Copy Number Analyses Identified a Novel Gene: APOBEC3A Related to Lipid Metabolism in the Pathogenesis of Preeclampsia
- 1School of Pharmacy, University of Chinese Academy of Sciences, Beijing, China
- 2State Key Laboratory of Drug Research, Center for Drug Safety Evaluation and Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China
- 3Department of Obstetrics, International Peace Maternity and Child Health Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 4Department of Biostatistics, Clinical Research Institute, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 5School of Renji Clinical Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 6Department of Obstetrics and Gynecology, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Background: Preeclampsia is a heterogeneous and complex disease with its pathogenesis mechanism not fully elucidated. A certain subset of patients with preeclampsia exhibit disturbances in lipid metabolism before clinical symptoms. Moreover, there is a tendency for preeclampsia to run in families. Whether genetic factors play a role in abnormal lipid metabolism during the incidence of preeclampsia has not been well investigated.
Methods: Preeclampsia patients (n = 110) and healthy age- and gravidity-matched pregnant women (n = 110) were enrolled in this study. Peripheral blood specimens were used for genomic analysis (n = 10/group) or laboratory validation (n = 100/group). We retrospectively obtained the baseline clinical characteristics of 68 preeclampsia patients and 107 controls in early pregnancy (12–14 gestational weeks). Correlation analyses between differential genes and baseline lipid profiles were performed to identify candidate genes. In vitro and in vivo gain-of-function models were constructed with lentivirus and adeno-associated virus systems, respectively, to investigate the role of candidate genes in regulating lipid metabolism and the development of preeclampsia.
Results: We observed that preeclampsia patients exhibited significantly elevated plasma TC (P = 0.037) and TG (P < 0.001) levels and increased body mass index (P = 0.006) before the disease onset. Within the region of 27 differential copy number variations, six genes potentially connected with lipid metabolism were identified. The aberrant copies of APOBEC3A, APOBEC3A_B, BTNL3, and LMF1 between preeclampsia patients and controls were verified by quantitative polymerase chain reaction. Especially, APOBEC3A showed a significant positive correlation with TC (P < 0.001) and LDL (P = 0.048) in early pregnancy. Then, our in vitro data revealed that overexpression of APOBEC3A disrupted lipid metabolism in HepG2 cells and affected both cholesterol and fatty acid metabolisms. Finally, in vivo study in a hepatic-specific overexpressed APOBEC3A mouse model revealed abnormal parameters related to lipid metabolism. Pregnant mice of the same model at the end of pregnancy showed changes related to preeclampsia-like symptoms, such as increases in sFlt-1 levels and sFlt-1/PLGF ratios in the placenta and decreases in fetal weight.
Conclusion: Our findings established a new link between genetics and lipid metabolism in the pathogenesis of preeclampsia and could contribute to a better understanding of the molecular mechanisms of preeclampsia.
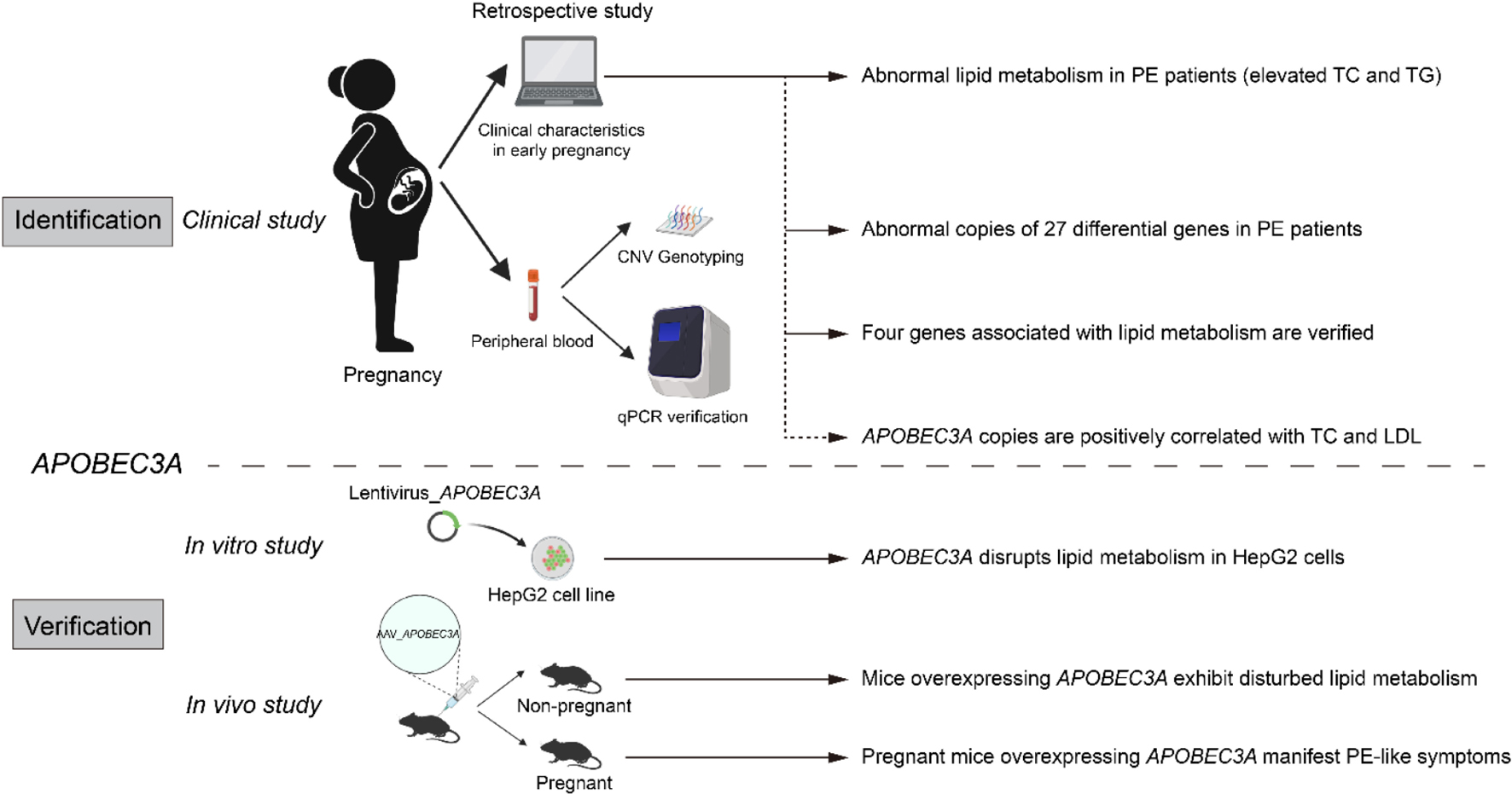
Graphical Abstract. Identification and validation of APOBEC3A’srole in regulating lipid metabolism in PE.
Introduction
Preeclampsia (PE) is a pregnancy-specific complication mainly characterized by hypertension and proteinuria and remains one of the leading causes of morbidity and mortality in pregnant women and fetuses (1). Currently, the incidence of PE in developed countries is about 1.3–6%, which is even higher in developing countries (2). PE is a heterogeneous and complex disease caused by multiple factors, including heredity and environment (3). However, the underlying mechanism remains elusive due to diverse races, geography, and other factors (4).
An emerging body of research has demonstrated that abnormalities in lipid metabolism may be connected with the etiology of PE, with certain patients manifesting lipid metabolism disturbances before the onset of clinical symptoms (5–7). The lipid profiles are clinically accessible parameters. We have previously reported that triglycerides (TG) and low-density lipoprotein cholesterol (LDL) are independent risk factors for PE, while high-density lipoprotein cholesterol (HDL) is an independent protective factor (8). Therefore, elucidating the relationship between abnormal lipid metabolism and the pathogenesis of PE can guide the early identification of high-risk individuals for PE.
Previous findings have revealed that PE is relatively common among daughters and sisters of women with PE (9, 10), indicating genetic factors play an essential role in the development of PE. Most studies on genetic factors in PE are based on expression profiles, including mRNA and protein levels, making it challenging to clarify the causal relationship between these factors and the onset of PE.
To elucidate the mechanistic link between abnormal lipid metabolism and genetics in the pathogenesis of PE, we analyzed the genomes of clinical specimens with DNA microarrays and performed laboratory validation. Additionally, we retrospectively collected clinical characteristics of pregnant women in early pregnancy (12–14 gestational weeks) and performed the correlation analysis of lipid profiles at baseline with the candidate genes. Furthermore, we constructed in vitro and in vivo gain-of-function models to explore the role of candidate genes in regulating lipid metabolism and participating in the development of PE.
