Carbapenemase genes distribution in clonal lineages of Acinetobacter baumannii: a comprehensive study on plasmids and chromosomes
- 1Department of Bacteriology, Pasteur Institute of Iran, Tehran, Iran
- 2Isfahan Endocrine and Metabolism Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
Background: The global spread of plasmids carrying carbapenemase genes within carbapenem resistant Acinetobacter baumannii (CRAB) strains poses a worldwide public health issue. In this study, we conducted a comprehensive genetic analysis of plasmids and chromosomes harboring the major carbapenemase genes (blaNDM, blaKPC, blaVIM, blaIMP, blaGES, blaOXA-58-like, blaOXA-24/40-like, blaOXA-143-like, and blaOXA-23-like) in CRAB strains using bioinformatic tools.
Methods: We retrieved plasmids and chromosomes carrying the major carbapenemase genes from GenBank. The size, replicon type, and conjugal apparatus of the plasmids were also determined. Furthermore, allele types, co-existence of other antimicrobial resistance genes alongside carbapenemases in plasmids or chromosomes, co-occurrence of carbapenemase genes, gene repetition, and sequence types (ST) of whole genomes were characterized.
Results: The database contained 113 plasmids and 38 chromosomes harboring carbapenemase genes. This investigation revealed that blaNDM and blaOXA-58-like were the predominant allele types in both the plasmids and chromosomes. Nine (7.96%) plasmids with blaNDM-1 were potentially conjugative. The most common replicon types of the plasmids were R3-T1, R3-T8, R3-T2, R3-T23, and RP-T1. The analysis revealed that blaNDM-1 and blaOXA-58-like genes possessed the highest variety of co-existence with other antibiotic resistance genes. The co-occurrence of dual carbapenemases was identified in 12 plasmids and 19 chromosomes. Carbapenemase gene repetitions were identified in 10 plasmids and one chromosome. Circular alignment revealed that the plasmids carrying the co-occurrence of blaNDM-1 and blaOXA-58 were more homogeneous. However, there was heterogeneity in certain regions of these plasmids. According to the minimum spanning tree (MST) results, the majority of the plasmids belonged to the genomes of ST2Pas, ST1Pas, ST422Pas, ST622Pas, and ST85Pas.
Conclusion: A. baumannii appears to have a strong ability for genome plasticity to incorporate carbapenemase genes on its plasmids and chromosomes to develop resistance against carbapenems. Mobilizable plasmids harboring carbapenemases significantly contribute to the dissemination of these genes. The genetic structure of the plasmids revealed a strong associations of class I integrons, ISAba-like structures, Tn4401 elements, and aac (6′)-Ib with carbapenemases. Furthermore, gene repetition may also be associated with carbapenem heteroresistance.
1 Introduction
Acinetobacter baumannii is a member of the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.). A. baumannii is an opportunistic pathogen that has been identified as an important cause of nosocomial infections, including ventilator-associated pneumonia in intensive care units, urinary tract, and bloodstream infections (Flores-Paredes et al., 2021; Nocera et al., 2021). These infections are associated with significant mortality rates owing to high levels of antimicrobial resistance (Alrahmany et al., 2022). Carbapenems are used as a last resort to treat severe infections caused by multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains (Zalegh et al., 2023). In addition, carbapenem-resistant A. baumannii (CRAB) isolates have been categorized by the World Health Organization (WHO) as one of the 12 top priority resistant bacteria that pose the greatest threat to public health (Hsieh et al., 2020). The main cause of carbapenem resistance in A. baumannii isolates is the acquisition of metallo-beta-lactamases (MBLs), class A carbapenemases (for example, KPC), and carbapenem-hydrolyzing class D beta-lactamase (CHDLs) enzymes (Kyriakidis et al., 2021).
Treatment of antibiotic-resistant A. baumannii isolates is becoming increasingly difficult due to the co-existence of carbapenemase-encoding and other antibiotic-resistance genes, including aminoglycoside-modifying genes and plasmid-mediated quinolone resistance (Wasfi et al., 2021). However, these resistance genes are often located in mobile genetic elements (MGEs), including conjugative plasmids, transposons, integrons, and insertion sequences. They can be easily expanded and transferred between various bacterial species (Horne et al., 2023). Conjugative plasmids, which include the origin of the transfer (oriT) region, relaxase enzyme, type IV coupling protein (T4CP), and a gene cluster for the bacterial type IV secretion system (T4SS) apparatus, are essential for the horizontal gene transfer and dissemination of carbapenemase genes (Li et al., 2018). Lam et al. (2022) classified A. baumannii plasmids into three categories based on the Pfam domains of the replication initiation protein including RP, R1, and R3. Plasmids that contain conjugation genes and are often associated with various acquired antibiotic resistance genes belong to the four most common types: RP-T1, R3-T1, R3-T2, and R3-T3 (Lam et al., 2023). Plasmids carrying resistance genes are strongly related to specific sequence types (STs), including ST2, ST1, and ST3, which can cause nosocomial infections and outbreaks (Frenk et al., 2022). Therefore, the genetic characterization of these plasmids in successful international clones is important for understanding the carbapenemase-harboring bacteria and for effective decision-making in infection prevention strategies.
In this study, we conducted an in silico analysis and comparative assessment of carbapenemase harboring plasmids (CHPs) and chromosomes (CHCs) in A. baumannii. The primary objective was to identify the genetic structures of various allele types of the major carbapenemase genes (blaIMP, blaVIM, blaGES, blaNDM, blaOXA-58-like, blaOXA24/40-like, blaOXA-143-like, and blaOXA-23-like) using bioinformatic tools. Additionally, this study characterized the genetic properties of both CHPs and CHCs, encompassing replicon types, conjugation ability, co-existence (linkage of carbapenemases with other antimicrobial resistance genes), co-occurrence (presence of at least two carbapenemase genes in one strain), gene repetition, and phylogenetic relatedness.
2 Materials and methods
2.1 Preparation of the initial dataset and characterization of carbapenemases
Nucleotide sequences of nine major carbapenemase genes (blaNDM, blaKPC, blaVIM, blaIMP, blaGES, blaOXA-58-like, blaOXA-24/40-like, blaOXA-143-like, and blaOXA-23-like) in A. baumannii strains were retrieved from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Benson et al., 2017). Two BLAST approaches including Microbial BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=MicrobialGenomes) and Standard Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLwwereeee_SPEC=GeoBlast&PAGcompleteSearch) was used to retrieve all completed plasmids and partial DNA fragments carrying carbapenemase genes, respectively. The criteria used to retrieve all carbapenemases were ≥ 80% identity and 100% coverage.
2.2 Allele types and genetic environment of carbapenemases
The allele types of the mentioned carbapenemase genes were determined (cut-off was 100% identity and 100% coverage) using the beta-lactamase database (http://bldb.eu/) (Naas et al., 2017). Additionally, the prevalence of each allele type was determined. Moreover, the genetic contexts of nine carbapenemase genes present on both plasmids and chromosomes were characterized. The most prevalent of genetic structure for each gene was depicted.
2.3 Detection of other AMR genes on the retrieved DNAs
The Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/home) was used to identify the presence of various antimicrobial resistance genes against beta-lactams, carbapenems, macrolides, aminoglycosides, and other antibiotics (Islam et al., 2023). The co-existence (gene linkage) of A. baumannii plasmids and chromosomes harboring major carbapenemase genes with various antimicrobial resistance-associated genes was investigated. Co-occurrence was determined based on the presence of at least two major carbapenemase genes in an isolate.
