicated in the glucose-response (69, 70). It has been reported in D. melanogaster (34), A. gambiae (63), A. pisum (22), and T. castaneum (16), but not in B. mori (17), A. mellifera, N. vitripennis, and P. humanus corporis. Here, C3 was identified as the orthologous gene to boss. These results indicated that boss has been randomly lost in insects during their evolutionary process. Moreover, three mGlu receptors (C4, C5, and C6) were found in A. lucorum, whereas there are only two mGlu receptors in D. melanogaster (34) and one mGlu receptor in A. pisum (22). Small expansions of A. lucorum mGlu receptors have been observed. There are some unclassified receptors in this family. C7 was the orthologous gene to smog. C8 and C9 showed 53 and 38% sequence identities with their counterparts in D. melanogaster, respectively. As shown in Figure S2, C9 showed a high expressed level in egg, adult head, and all nymph tissues, except leg, while the function of its orthologous gene is unclear. C10 was an orphan receptor that has not been reported in D. melanogaster and A. pisum (22, 34).
3.4 Family-F GPCRs
Family-F GPCRs comprise the frizzled gene family and the smoothened gene (34, 71). In this study, four putative A. lucorum GPCRs were identified in the frizzled/smoothened GPCR family, which are orthologous to D. melanogaster fz, fz2, fz3, and smo (Table S1 and Figure S3). Our results indicated that orthologs for D. melanogaster fz4 were missing in A. lucorum. Among family-F, F4 (smo) showed the highest expression in the egg with a TPM of 61.
4 Discussion
In this study, we systematically identified 133 GPCRs from A. lucorum. Compared with other model insects, we also found the GPCR genes remarkably expanded among the biogenic amine receptors, neuropeptide and protein hormone receptors, and the B1 subfamily (Table 4). Some of them had been reported in R. prolixus or C. lectularius (24, 27), such as the duplications of 5-HT7, Lkr, PK1-R, ETHR, CCAP-R, NPFR, SIFaR, and DH31-R. However, missing TyrR, Tre1, TrissinR, MsR, and some orphan receptors has also been observed in the genome of A. lucorum (Tables S2, S3). All these predicted GPCRs were quantified by transcriptome data. Although most GPCR genes showed a low expression level in A. lucorum, there were a few highly expressed GPCR genes, such as the long-wavelength opsin and Cirl. By comparative analysis, we also found C2 LGR types were widely distributed in Hemiptera. All these aspects will be discussed in detail below.
4.1 GPCRs Gene Expansion Occurred in A. lucorum
Compared with other well-studied insects, we noticed that the number of genes coding for GPCRs is obviously larger than for other insects, especially expanded among the biogenic amine receptors, neuropeptide and protein hormone receptors, and B1 subfamily (Table 4). There were 26 GPCRs duplicated in A. lucorum. Twenty-three of them were classified into the three subfamilies mentioned above. By MCScanX analysis, we found six tandem duplication events occurred among Rh6, Rh7, AstA-R, ETHR, FMRFaR, and AdoR, while most GPCR genes duplicated dispersedly. Considering the location of GPCR genes on chromosomes, it suggested that the duplication of GPCR genes mainly occurred as independent duplications and transitions (Figure 4).
The duplicate biogenic amine receptors in A. lucorum included 5-HT1A, 5-HT1B, 5-HT2A, 5-HT2B, 5-HT7, Dop2R, mAChR-B, Oamb, Octbeta2R, and Octbeta3R. These biogenic amine receptors can regulate many behaviors including flight and fight, learning and memory, sleep and wakefulness, feeding, and social and reproductive behaviors (72–74). For example, 5-HT1A was related to locomotor activity in B. mori. Injecting the antagonist of the Bm5-HT1A receptor into larvae caused slow or weak motility, and adults had lowered courtship vitality or moving speed (75). mAChRs have also been reported to be critical in regulation of locomotory behavior in Drosophila (76). In addition, 5-HT1B mediates hemocyte phagocytosis and serotonergic signaling performs critical modulatory functions in immune systems. Moreover, 5-HT7 and Dop2R were shown to be associated with learning ability (77, 78) and octopamine receptors were required for ovulation in D. melanogaster (79). The ligands of neuropeptide and protein hormone receptors and the B1 subfamily belong to neuropeptides, which also play an important role in the regulation of development, reproduction, feeding, courtship, aggression, olfaction, locomotor activity, circadian rhythm, and many other physiological processes in insects (21, 80, 81). The gene expansion of these three subfamilies indicated that A. lucorum had a more complex peptidergic signaling system.
Expansions of genes associated with omnivorousness and mesophyll feeding, such as those related to digestion, chemosensory perception, and detoxification, were also observed in A. lucorum (32). Gustatory receptors (Grs) and odorant receptors (Ors) are thought to be the most important chemosensory receptors. Like GPCRs, Ors, and Grs are seven-transmembrane domain receptors but belong to the chemosensory 7tm receptor superfamily (82, 83). It has been suggested that the cause of gene expansion in GPCRs might be similar to that of chemosensory receptors, also to better adapt to the environment (32). A. lucorum is found in natural and agricultural ecosystems throughout the world (30), and many of them are generalists, exhibiting diverse feeding habits or preferences (e.g., feeding on leaf, stem, inflorescences, nectar, pollen, and fruit) (32). These results indicated the complex peptidergic signaling system is more favorable for A. lucorum to adapt to multiple living environments and multiple hosts.