Materials and Methods
Clinical Study Design and Oversight
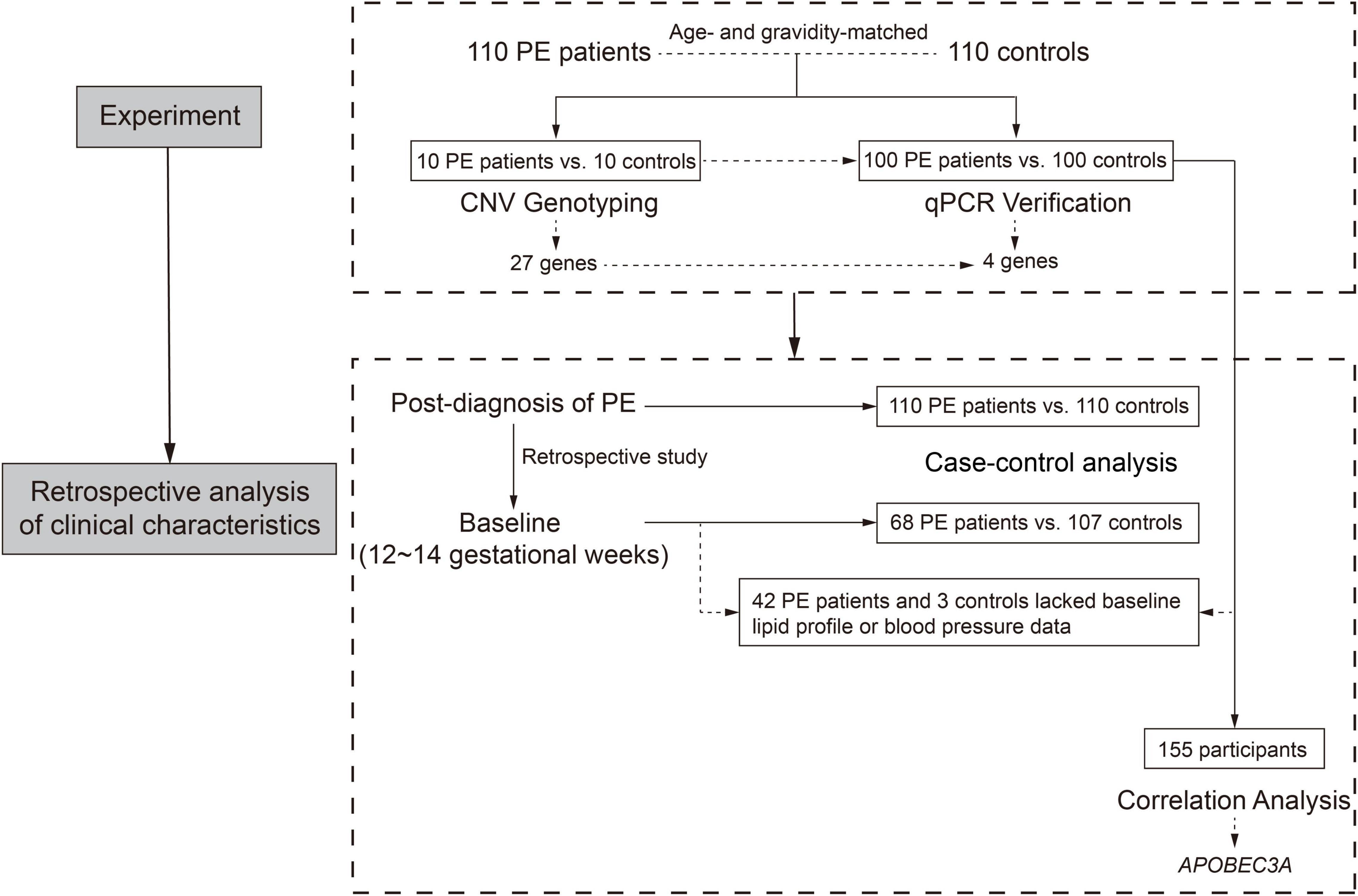
From October 2013 to December 2017, we recruited 220 participants, including 110 PE patients and 110 controls, from two institutes in Shanghai, China (Department of Obstetrics, International Peace Maternity and Child Health Hospital, and Department of Obstetrics, Renji Hospital, Shanghai Jiao Tong University School of Medicine). The case-control study flow chart is displayed in Figure 1. Fasting peripheral blood specimens were collected before delivery. The inclusion criteria for the PE group were pregnant women with hypertension (≥ 140 mmHg systolic and ≥ 90 mmHg diastolic blood pressure), proteinuria higher than 0.3 g/d or qualitatively + above. In contrast, healthy age- and gravidity-matched pregnant women with normotensive pre-pregnancy and pregnancy and without obstetric complications as the control group.
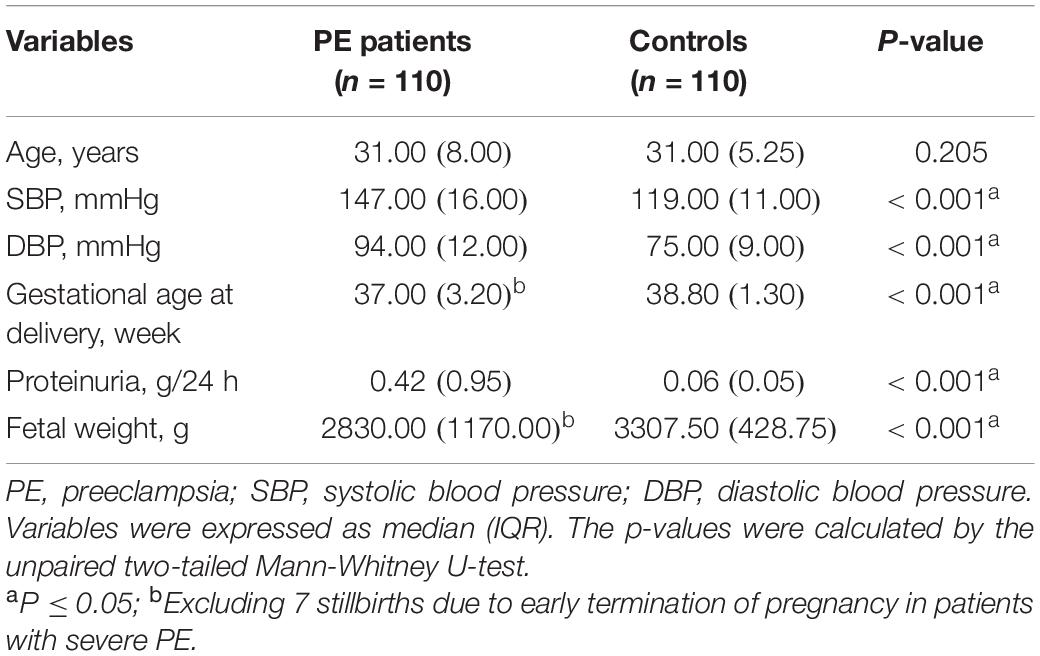
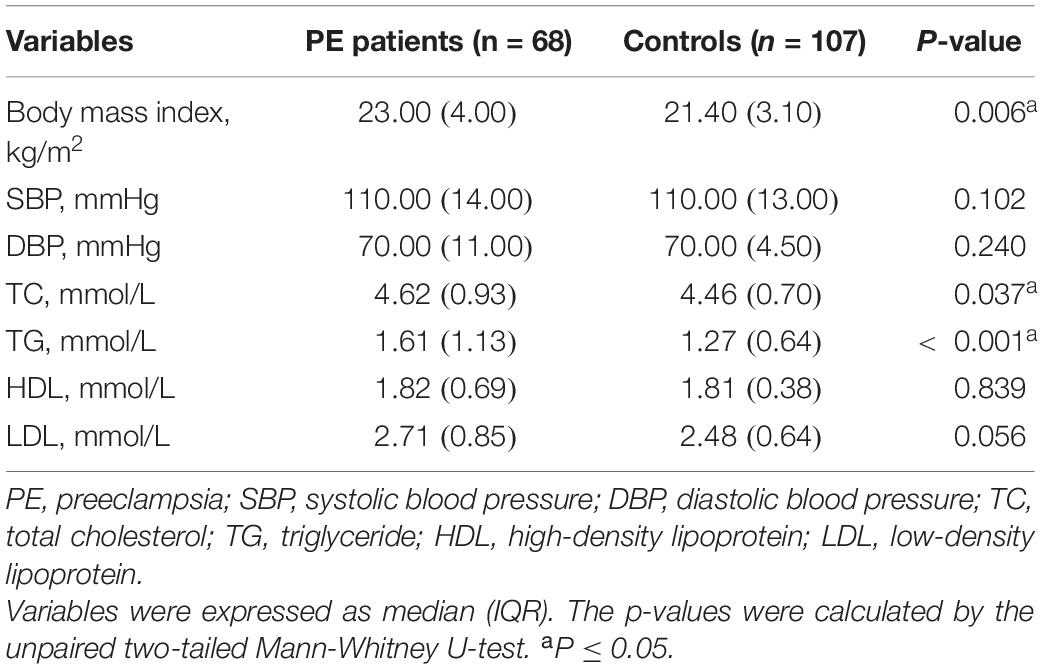
Table 1 lists clinical features at the patients’ initial diagnosis of PE. We retrospectively collected clinical characteristics of subjects at their initial routine prenatal examination in early pregnancy (12–14 gestational weeks), which is listed in Table 2. Since the Department of Obstetrics at Renji Hospital is a consultation and resuscitation center for maternal near-miss in Shanghai, some PE patients in this hospital were referred from other institutions, so the baseline data of some subjects in early pregnancy (12–14 weeks of gestation) are missing. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Medical Research Ethics Committee of International Peace Maternity and Child Health Hospital (the Ethics Committee Approval Protocol No. 5 of 28 February 2014). All subjects provided broad informed consent for the research use of their biological samples.
