- 1Department of Urology, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Department of Research Management and International Cooperation, The Affiliated Hospital of Qingdao University, Qingdao, China
Background: Adrenocortical carcinoma (ACC) is an aggressive and rare neoplasm that originates from the cortex of the adrenal gland. N6-methyladenosine (m6A) RNA methylation, the most common form of mRNA modification, has been reported to be correlated with the occurrence and development of the malignant tumor. This study aims to identify the significance of m6A RNA methylation regulators in ACC and construct a m6A based signature to predict the prognosis of ACC patients.
Materials and methods: RNA-seq data from The Cancer Genome Atlas (TCGA) database was used to identify the expression level of m6A RNA methylation regulators in ACC. An m6A based signature was further constructed and its prognostic and predictive values were assessed by survival analysis and nomogram.
Results: 11 m6A RNA regulators were differentially expressed in ACC and three m6A RNA regulators were finally selected in a signature to predict the prognosis of ACC patients. Survival analysis indicated that high risk scores were closely related to poor survival outcomes in ACC patients. Univariate and multivariate Cox regression analyses demonstrated that the m6A based signature was an independent prognostic factor for ACC patients. A nomogram with clinical factors and the m6A based signature was also constructed to superiorly predict the prognosis of ACC patients. The expression levels of m6A RNA methylation regulators, which were contained in the signature, were also verified in human ACC tissues and normal tissues by using vitro experiments.
Conclusion: We identified and validated an m6A based signature, which can be used as an independent prognostic factor in evaluating the prognosis of ACC patients. Further clinical trials and experimental explorations are needed to confirm our observations and mechanisms underlying prognostic values of these m6A RNA methylation regulators in ACC.
Introduction
Adrenocortical carcinoma (ACC) is a rare disease of the adrenal cortex with aggressive behavior, and only 16% to 47% of patients are still alive after five years (1). The significant progress in the treatment of ACC, the molecular mechanisms, drug resistance, and reliable prognostic biomarkers in ACC remain poorly understood. Over the past decade, methods in genomic study have multiplied our knowledge about gene expression, genetic, and epigenetic alterations at the pan-genomic level in ACC. This highlights the importance of genetic counseling to identify potential prognostic biomarkers in patients with ACC. N6-methyladenosine (m6A) is the most abundant internal modification of mRNAs in higher eukaryotes and mainly influences mRNA stability, translation efficiency, variable splicing, and localization by dynamic regulation of methyltransferases (“writers”), m6A-binding proteins (“readers”), and demethylases (“erasers”) (2–4). Previous studies also demonstrated that m6A RNA regulators were significantly associated with the prognosis, chemoresistance, and radioresistance of cancers, such as breast cancer, bladder cancer, gastric cancer, and pancreatic cancer (5–8). In addition, m6A RNA methylation regulators can promote the occurrence and progression of cancers by multiple signaling pathways, such as regulating the epithelial mesenchymal transition of cancer, Wnt/PI3K-Akt signaling, and modulating genes expression levels (9–11). However, the expression levels and prognostic values of m6A RNA methylation regulators in ACC have not been elucidated.
In the present study, we first utilized the transcriptome data from The Cancer Genome Atlas (TCGA) to identify the significance of 13 m6A RNA methylation regulators in ACC patients. We evaluated the interaction and correlation among m6A RNA methylation regulators and identified two clusters of ACC patients with different survival outcomes. Subsequently, an m6A based signature was further developed and was closely related to the prognosis and clinicopathologic characteristics of ACC patients. Furthermore, a nomogram was constructed to validate its predictive value in ACC.
Materials and Methods
Data Collection
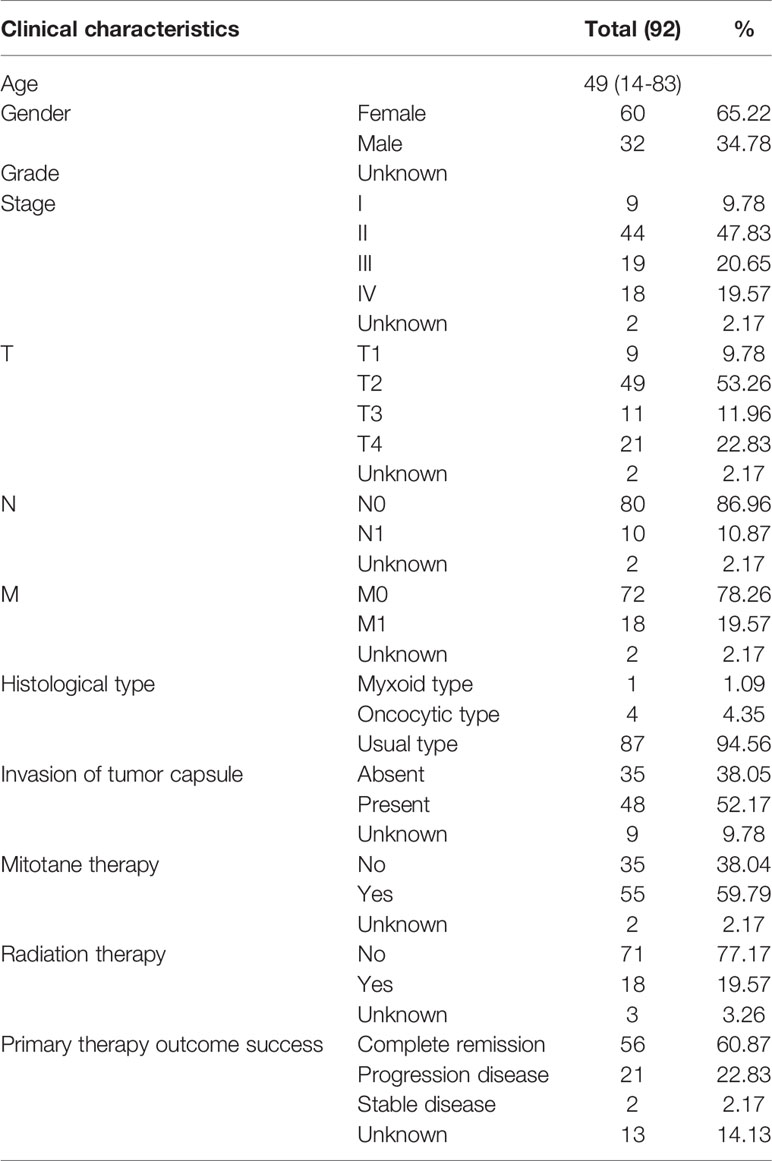
RNA-sequencing (RNA-seq) expression profiles of the ACC cohort were obtained from TCGA official website (https://tcga‐data.nci.nih.gov/tcga/). A total of 92 patients were enrolled in our study and detailed clinical characteristics are described in Table 1. RNA-sequencing (RNA-seq) expression profiles of normal cohort contained 128 normal samples, which were download from the Genotype-Tissue Expression (GTEx) database (https://xenabrowser.net/datapages/). Thirteen widely recognized m6A RNA regulators were retrieved from published literature, including METTL3 (methyltransferase-like 3), METTL14 (methyltransferase-like 14), WTAP (Wilms tumor 1-associated protein), KIAA1429 (VIRMA, vir like m6A methyltransferase associated), RBM15 (RNA binding motif protein 15), FTO (fat mass and obesity-associated gene), ALKBH5 (alkB homolog 5, RNA demethylase), YTHDC1 (YTH domain containing 1), YTHDC2 (YTH domain containing 2), YTHDF1 (YTH N6-methyladenosine RNA binding protein 1), YTHDF2 (YTH N6-methyladenosine RNA binding protein 2), HNRNPC (heterogeneous nuclear ribonucleoprotein C), and ZC3H13 (zinc finger CCCH-type containing 13).
Differential Expression Analysis of m6A RNA Methylation Regulators in ACC
The expression levels of 13 m6A RNA regulators between ACC and normal samples were measured using edgeR (version R 3.5.1, https://bioconductor.org/packages/release/bioc/) (12). P < 0.05 was recognized as the cut-off value. Heatmap and violin plot were used to display the expression levels of m6A RNA methylation regulators in ACC and normal samples.
Consensus Clustering Analysis
To determine the relationship between the expression levels of m6A RNA regulators and the prognosis of ACC patients, we classified ACC patients into different groups by consensus clustering analysis with “ConsensusClusterPlus” in R. Kaplan-Meier method was performed to investigate the survival differences between two clusters.