4.2 A. lucorum Appears to Have Evolved From a Novel Large Clade of Known FMRFaRs
FMRFamide (FMRFa) is a cardioexcitatory peptide that was first isolated from the nervous system of the clam, Macrocallista nimbosa (84), and is active as a tetrapeptide only in mollusks and annelids. Since the discovery of FMRFa, peptides with extended length at the N-terminal portion have been reported, such as myosuppressin (Ms) (85). Here, nine receptors (A54–62) displayed a certainly evolutionary kinship with the FMRFaR of D. melanogaster, A. pisum, C. lectularius, and R. prolixus, while the MsR is missing in A. lucorum (Figure 2). By reconstruction of the phylogenetic tree of MsR and FMRFaR with more species (17, 86), we found these receptors were closer to the known FMRFaRs (Figure 7). Among these, A54 and A55 were clustered in a single clade with the FMRFaRs that had been identified in other insects, whereas the other seven receptors are clustered on the other single clade. The most similar proteins in UniProtKB/Swiss-Prot of these receptors are both D. melanogaster FMRFaRs (CG2114). Among these receptors, A54 was the ortholog to the insect FMRFaRs with the smallest e-value of 1.75E−130, and A55, which was adjacent located on chromosome 9, is a tandem duplication that occurred at the beginning of this receptor expansion. However, the other seven receptors (A56–62) were scattered across six chromosomes, which indicated these seven receptors might have arisen from transposition.
By searching in the genome of other heteropteran insects, we found there are only one or two FMRFaRs in each heteropteran species. We suggest A54 and A55 should be classified as FMRFaRs, while the others (A56–62) were named as FMRFaR-like for the moment. This branch might be another unknown GPCR or even contain the MsR. Recent research had found that FMRFaR stimulates intracellular calcium signaling through the IP3R and helps maintain neuronal excitability in a subset of dopaminergic neurons for positive modulation of flight bout durations (87), and the ligand can reduce spontaneous muscle contractions, such as in the intestinal muscle and the heart rate, which also have an impact on movements (88, 89). A. lucorum has great flight capacity and its adults can fly 151.3 km within 48 h (90). A large FMRFaR-like branch evolved in A. lucorum may help it maintain strong flight capability.
4.3 Only a Few GPCR Genes Showed High Expression Levels in A. lucorum
In our research, only 23 GPCR genes (17.3% of all GPCR genes) were expressed highly (TPM >10) in at least one tissue, while most GPCR genes showed a low expression level in A. lucorum. This result is consistent with previous studies in which most GPCRs showed a low endogenous expression level, even at the mRNA level (91–93). Certainly, a low expression level of GPCRs does not necessarily equate to functional insignificance (94).
Here, through transcriptome analysis, we found the expression levels of opsins were higher than in other subfamilies. Opsins that originated early in metazoan evolution mediate the response to visual stimuli primarily. When stimulated by light, opsins can activate a downstream signaling cascade by conformational change (95). Most opsin genes are expressed in photoreceptors, but there are opsins expressed in other tissues, suggesting some nonvisual functions (96, 97). In A. lucorum, opsin genes (A1–3) were expressed highly not only in head but also in leg, wing, and mouthpart, indicating these opsins may execute some nonvisual functions (Figure 1). Among these, the LW opsin (A1) showed the highest expression levels in the head tissue of adults (TPM = 28,787, at least 20 times more than other GPCR; Figure 5). The peak absorbance of the LW opsin is 500–600 nm, which corresponds to yellow-green light. As night sets in, the natural ambient light is increasingly dominated by longer wavelengths (98, 99). The importance of LW opsin had been reported in many nocturnal insects (100, 101). The adults of the A. lucorum were mainly active from dusk to early morning (102). High expression levels of LW opsin may help the organism adapt to a low light environment. We found B10, orthologous to Cirl, is the most widely expressed GPCR gene, which can be tested in all tissues (Figure 5). Cirl belongs to a unique branch of GPCRs and, specifically, is an adhesion GPCR (103, 104). The orthologs of Cirl have been discovered in almost all animals from invertebrates to vertebrates, including humans (105). There are three homologs of Cirl in most vertebrates (Cirl‐1, Cirl‐2, and Cirl‐3) and two in birds and worms, whereas there is only one homolog in insects—which is most homologous to vertebrate Cirl-2 (103, 106). The expression pattern of insects Cirl, which had been reported to be expressed in multiple tissues (103, 107), was also like vertebrate Cirl-2 (108). Although there is only one Cirl member in insect species, Cirl is still involved in multiple physiological processes, which can regulate sensory, developmental, reproductive, and immune functions in insects (104, 109). Here, B10 had been detected in all transcriptomic samples and its distribution range is wider than in T. castaneum and D. melanogaster (103, 106). This kind of expression pattern suggested B10 is crucial in the development of A. lucorum.
4.4 Type C2 LGRs Are Mainly Distributed in Hemiptera and Phthiraptera Insects
Within the neuropeptide and protein hormone receptor subfamily, LGRs are a distinct subgroup with important functions in development and reproduction (110). Three distinct types of LGRs have been defined based on their structural characteristics and they are distinguished by the number of leucine-rich repeat (LRR) motifs, the absence or presence of a low density lipoprotein receptor domain class A (LDLa) motif, and their type-specific hinge region. Generally, type B LGRs have about twice the number of LRRs compared to the other two types. An exclusive feature of the type C LGRs is the presence of at least one LDLa motif in the ectodomain. The more general type containing only one LDLa will be referred to as type C1, whereas type C2 contained more than one LDLa (111).