2.4 Genetic characterization and replicon typing of plasmids carrying carbapenemases
The conjugation apparatus of the plasmids was identified using oriTfinder (https://bioinfo-mml.sjtu.edu.cn/oriTfinder/) (Li et al., 2018). The presence or absence of conjugation elements, including oriT, relaxase-encoding genes, T4CP, and T4SS in the CHPs was determined using this web tool. The replicon type for each plasmid was determined using local BLASTn. In addition, carbapenemase gene repetition in the plasmids was performed. Moreover, information on the geographical regions, isolation sources, collection date, host disease, and hosts of all isolates harboring crabapenemase genes were extracted.
2.5 Phylogenetic analysis and circular alignment of carbapenemase-harboring plasmids
We conducted a distribution analysis of bacterial accessory genomic elements using ClustAGE (https://bmcbioinformatics.biomedcentral.com) (Ozer, 2018). This tool creates a presence/absence matrix, which is subsequently converted into an unweighted pair-group method with arithmetic mean (UPGMA) dendrogram using the Anvi’o tool. UPGMA was used to determine the phylogenetic relationships among all CHPs (Xu et al., 2023). The results were visualized using the Interactive Tree Of Life (iTOL) platform (https://itol.embl.de) (Letunic and Bork, 2021). In addition, multiple circular alignments were applied to compare the similarity and heterogeneity of plasmids carrying blaNDM-1/blaOXA58-like using the BLAST Ring Image Generator (BRIG) version 0.95 (https://brig.sourceforge.net/) (Yao et al., 2022). Differences in the gene content of these plasmids were determined through pan-genome analysis using Roary (Rapid large-scale prokaryote pan-genome analysis) version 3.11.2 (http://sanger-pathogens.github.io/Roary) (Page et al., 2015), which operates on annotated assemblies in GFF3 format produced by Prokka (rapid prokaryotic genome annotation) version 1.14.5 (http://www.vicbioinformatics.com/software.prokka.shtml) (Seemann, 2014).
2.6 Clonal relatedness of strains harboring carbapenemase genes
The distribution of major carbapenemase genes among the different STs was assessed. For each plasmid, ST of the related chromosome was determined using seven housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) from the PubMLST database (https://pubmlst.org). The clonal relatedness of STs was characterized using PHYLOViZ version 2.0 (http://www.phyloviz.net) to generate a minimum spanning tree (MST) for all STs (Nascimento et al., 2017).
3 Results
3.1 The prevalence and the allele types of the various carbapenemase genes
A total of 415 DNA fragments containing partial DNA plasmids (n= 264), complete plasmids (n= 113), and chromosomes (n= 38) carrying carbapenemase genes were extracted from the GenBank database using microbial and standard BLAST. The most prevalent alleles of carbapenemase genes found on the all retrieved DNAs were blaNDM-1 (77/96), blaOXA-58 (64/81), blaOXA-72 (42/70), blaOXA-366 (11/53), blaIMP-1 (13/47), blaGES-11 (12/32), blaOXA-253 (4/17), blaVIM-2 (6/13), and blaKPC-3 (3/6) (Figure 1).
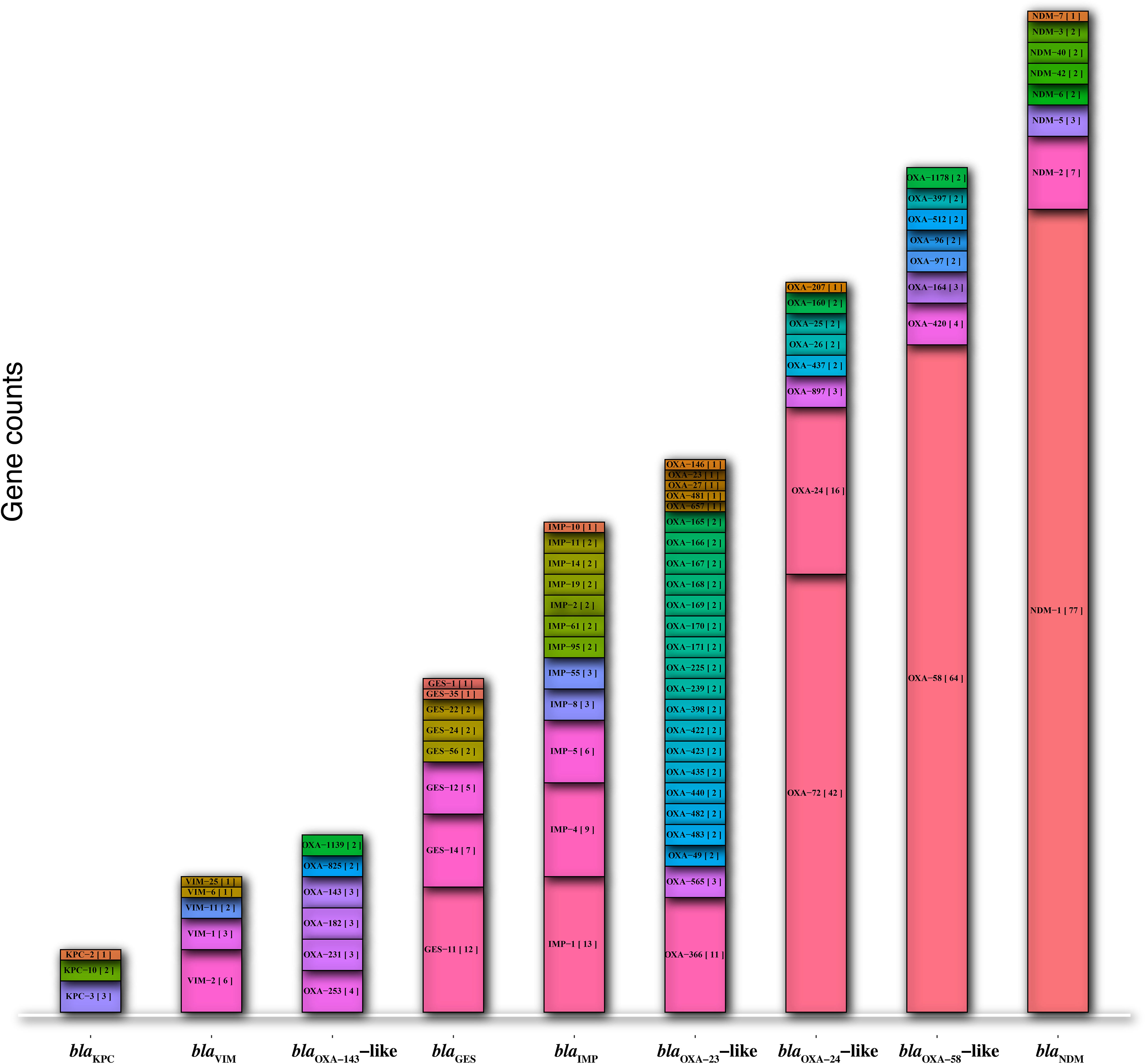
Figure 1 The prevalence of allele types in the nine major carbapenemase genes (blaNDM, blaKPC, blaVIM, blaIMP, blaGES, blaOXA-58-like, blaOXA-24/40-like, blaOXA-143-like, and blaOXA-23-like).