Construction and Evaluation of an m6A Based Signature
Univariate Cox regression analysis was performed using the R package “survival” to identify m6A RNA methylation regulators that were associated with the OS of ACC patients. A least absolute shrinkage and selection operator (LASSO) Cox regression method was adopted to establish an optimal m6A based signature for predicting the prognosis of ACC patients by using the R package called “glmnet” and the m6A based signature risk score = Σ(βi×Expi) (i = the number of m6A RNA regulators) (13, 14). After calculating the risk scores of ACC patients, patients were further stratified into high-risk and low-risk groups according to the median risk score. Kaplan-Meier curves were used to evaluate the survival outcome differences between the high-risk and low-risk groups. The survival ROC package was used to conduct the receiver operating characteristic curve (ROC) to evaluate the prediction accuracy of the m6A based signature. Univariate and multivariate Cox regression analyses were used to explore prognostic values of the m6A based signature and various clinical characteristics. In addition, the Wilcoxon signed-rank test was applied to identify the relationship between the m6A based signature risk score and clinicopathological characteristics.
Development of a Nomogram Based on the Signature Risk Score and Clinicopathological Characteristics
Clinical factors (age, gender, stage, T, N, M, invasion of tumor capsule, mitotane therapy, radiation therapy) and the m6A based signature risk score were used to construct a prognostic nomogram to assess the probability of 1-, 2-, and 3-year OS for ACC patients via the R package (https://cran.r-project.org/web/packages/rms/) (15).
RNA Isolation and Reverse Transcription−Quantitative PCR
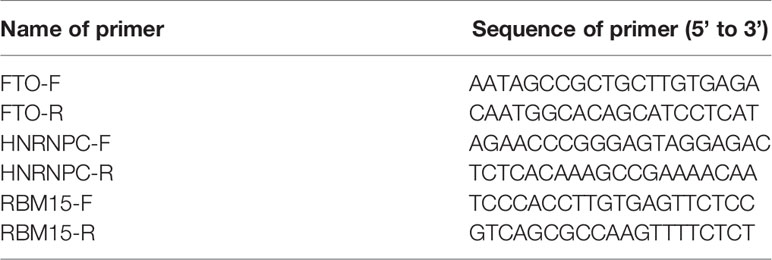
To further validate the expression levels of three m6A RNA regulators in our ACC tissues and normal tissues, RNA isolation and reverse transcription−quantitative PCR (RT-qPCR) were performed. The RNAprep pure FFPE kit (DP439, TIANGEN Biotech(Beijing)Co, Ltd, CHN) was used to extract the total RNA from tissue specimens based on the manufacturer’s instructions. For the detection of mRNA levels, the total RNA (500 ng) was transcribed into cDNA using a PrimeScript™ RT reagent kit (Perfect Real Time) (Takara, code no RR037A). All the primers were synthesized by Huada Gene (Beijing, China) and the sequences are shown in Table 2. The amplification of cDNAs was conducted with Roche LightCycler 480II real-time PCR detection system (Roche, Basel, Switzerland). Gene expression was normalized against GADPH and relative expression levels of RBM15, FTO, and HNRNPC were determined by the comparative threshold cycle (Ct) method using the formula 2−(ΔΔCt).
Results
Expression Patterns of 13 m6A RNA Regulators in ACC
A heatmap was generated to screen the expression levels of 13 m6A RNA regulators between ACC and normal samples. Red represents high expression, while green represents low expression. KIAA1429 (p < 0.001), HNRNPC (p < 0.001), RBM15 (p < 0.001), METTL3 (p < 0.001), ZC3H13 (p < 0.001), WTAP (p < 0.001), YTHDF1 (p < 0.001), FTO (p < 0.001), YTHDF2 (p < 0.001), and ALKBH5 (p < 0.001) were differentially expressed in ACC samples (Figure 1A). Compared to normal tissue samples, KIAA1429, HNRNPC, METTL3, WTAP, YTHDC1, and FTO were down-regulated in ACC, and RBM15, ZC3H13, YTHDF1, YTHDF2, and ALKBH5 were up-regulated in ACC (Figure 1B).
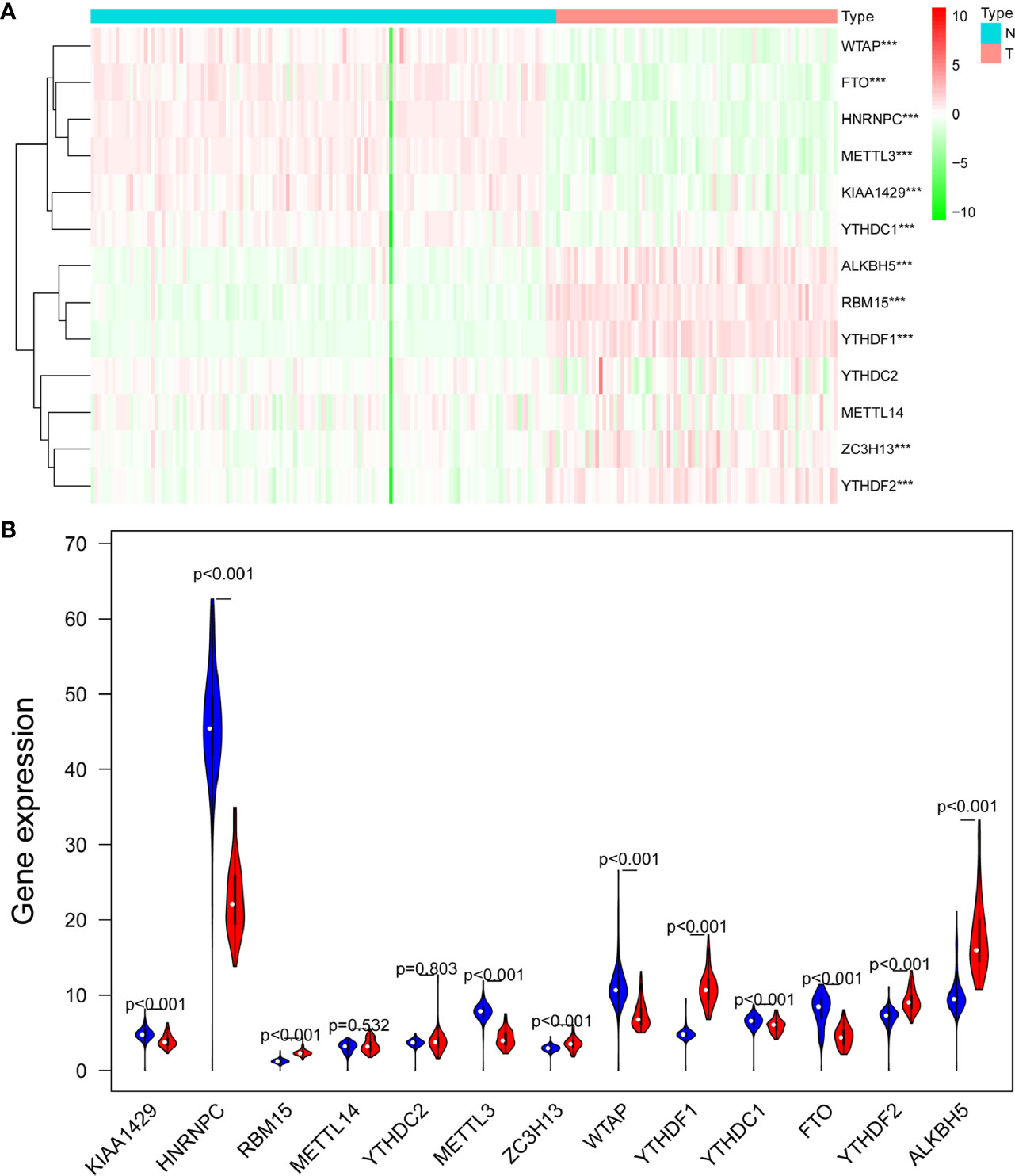
Figure 1 The expression levels of 13 m6A RNA methylation regulators between ACC and normal samples. (A) The heatmap was used to screen the expression levels of m6A RNA methylation regulators in each clinical sample. (B) The violin plot showed the significantly differentially expressed m6A RNA methylation regulators between ACC and the normal samples. ***p < 0.005.