Type C2 LGRs were first discovered in echinoderms, mollusks, and in one insect species (Pediculus humanis corporis). In our study, we found it existed in all hemipteran insects that we studied. Combining recent work, we mentioned that type C2 LGRs are reported in many hemipteran insects and P. h. humanus (18, 23, 25, 26), and are lost in other orders of insects (16, 17, 63, 86) (Figure 6). Until now, the presence of type C2 LGRs have been found in all Hemiptera insects in which their GPCRs have been identified (22, 23, 25, 26). Among insects, Phthiraptera is one of the orders most closely related to Hemiptera (112). Type C2 LGRs may be present in the common ancestor of these two orders. To clarify the distribution of type C2 LGRs in insects, we checked all protein sequences in the non-redundant protein sequences database (nr) of NCBI. The result showed, except for Hemiptera and Phthiraptera, type C2 LGRs were also founded in Zootermopsis nevadensis of Blattodea. In terms of functionality, LGRs have important functions in development and reproduction. Type A LGRs and type B LGRs are stimulated by large dimeric protein hormones (110), regulating the adult eclosion of insects, and cuticle tanning (113, 114), while type C1 LGRs are the receptors of insulin-like peptide 7 and insulin-like peptide 8 and they coordinate organ growth in D. melanogaster (115–117). At present, the function of type C2 LGRs is undetermined. The function of type C2 LGRs and the existence of type C2 LGRs in Phthiraptera need to be explored in future work.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
The majority of the work described here was carried out by HG. This work was also assisted by YL, MW, XS, JT, and FF. BL designed the study and crucially revised the manuscript for important intellectual content and data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31872970 & 3217030192), and the Graduate Research and Innovation Projects of Jiangsu Province (No. KYCX21_1361).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.773669/full#supplementary-material
References
1. Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of Molecular Switches in GPCRs–Theoretical and Experimental Studies. Curr Med Chem (2012) 19:1090–109. doi: 10.2174/092986712799320556
2. Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular Signatures of G-Protein-Coupled Receptors. Nature (2013) 494:185–94. doi: 10.1038/nature11896
3. King N, Hittinger CT, Carroll SB. Evolution of Key Cell Signaling and Adhesion Protein Families Predates Animal Origins. Science (2003) 301:361–3. doi: 10.1126/science.1083853
4. Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin J-P, Davenport AP, et al. International Union of Pharmacology. XLVI. G Protein-Coupled Receptor List. Pharmacol Rev (2005) 57:279–88. doi: 10.1124/pr.57.2.5
5. Attwood TK, Findlay JBC. Fingerprinting G-Protein-Coupled Receptors. Protein Engineering Design Selection (1994) 7:195–203. doi: 10.1093/protein/7.2.195
6. Ilter M, Mansoor S, Sensoy O. Utilization of Biased G Protein-Coupled Receptor Signaling Towards Development of Safer and Personalized Therapeutics. Molecules (2019) 24:e2052. doi: 10.3390/molecules24112052
7. Kowalik L, Chen JK. Illuminating Developmental Biology Through Photochemistry. Nat Chem Biol (2017) 13:587–98. doi: 10.1038/nchembio.2369
8. Ma Z, Taruno A, Ohmoto M, Jyotaki M, Lim JC, Miyazaki H, et al. CALHM3 Is Essential for Rapid Ion Channel-Mediated Purinergic Neurotransmission of GPCR-Mediated Tastes. Neuron (2018) 98:547–561.e10. doi: 10.1016/j.neuron.2018.03.043
9. Michaloski JS, Galante PAF, Nagai MH, Armelin-Correa L, Chien M-S, Matsunami H, et al. Common Promoter Elements in Odorant and Vomeronasal Receptor Genes. PloS One (2011) 6:e2906. doi: 10.1371/journal.pone.0029065
10. Panossian A, Hamm R, Kadioglu O, Wikman G, Efferth T. Synergy and Antagonism of Active Constituents of ADAPT-232 on Transcriptional Level of Metabolic Regulation of Isolated Neuroglial Cells. Front Neurosci (2013) 7:16. doi: 10.3389/fnins.2013.00016
11. Saroz Y, Kho DT, Glass M, Graham ES, Grimsey NL. Cannabinoid Receptor 2 (CB2) Signals via G-Alpha-s and Induces IL-6 and IL-10 Cytokine Secretion in Human Primary Leukocytes. ACS Pharmacol Transl Sci (2019) 2:414–28. doi: 10.1021/acsptsci.9b00049
12. Sharma N, Akhade AS, Qadri A. Sphingosine-1-Phosphate Suppresses TLR-Induced CXCL8 Secretion From Human T Cells. J Leukoc Biol (2013) 93:521–8. doi: 10.1189/jlb.0712328