3.2 The prevalence of carbapenemase genes carrying on plasmids and chromosomes
The prevalence of plasmids harboring carbapenemase genes was as follows: blaOXA-24 (46/113), blaOXA-58 (34/113), blaNDM (25/113), and blaGES (8/113). In contrast, the prevalence of chromosomes bearing these genes included blaNDM (32/38). The detailed information is as follows: 12 blaNDM-1, one blaNDM-2, one blaNDM-6, one blaNDM-1/blaOXA-58-like, 17 blaNDM-1/blaOXA-23-like, blaOXA-23-like (3/38), blaOXA-58-like (2/38), and blaKPC-3 (1/38) (Tables 1, 2).
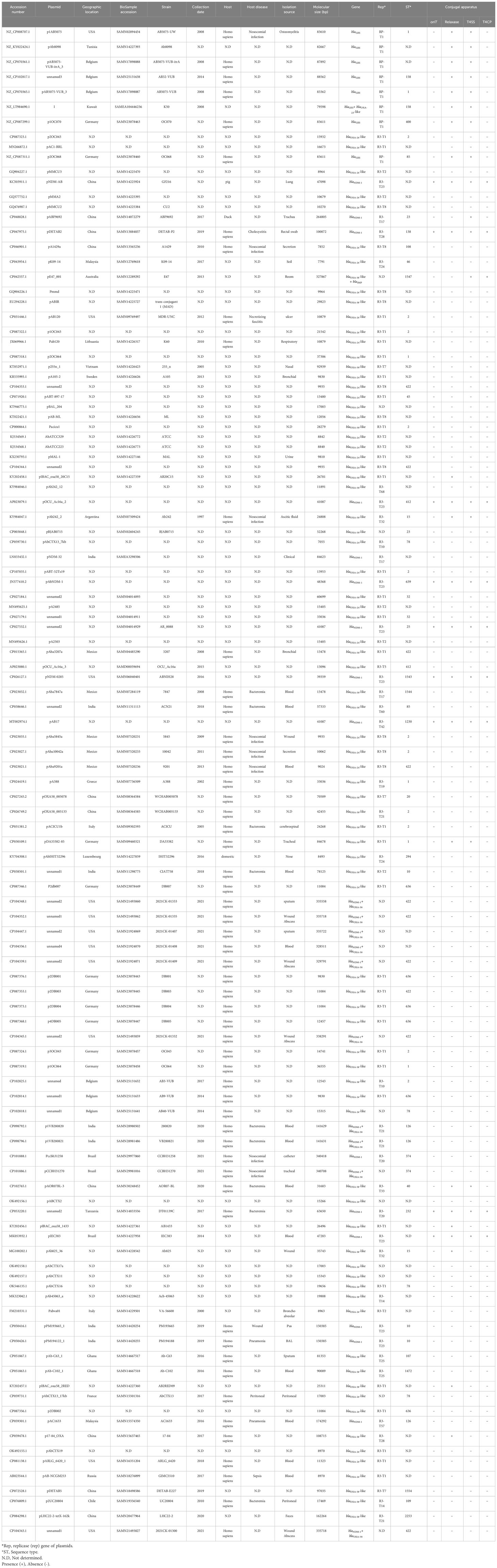
Table 1 The data on the geographical regions, isolation sources, years, hosts, genetic characteristics and Rep types of the plasmids harboring carbapenemases isolated from A. baumannii strains.
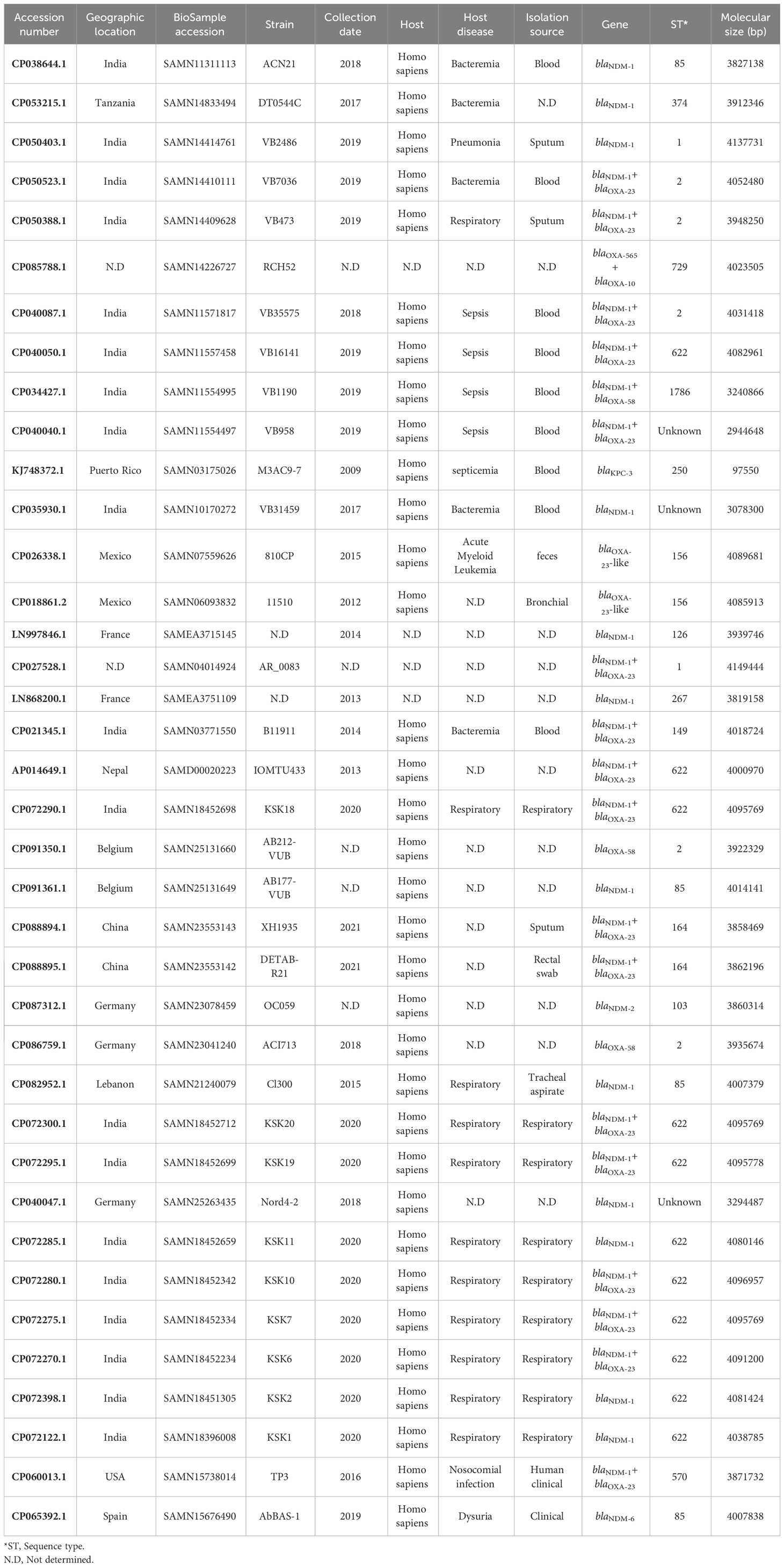
Table 2 The data on the geographical regions, isolation sources, years, hosts, and clonal relatedness of the chromosomes of A. baumannii strains harboring carbapenemases.