Consensus Clustering of m6A RNA Methylation Regulators Identified Two Clusters of ACC Patients With Different Survival Outcomes
Based on the expression similarity of m6A RNA regulators, k=2 was the most optimal approach to divide ACC patients into two clusters, namely cluster 1 and cluster 2 (Figures 2A-C). Compare with cluster 1, most m6A RNA methylation regulators were highly expressed in cluster 2 (Supplementary 1). Survival analysis indicated that a significantly shorter overall survival (OS) (p = 3.311e-02), disease-specific survival (DSS) (p = 3.589e-02), disease-free interval (DFI) (p = 2.817e-04), and progression-free interval (PFI) (p = 2.957e-04) were observed in ACC patients in cluster 2 compared to those in cluster 1 (Figures 3A-D).
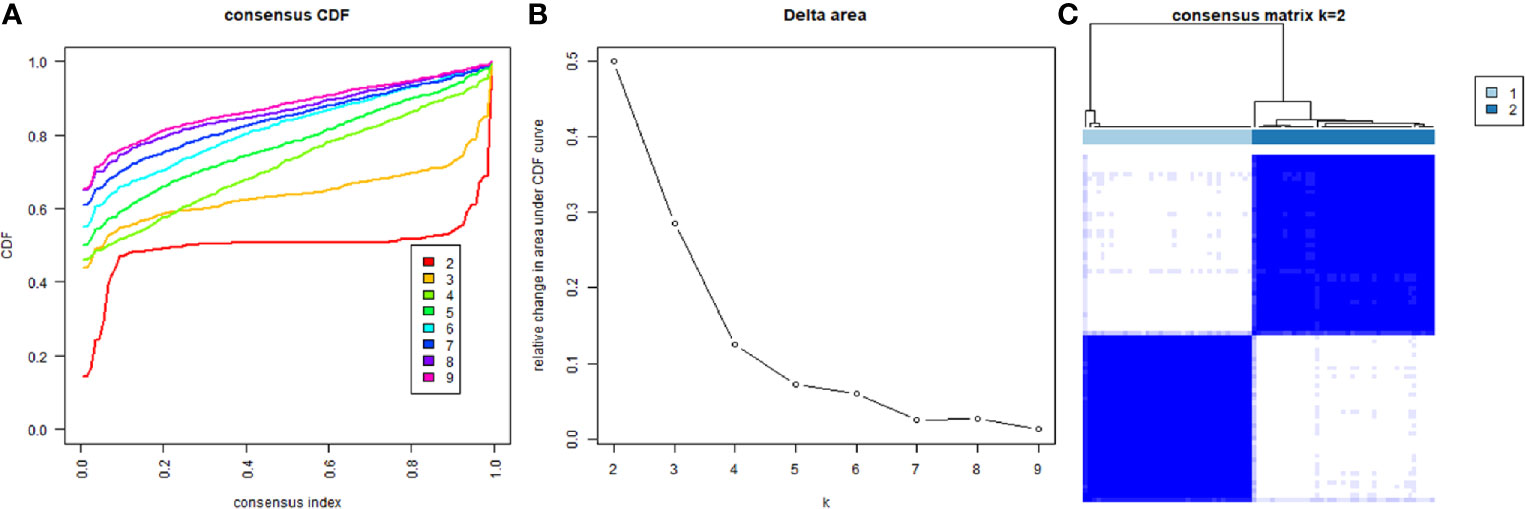
Figure 2 Divided ACC patients into different groups according to the expression of m6A RNA methylation regulators. (A) Consensus clustering cumulative distribution function CDF for k = 2 to 9. (B) Relative change in area under CDF curve for k = 2 to 9. (C) The TCGA ACC cohort was divided into two distinct clusters when k = 2.
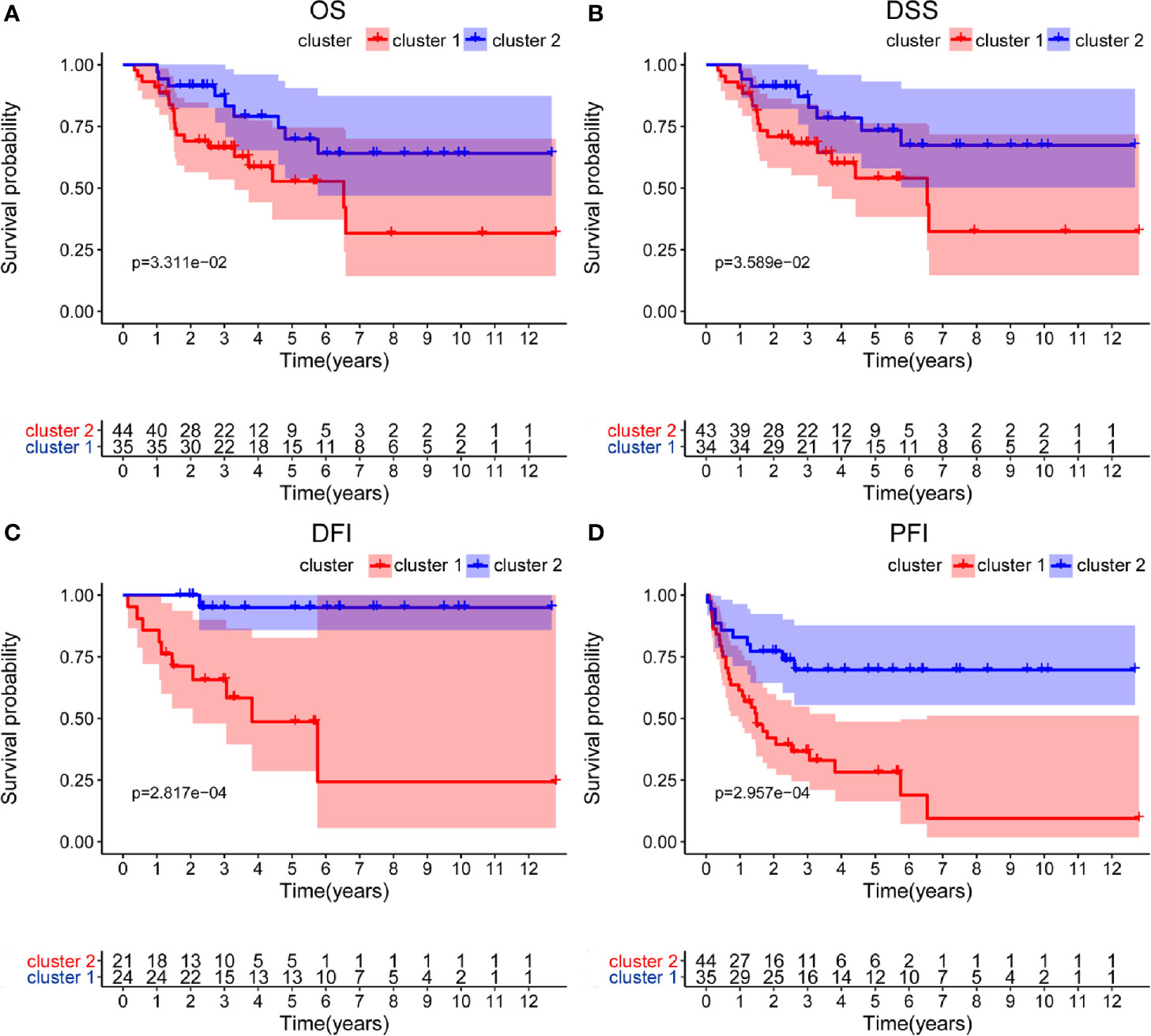
Figure 3 Differential survival outcomes of ACC patients in the two different clusters. (A–D) Survival analysis indicated that a significant shorter overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) were observed in ACC patients in cluster 2 than those in cluster 1.