13. Kim KH, Kim JU, Kim BH, Shin JH, Hong SJ, Lee SR, et al. An Effect of Thermo-Stimulation of Lower Abdomen on Autonomic Nervous System: An Experimental Study. J Pharmacopuncture (2018) 21:98–103. doi: 10.3831/KPI.2018.21.012
14. Strachan RT, Ferrara G, Roth BL. Screening the Receptorome: An Efficient Approach for Drug Discovery and Target Validation. Drug Discov Today (2006) 11:708–16. doi: 10.1016/j.drudis.2006.06.012
15. Hauser AS, Chavali S, Masuho I, Jahn LJ, Martemyanov KA, Gloriam DE, et al. Pharmacogenomics of GPCR Drug Targets. Cell (2018) 172:41–54.e19. doi: 10.1016/j.cell.2017.11.033
16. Hauser F, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, et al. A Genome-Wide Inventory of Neurohormone GPCRs in the Red Flour Beetle Tribolium Castaneum. Front Neuroendocrinol (2008) 29:142–65. doi: 10.1016/j.yfrne.2007.10.003
17. Fan Y, Sun P, Wang Y, He X, Deng X, Chen X, et al. The G Protein-Coupled Receptors in the Silkworm, Bombyx Mori. Insect Biochem Mol Biol (2010) 40:581–91. doi: 10.1016/j.ibmb.2010.05.005
18. Li C, Song X, Chen X, Liu X, Sang M, Wu W, et al. Identification and Comparative Analysis of G Protein-Coupled Receptors in Pediculus Humanus Humanus. Genomics (2014) 104:58–67. doi: 10.1016/j.ygeno.2014.06.002
19. Zhang H, Bai J, Huang S, Liu H, Lin J, Hou Y. Neuropeptides and G-Protein Coupled Receptors (GPCRs) in the Red Palm Weevil Rhynchophorus Ferrugineus Olivier (Coleoptera: Dryophthoridae). Front Physiol (2020) 11:159. doi: 10.3389/fphys.2020.00159
20. Birgül Iyison N, Shahraki A, Kahveci K, Düzgün MB, Gün G. Are Insect GPCRs Ideal Next-Generation Pesticides: Opportunities and Challenges. FEBS J (2021) 288:2727–45. doi: 10.1111/febs.15708
21. Nässel DR, Zandawala M. Recent Advances in Neuropeptide Signaling in Drosophila, From Genes to Physiology and Behavior. Prog Neurobiol (2019) 179:101607. doi: 10.1016/j.pneurobio.2019.02.003
22. Li C, Yun X, Hu X, Zhang Y, Sang M, Liu X, et al. Identification of G Protein-Coupled Receptors in the Pea Aphid, Acyrthosiphon Pisum. Genomics (2013) 102:345–54. doi: 10.1016/j.ygeno.2013.06.003
23. Li X, Du L, Jiang X-J, Ju Q, Qu C-J, Qu M-J, et al. Identification and Characterization of Neuropeptides and Their G Protein-Coupled Receptors (GPCRs) in the Cowpea Aphid Aphis Craccivora. Front Endocrinol (Lausanne) (2020) 11:640. doi: 10.3389/fendo.2020.00640
24. Benoit JB, Adelman ZN, Reinhardt K, Dolan A, Poelchau M, Jennings EC, et al. Unique Features of a Global Human Ectoparasite Identified Through Sequencing of the Bed Bug Genome. Nat Commun (2016) 7:10165. doi: 10.1038/ncomms10165
25. Wang Z, Zhou W, Hameed MS, Liu J, Zeng X. Characterization and Expression Profiling of Neuropeptides and G-Protein-Coupled Receptors (GPCRs) for Neuropeptides in the Asian Citrus Psyllid, Diaphorina Citri (Hemiptera: Psyllidae). Int J Mol Sci (2018) 19:e3912. doi: 10.3390/ijms19123912
26. Tanaka Y, Suetsugu Y, Yamamoto K, Noda H, Shinoda T. Transcriptome Analysis of Neuropeptides and G-Protein Coupled Receptors (GPCRs) for Neuropeptides in the Brown Planthopper Nilaparvata Lugens. Peptides (2014) 53:125–33. doi: 10.1016/j.peptides.2013.07.027
27. Ons S, Lavore A, Sterkel M, Wulff JP, Sierra I, Martínez-Barnetche J, et al. Identification of G Protein Coupled Receptors for Opsines and Neurohormones in Rhodnius Prolixus. Genomic and Transcriptomic Analysis. Insect Biochem Mol Biol (2016) 69:34–50. doi: 10.1016/j.ibmb.2015.05.003
28. Xu P, Lu B, Chao J, Holdbrook R, Liang G, Lu Y. The Evolution of Opsin Genes in Five Species of Mirid Bugs: Duplication of Long-Wavelength Opsins and Loss of Blue-Sensitive Opsins. BMC Ecol Evol (2021) 21:66. doi: 10.1186/s12862-021-01799-5
29. Cassis G, Schuh RT. Systematics, Biodiversity, Biogeography, and Host Associations of the Miridae (Insecta: Hemiptera: Heteroptera: Cimicomorpha). Annu Rev Entomol (2012) 57:377–404. doi: 10.1146/annurev-ento-121510-133533
30. Pan H, Liu B, Lu Y, Wyckhuys KAG. Seasonal Alterations in Host Range and Fidelity in the Polyphagous Mirid Bug, Apolygus Lucorum (Heteroptera: Miridae). PloS One (2015) 10:e0117153. doi: 10.1371/journal.pone.0117153
31. Lu Y, Wu K, Jiang Y, Xia B, Li P, Feng H, et al. Mirid Bug Outbreaks in Multiple Crops Correlated With Wide-Scale Adoption of Bt Cotton in China. Science (2010) 328:1151–4. doi: 10.1126/science.1187881
32. Liu Y, Liu H, Wang H, Huang T, Liu B, Yang B, et al. Apolygus Lucorum Genome Provides Insights Into Omnivorousness and Mesophyll Feeding. Mol Ecol Resour (2021) 21:287–300. doi: 10.1111/1755-0998.13253