Copy Number Variation Genotyping and Data Analysis
Genomic DNA was extracted from the peripheral blood (n = 10/group) of pregnant women and detected using Affymetrix CytoScan HD Cytogenetics Chips according to the manufacturer’s recommended protocol. Quality control analysis indicated that all microarrays were qualified, except one was judged as male by the Chromosome Analysis Suite software for unclear reasons (Supplementary Table 1). Therefore, we excluded this sample from the analysis of the microarray data.
The raw data was processed by Chromosome Analysis Suite analysis software from Affymetrix to obtain all the CNVs, including duplications and deletions. Given the small sample size for microarray analysis, we opted for CNVs that occurred with a frequency difference of more than 30% between the two groups. These CNVs did not overlap with the high-frequency CNV regions of ordinary individuals in the Database of Genomic Variants1 as differential CNVs. The differential genes within the region of differential CNVs were given gene annotation by the Gene Ontology (GO) database. The GO function enrichment analysis was employed to obtain the special functions for these genes using Fisher’s exact test and χ2 test with the threshold of P < 0.05 and FDR < 0.25.
Verification of Gene Copy Numbers and Clinical Correlation Analysis
Total genomic DNA was extracted from peripheral blood specimens (n = 100/group) of pregnant women using the AxyPrep Multisource Genomic DNA Miniprep Kit (Axygen Scientific) following the manufacturer’s instructions. The genes linked to lipid metabolism were verified by quantitative polymerase chain reaction (qPCR), and GAPDH was used as an endogenous control at the DNA level. Details of the primer sequences are shown in Supplementary Table 2. In this study, correlations between candidate genes and clinical lipid profiles were analyzed by SPSS Statistics 25, using Pearson’s correlation test.
Establishment of Lipid Accumulation Model in vitro
HepG2 (ATCC) cell line was cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 1% Penicillin-Streptomycin (Thermo Fisher Scientific) at 37°C in a 5% CO2 atmosphere. We infected cells with lentivirus carrying APOBEC3A _Flag or negative control and screened the cells using puromycin (Thermo Fisher Scientific) to obtain a pool of cells overexpressing the APOBEC3A gene (or negative control). DMEM medium with 1 mM oleic acid (Sigma) served as the fat accumulation inducer for the cells. Cell cultures were collected after being starved overnight and treated with the fat accumulation inducer for 24 h. The intracellular lipid content was determined by a biochemical analyzer (Roche). The results were normalized to the total protein concentration.
Adeno-Associated Virus-Mediated Overexpression of APOBEC3A in Mice
The coding sequence of human APOBEC3A, the 3 × flag tag, and a green fluorescent protein were simultaneously subcloned into the AAV2/8 plasmid (AAV_APOBEC3A) under the control of the thyroxine-binding globulin promoter, a hepatic-specific promoter. This serotype is known to have a stable expression in the liver after injection (11). A virus containing green fluorescent protein alone under control of the thyroxine-binding globulin promoter was used as a control virus (AAV_Mock).
Separate groups of 6- to 8-week-old wild-type, C57BL/6J female mice (n = 14/group) were injected intraperitoneally with 1.5 × 1010 vector genomes/mouse of the relevant vectors. Mice were fed conventional chow throughout the study. They were sacrificed to investigate the impact of APOBEC3A on lipid metabolism (n = 6/group) or mated to explore the effect of the APOBEC3A gene on pregnant mice (n = 8/group, a positive vaginal plug was marked as gestation day 0, pregnant mice were sacrificed on gestation day 18) at 5 weeks (35 days) after AAV administration. Mice were fasted for 4 h, anesthetized with isoflurane, and whole blood was collected by cardiac puncture into BD Microtainer EDTA collection tubes. Plasma samples were used for lipid analysis and assayed using a biochemical analyzer (Roche). Liver and placenta specimens were obtained and immediately fast-frozen in nitrogen and stored at −80°C for subsequent testing. Fetuses were delivered by cesarean section, and their weights were measured. We performed all animal procedures according to protocols approved by the Institutional Animal Care and Use Committee of Shanghai Institute of Materia Medica, Chinese Academy of Sciences.
RNA-Seq and Cluster Analysis
Total RNA was extracted from HepG2 cells (n = 3/group) and used for RNA-seq analysis. cDNA library construction and sequencing were performed by the Shanghai Majorbio Bio-Pharm Technology Co., Ltd. using the majorbio cloud platform. High-quality reads were aligned to the human reference genome (GRCh38) using Bowtie2. We normalized the expression level of each gene to the fragment of the exon model per million mapped reads (FPKM) based on the expectation-maximization method. NOISeq method was used to screen out differentially expressed genes between the two groups with a fold change ≥ of 1.5. Function and pathway annotation and enrichment analysis were based on the GO database and the Kyoto Encyclopedia of Genes and Genomes database.
We obtained the list of genes connected with cholesterol homeostasis and fatty acid metabolism from the Gene Set Enrichment Analysis website. The FPKM of these genes were logarithmically (fold-change) converted, and we visualized the differences between the two groups through heatmap, which was performed by GraphPad Prism V.8 software.
Reverse Transcription-Polymerase Chain Reaction and Immunoblot Analysis
Cell cultures (n = 3/group) or liver tissues (n = 6/group) were lysed in TRIzol (Invitrogen), and total RNA was extracted according to the manufacturer’s instructions. cDNA was synthesized using PrimeScript™ RT Master Mix (Takara). A reverse transcription-polymerase chain reaction was performed using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific) on an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific). Primer sequences for human cells are listed in Supplementary Table 3 (Supplementary Table 4 is for mouse tissues). All results were normalized to GAPDH expression and calculated using the comparative CT (ΔΔCT) method.
Cell cultures (n = 3/group) were lysed with RIPA lysis buffer (Thermo Fisher Scientific). Protein lysates were separated on 10% SDS-PAGE gels and electrophoretically transferred to polyvinylidene fluoride membranes (Millipore). The membranes were blocked with 5% skim milk and incubated overnight at 4°C with mouse monoclonal anti-FLAG® M2 (Sigma) as primary antibodies. Then, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (Goat anti-rabbit, or -mouse IgG, Jackson ImmunoResearch). Protein band chemiluminescence was detected using an ECL Plus immunoblot detection system (Millipore). We performed densitometric analysis on low-exposure images with ImageJ (National Institutes of Health). Rabbit polyclonal anti-β-Tubulin antibody (Proteintech) was used as a loading control to confirm equivalent protein loading on the same membrane.
Immunohistochemistry Analysis
Fresh liver tissue samples were fixed in 4% paraformaldehyde and embedded in paraffin. We then sectioned them into 4-μm-thick sections for immunohistochemical staining. The primary antibodies used in this assay were rabbit monoclonal IgG [EPR25A]-isotype control (Abcam) and rabbit polyclonal to APOBEC3A (Abcam). Immunostaining was performed according to the standard protocol. Then, labeling was visualized using the diaminobenzidine method. Counterstaining was performed using Mayer’s hematoxylin.
Enzyme-Linked Immunosorbent Assay
Approximately 25 mg of the liver (n = 6/group) or placental tissue (n = 8/group) was weighed and made into a 10% saline tissue homogenate, centrifuged at 4°C for 10 min at 3,000 g to obtain the supernatant for subsequent assays. TG or total cholesterol (TC) levels in the liver were measured by a biochemical analyzer (Roche). Placental growth factor (PLGF) or soluble fms-like tyrosine kinase-1 (sFlt-1) concentrations in plasma or placenta were measured by mouse PLGF or sFlt-1 Enzyme-linked immunosorbent assay kits (R&D Systems). The results were normalized by tissue mass.