Identification of Survival-Related m6A RNA Regulators and Construction of a m6A Based Signature
Univariate Cox regression analysis was applied to identify survival-related m6A RNA regulators in ACC patients. The result demonstrated that RBM15 (p < 0.001), HNRNPC (p < 0.001), and YTHDF2 (p = 0.028) were significantly related to OS and these three m6A regulators were risk genes with HR larger than 1 (Figure 4). Subsequently, LASSO Cox regression analysis was performed to construct an m6A based signature that contained three m6A RNA regulators, including RBM15, HNRNPC, and FTO. We further calculated the risk score for ACC patients as follows: risk score = (0.3070 × expression value of RBM15) + (0.0235 × expression value of HNRNPC) + (-0.0673 × expression value of FTO), and divided ACC patients into high-risk and low-risk groups following the cut-off of the median risk score. Survival analysis revealed that the high-risk group had shorter OS (p = 1.396e-05), DSS (p = 2.96e-05), DFI (p = 9.582e-03), and PFI (p = 2.088e-06) when compared with the low-risk group (Figures 5A-D). In addition, high risk scores also indicate poor OS in ACC patients, which received mitotane therapy (p = 2.908e-03, Figure 5E). Furthermore, ROC curves were used to validate the prediction accuracy of the signature and the areas under the curves (AUCs) of risk score, age, gender, stage, T, N, M, invasion, mitotane therapy, and radiation therapy were 0.747, 0.538, 0.659, 0.679, 0.745, 0.455, 0.583, 0.727, 0.689, and 0.402, which indicated superior prediction accuracy of the m6A based signature in survival outcomes (Figure 5F).
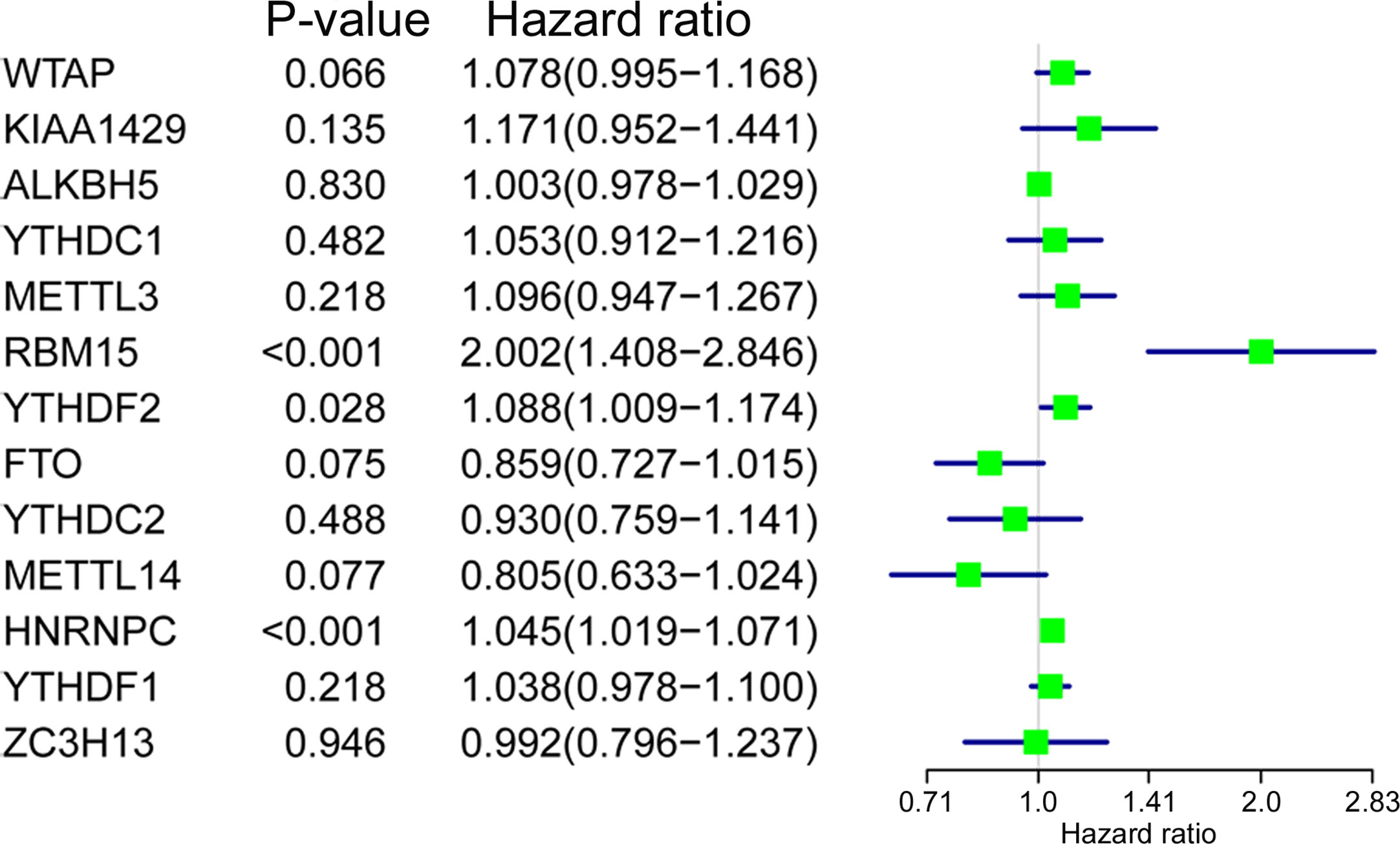
Figure 4 The relationship between m6A RNA methylation regulators and OS of ACC patients. Univariate Cox regression analysis showed that RBM15, YTHDF2, and HNRNPC are closely associated with the OS of BC patients (p < 0.05).
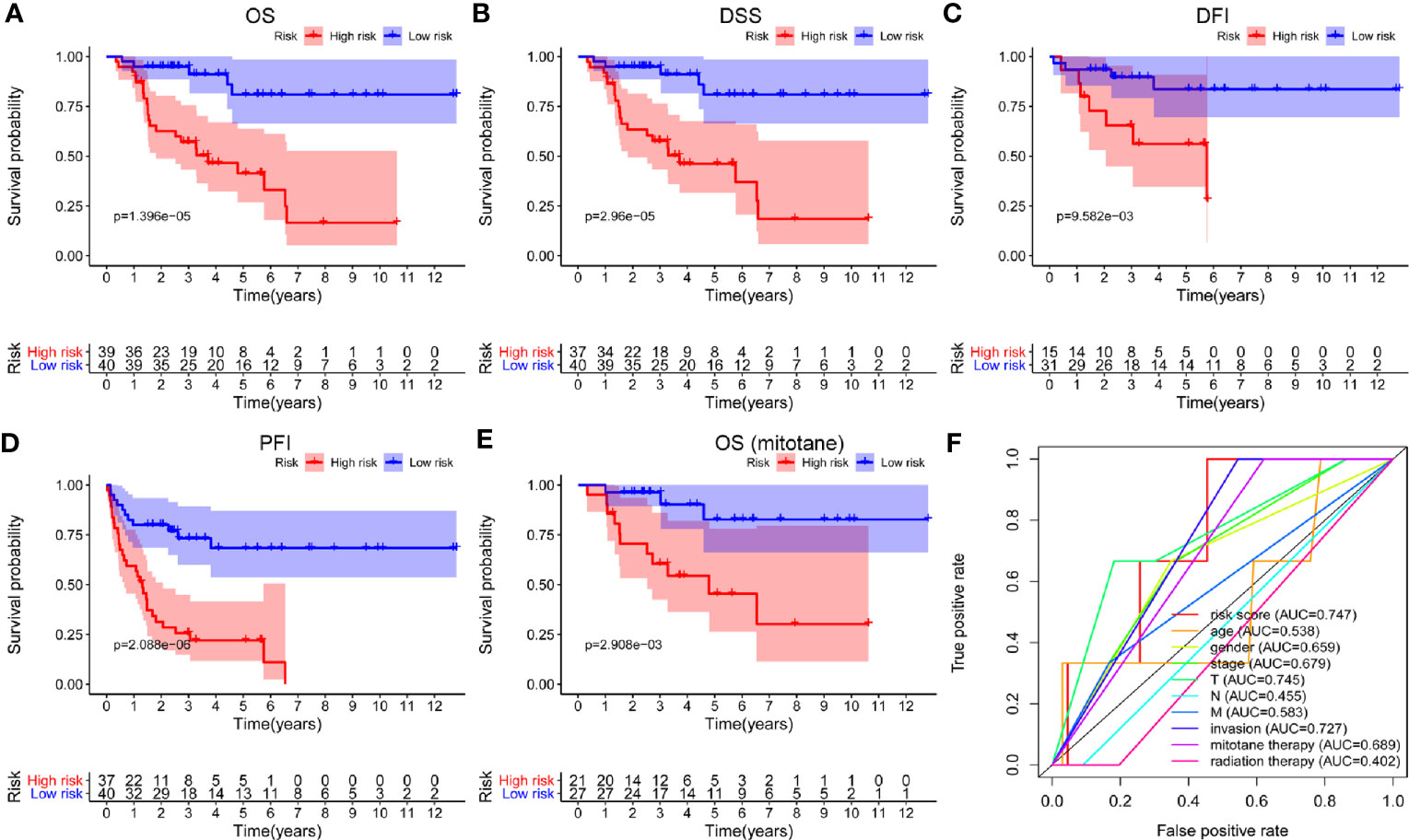
Figure 5 Survival analysis of the m6A based signature in ACC. (A-D) Kaplan-Meier curves revealed that the high-risk group had significantly shorter overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) compared with the low-risk group. (E) In addition, a high risk score also indicated poor OS in ACC patients who received mitotane therapy. (F) ROC curves showed that the area under the curves (AUCs) of risk score, age, gender, stage, T, N, M, invasion, mitotane therapy, and radiation therapy were 0.747, 0.538,0.659, 0.679,0.745, 0.455, 0.583, 0.727, 0.689, and 0.402.