33. Gelbart WM, Crosby M, Matthews B, Rindone WP, Chillemi J, Russo Twombly S, et al. FlyBase: A Drosophila Database. The FlyBase Consortium. Nucleic Acids Res (1997) 25:63–6. doi: 10.1093/nar/25.1.63
34. Brody T, Cravchik A. Drosophila Melanogaster G Protein-Coupled Receptors. J Cell Biol (2000) 150:F83–8. doi: 10.1083/jcb.150.2.F83
35. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: A Better Web Interface. Nucleic Acids Res (2008) 36:W5–9. doi: 10.1093/nar/gkn201
36. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting Transmembrane Protein Topology With a Hidden Markov Model: Application to Complete Genomes. J Mol Biol (2001) 305:567–80. doi: 10.1006/jmbi.2000.4315
37. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: Protein Domains Identifier. Nucleic Acids Res (2005) 33:W116–20. doi: 10.1093/nar/gki442
38. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res (2002) 30:3059–66. doi: 10.1093/nar/gkf436
39. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol (2015) 32:268–74. doi: 10.1093/molbev/msu300
40. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat Methods (2017) 14:587–9. doi: 10.1038/nmeth.4285
41. Leinonen R, Sugawara H, Shumway M. International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res (2011) 39:D19–21. doi: 10.1093/nar/gkq1019
42. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
43. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-Based Genome Alignment and Genotyping With HISAT2 and HISAT-Genotype. Nat Biotechnol (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
44. Zhao S, Ye Z, Stanton R. Misuse of RPKM or TPM Normalization When Comparing Across Samples and Sequencing Protocols. RNA (2020) 26:903–9. doi: 10.1261/rna.074922.120
45. Liao Y, Smyth GK, Shi W. Featurecounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
46. Smid M, Coebergh van den Braak RRJ, van de Werken HJG, van Riet J, van Galen A, de Weerd V, et al. Gene Length Corrected Trimmed Mean of M-Values (GeTMM) Processing of RNA-Seq Data Performs Similarly in Intersample Analyses While Improving Intrasample Comparisons. BMC Bioinf (2018) 19:236. doi: 10.1186/s12859-018-2246-7
47. Letunic I, Bork P. Interactive Tree Of Life (iTOL) V4: Recent Updates and New Developments. Nucleic Acids Res (2019) 47:W256–9. doi: 10.1093/nar/gkz239
48. Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res (2012) 40:e49. doi: 10.1093/nar/gkr1293
49. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant (2020) 13:1194–202. doi: 10.1016/j.molp.2020.06.009
50. Velarde RA, Sauer CD O, Walden KK, Fahrbach SE, Robertson HM. Pteropsin: A Vertebrate-Like Non-Visual Opsin Expressed in the Honey Bee Brain. Insect Biochem Mol Biol (2005) 35:1367–77. doi: 10.1016/j.ibmb.2005.09.001
51. Salcedo E, Huber A, Henrich S, Chadwell LV, Chou W-H, Paulsen R, et al. Blue- and Green-Absorbing Visual Pigments Ofdrosophila: Ectopic Expression and Physiological Characterization of the R8 Photoreceptor Cell-Specific Rh5 and Rh6 Rhodopsins. J Neurosci (1999) 19:10716–26. doi: 10.1523/JNEUROSCI.19-24-10716.1999
52. Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, et al. Genomics, Transcriptomics, and Peptidomics of Neuropeptides and Protein Hormones in the Red Flour Beetle Tribolium Castaneum. Genome Res (2008) 18:113–22. doi: 10.1101/gr.6714008
53. Dolezelova E, Nothacker H-P, Civelli O, Bryant PJ, Zurovec M. A Drosophila Adenosine Receptor Activates cAMP and Calcium Signaling. Insect Biochem Mol Biol (2007) 37:318–29. doi: 10.1016/j.ibmb.2006.12.003
54. Abbracchio MP, Boeynaems J-M, Barnard EA, Boyer JL, Kennedy C, Miras-Portugal MT, et al. Characterization of the UDP-Glucose Receptor (Re-Named Here the P2Y14 Receptor) Adds Diversity to the P2Y Receptor Family. Trends Pharmacol Sci (2003) 24:52–5. doi: 10.1016/S0165-6147(02)00038-X
55. Abbracchio MP, Burnstock G. Purinoceptors: Are There Families of P2X and P2Y Purinoceptors? Pharmacol Ther (1994) 64:445–75. doi: 10.1016/0163-7258(94)00048-4
56. Yu R, Liu H, Peng X, Cui Y, Song S, Wang L, et al. The Palmitoylation of the N-Terminal Extracellular Cys37 Mediates the Nuclear Translocation of VPAC1 Contributing to Its Anti-Apoptotic Activity. Oncotarget (2017) 8:42728–41. doi: 10.18632/oncotarget.17449
57. Li C, Chen M, Sang M, Liu X, Wu W, Li B. Comparative Genomic Analysis and Evolution of Family-B G Protein-Coupled Receptors From Six Model Insect Species. Gene (2013) 519:1–12. doi: 10.1016/j.gene.2013.01.061
58. Harmar AJ. Family-B G-Protein-Coupled Receptors. Genome Biol (2001) 2:reviews3013.1. doi: 10.1186/gb-2001-2-12-reviews3013