Untargeted Relative Quantitative Lipidomics
Lipids from liver specimens (n = 6/group) were extracted according to the methyl tert-butyl ether method (12). Samples were separated and analyzed using an ultra-performance liquid chromatography system (Waters) equipped with a Q-Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Lipid lists were obtained by annotation using Lipid Search software, and differential lipids were identified by t-test and orthogonal partial least squares discrimination analysis test. The Beijing Genomics Institute performed this assay.
Statistical Analysis
Data were expressed as mean (SD) or median (IQR) when appropriate. Comparisons of continuous variables between two groups were made by unpaired two-tailed Student’s t-test or Mann-Whitney U-test when variables were normally or non-normally distributed, respectively. Statistical tests were performed by GraphPad Prism V.8. Correlation analysis was conducted by SPSS Statistics 25. P ≤ 0.05 (*) was considered statistically significant. Other statistical analysis methods used have been described separately in the respective methods section.
Graphical abstract was created using graphics from www.Biorender.com.
Results
Preeclampsia Patients Showed a Trend of Abnormal Lipid Metabolism Before the Onset of the Disease
Compared with the control group, patients with PE presented with elevated blood pressure, increased proteinuria, decreased gestational age at delivery, and fetal growth restriction (Table 1), consistent with previous reports (13). The clinical characteristics of 68 PE patients and 107 controls in early pregnancy (12–14 gestational weeks) were retrospectively obtained, as shown in Figure 1 and Table 2. The findings showed that those pregnant women who subsequently developed PE exhibited distinctly elevated plasma TC (P = 0.037) and TG (P < 0.001) levels before the disease onset and had significantly increased body mass index (P = 0.006). However, their blood pressure (systolic blood pressure, P = 0.102; diastolic blood pressure, P = 0.240) did not differ substantially in early pregnancy compared to controls. These findings indicate that PE patients tend to have abnormal lipid metabolism before the onset of the disease, but the causative mechanisms have not been elucidated.
Differential Genes Between Two Groups Were Enriched in Functions Related to Metabolic Processes
From the genomic analysis of 10 PE patients and 10 controls (clinical characteristics are shown in Supplementary Table 5), we obtained 27 differential CNVs, including 22 deletions and 5 duplications (Figure 2A). The identified variants ranged in size from 1,409 base pairs to nearly 151.7 kb (median = 10.8 kb), and these CNV regions contained 27 differential genes (Figure 2B and Supplementary Table 6). GO functional enrichment analysis revealed that the biological functions of these differential genes were enriched in response to nutrient levels, protein hydrolysis, developmentally programmed cell death, and regulation of triglyceride/cholesterol/lipoprotein metabolism (Figure 2C).
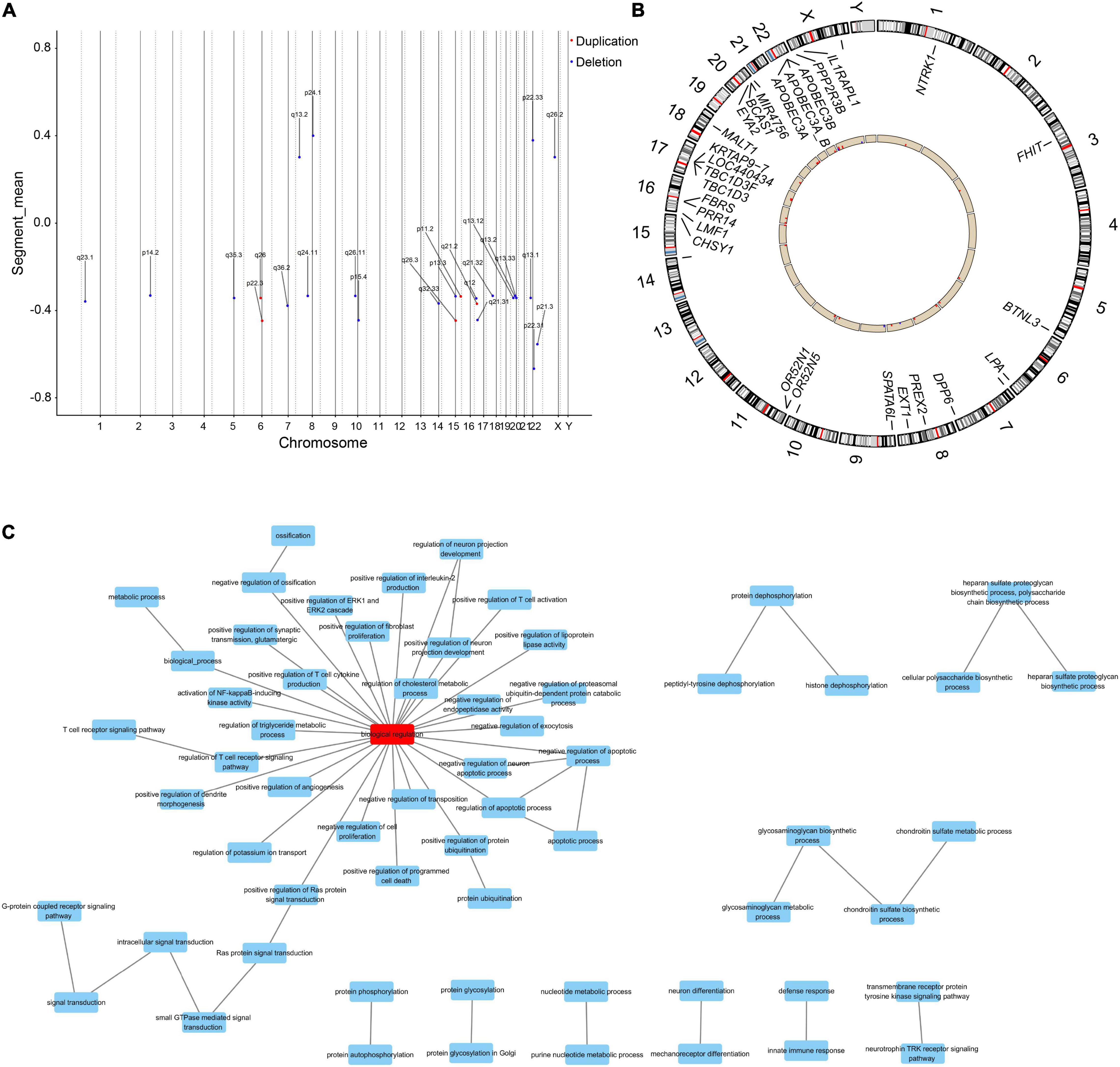
Figure 2. The differential genes linked to aberrant CNVs are enriched in metabolism-related biological functions. (A) Distributions of aberrant CNVs on each chromosome. Red and blue dots indicate whether the type of the CNV is duplication or deletion, respectively. (B) Distribution of differential genes within the region of differential CNVs in the genome. Red columns indicate that the copy number of this gene is increased in PE patients compared to controls, and blue columns indicate a decrease. (C) Differential genes were enriched in functions associated with lipid and cholesterol metabolism, immunity, and development.
Aberrant Copies of Genes Linked to Lipid Metabolism Were Verified in 100 Preeclampsia Patients and 100 Controls
We identified six genes in the differential CNV regions that may be connected with lipid metabolism. The LPA gene located in the 6q26 region from 161.032 to 161.047 Mb was reported in previous research on PE (14–17), and we detected this duplication in one case (10%) and four controls (44.4%). APOBEC3A, APOBEC3A_B, and APOBEC3B in 22q13.1 from 39.350 to 39.390 Mb were detected in one case (10%) and four controls (44.4%). The BTNL3 gene in the 5q35.3 region from 180.379 to 180.431 Mb, with this deletion detected in one case (10%) and four controls (44. 4%). The LMF1 gene in the 16p13.3 region from 1.002 to 1.003 Mb, with this duplication detected in no one case and three controls (33.3%). These four regions of differential CNVs were confirmed by qPCR with significantly increased copy numbers of APOBEC3A, APOBEC3A_B, BTNL3, and LMF1 in PE patients compared with controls (Figures 3A–D). In contrast, the decrease in copies of LPA and the increase in copies of APOBEC3B were not statistically significant (Figures 3E,F).