3.3 Genetic characterization of plasmids harboring carbapenemases
The size of the complete plasmids harboring carbapenemase genes varied from 83,610 to 335,718 bp. The majority of plasmids carrying carbapenemase genes, such as blaNDM, blaOXA-58-like, and blaOXA-24-like, fell within the range of 10,000 to 50,000 bp. In contrast, plasmids housing blaGES genes tended to be larger, typically in the range of 50,000 to 100,000 bp. Nine (7.96%, 9/113) plasmids were identified as potentially conjugative, as they carried all four essential conjugal components including oriT, relaxase, type IV coupling protein (T4CP), and type IV secretion system (T4SS). The presence of blaGES was exclusively confined to plasmids with RP-T1 replicon type (100%, 8/8). On the other hand, the blaNDM genes were primarily related to plasmids with the R3-T23 replicon type (32%, 8/25). Similarly, blaOXA-24 and blaOXA-58 were predominantly associated of plasmids with the R3-T1 replicon type (45.65%, 21/46 and 32.35%, 11/34) (Table 1).
3.4 Epidemiological data on A. baumannii harboring carbapenemases in plasmids and chromosomes
In this study, a notable proportion of A. baumannii strains harboring carbapenemases in plasmids and chromosomes were derived from blood samples. Furthermore, it is important to highlight that all hosts of these isolates were Homo sapiens. Respiratory and nosocomial infections were the predominant diseases associated with the isolates. Notably, plasmids carrying the blaGES (25%, 2/8), blaOXA-24-like (13.04%, 6/46), blaNDM (32%, 8/25), and blaOXA-58-like (17.6%, 6/34) genes were identified in Germany, the United States, and China, respectively. Additionally, a substantial percentage of chromosomes harboring carbapenemases was identified in India (50%, 19/38). Detailed information on the geographical regions, isolation sources, collection date, host disease, and hosts of plasmids and chromosomes carrying carbapenemases is shown in Tables 1, 2.
3.5 The co-existence of other antimicrobial resistance genes in the plasmids and chromosomes harboring carbapenemases
Antimicrobial resistance genes against various classes of antibiotics, including beta-lactams, carbapenems, macrolides, aminoglycosides, sulfonamides, trimethoprim (TMP), tetracyclines, rifampin, fluoroquinolones, and phenicols, along with genes encoding efflux pump proteins and antiseptic resistance genes, were found in plasmids and chromosomes carrying carbapenemase genes. Carbapenemase gene carriage is a prerequisite for plasmid selection. Co-existence of blaIMP and resistance genes against macrolides, aminoglycosides, beta-lactams, carbapenems, sulfonamides, TMP, phenicols, and a disinfectant (qacEdelta1) was observed. Additionally, blaGES co-existed with resistance genes against aminoglycosides, beta-lactams, carbapenems, sulfonamides, TMP, and a disinfectant (qacEdelta1). The blaOXA-24/40-like-resistant genes related to macrolides. The blaNDM genes were associated with resistance genes against beta-lactams, carbapenems, macrolides, aminoglycosides, sulfonamides, TMP, tetracyclines, rifampin, fluoroquinolones, phenicols, and disinfectants (qacG and qacE). Similarly, blaOXA-58-like was observed in co-existence with resistance genes against beta-lactams, carbapenems, macrolides, aminoglycosides, sulfonamides, TMP, tetracyclines, rifampin, fluoroquinolones, phenicols, and disinfectants (qacEDelta1, and qacG). Moreover, blaOXA-24/40-like was linked to macrolide resistance genes (Figure 2).
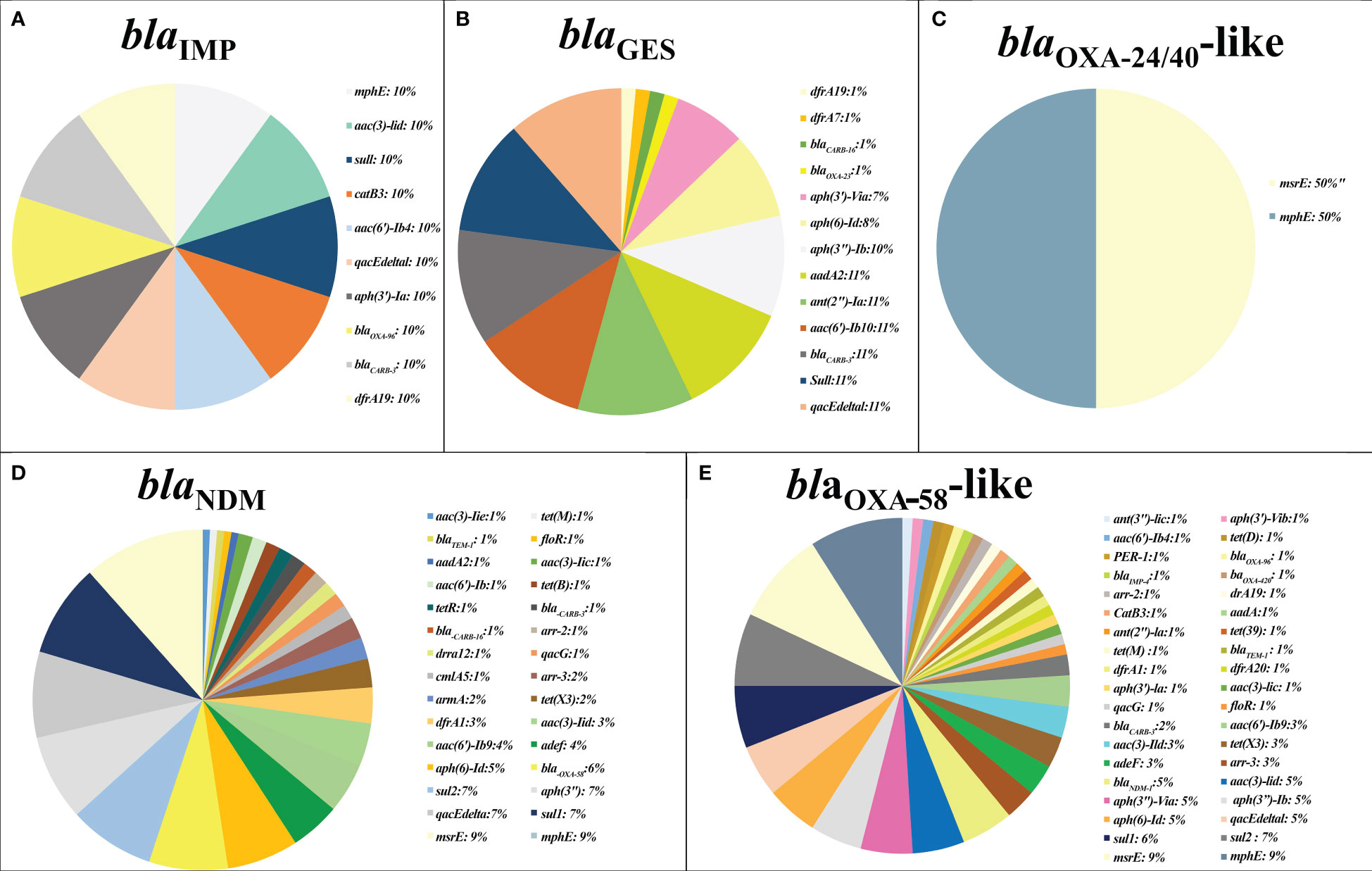
Figure 2 The prevalence of other antimicrobial resistance in plasmids containing major carbapenemase genes. The co-existence rate of other antibiotic resistance genes detected in plasmids harboring blaIMP gene (A), blaGES gene (B), blaOXA-24/40-like gene (C), blaNDM gene (D), and blaOXA-58-like (E).