59. Liapakis G, Matsoukas M-T, Karageorgos V, Venihaki M, Mavromoustakos T. Family B G Protein-Coupled Receptors and Their Ligands: From Structure to Function. Curr Med Chem (2017) 24:3323–55. doi: 10.2174/0929867324666170303162416
60. Zandawala M, Li S, Hauser F, Grimmelikhuijzen CJP, Orchard I. Isolation and Functional Characterization of Calcitonin-Like Diuretic Hormone Receptors in Rhodnius Prolixus. PloS One (2013) 8:e82466. doi: 10.1371/journal.pone.0082466
61. Nagasaki K, Asami T, Kikuchi T, Uchiyama M. Longitudinal Growth of the Short Bones of the Hand in a Girl With Pseudohypoparathyroidism Type Ia. Clin Pediatr Endocrinol (2007) 16:23–9. doi: 10.1297/cpe.16.23
62. Xie J, Sang M, Song X, Zhang S, Kim D, Veenstra JA, et al. A New Neuropeptide Insect Parathyroid Hormone iPTH in the Red Flour Beetle Tribolium Castaneum. PloS Genet (2020) 16:e1008772. doi: 10.1371/journal.pgen.1008772
63. Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, et al. G Protein-Coupled Receptors in Anopheles Gambiae. Science (2002) 298:176–8. doi: 10.1126/science.1076196
64. Vogel KJ, Brown MR, Strand MR. Phylogenetic Investigation of Peptide Hormone and Growth Factor Receptors in Five Dipteran Genomes. Front Endocrinol (2013) 4:193. doi: 10.3389/fendo.2013.00193
65. Friedrich M, Jones JW. Gene Ages, Nomenclatures, and Functional Diversification of the Methuselah/Methuselah-Like GPCR Family in Drosophila and Tribolium. J Exp Zool B Mol Dev Evol (2016) 326:453–63. doi: 10.1002/jez.b.22721
66. Mitri C, Parmentier M-L, Pin J-P, Bockaert J, Grau Y. Divergent Evolution in Metabotropic Glutamate Receptors: A New Receptor Activated By an Endogenous Ligand Different From Glutamte in Insects*. J Biol Chem (2004) 279:9313–20. doi: 10.1074/jbc.M310878200
67. Mezler M, Müller T, Raming K. Cloning and Functional Expression of GABA(B) Receptors From Drosophila. Eur J Neurosci (2001) 13:477–86. doi: 10.1046/j.1460-9568.2001.01410.x
68. Josefsson LG. Evidence for Kinship Between Diverse G-Protein Coupled Receptors. Gene (1999) 239:333–40. doi: 10.1016/s0378-1119(99)00392-3
69. Papagiannouli F, Lohmann I. Stage-Specific Control of Stem Cell Niche Architecture in the Drosophila Testis by the Posterior Hox Gene Abd-B. Comput Struct Biotechnol J (2015) 13:122–30. doi: 10.1016/j.csbj.2015.01.001
70. Kohyama-Koganeya A, Kurosawa M, Hirabayashi Y. Differential Effects of Tissue-Specific Deletion of BOSS on Feeding Behaviors and Energy Metabolism. PloS One (2015) 10:e0133083. doi: 10.1371/journal.pone.0133083
71. Huang H-C, Klein PS. The Frizzled Family: Receptors for Multiple Signal Transduction Pathways. Genome Biol (2004) 5:234. doi: 10.1186/gb-2004-5-7-234
72. Tierney AJ. Invertebrate Serotonin Receptors: A Molecular Perspective on Classification and Pharmacology. J Exp Biol (2018) 221:jeb184838. doi: 10.1242/jeb.184838
73. Verlinden H. Dopamine Signalling in Locusts and Other Insects. Insect Biochem Mol Biol (2018) 97:40–52. doi: 10.1016/j.ibmb.2018.04.005
74. Karam CS, Jones SK, Javitch JA. Come Fly With Me: An Overview of Dopamine Receptors in Drosophila Melanogaster. Basic Clin Pharmacol Toxicol (2020) 126 Suppl 6:56–65. doi: 10.1111/bcpt.13277
75. Xiong R, Zhao W, Chen X, Li T, Li H, Li Y, et al. Pharmacological Characterization of the 5-HT1A Receptor of Bombyx Mori and Its Role in Locomotion. Comp Biochem Physiol A Mol Integr Physiol (2019) 231:56–65. doi: 10.1016/j.cbpa.2019.01.020
76. Malloy CA, Somasundaram E, Omar A, Bhutto U, Medley M, Dzubuk N, et al. Pharmacological Identification of Cholinergic Receptor Subtypes: Modulation of Locomotion and Neural Circuit Excitability in Drosophila Larvae. Neuroscience (2019) 411:47–64. doi: 10.1016/j.neuroscience.2019.05.016
77. Sun R, Delly J, Sereno E, Wong S, Chen X, Wang Y, et al. Anti-Instinctive Learning Behavior Revealed by Locomotion-Triggered Mild Heat Stress in Drosophila. Front Behav Neurosci (2020) 14:41. doi: 10.3389/fnbeh.2020.00041
78. Ganguly A, Qi C, Bajaj J, Lee D. Serotonin Receptor 5-HT7 in Drosophila Mushroom Body Neurons Mediates Larval Appetitive Olfactory Learning. Sci Rep (2020) 10:21267. doi: 10.1038/s41598-020-77910-5
79. Lee H-G, Seong C-S, Kim Y-C, Davis RL, Han K-A. Octopamine Receptor OAMB Is Required for Ovulation in Drosophila Melanogaster. Dev Biol (2003) 264:179–90. doi: 10.1016/j.ydbio.2003.07.018
80. Terhzaz S, Cabrero P, Robben JH, Radford JC, Hudson BD, Milligan G, et al. Mechanism and Function of Drosophila Capa GPCR: A Desiccation Stress-Responsive Receptor With Functional Homology to Human neuromedinU Receptor. PloS One (2012) 7:e29897. doi: 10.1371/journal.pone.0029897