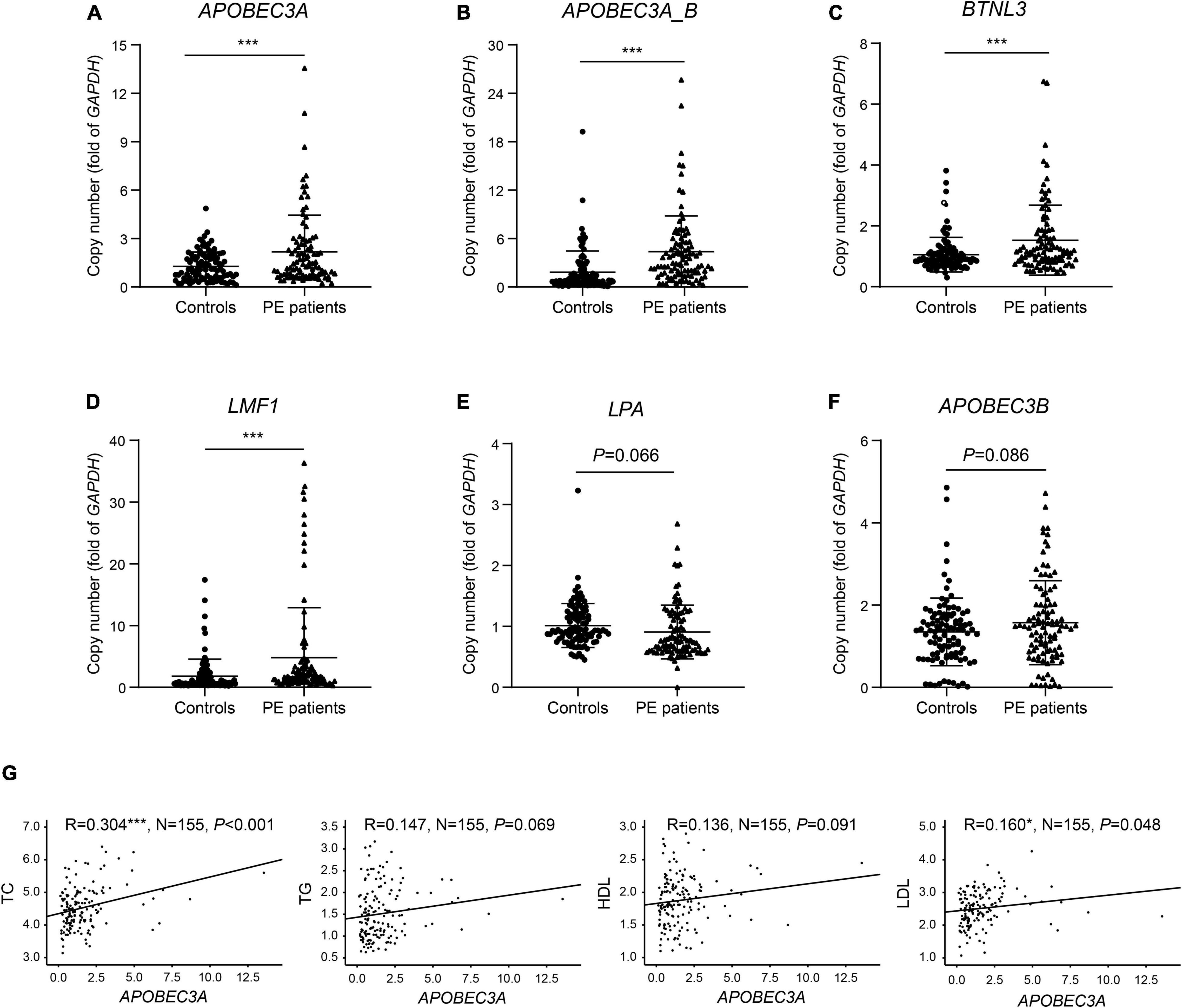
Figure 3. Verification of copies for lipid metabolism-related genes and correlation of copies of APOBEC3A with lipid profiles. The copy numbers of (A) APOBEC3A, (B) APOBEC3A_B, (C) BTNL3, (D) LMF1, (E) LPA, (F) APOBEC3B in PE patients and controls (n = 100/group). (G) Correlation of plasma lipid profiles with APOBEC3A copies (n = 155). The unpaired two-tailed Student’s t-test determined statistical significances between two groups. The Pearson correlation coefficient analyzed the correlations. Data were represented as the mean ± SEM. ***P < 0.001, *P < 0.05. PE, preeclampsia.
APOBEC3A Showed a Significant Positive Correlation With Total Cholesterol and Low-Density Lipoprotein Cholesterol in Human Subjects
To identify the predisposing genes causing lipid metabolism disturbance in PE patients, we collected clinical lipid profiles from these participants’ initial prenatal examination (12–14 gestational weeks) and analyzed the correlation between lipid profiles and the copy number of these candidate genes. The results indicated significant positive correlations between the copy number of APOBEC3A and both TC (P < 0.001) and LDL (P = 0.048) levels in serum (Figure 3G). In contrast, we did not find significant correlations between other candidate genes and clinical lipid profiles here, except for copies of both BTNL3 (P = 0.012) and LPA (P = 0.006), which were significantly positively correlated with HDL levels (Supplementary Figures 1A–E). For the first time, we report an interesting finding that the APOBEC3A gene may play a role in regulating lipid metabolism and be related to the pathogenesis of PE. Subsequently, we investigated how the novel gene APOBEC3A participates in regulating lipid metabolism and the pathogenesis of PE.
APOBEC3A Disrupted Lipid Metabolism in HepG2 Cells in vitro
The liver is recognized as an essential metabolic organ in humans (18). To explore the underlying mechanism of APOBEC3A in lipid metabolism regulation, we constructed an in vitro model of HepG2 (HepG2APOBEC3AOE), a human hepatocellular carcinoma cell line overexpressing APOBEC3A (Figure 4A). Both TC and TG levels were distinctly upregulated in HepG2APOBEC3AOE cells compared to mock cells treated with or without fat accumulation inducer (Figure 4B). RNA-seq analysis revealed that the differentially expressed genes in HepG2APOBEC3AOE were enriched in the biological function of response to lipid (Figure 4C) and the pathway of regulation of lipolysis in the adipocytes (Figure 4D). Furthermore, hallmark gene sets related to cholesterol homeostasis and metabolism of fatty acids in HepG2APOBEC3AOE (Figure 4E) showed distinct differences compared to the mock cells. The genes linked to cholesterol biosynthesis (FDPS, DGAT2) and fatty acid biosynthesis (SCD, FASN), as well as fatty acid absorption (CD36), were upregulated in HepG2APOBEC3AOE. On the other hand, the genes associated with cholesterol catabolism (GPD1) and fatty acid β-oxidation (CPT1A) were downregulated (Figure 4F). These findings indicate that APOBEC3A may be involved in both cholesterol and fatty acid metabolic processes.
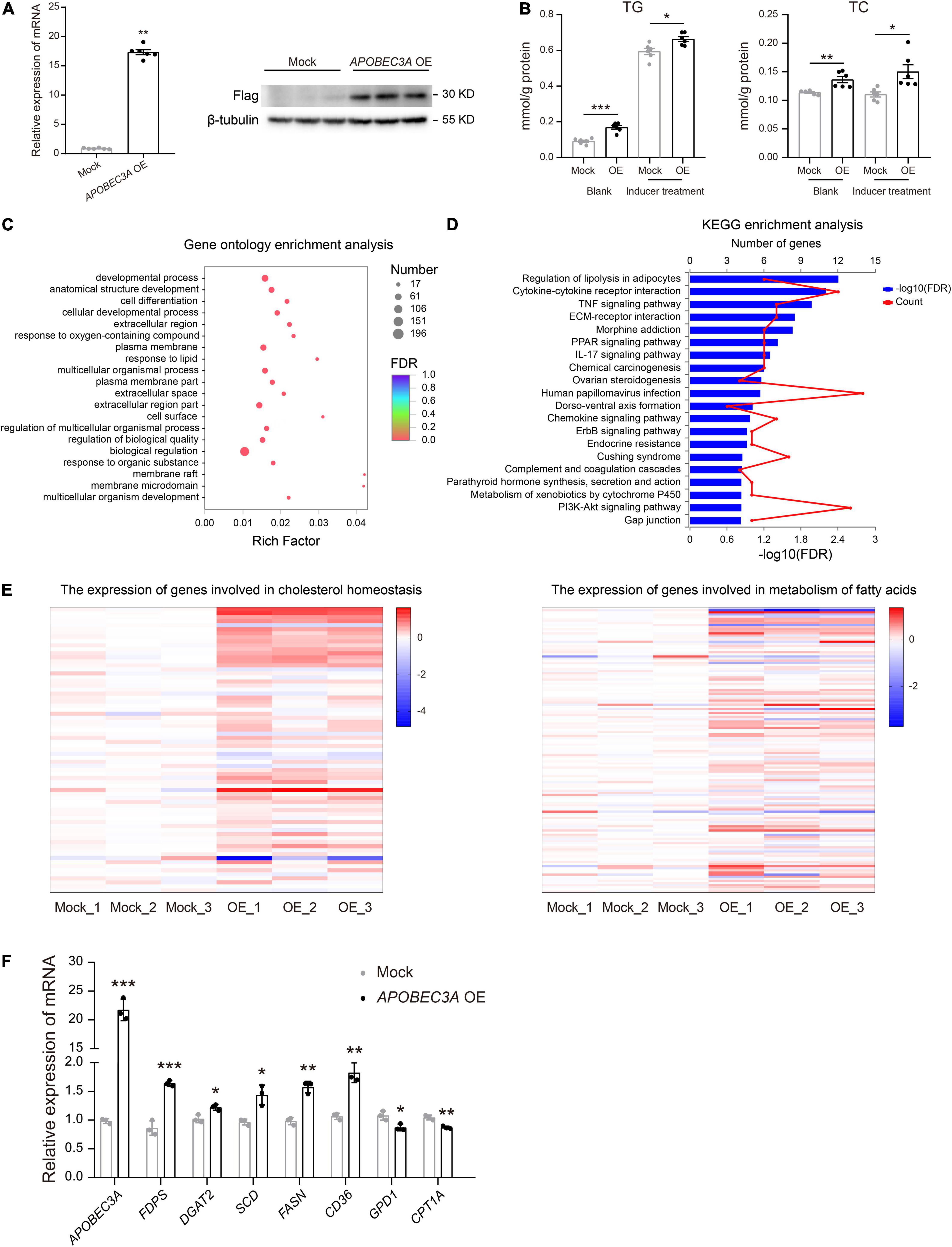
Figure 4. HepG2APOBEC3AOE exhibited perturbed cholesterol and fatty acid metabolism. (A) HepG2 cell lines over-expressing APOBEC3A were constructed. (B) TG and TC levels in the lysates of HepG2APOBEC3AOE and mock cells (n = 6/group). (C) Differential genes between HepG2APOBEC3AOE and mock cells were enriched in functions of respond to lipid. (D) Differential genes between HepG2APOBEC3AOE and mock cells were enriched in pathways of the regulation of lipolysis in adipocytes. (E) The expression of genes connected with cholesterol homeostasis and metabolism of fatty acids in HepG2APOBEC3AOE and mock cells (n = 3/group). (F) The mRNA expression of multiple genes related to cholesterol and fatty acid metabolism in HepG2APOBEC3AOE and mock cells (n = 3/group). The unpaired two-tailed Student’s t-test determined statistical significances between two groups. Data were represented as the mean ± SEM. ***P < 0.001, **P < 0.01, *P < 0.05. OE, HepG2APOBEC3AOE cells; Mock, HepG2 mock cells.