The m6A Based Signature Is an Independent Prognostic Factor for ACC
To evaluate the prognostic significance of the m6A based signature and various clinical factors, univariate and multivariate Cox regression analyses were performed. Univariate Cox regression analysis showed that stage (p < 0.001), T (p < 0.001), M (p < 0.001), invasion of tumor capsule (p = 0.030), mitotane therapy (p = 0.037), and risk score (p < 0.001) were significantly associated with the OS of ACC patients (Figure 6A). Multivariate Cox regression analysis demonstrated that the m6A based signature could serve as an independent prognostic factor in ACC (p = 0.016, Figure 6B).
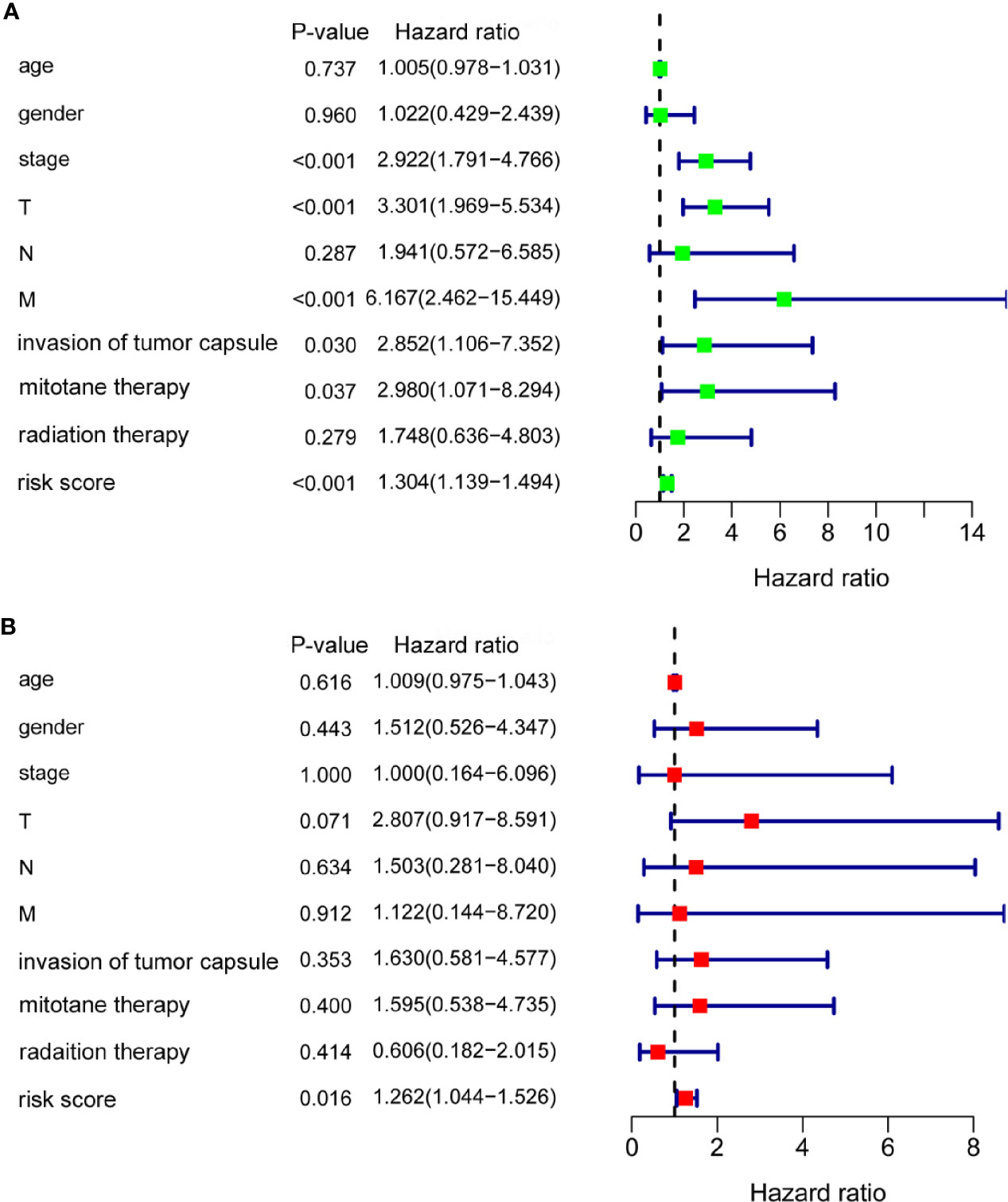
Figure 6 The m6A based signature was an independent prognostic factor for ACC. (A) Univariate Cox regression analysis showed that stage, T, M, invasion of tumor capsule, mitotane therapy, and risk score were significantly associated with the OS of ACC patients. (B) Multivariate Cox regression analysis demonstrated that the m6A based signature could serve as an independent prognostic factor in ACC.
Association Between m6A Based Signature Risk Score and Clinical Factors
The treatment strategies for ACC patients depend largely on clinicopathologic characteristics. Therefore, we analyzed the relationship between the m6A based signature risk score and clinical factors in ACC patients. Our study showed that the risk score was closely related to histological type (p = 0.008), stage (p = 1.855e-04), T (p = 0.002), N (p = 0.037), M (p = 0.004), and progression (p = 0.002), but not associated with age (p = 0.231), gender (p = 0.723), and invasion of tumor capsule (p = 0.377) in ACC patients (Figure 7).
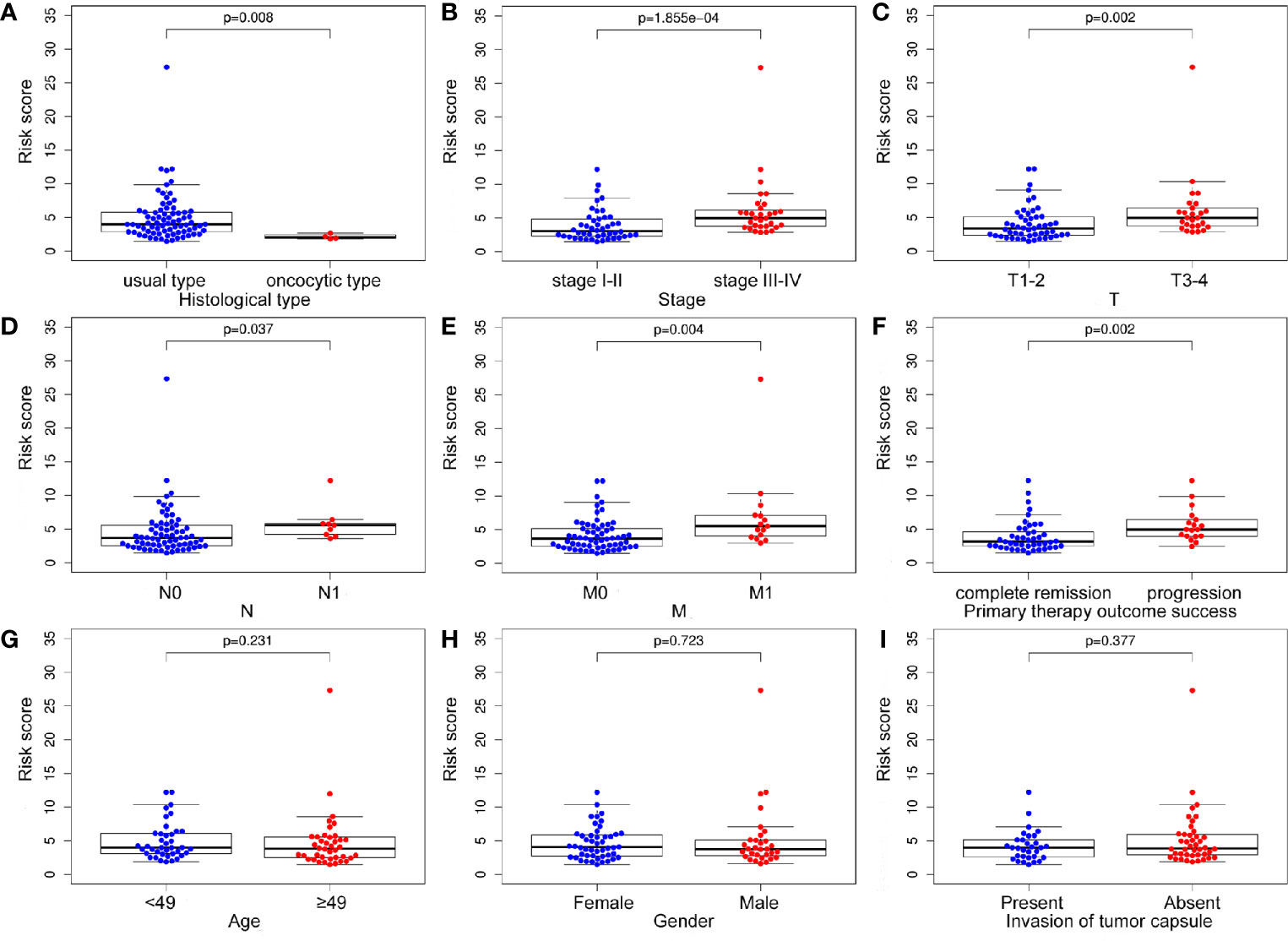
Figure 7 The relationship between the m6A based signature and clinical characteristics. (A-F) The risk score was closely related to histological type, stage, T, N, M, and progression of ACC. (G-I) However, the signature was not associated with age, gender, and invasion of tumor capsule in ACC patients.