3.6 The co-occurrence of plasmids and chromosomes harboring carbapenemase genes
Analysis of the data retrieved from the GenBank database revealed the co-occurrence of predominant allele types of carbapenemase genes in the plasmids and chromosomes. This co-occurrence was observed in 12 plasmids, among which 10 harbored both blaNDM-1 and blaOXA-58-like genes, one contained both blaOXA-58-like and blaIMP genes, and one harbored both blaGES and blaOXA-23-like genes. Notably, there was no co-occurrence of blaOXA-24-like and other carbapenemase genes in the plasmids dataset (Table 1 and Figure 3). Furthermore, co-occurrence was found in 19 chromosomes, which can be categorized as follows:17 chromosomes carried both blaNDM-1 and blaOXA-23-like genes, one chromosome contained both blaNDM-1 and blaOXA-58-like genes, and one chromosome exhibited co-occurrence of blaOXA-23-like and blaOXA-10 genes. Notably, there was no co-occurrence of blaKPC-3 with other carbapenemase genes in the chromosomes (Table 2).
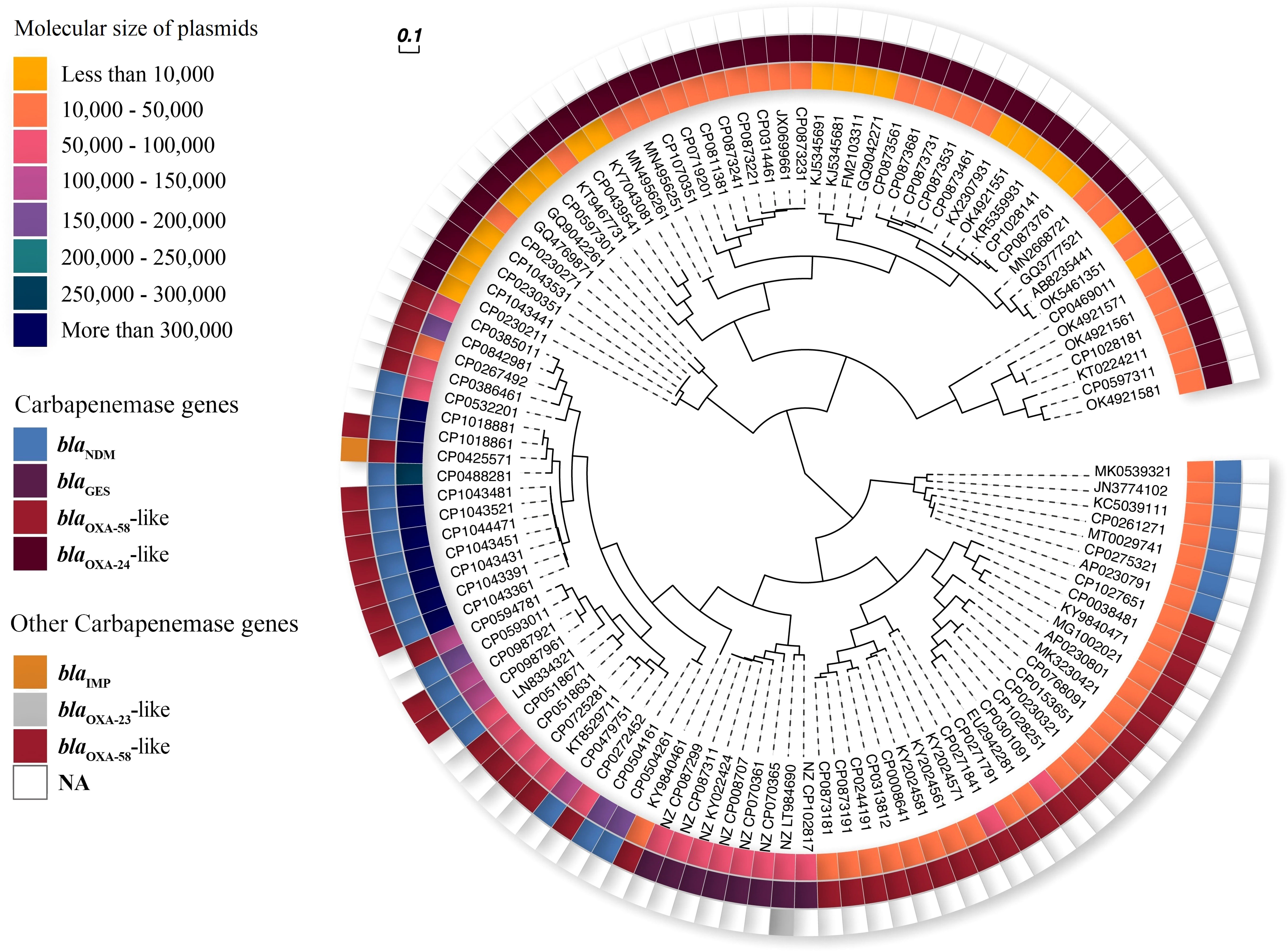
Figure 3 Phylogenetic UPGMA of carbapenemase-harboring plasmids from A. baumannii. Inner ring: 8 ranges for size of plasmids (yellow to navy blue color spectrum). Middle ring: carbapenemase genes (blue-blaNDM, purple-blaGES, red-blaOXA-58-like, and crimson-blaOXA-24-like). Outer ring: other carbapenemase genes (orange-blaIMP, gray-blaOXA-23-like, red-blaOXA-58-like).
3.7 The gene repetition in the plasmids and chromosomes
In this study, carbapenemase gene repetitions were identified in seven plasmids harboring blaOXA-24-like (CP107035.1, CP087324.1, CP087323.1, CP081138.1, CP071920.1, CP031446.1, and JX069966.1), three plasmids harbored blaOXA-58-like (CP027184.1, CP027179.1, and CP000864.1), and one chromosome (CP091350.1) had blaOXA-58-like. Carbapenemase gene repetitions were not detected within the plasmids or chromosomes carrying other carbapenemase gene variants.
3.8 Genetic environment of carbapenemase genes
Two copies of ISAba125 flanked blaNDM-1. The blaNDM cluster was sequentially embedded in the ISAba125-IS30- blaNDM-bleMBL-tat-cutA structure. blaOXA-58 is located in the ISAba3-blaOXA-58-ISAba3-like structure. This was followed by araC1 (a putative transcriptional regulator) and lysE (a putative threonine efflux protein). The blaKPC gene was flanked by two copies of Tn4401. This transposon also harbors the ISKpn6, ISKpn7, transposase, and resolvase genes. The blaGES gene is located in a class 1 integron. The blaGES gene cassette is downstream of aacA4 gene, which encodes AAC (6′)-Ib aminoglycoside acetyltransferase. This was followed by the dfrA7 gene cassette, trimethoprim resistance gene, and qacEdelta1. The blaIMP gene was found in the blaIMP-qacG2-aacA4-catB3-sul cassette array and was the most abundant gene in the class 1 integrons. This array contained genes that confer resistance to quaternary ammonium compounds (qacG), aminoglycosides (aacA4), and chloramphenicol (catB3). XerC/XerD-like binding sites flanked the blaOXA-24/40-like genes. In addition, blaOXA-23 was flanked by two copies of ISAba1. This arrangement was followed by the ATPase and helicase genes (Figure 4).