Mice With Hepatic-Specific Overexpression of APOBEC3A Exhibit Disturbed Lipid Metabolism
To explore the potential roles of the APOBEC3A gene in regulating lipid metabolisms and participation in the pathogenesis of PE in vivo, we injected AAV_APOBEC3A and AAV_Mock intraperitoneally in adult C57BL/6J female mice (Figures 5A,B and Supplementary Figure 2). The first important finding was that TC and TG levels were elevated in both plasma and liver of APOBEC3A over-expressing mice (Figure 5C). Moreover, the expression of genes related to cholesterol biosynthesis (Hmgcr, Hmgcs2, Fdps) and fatty acid biosynthesis (Srebp1, Fasn, Acc1) were increased. In contrast, the expression of genes linked to fatty acid β-oxidation (Adiporq, Acoxl) was decreased in the liver of APOBEC3A over-expressing mice (Figure 5D). Lipidomics indicated that fatty acids, (O-acyl)-1-hydroxy fatty acids, triglycerides, and diacylglycerol were elevated in mice over-expressing APOBEC3A (Figure 5E and Supplementary Figure 3A). Moreover, pathway enrichment analysis indicated that differential lipids were enriched in glycerophospholipid metabolism, adipocytokine signaling pathway, fat digestion and absorption, regulation of lipolysis in adipocytes, and cholesterol metabolism (Supplementary Figure 3B). These results imply that APOBEC3A may play a significant role in the biosynthesis of fatty acids and triglycerides.
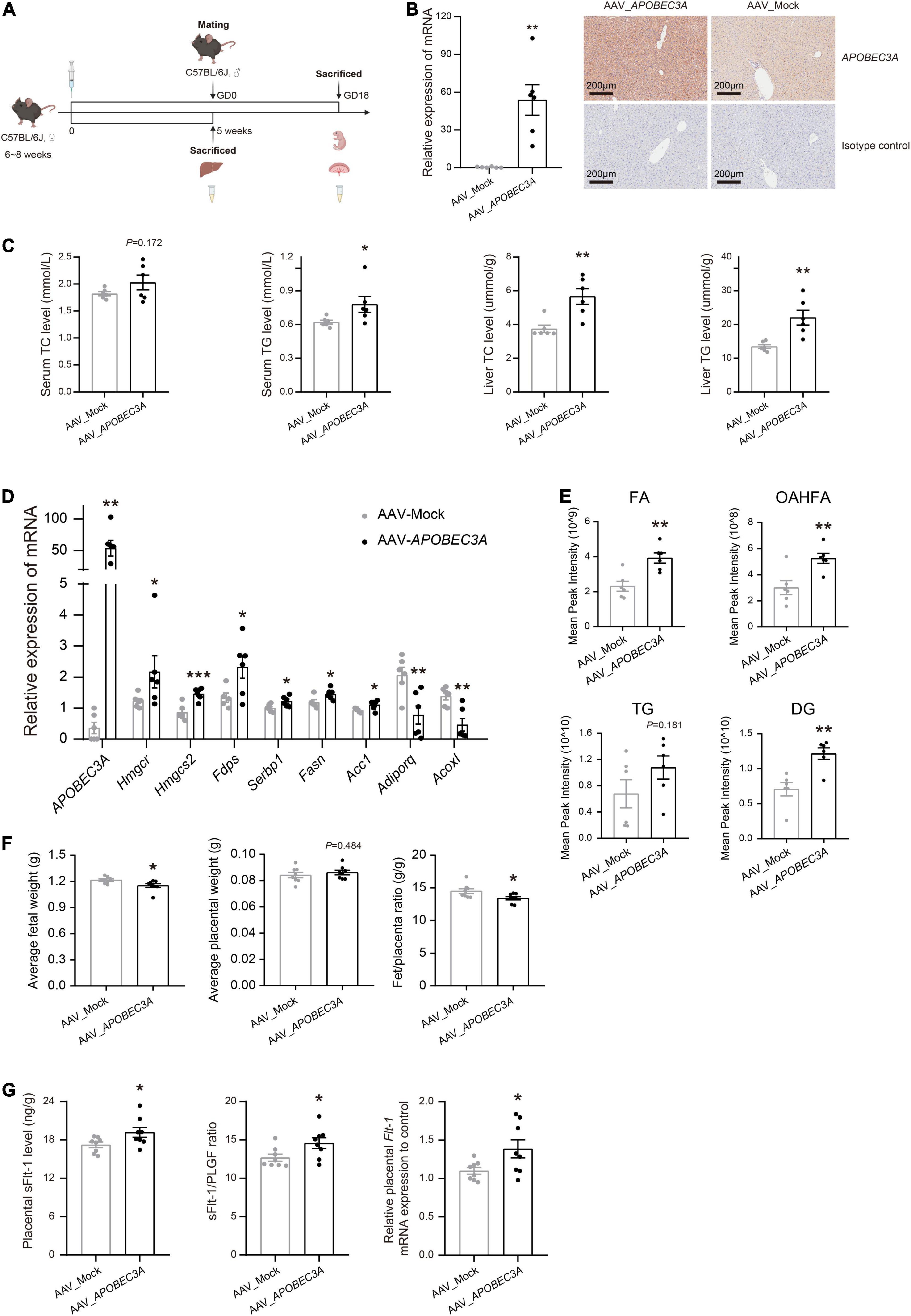
Figure 5. Hepatic-specific overexpression of APOBEC3A in mice manifested abnormal lipid metabolism and PE-like symptoms. (A) Experimental design and timeline for the in vivo experiment in mice. (B) The mRNA and protein expression of APOBEC3A gene in the liver of non-pregnant mice injected with AAV_APOBEC3A or AAV_Mock (n = 6/group). (C) TC and TG levels in plasma and liver of mice injected with AAV_APOBEC3A or AAV_Mock at 5 weeks after AAV administration (n = 6/group). (D) The mRNA expression of multiple genes related to cholesterol and fatty acid metabolism in mice injected with AAV_APOBEC3A or AAV_Mock (n = 3–6/group). (E) Levels of FA, OAHFA, TG, and DG metabolites in mice injected with AAV_APOBEC3A or AAV_Mock (n = 6/group). (F) The average weight of fetuses and placentas in pregnant mice injected with AAV_APOBEC3A or AAV_Mock (n = 8/group). (G) The protein and mRNA levels of sFlt-1 in the placenta of pregnant mice injected with AAV_APOBEC3A or AAV_Mock (n = 8/group). The unpaired two-tailed Student’s t-test determined statistical significances between two groups. Data were represented as the mean ± SEM. ***P < 0.001, **P < 0.01, *P < 0.05. FA, fatty acids; OAHFA, (O-acyl)-1-hydroxy fatty acids; TG, triglycerides; DG, diacylglycerol. GD, day of gestation.