Construction of a Prognostic Nomogram for ACC
To establish a clinically applicable method for monitoring the prognosis of ACC patients, we constructed a prognostic nomogram by combining clinical factors (age, gender, stage, T, N, M, invasion of tumor capsule, mitotane therapy, and radiation therapy) with the m6A based signature risk score. The result indicated that the prognostic nomogram could superiorly predict the 1-, 2-, and 3-year OS of ACC patients (Figure 8).
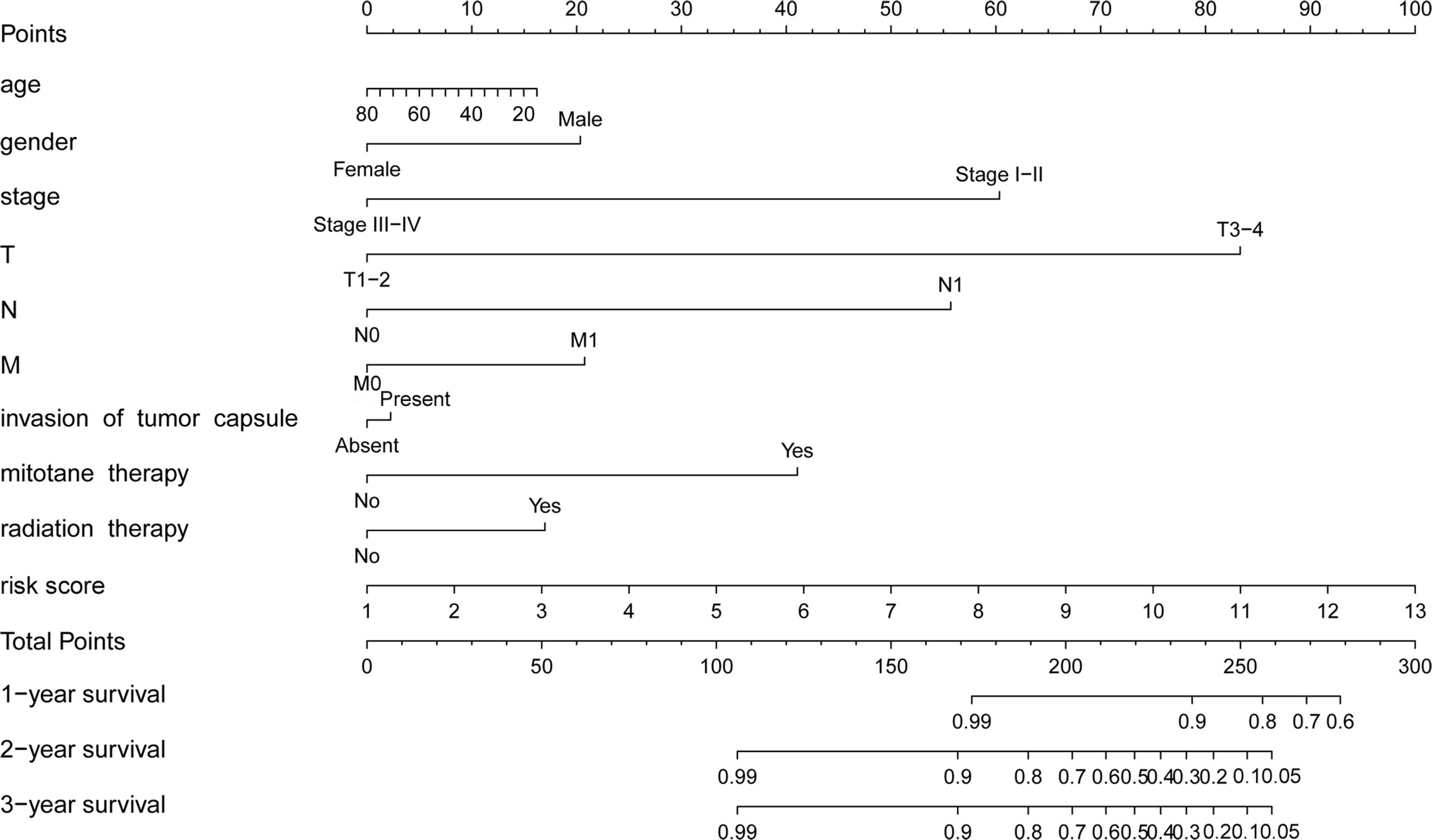
Figure 8 Prognostic nomogram with clinical features and m6A based signature for ACC. The nomogram could superiorly predict 1-, 2-, and 3-year OS of ACC patients.
Validation of the Expression Levels of Three m6A RNA Methylation Regulators by in Vitro Experiments
RNA isolation and reverse transcription−quantitative PCR (RT-qPCR) were further performed to validate the expression levels of the three selected m6A RNA methylation regulators in 3 newly diagnosed ACC patients. Retroperitoneal laparoscopic resections of the adrenal tumor were performed on all patients. We applied the paired t-test to evaluate the differences between the ACC tissues and the adjacent non-tumor adrenal tissues. The results demonstrated significant differences in the expression levels of three m6A RNA methylation regulators (RBM15, HNRNPC, and FTO) between ACC and normal tissues (Figure 9). Compared to normal samples, HNRNPC and FTO were down-regulated and RBM15 was up-regulated in ACC samples. The results from this qRT-PCR validation in three newly diagnosed ACC patients were consistent with the above bioinformatics results.