81. Williams MJ, Akram M, Barkauskaite D, Patil S, Kotsidou E, Kheder S, et al. CCAP Regulates Feeding Behavior via the NPF Pathway in Drosophila Adults. Proc Natl Acad Sci USA (2020) 117:7401–8. doi: 10.1073/pnas.1914037117
82. Antony B, Soffan A, Jakše J, Abdelazim MM, Aldosari SA, Aldawood AS, et al. Identification of the Genes Involved in Odorant Reception and Detection in the Palm Weevil Rhynchophorus Ferrugineus, an Important Quarantine Pest, by Antennal Transcriptome Analysis. BMC Genomics (2016) 17:69. doi: 10.1186/s12864-016-2362-6
83. Chen H, Lahey Z, Talamas EJ, Johnson NF. Identification and Expression of Chemosensory Receptor Genes in the Egg Parasitoid Trissolcus Basalis. Comp Biochem Physiol Part D Genomics Proteomics (2021) 37:100758. doi: 10.1016/j.cbd.2020.100758
84. Price DA, Greenberg MJ. Purification and Characterization of a Cardioexcitatory Neuropeptide From the Central Ganglia of a Bivalve Mollusc. Prep Biochem (1977) 7:261–81. doi: 10.1080/00327487708061643
85. Holman GM, Cook BJ, Nachman RJ. Primary Structure and Synthesis of a Blocked Myotropic Neuropeptide Isolated From the Cockroach, Leucophaea Maderae. Comp Biochem Physiol Part C: Comp Pharmacol (1986) 85:219–24. doi: 10.1016/0742-8413(86)90077-0
86. Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJP. A Review of Neurohormone GPCRs Present in the Fruitfly Drosophila Melanogaster and the Honey Bee Apis Mellifera. Prog Neurobiol (2006) 80:1–19. doi: 10.1016/j.pneurobio.2006.07.005
87. Ravi P, Trivedi D, Hasan G. FMRFa Receptor Stimulated Ca2+ Signals Alter the Activity of Flight Modulating Central Dopaminergic Neurons in Drosophila Melanogaster. PloS Genet (2018) 14:e1007459. doi: 10.1371/journal.pgen.1007459
88. Orchard I, Lange AB, Bendena WG. FMRFamide-Related Peptides: A Multifunctional Family of Structurally Related Neuropeptides in Insects. In: Advances in Insect Physiology. London: Academic Press (2001). p. 267–329. doi: 10.1016/S0065-2806(01)28012-6
89. Hewes RS, Snowdeal EC, Saitoe M, Taghert PH. Functional Redundancy of FMRFamide-Related Peptides at the Drosophila Larval Neuromuscular Junction. J Neurosci (1998) 18:7138–51. doi: 10.1523/JNEUROSCI.18-18-07138.1998
90. Lu Y, Wu K, Guo Y. Flight Potential of Lygus Lucorum (Meyer-Dür) (Heteroptera: Miridae). Environ Entomol (2007) 36:1007–13. doi: 10.1603/0046-225x(2007)36[1007:fpollm]2.0.co;2
91. Sriram K, Wiley SZ, Moyung K, Gorr MW, Salmerón C, Marucut J, et al. Detection and Quantification of GPCR mRNA: An Assessment and Implications of Data From High-Content Methods. ACS Omega (2019) 4:17048–59. doi: 10.1021/acsomega.9b02811
92. Su D, Wang M, Ye C, Fang J, Duan Y, Zhang Z, et al. One-Step Generation of Mice Carrying a Conditional Allele Together With an HA-Tag Insertion for the Delta Opioid Receptor. Sci Rep (2017) 7:44476. doi: 10.1038/srep44476
93. Chung KY, Day PW, Vélez-Ruiz G, Sunahara RK, Kobilka BK. Identification of GPCR-Interacting Cytosolic Proteins Using HDL Particles and Mass Spectrometry-Based Proteomic Approach. PloS One (2013) 8:e54942. doi: 10.1371/journal.pone.0054942
94. Wise H. The Roles Played by Highly Truncated Splice Variants of G Protein-Coupled Receptors. J Mol Signal (2012) 7:13. doi: 10.1186/1750-2187-7-13
95. Feuda R, Marlétaz F, Bentley MA, Holland PWH. Conservation, Duplication, and Divergence of Five Opsin Genes in Insect Evolution. Genome Biol Evol (2016) 8:579–87. doi: 10.1093/gbe/evw015
96. Shichida Y, Matsuyama T. Evolution of Opsins and Phototransduction. Philos Trans R Soc Lond B Biol Sci (2009) 364:2881–95. doi: 10.1098/rstb.2009.0051
98. Hu K, De Rosa E, Anderson AK. Differential Color Tuning of the Mesolimbic Reward System. Sci Rep (2020) 10:10223. doi: 10.1038/s41598-020-66574-w
99. Hall DW, Sander SE, Pallansch JC, Stanger-Hall KF. The Evolution of Adult Light Emission Color in North American Fireflies. Evolution (2016) 70:2033–48. doi: 10.1111/evo.13002
100. Zhang S, Kong X, Liu F, Zhang Z. Identification and Expression Patterns of Opsin Genes in a Forest Insect, Dendrolimus Punctatus. Insects (2020) 11:e116. doi: 10.3390/insects11020116
101. Liu Y-J, Yan S, Shen Z-J, Li Z, Zhang X-F, Liu X-M, et al. The Expression of Three Opsin Genes and Phototactic Behavior of Spodoptera Exigua (Lepidoptera: Noctuidae): Evidence for Visual Function of Opsin in Phototaxis. Insect Biochem Mol Biol (2018) 96:27–35. doi: 10.1016/j.ibmb.2018.03.006