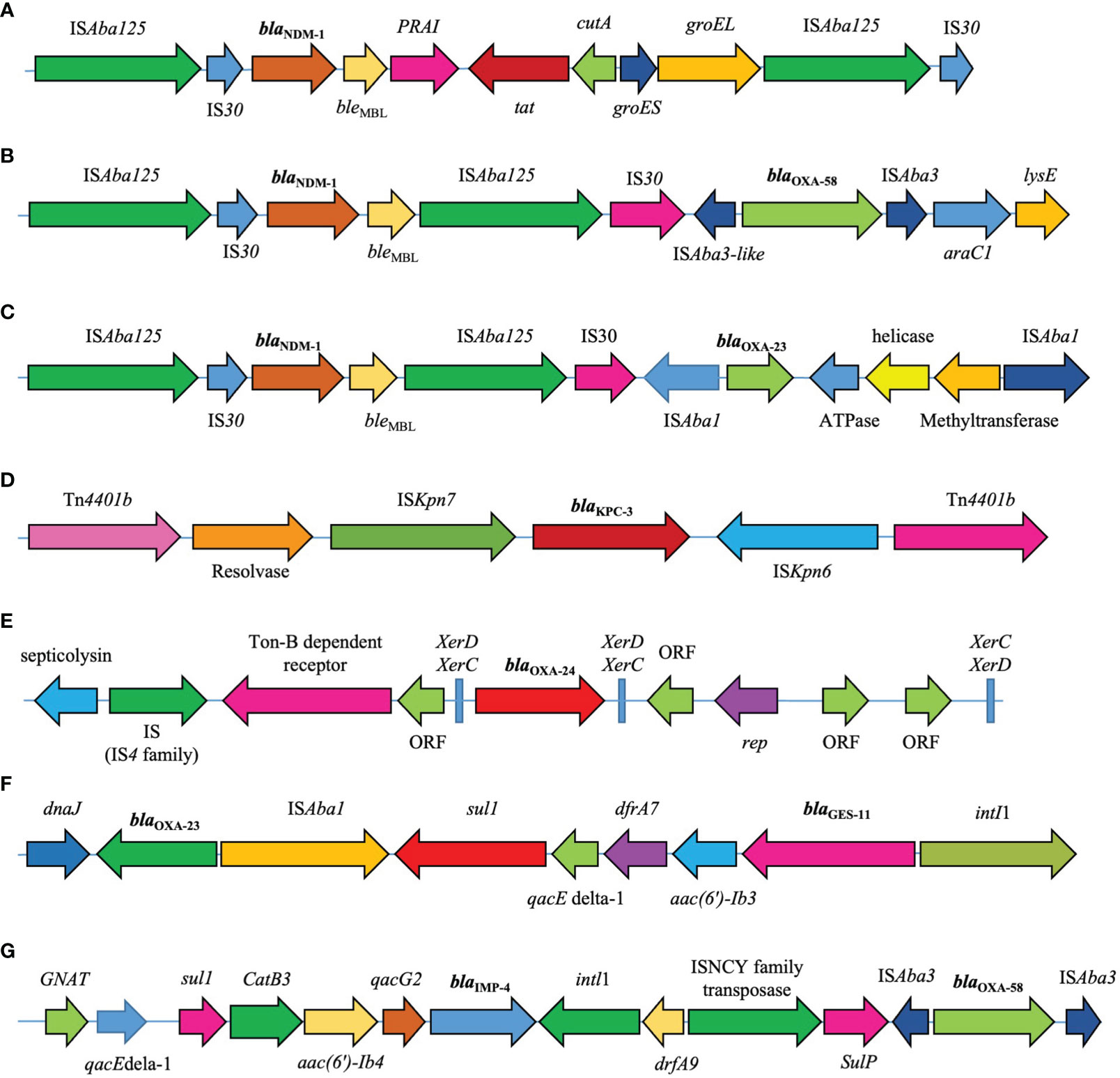
Figure 4 Genetic environments of major carbapenemase genes (A) blaNDM-1 (accession number: KC503911.1). (B) blaNDM-1/blaOXA-58 (CP104348.1). (C) blaNDM-1/blaOXA-23 (CP050523.1). (D) blaKPC (KJ748372.1). (E) blaOXA-24/40-like (CP043954.1). (F) The blaOXA-23/blaGES (LT984690.1). (G) The blaOXA-58/blaIMP (CP042557.1).
3.9 Comparative analysis of plasmids carrying blaNDM-1 and blaOXA-58
Multiple circular alignments of the plasmids carrying the co-occurrence of blaNDM-1 and blaOXA-58 were performed, including p1VB280820 (CP098792.1), p1VB280821 (CP098796.1), pAC1633-1 (CP059301.1), pABF9692 (CP048828.1), unnamed4 (CP104336.1), unnamed2 (CP104339.1), unnamed2 (CP104348.1), unnamed1 (CP104352.1), unnamed1 (CP104343.1), unnamed2 (CP104345.1), pccbh31258 (CP101888.1), and pCCBH31270 (CP101886.1). In this study, plasmid unnamed2 (CP104447.1) was used as a reference (Figure 5). These plasmids appeared to be more homogeneous in their genetic structure, marked by the presence of a set of common genes, including acrA, acrE, actP, adh, aes, aphA, betI, bin3, aacA4, and acoR, as well as the antibiotic resistance genes, blaNDM-1 and blaOXA-58, which were consistently present across all plasmids.