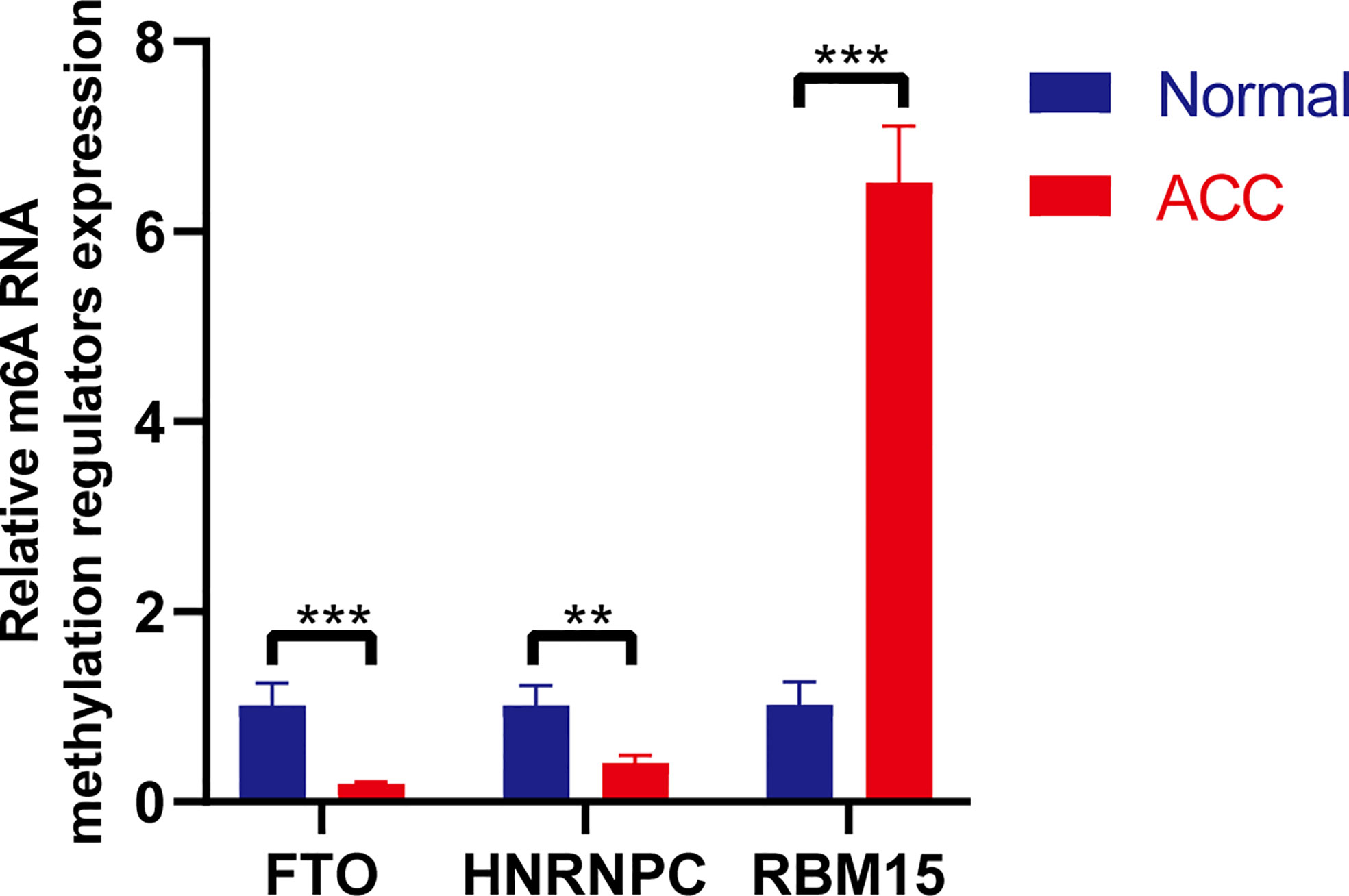
Figure 9 RT-PCR validation of three m6A RNA regulators in ACC and normal tissues. The results indicated that significant differences in the expression levels of three m6A RNA methylation regulators (RBM15, HNRNPC, and FTO) between ACC and normal tissues. **p < 0.01; ***p < 0.005.
Discussion
ACC is a rare malignant tumor in humans, and its pathogenesis is a complex process, involving a great number of aberrant gene expression profiles in multiple signaling pathways. There are no effective treatments for ACC patients with advanced stage or metastasis and lack of robust biomarkers to predict the prognosis and metastasis of ACC patients. Considering the importance of m6A RNA methylation regulators in cancer progression, it is crucial to explore biomarkers of prevention and treatment to ACC from m6A RNA regulators. In the present study, we first systematically evaluated the expression levels of 13 m6A RNA methylation regulators and further constructed an m6A based prognostic signature in ACC by using the TCGA database. 11 m6A RNA methylation regulators were differentially expressed in ACC, and RBM15, YTHDF2, and HNRNPC were associated with the prognosis of ACC patients. In addition, based on the expression levels of 13 m6A RNA regulators, we classified ACC patients into two groups, clusters 1/2, through consensus cluster analysis. Survival analysis indicated that clusters 1/2 were significantly related to the survival outcomes of ACC patients. Subsequently, three m6A RNA regulators (RBM15, HNRNPC, and FTO) were finally screened out from a signature and survival analysis revealed a positive correlation between high risk scores and poor survival outcomes in ACC. In addition, the m6A based signature was associated with the OS of ACC patients who received mitotane therapy. Therefore, the m6A based signature may be applied as a novel predictive biomarker for ACC patients after mitotane therapy. Univariate and multivariate analyses demonstrated that the m6A based signature was an independent prognostic factor for ACC. Moreover, the m6A based signature risk score was correlated with histological type, advanced stage, and progression of ACC. These results indicated that the m6A based signature may provide a valuable tool for predicting the prognosis and progression of ACC. Furthermore, a nomogram integrated with m6A based signature and clinical features was constructed, which can superiorly monitor the survival of patients with ACC. Finally, RT-qPCR demonstrated significant differences in the expression levels of three m6A RNA methylation regulators (RBM15, HNRNPC, and FTO) between ACC and normal tissues.
Among these m6A RNA methylation regulators, FTO, as a genuine demethylase of m6A modification, negatively regulates the expression of its critical target RNAs, such as ASB2 and RARA. FTO plays its major role through reducing the abundance of internal m6A modification, especially in the 3′ untranslated regions (3′-UTRs), which in turn leads to decreased stability of the target mRNA transcripts and influences the occurrence and progression of acute myeloid leukemia (AML) (16). MYC is a direct and functionally essential target of FTO, and FTO knockdown enhances the expression level of m6A on MYC mRNA (especially at the 5′ UTR and middle exons), leading to mRNA decay and MYC down-regulation, and thereby inhibition of MYC signaling, which plays an important role in the tumor (17). Previous studies showed that the abnormally high expression of FTO was associated with worse outcomes in cancers, such as gastric cancer, endometrial carcinoma, and lung squamous cell carcinoma (18–20). Moreover, FTO influences the progression of cancer by regulating multiple signaling pathways, such as FTO-PGC-1 α signaling axis, mediating PKM2 demethylation, impairing the translation efficiency of E2F1 and Myc (21–24). ALKBH5 also functions as an m6A demethylase, and increases the expression of its key target, FOXM1 (forkhead box M1), to promote the development of cancers by reducing m6A abundance on target mRNA transcripts (especially at the 3′ UTR) (25). METTL14 was defined as an m6A writer and exerts its oncogenic role through m6A-dependent post-transcriptional regulation of its critical mRNA targets such as MYB and MYC. METTL14 mainly promotes the expression of MYB and MYC by increasing m6A abundance and enhancing the stability of the target mRNA transcripts (26). The biological function of METTL3 is likely attributed to the promotion of the translation of its mRNA targets, such as MYC, BCL-2, and PTEN, in an m6A-dependent manner (27). Overexpression of METTL3 significantly promoted tumorigenesis and metastasis of hepatocellular carcinoma, mostly through regulation of SOCS2 expression by an m6qa- and YTHDF2-dependent mechanisms (28). RBM15, an RNA binding protein, was categorized as the component of writers and its target is the lncRNA XIST, which mediates X-inactivation and gene silencing during development (29). RBM15-assisted XIST methylation is necessary for XIST-mediated silencing, thus revealing the first functional role for transcript-specific methylation directed by RBM15. RBM15 was also significantly upregulated in cancers and was associated with the prognosis of cancers, such as gastric cancer and chronic myelogenous leukemia (CML) (30, 31). Furthermore, RBM15 can modulate the growth, proliferation, cell cycle, and apoptosis of CML cells via Notch signaling (31). The fates of m6A-modified mRNAs rely on the functions of distinct proteins that recognize them (i.e., readers), which may affect the stability, splicing, and/or translation of target mRNAs. Dysregulation of m6A modification and the related proteins also contributes to the initiation and progression of cancers. Members of the YT521-B homology (YTH) domain family of proteins, including YTHDF1, YTHDF2, YTHDC1, and YTHDC2, have been identified as direct m6A readers. While YTHDF2 and YTHDC2 may promote decay of target mRNAs, YTHDF1 and YTHDC2 can promote the translation of target mRNAs. Besides, YTHDC1 likely impacts splicing and nuclear export of target mRNAs (32–38). HNRNPC, a regulator in RNA splicing, has been upregulated in various cancers and tumor cell lines, such as glioblastoma, hepatocellular carcinoma, and lung cancer (39–41). HNRNPC may induce human lung cancer cell invasion and metastasis by activating the IFN-α-JAK-STAT1 signaling pathway (42). Wu et al. indicated that inhibition of HNRNPC can suppress the proliferation and tumor growth by mediating the cascade of interferon responses (43). In our study, a prognostic signature incorporating three m6A RNA regulators was established based on TCGA data. The AUC of the m6A based signature was 0.747, which showed satisfactory accuracy in predicting the prognosis of ACC patients. Our results indicated that the m6A based signature can be used to identify ACC patients at high risk and provide early interventions to improve the prognosis. However, as a retrospective study, our research still has a bias due to heterogeneity, although almost all the clinical factors in ACC cohorts available from the TCGA database have been included. Second, as a bioinformatic study based on a public database, further experimental studies are needed to explore the potential effect and mechanism of these m6A RNA regulators in the development of ACC.