Pregnant Mice With Hepatic-Specific Overexpression of APOBEC3A Exhibit Preeclampsia-Like Symptoms
Fetuses in the AAV_APOBEC3A group had decreased fetuses/placenta ratios, which appeared to be driven by a significant reduction in fetal weight (Figure 5F), showing intrauterine growth restriction, a common clinical manifestation of PE. The sFlt-1/PLGF ratio is a well-known marker for the short-term prediction of PE (19, 20). We measured the levels of PLGF and sFlt-1 in mouse placenta homogenates and serum by ELISA. The protein and mRNA levels of sFlt-1 and the sFlt-1/PLGF ratio were significantly increased in the placenta of the AAV_APOBEC3A group compared with the AAV_Mock group (Figure 5G). In contrast, there was no substantial change in both protein and mRNA levels of PLGF in placental tissues (Supplementary Figure 4A). Besides, these changes mentioned above were not observed in serum samples (Supplementary Figure 4B). These findings indicate that APOBEC3A may be involved in the pathogenesis of PE.
Discussion
Overall, our study identified 27 aberrant CNVs related to PE, including 22 deletions and 5 duplications. The differential genes in these CNV regions were enriched mainly in metabolism-related biological functions. Moreover, aberrant copies of APOBEC3A, APOBEC3A_B, BTNL3, and LMF1 were validated in our cohort, especially the copy number of APOBEC3A was markedly correlated with clinical lipid parameters of human subjects in early pregnancy. To our best knowledge, we are the first to report that APOBEC3A plays a role in regulating lipid metabolism and participating in the pathogenesis of PE.
Despite the growing attention to the pathogenesis of PE, there are still few reports investigating its pathogenesis related to CNV, i.e., duplications or deletions of genomic segments greater than 1 kb in length (21). Although most genome studies have focused on single nucleotide variants (SNVs), CNVs cover more bases than SNVs and may have more potent effects on gene expression and structure (22, 23). Zhao et al. (24) analyzed CNVs from a US cohort and identified two relevant genes, PDXDC1 and PSG11. However, the hypothesis of them being involved in the pathogenesis of PE was not fully supported by further laboratory studies. The above two genes have no overlap with our findings, which may be interpreted as a difference in the genetic background of the two study cohorts.
Abnormal lipid metabolism in pregnant women has been reported to be possibly connected with the pathogenesis of PE (6). PE-related endothelial dysfunction may be associated with changes in lipid structure and oxidative stress caused by abnormal lipid metabolism (25, 26). Physiological hyperlipidemia, which occurs during normal pregnancy, increases the energy supply to both mothers and fetuses. Once the concentrations of TG and free fatty acid in pregnant women gradually increase beyond the physiological tolerance ranges of normal pregnancy, lipid molecules could accumulate and cause extensive vascular endothelial damage and systemic inflammation, eventually leading to PE (25). Consistent with previous reports, we found a significant increase of TC and TG in PE patients in early pregnancy compared with controls, which provides further evidence for abnormal lipid metabolism as a susceptibility factor for PE.
APOBEC3A belongs to the apolipoprotein B mRNA editing enzyme and catalytic polypeptide-like protein family, catalyzing cytidine deamination in single-stranded DNA/RNA to uridine (27). It plays an essential role in the host’s innate immune by inducing viral mutations to protect cells from viral infection (28). Apart from this, APOBEC3A-induced genomic mutations may drive the progression of multiple cancers (29). In this study, we identified a deletion CNV including APOBEC3A, APOBEC3A_B, and APOBEC3B in the genomes of controls (44%). In contrast, the incidence of this CNV is lower in PE patients (10%), which may lead to elevated levels of both APOBEC3A and APOBEC3B in PE patients. APOBEC3A_B has the promoter and coding region of APOBEC3A and the 3’UTR of APOBEC3B. The resulting chimeric APOBEC3A transcription product would lead to a further increase in intracellular APOBEC3A levels (30). Subsequently, we also confirmed by qPCR that the copy numbers of APOBEC3A and APOBEC3A_B genes were markedly higher in PE patients than in controls. Notably, our findings showed that the copy number of APOBEC3A was statistically positively correlated with serum concentrations of TC and LDL. LPA is known to be associated with metabolic diseases (31) and may be involved in the pathogenesis of PE (14–17). BTNL3 has also been reported to have a possible role in intestinal metabolism and inflammation in neonates with intrauterine growth restriction (32). Our study found that copies of both LPA and BTNL3 were significantly and positively associated with HDL levels. Moreover, in vivo and in vitro studies suggested that APOBEC3A may regulate cholesterol and fatty acid metabolisms. These findings filled the gap in the role of APOBEC3A in regulating lipid metabolisms. Of note, we found that APOBEC3A expression was also increased in patients with non-alcoholic steatohepatitis (NASH), especially in NASH in combination with fibrosis (unpublished data), suggesting that APOBEC3A may play a role in other metabolic diseases as well. Overall, elevated APOBEC3A copies may lead to increased cholesterol and fatty acid biosynthesis and decreased catabolism, which increases the risks of various metabolic diseases. These findings may guide the dietary management and clinical use of drugs in such high-risk groups and have good clinical application prospects, although the specific mechanism by which APOBEC3A regulates lipid metabolism needs in-depth study.
PE is a gestational disease, and the main manifestations in animal models include elevated blood pressure and urinary protein, restricted fetal growth, elevated sFlt-1, decreased PLGF, and increased sFlt-1/PLGF ratio (33). In the present study, we found that the average fetal weight per pregnant mice overexpressing APOBEC3A was distinctly reduced (Figure 5F), a manifestation of growth restriction. Both sFlt-1 levels and sFlt-1/PLGF ratios were distinctly increased in the placenta (Figure 5G). However, changes in plasma parameters were not evident (Supplementary Figure 4), which led us to speculate that to be due to vascular endothelial damages may be only related to local (placenta) changes in PE rather than systemic ones. On the other hand, PE is a complex disease induced by multiple factors where a single element may be difficult to interpret the pathogenesis of PE. Our molecular and clinical data suggest that aberrant APOBEC3A copies could at least partly explain the pathogenesis of PE.
In terms of limitations, we did not conduct a subgroup analysis of the collected cases as early-onset and late-onset PE due to limited recruitment, despite the pathogenesis of PE may differ among patients in those two subtypes. Further, our recruited participants were all from one city in China, as PE patients from other regions were not available at the time of this study. This study retrospectively collected information on the lipid profile of pregnant women in early gestation, which resulted in a certain percentage of missing data. We did not account for the effect of possible confounding factors, such as familial dyslipidemia and the use of lipid-lowering drugs, on this study. In addition, as the blood pressure of mice may be affected by various conditions such as sounds, light, and temperature, we did not measure the blood pressure of mice in this study. Finally, in vivo and in vitro knockout models need to be constructed to explore further the more detailed mechanisms that APOBEC3A may modulate lipid metabolism and participate in the pathogenesis of PE.
Collectively, this study established a novel link between genetics and lipid metabolism in the development of PE. That is, aberrant copies of APOBEC3A may be involved in the pathogenesis of PE by regulating lipid metabolism. This critical finding will likely facilitate a better understanding of the molecular mechanisms of PE. It is also our understanding that APOBEC3A copies and lipid profiles may potentially have clinical applications to benefit the early identification of pregnant women at high risk of PE, leading to early clinical intervention to reduce the risk of developing PE.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by The Medical Research Ethics Committee of International Peace Maternity and Child Health Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by The Institutional Animal Care and Use Committee of Shanghai Institute of Materia Medica, Chinese Academy of Sciences.
Author Contributions
NL, L-KG, and B-SW conceived and designed the experiments. NL, ML, and J-HS performed the experiments. Y-NG, X-JW, JM, XZ, YC, and J-HL provided clinical materials. NL, Y-TH, FZ, HH, and J-LX collected and analyzed the data. NL wrote the manuscript. L-KG and B-SW reviewed and edited the manuscript. All authors read and approved the submitted version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81573162).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We express gratitude to all patients and healthy individuals that participated in this study. We wish to acknowledge the support of the Department of Obstetrics, International Peace Maternity and Child Health Hospital, and the Department of Obstetrics, Renji Hospital, Shanghai Jiao Tong University School of Medicine. We also thank Shanghai Qiming Information Technology Co, Ltd., Shanghai Benegene Biotechnology Co, Ltd., Shanghai Majorbio Bio-Pharm Technology Co., Ltd., and Beijing Genomics Institute for their excellent technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.841249/full#supplementary-material
Abbreviations
AAV, Adeno-associated Virus; CNVs, copy number variations; FPKM, fragment of the exon model per million mapped reads; GO, Gene Ontology; HDL, high-density lipoprotein; LDL, low-density lipoprotein; PE, preeclampsia; PLGF, placental growth factor; qPCR, quantitative polymerase chain reaction; sFlt-1, soluble fms-like tyrosine kinase-1; SNVs, single nucleotide variants; TC, total cholesterol; TG, triglyceride.