102. Geng H, Lu Y, Yang Y. Activity of Adult Apolygus Lucorum in Cotton Fields. Chin J Appl Entomol (2012) 49:601–4.
103. Gao S, Liu X, Liu J, Xiong W, Song X, Wu W, et al. Identification and Evolution of Latrophilin Receptor Gene Involved in Tribolium Castaneum Devolopment and Female Fecundity. Genesis (2017) 55:e23081. doi: 10.1002/dvg.23081
104. Xiong W, Gao S, Lu Y, Wei L, Mao J, Xie J, et al. Latrophilin Participates in Insecticide Susceptibility Through Positively Regulating CSP10 and Partially Compensated by OBPC01 in Tribolium Castaneum. Pestic Biochem Physiol (2019) 159:107–17. doi: 10.1016/j.pestbp.2019.06.005
105. Nordström KJV, Lagerström MC, Wallér LMJ, Fredriksson R, Schiöth HB. The Secretin GPCRs Descended From the Family of Adhesion GPCRs. Mol Biol Evol (2009) 26:71–84. doi: 10.1093/molbev/msn228
106. Silva J-P, Ushkaryov YA. The Latrophilins, “Split-Personality” Receptors. Adv Exp Med Biol (2010) 706:59–75. doi: 10.1007/978-1-4419-7913-1_5
107. van der Voet M, Harich B, Franke B, Schenck A. ADHD-Associated Dopamine Transporter, Latrophilin and Neurofibromin Share a Dopamine-Related Locomotor Signature in Drosophila. Mol Psychiatry (2016) 21:565–73. doi: 10.1038/mp.2015.55
108. Ichtchenko K, Bittner MA, Krasnoperov V, Little AR, Chepurny O, Holz RW, et al. A Novel Ubiquitously Expressed Alpha-Latrotoxin Receptor is a Member of the CIRL Family of G-Protein-Coupled Receptors. J Biol Chem (1999) 274:5491–8. doi: 10.1074/jbc.274.9.5491
109. Wei L, Gao S, Xiong W, Liu J, Mao J, Lu Y, et al. Latrophilin Mediates Insecticides Susceptibility and Fecundity Through Two Carboxylesterases, Esterase4 and Esterase6, in Tribolium Castaneum. Bull Entomol Res (2019) 109:534–43. doi: 10.1017/S0007485318000895
110. Van Loy T, Vandersmissen HP, Van Hiel MB, Poels J, Verlinden H, Badisco L, et al. Comparative Genomics of Leucine-Rich Repeats Containing G Protein-Coupled Receptors and Their Ligands. Gen Comp Endocrinol (2008) 155:14–21. doi: 10.1016/j.ygcen.2007.06.022
111. Van Hiel MB, Vandersmissen HP, Van Loy T, Vanden Broeck J. An Evolutionary Comparison of Leucine-Rich Repeat Containing G Protein-Coupled Receptors Reveals a Novel LGR Subtype. Peptides (2012) 34:193–200. doi: 10.1016/j.peptides.2011.11.004
112. Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics Resolves the Timing and Pattern of Insect Evolution. Science (2014) 346:763–7. doi: 10.1126/science.1257570
113. Vandersmissen HP, Van Hiel MB, Van Loy T, Vleugels R, Vanden Broeck J. Silencing D. Melanogaster Lgr1 Impairs Transition From Larval to Pupal Stage. Gen Comp Endocrinol (2014) 209:135–47. doi: 10.1016/j.ygcen.2014.08.006
114. Bai H, Palli SR. Functional Characterization of Bursicon Receptor and Genome-Wide Analysis for Identification of Genes Affected by Bursicon Receptor RNAi. Dev Biol (2010) 344:248–58. doi: 10.1016/j.ydbio.2010.05.003
115. Colombani J, Andersen DS, Boulan L, Boone E, Romero N, Virolle V, et al. Drosophila Lgr3 Couples Organ Growth With Maturation and Ensures Developmental Stability. Curr Biol (2015) 25:2723–9. doi: 10.1016/j.cub.2015.09.020
116. Vallejo DM, Juarez-Carreño S, Bolivar J, Morante J, Dominguez M. A Brain Circuit That Synchronizes Growth and Maturation Revealed Through Dilp8 Binding to Lgr3. Science (2015) 350:aac6767. doi: 10.1126/science.aac6767
Keywords: identification, GPCRs, Apolygus lucorum, expansion, phylogenetic analysis
Citation: Gao H, Li Y, Wang M, Song X, Tang J, Feng F and Li B (2021) Identification and Expression Analysis of G Protein-Coupled Receptors in the Miridae Insect Apolygus lucorum. Front. Endocrinol. 12:773669. doi: 10.3389/fendo.2021.773669
Received: 10 September 2021; Accepted: 08 November 2021;
Published: 26 November 2021.
Edited by:
Maurice Richard Elphick, Queen Mary University of London, United KingdomReviewed by:
Christian Wegener, Julius Maximilian University of Würzburg, GermanyMeet Zandawala, Brown University, United States
Copyright © 2021 Gao, Li, Wang, Song, Tang, Feng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Li, libin@njnu.edu.cn
 Han Gao
Han Gao Bin Li
Bin Li