In conclusion, we identified and validated an m6A based signature, which can be used as an independent prognostic signature in predicting the prognosis of ACC patients. Further clinical trials and experimental explorations are needed to validate our observations and mechanisms underlying the prognostic value of m6A RNA methylation regulators in ACC.
Data Availability Statement
Publicly available datasets were analyzed for this study. These can be found here: https://tcga‐data.nci.nih.gov/tcga/ and https://xenabrowser.net/datapages/.
Ethics Statement
The studies involving human participants were reviewed and approved by The Affiliated Hospital of Qingdao University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CS: Conceptualization, methodology, and writing—original draft. JL: Data curation. XY and WJ: Supervision. YW: Writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81972378, 81101932).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.568397/full#supplementary-material
Supplementary Figure 1 | The violin plot showed the differentially expressed m6A RNA methylation regulators between cluster 1 and cluster 2.
References
1. Stigliano A, Chiodini I, Giordano R, Faggiano A, Canu L, Della Casa S, et al. Management of adrenocortical carcinoma: a consensus statement of the Italian Society of Endocrinology (SIE). J Endocrinol Invest (2016) 39(1):103–21. doi: 10.1007/s40618-015-0349-9
2. Batista PJ. The RNA modification N(6)-methyladenosine and its implications in human disease. Genomics Proteomics Bioinformatics (2017) 15(3):154–63.
3. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res (2018) 28(6):616–24. doi: 10.1038/s41422-018-0040-8
4. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet (2014) 15(5):293–306. doi: 10.1038/nrg3724
5. Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6-methyladenosine-related Genomic Targets are Altered in Breast Cancer Tissue and Associated with Poor Survival. J Cancer (2019) 10(22):5447–59. doi: 10.7150/jca.35053
6. Chen M, Nie ZY, Wen XH, Gao YH, Cao H, Zhang SF. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep (2019) 39(12):BSR20192892. doi: 10.1042/BSR20192892
7. Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig Dis Sci (2019) 64(6):1503–13. doi: 10.1007/s10620-018-5452-2
8. Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol (2018) 52(2):621–9. doi: 10.3892/ijo.2017.4219
9. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun (2019) 10(1):2065. doi: 10.1038/s41467-019-09865-9
10. Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med (2019) 8(10):4766–81. doi: 10.1002/cam4.2360
11. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer (2019) 18(1):127. doi: 10.1186/s12943-019-1053-8
12. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26(1):139–40. doi: 10.1093/bioinformatics/btp616
13. Wan B, Liu B, Yu G, Huang Y, Lv C. Differentially expressed autophagy-related genes are potential prognostic and diagnostic biomarkers in clear-cell renal cell carcinoma. Aging (Albany NY) (2019) 11(20):9025–42. doi: 10.18632/aging.102368
14. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol (2016) 17(1):174. doi: 10.1186/s13059-016-1028-7
15. Shen C, Liu J, Wang J, Zhong X, Dong D, Yang X, et al. Development and validation of a prognostic immune-associated gene signature in clear cell renal cell carcinoma. Int Immunopharmacol (2020) 81:106274. doi: 10.1016/j.intimp.2020.106274
16. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell (2017) 31(1):127–41. doi: 10.1016/j.ccell.2016.11.017
17. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell (2018) 172(1-2):90–105.e23. doi: 10.1016/j.cell.2017.11.031
18. Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep (2017) 38(4):2285–92. doi: 10.3892/or.2017.5904
19. Zhu Y, Shen J, Gao L, Feng Y. Estrogen promotes fat mass and obesity-associated protein nuclear localization and enhances endometrial cancer cell proliferation via the mTOR signaling pathway. Oncol Rep (2016) 35(4):2391–7. doi: 10.3892/or.2016.4613
20. Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun (2018) 502(4):456–64. doi: 10.1016/j.bbrc.2018.05.175
21. Li J, Zhu L, Shi Y, Liu J, Lin L, Chen X. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am J Transl Res (2019) 11(9):6084–92.
22. Zou D, Dong L, Li C, Yin Z, Rao S, Zhou Q. The m6A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int (2019) 19:321. doi: 10.1186/s12935-019-1045-1
23. Singh B, Kinne HE, Milligan RD, Washburn LJ, Olsen M, Lucci A. Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PloS One (2016) Jul 811(7):e0159072. doi: 10.1371/journal.pone.0159072
24. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell (2019) Apr 1535(4):677–691.e10. doi: 10.1016/j.ccell.2019.03.006
25. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31(4):591–606.e6. doi: 10.1016/j.ccell.2017.02.013
26. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell (2018) 22(2):191–205.e9. doi: 10.1016/j.stem.2017.11.016
27. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med (2017) 23(11):1369–76. doi: 10.1038/nm.4416
28. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology (2018) 67(6):2254–70. doi: 10.1002/hep.29683
29. Meyer KD, Jaffrey SR. Rethinking m(6)a readers, writers, and erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
30. Su Y, Huang J, Hu J. m6A RNA Methylation Regulators Contribute to Malignant Progression and Have Clinical Prognostic Impact in Gastric Cancer. Front Oncol (2019) 9:1038:1038. doi: 10.3389/fonc.2019.01038
31. Yang Y, Wang S, Zhang Y, Zhu X. Biological effects of decreasing RBM15 on chronic myelogenous leukemia cells. Leuk Lymphoma (2012) 53(11):2237–44. doi: 10.3109/10428194.2012.684350
32. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
33. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
34. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
35. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
36. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res (2017) 27(3):444–7. doi: 10.1038/cr.2017.10
37. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
38. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife (2017) 6:e31311. doi: 10.7554/eLife.31311
39. Park YM, Hwang SJ, Masuda K, Choi KM, Jeong MR, Nam DH, et al. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol Cell Biol (2012) 32(20):4237–44. doi: 10.1128/MCB.00443-12
40. Sun W, Xing B, Sun Y, Du X, Lu M, Hao C, et al. Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol Cell Proteomics (2007) 6(10):1798–808. doi: 10.1074/mcp.M600449-MCP200
41. Pino I, Pío R, Toledo G, Zabalegui N, Vicent S, Rey N, et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer (2003) 41(2):131–43. doi: 10.1016/s0169-5002(03)00193-4
42. Yan M, Sun L, Li J, Yu H, Lin H, Yu T, et al. RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer. J Exp Clin Cancer Res (2019) 38(1):478. doi: 10.1186/s13046-019-1479-2
Keywords: adrenocortical carcinoma, m6A RNA methylation regulators, prognosis, signature, The Cancer Genome Atlas
Citation: Shen C, Liu J, Yang X, Jiao W and Wang Y (2021) Development and Validation of an m6A RNA Methylation Regulators-Based Signature for Predicting the Prognosis of Adrenocortical Carcinoma. Front. Endocrinol. 12:568397. doi: 10.3389/fendo.2021.568397
Received: 01 June 2020; Accepted: 13 January 2021;
Published: 22 February 2021.
Edited by:
Antonino Belfiore, University of Catania, ItalyReviewed by:
Naoyuki Kataoka, The University of Tokyo, JapanMarco Volante, University of Turin, Italy
Rosamaria Lappano, University of Calabria, Italy
Copyright © 2021 Shen, Liu, Yang, Jiao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Jiao, doctorjiaowei@sina.com; Yonghua Wang, doctoryonghua123@sina.com
†These authors have contributed equally to this work
 Chengquan Shen1†
Chengquan Shen1† Yonghua Wang
Yonghua Wang