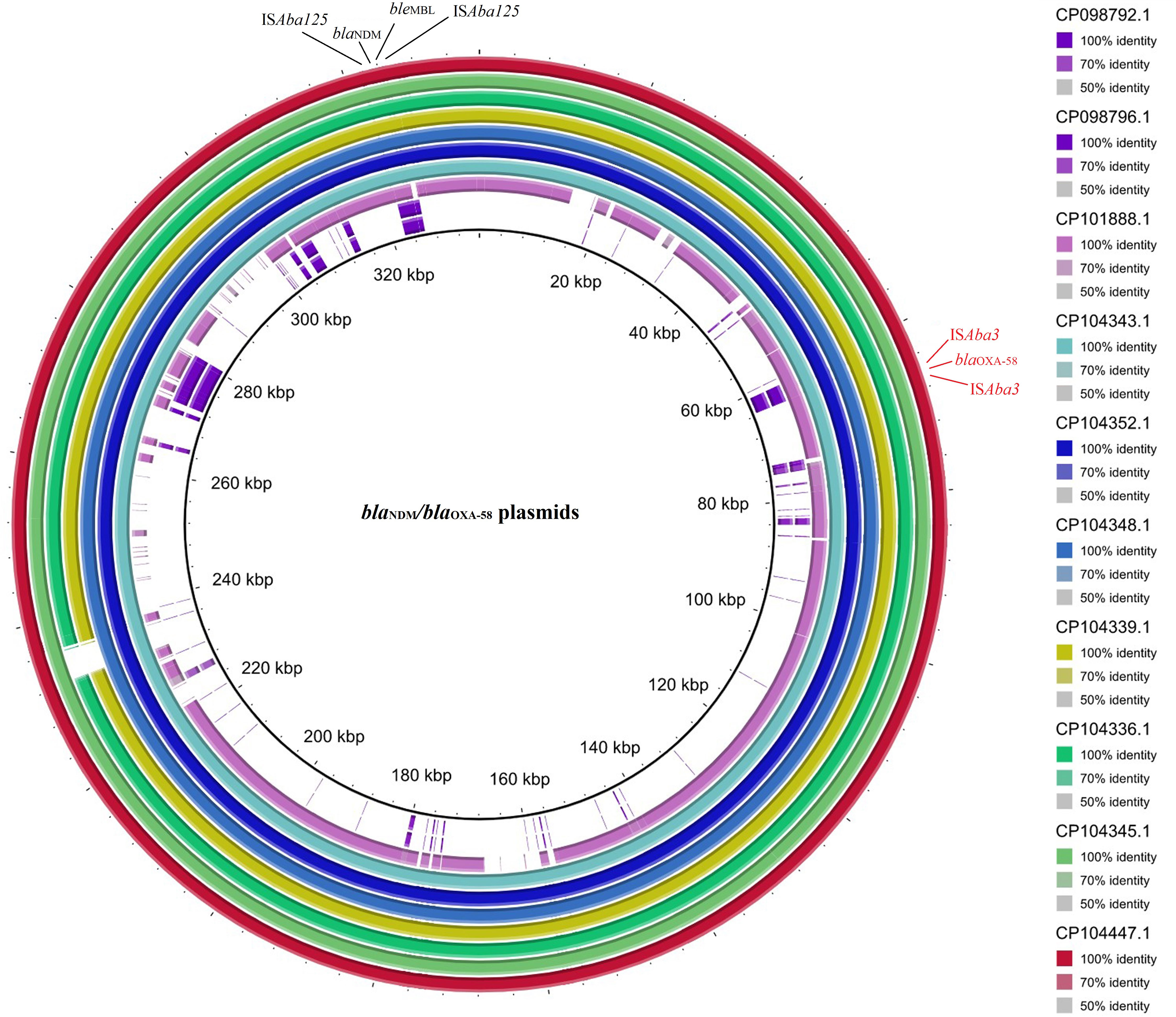
Figure 5 The multiple circular sequence alignment and genetic environment of plasmids harboring blaNDM-1/blaOXA-58 in A. baumannii. Inner ring: size of plasmids with co-occurrence of blaNDM-1 and blaOXA-58, and outer ring: the reference strain. Color spectrum: identity percent of the plasmids with the reference strain.
Despite their homogeneity, certain regions of the plasmids displayed heterogeneity. For example, plasmid pccbh31258 (CP101888.1), featured unique genes such as camA and cydA, and plasmid p1VB280820 (CP098792.1), had rep and sasA genes. Additionally, plasmids unnamed4 (CP104336.1) and unnamed2 (CP104345.1) were identified the unique genes noc and recD2, respectively.
3.10 Clonal relatedness of A. baumannii strains carrying carbapenemase genes
The clonal relatedness of A. baumannii strains carrying carbapenemase genes can be classified as follows. Most blaOXA-58-like and blaOXA-24/40-like genes were associated with ST2pas. The blaNDM gene were predominantly associated with ST622pas. In contrast, the blaIMP, blaKPC, and blaOXA-23-like genes were primarily correlated with ST1547pas, ST250pas, ST156pas, and ST729pas, respectively. Some STs were linked to only one specific carbapenemase gene, such as ST1547pas with blaIMP, ST250pas with blaKPC, ST156pas,or ST729pas with blaOXA-23-like. Additional STs, including ST2pas, ST422pas, ST1pas, ST85pas, ST1786pas, ST23pas, ST10pas, ST412pas, ST374pas, ST1554pas, ST138pas, and ST126pas, displayed a multi-harboring pattern with carbapenemase genes, including blaNDM, blaOXA-58-like, blaOXA-23-like, blaOXA-24/40-like, and blaGES. In a separate analysis of the A. baumannii chromosomes, thirty-eight non-redundant chromosomes were identified, revealing 15 different STs. Notably, the co-occurrence of the blaNDM and blaOXA-23-like genes was predominantly associated with ST622pas. In contrast, chromosomes carrying blaOXA-58-like genes were exclusively linked to ST2pas. Moreover, chromosomes associated with ST103pas, ST85pas, and ST250pas carried various carbapenemase genes, including blaNDM-2, blaNDM-6, and blaKPC-3, respectively. Chromosomes harboring blaOXA-23-like genes were exclusively identified in association with ST156pas (Figure 6).
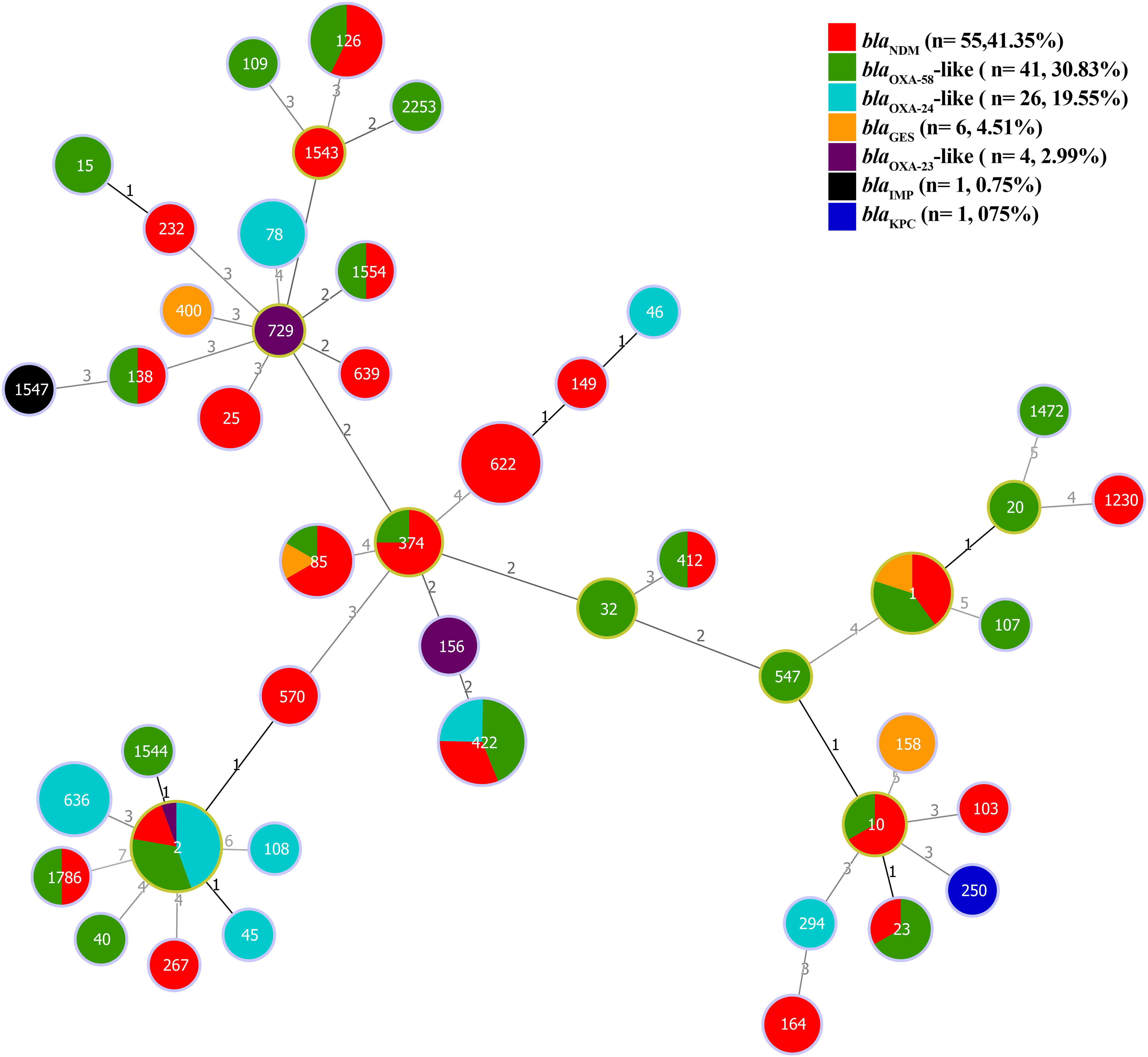
Figure 6 The minimum spanning tree (MST) of STs carrying plasmids containing carbapenemase genes (blaNDM, blaKPC, blaIMP, blaGES, blaOXA-58-like, blaOXA-24/40-like, and blaOXA-23-like) with a similarity cut-off > 4 allelic types based on multi-locus sequence typing (MLST) scheme. The numbers on the lines indicate allelic differences between the two separate sequence types. Red: blaNDM, Green: blaOXA-58-like, Blue: blaOXA-24-like, Orange: blaGES, Purple: blaOXA-23-like, Black: blaIMP, and Blue Navy: blaKPC genes.