Footnotes
References
1. Phipps EA, Thadhani R, Benzing T, Karumanchi SA. Pre-eclampsia: pathogenesis, novel diagnostics and therapies. Nat Rev Nephrol. (2019) 15:275–89. doi: 10.1038/s41581-019-0119-6
2. Irianti VN, Rasipin SB. Preeclampsia tree educational model for pregnant women as an effort to change preeclampsia prevention behavior. Int J Innov Sci Res Technol. (2021) 6:683–7.
3. Nejatizadeh A, Stobdan T, Malhotra N, Pasha MA. The genetic aspects of pre-eclampsia: achievements and limitations. Biochem Genet. (2008) 46:451–79. doi: 10.1007/s10528-008-9163-9
4. Thakoordeen S, Moodley J, Naicker T. Candidate gene, genome-wide association and bioinformatic studies in pre-eclampsia: a review. Curr Hypertens Rep. (2018) 20:91. doi: 10.1007/s11906-018-0891-x
5. Jeyabalan A. Epidemiology of preeclampsia: impact of obesity. Nutr Rev. (2013) 71:S18–25. doi: 10.1111/nure.12055
6. Ghio A, Bertolotto A, Resi V, Volpe L, Cianni GD. Triglyceride metabolism in pregnancy. Adv Clin Chem. (2011) 55:133–53. doi: 10.1016/b978-0-12-387042-1.00007-1
7. Kaaja R. Lipid abnormalities in pre-eclampsia: implications for vascular health. Clin Lipidol. (2011) 6:71–8.
8. Zhang F, Wang X-J, Ma J, He Y-T, He H, Zhai J, et al. Association between blood lipid profile in early pregnancy and risk of preeclampsia: a study based on real world data. J Shanghai Jiaotong Univ. (2021) 41:483–8.
9. Chesley LC, Cooper DW. Genetics of hypertension in pregnancy: possible single gene control of pre-eclampsia and eclampsia in the descendants of eclamptic women. Br J Obstet Gynaecol. (1986) 93:898–908. doi: 10.1111/j.1471-0528.1986.tb08006.x
10. Esplin MS, Fausett MB, Fraser A, Kerber R, Mineau G, Carrillo J, et al. Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med. (2001) 344:867–72. doi: 10.1056/nejm200103223441201
11. Yan Z, Yan H, Ou H. Human thyroxine binding globulin (TBG) promoter directs efficient and sustaining transgene expression in liver-specific pattern. Gene. (2012) 506:289–94. doi: 10.1016/j.gene.2012.07.009
12. Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. (2008) 49:1137–46. doi: 10.1194/jlr.D700041-JLR200
13. Roberts JM, August PA, Bakris G, Barton JR, Bernstein IM, Druzin M, et al. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet Gynecol. (2013) 122:1122–31.
14. Wang J, Mimuro S, Lahoud R, Trudinger B, Wang XL. Elevated levels of lipoprotein(a) in women with preeclampsia. Am J Obstet Gynecol. (1998) 178:146–9. doi: 10.1016/s0002-9378(98)70642-8
15. Baksu B, Baksu A, Davas I, Akyol A, Gulbaba G. Lipoprotein(a) levels in women with pre-eclampsia and in normotensive pregnant women. J Obstet Gynaecol Res. (2005) 31:277–82. doi: 10.1111/j.1447-0756.2005.00276.x
16. Manten GT, van der Hoek YY, Marko Sikkema J, Voorbij HA, Hameeteman TM, Visser GH, et al. The role of lipoprotein (a) in pregnancies complicated by pre-eclampsia. Med Hypotheses. (2005) 64:162–9. doi: 10.1016/j.mehy.2004.04.026
17. Fanshawe AE, Ibrahim M. The current status of lipoprotein (a) in pregnancy: a literature review. J Cardiol. (2013) 61:99–106. doi: 10.1016/j.jjcc.2012.09.009
18. Campbell I. Liver: metabolic functions. Anaesth Intensive Care Med. (2006) 7:51–4. doi: 10.1383/anes.2006.7.2.51
19. Levine RJ, Maynard SE, Qian C, Lim K-H, England LJ, Yu KF, et al. Circulating angiogenic factors and the risk of preeclampsia. N Engl J Med. (2004) 350:672–83.
20. Zeisler H, Llurba E, Chantraine F, Vatish M, Staff AC, Sennstrom M, et al. Predictive value of the sFlt-1:PlGF ratio in women with suspected preeclampsia. N Engl J Med. (2016) 374:13–22. doi: 10.1056/NEJMoa1414838
21. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. (2006) 444:444–54. doi: 10.1038/nature05329
22. Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. (2015) 349:aab3761. doi: 10.1126/science.aab3761
23. Gamazon ER, Stranger BE. The impact of human copy number variation on gene expression. Brief Funct Genomics. (2015) 14:352–7. doi: 10.1093/bfgp/elv017
24. Zhao LL, Triche EW, Walsh KM, Bracken MB, Saftlas AF, Hoh J, et al. Genome-wide association study identifies a maternal copy-number deletion in PSG11 enriched among preeclampsia patients. BMC Pregnancy Childbirth. (2012) 12:61. doi: 10.1186/1471-2393-12-61
25. Sattar N, Petrie JR, Jaap AJ. The atherogenic lipoprotein phenotype and vascular endothelial dysfunction. Atherosclerosis. (1998) 138:229–35. doi: 10.1016/s0021-9150(98)00037-9
26. Lorentzen B, Henriksen T. Plasma lipids and vascular dysfunction in preeclampsia. Semin Reprod Endocrinol. (1998) 16:33–9. doi: 10.1055/s-2007-1016250
27. Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. (2002) 79:285–96. doi: 10.1006/geno.2002.6718
28. Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. (2008) 9:229. doi: 10.1186/gb-2008-9-6-229
29. Law EK, Levin-Klein R, Jarvis MC, Kim H, Argyris PP, Carpenter MA, et al. APOBEC3A catalyzes mutation and drives carcinogenesis in vivo. J Exp Med. (2020) 217:e20200261. doi: 10.1084/jem.20200261
30. Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat Commun. (2014) 5:5129. doi: 10.1038/ncomms6129
31. Schmidt K, Noureen A, Kronenberg F, Utermann G. Structure, function, and genetics of lipoprotein (a). J Lipid Res. (2016) 57:1339–59. doi: 10.1194/jlr.R067314
32. Huang S, Wu Z, Yuan X, Li N, Li T, Wang J, et al. Transcriptome differences suggest novel mechanisms for intrauterine growth restriction mediated dysfunction in small intestine of neonatal piglets. Front Physiol. (2020) 11:561. doi: 10.3389/fphys.2020.00561
Keywords: preeclampsia, APOBEC3A, lipid metabolism, copy number variation, genetics
Citation: Liu N, Guo Y-N, Wang X-J, Ma J, He Y-T, Zhang F, He H, Xie J-L, Zhuang X, Liu M, Sun J-H, Chen Y, Lin J-H, Gong L-K and Wang B-S (2022) Copy Number Analyses Identified a Novel Gene: APOBEC3A Related to Lipid Metabolism in the Pathogenesis of Preeclampsia. Front. Cardiovasc. Med. 9:841249. doi: 10.3389/fcvm.2022.841249
Received: 22 December 2021; Accepted: 26 April 2022;
Published: 16 May 2022.
Edited by:
Rossana Orabona, Obstetrics and Gynaecology, ASST Spedali Civili, ItalyReviewed by:
Tlili Barhoumi, King Abdullah International Medical Research Center (KAIMRC), Saudi ArabiaSerena Migliarino, University of Magna Graecia, Italy
Copyright © 2022 Liu, Guo, Wang, Ma, He, Zhang, He, Xie, Zhuang, Liu, Sun, Chen, Lin, Gong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Kun Gong, lkgong@cdser.simm.ac.cn; Bing-Shun Wang, wangbingshun@sjtu.edu.cn
 Nan Liu
Nan Liu Yu-Na Guo3
Yu-Na Guo3  Yun-Ting He
Yun-Ting He Bing-Shun Wang
Bing-Shun Wang