4 Discussion
Carbapenem resistant A. baumannii poses a significant challenge because it is a leading cause of community-acquired and nosocomial infections, contributing to elevated morbidity and mortality rates (Mirzaei et al., 2020). Carbapenems are last-line antibiotics against these infections. However, MDR and XDR A. baumannii isolates complicate treatment (Katip et al., 2022).
The presence of carbapenemase genes, including blaNDM, blaKPC, blaVIM, blaIMP, blaGES, blaOXA-58-like, blaOXA-24/40-like, and blaOXA-23-like, could be a public health concern. In addition, carbapenemase genes on conjugative plasmids contribute to higher dissemination rates of these genes between bacteria (Benson et al., 2017). In the current study, bioinformatic tools were used to present more information on the genetic characteristics of A. baumannii plasmids and chromosomes harboring carbapenemase genes.
According to the results of this study, blaNDM and blaOXA-58 were the most prevalent carbapenemase genes in the plasmids and chromosomes. Consistent with the current study, Monnheimer et al. showed that the most abundant carbapenemases in A. baumannii were belonged to these genes (Monnheimer et al., 2021).
Our study revealed significant findings regarding the co-existence of various aminoglycoside resistance genes, such as aac(3)-Iid, aac(6’)-Ib4, aph(3’)-Ia, aadA2, ant(2’’)-Ia, aac(6’)-Ib1, aph(6)-Id, aph(3’)-Via, aph(3’’)-Ib, aph(3’’)-Ib, aph(6)-Id, aac(6’)-Ib9, aac(3)-IId, aph(3’)-Ia, aac(3)-Iic, ant(3’’)-Iic, aph(3’)-Vib, arr-2, aph(3’)-VI, aac(6’)-Ib9, and aac(3)-Iie with carbapenemases. Furthermore, Nowak et al. (2014) have previously reported similar observations regarding the co-existence of carbapenemases and aminoglycoside resistance genes (Nowak et al., 2014).
Any action to combat the spread of antibiotic resistance genes requires the identification of the potential sources and genetic environment of these genes. The study of MGEs associated with antibiotic resistance genes could provide valuable epidemiological information to identify potential sources. The genetic environment of the carbapenemase genes is consistent with the results of many studies conducted on this topic. Among the MBLs, blaKPC and blaNDM were associated with Tn4401/non-Tn4401 elements and ISAba125/Tn125, respectively. Whereas, the blaVIM and blaIMP were associated with class 1 integron (Reyes et al., 2020; Wang et al., 2023). Nguye et al. showed that the blaOXA-58 is located in an ISAba3-blaOXA-58-ISAba3-like structure (Nguyen et al., 2020).
In parallel with our study findings, Lasarte-Monterrubio et al. revealed that the blaOXA-24/40-like was associated with the XerC/XerD-like binding site, while blaOXA-23 was flanked by ISAba (Lasarte-Monterrubio et al., 2022). Transposons and integrons, found in different plasmids and bacterial clones, are indicative of their transmissibility and mobilization within various genetic elements (Reyes et al., 2020). MGEs, including conjugative plasmids, transposons, integrons, and bacteriophages, act as carriers for the acquisition and transfer of antibiotic resistance genes. They play an important role in transferring resistance genes among bacteria (Nadella et al., 2022). In the current study, the majority of conjugative plasmids were linked to blaNDM, which may be the primary reason for their high prevalence co-existence with other antibiotic-resistance genes. In the present study, the most prevalent replicon types were R3-T1 and R3-T2. Previous studies have shown that blaNDM-1 in A. baumannii isolates may have chromosomal or plasmid origin (Sánchez-Urtaza et al., 2023). According to the results of current study, Chen et al., demonstrated the co-existence of blaOXA-58-like with blaNDM, blaOXA-58-like with blaIMP, and blaGES with blaOXA-23-like. This phenomenon is related to the fact that these genes are located on the same conjugated plasmid (Chen et al., 2019). In a recent study, blaOXA-24/40-like genes were not found to co-occur with other carbapenemase genes. Moreover, CHCs analysis revealed the co-occurrence of blaNDM-1, blaOXA-23, and blaOXA-58. This finding was consistent with the results of Ramoul et al. (2016). Additionally, we identified the co-occurrence of blaOXA-565 and blaOXA-10.
Carbapenemase gene repetition is a significant evolutionary process that affects environmental adaptation of bacteria. Repetition of antimicrobial genes is a prevalent metabolic factor that gradually changes during evolution, and plays an important role in antimicrobial resistance. Specifically, we found carbapenemase gene repetitions in blaOXA-24 and blaOXA-58, which were located on both the plasmids and chromosomes. These duplications might have contributed to the development of carbapenems heteroresistance.
MST results revealed that the predominant STs associated with various carbapenemase genes were ST622pas for blaNDM, ST2pas for blaOXA-58-like, and blaOXA-24/40-like, ST1547pas for blaIMP, ST250pas for blaKPC, ST156pas for blaOXA-23-like, and ST158pas for blaGES. In addition, there were several other multi-harbor carbapenemase STs, including ST422pas, ST1pas, ST85pas, ST1786pas, ST23pas, ST10pas, ST412pas, ST374pas, ST1554pas, ST138pas, and ST126pas. These multi-harbor STs were associated with plasmids containing the major carbapenemase genes, including blaNDM, blaOXA-58-like, blaOXA-23-like, blaOXA-24/40-like, and blaGES.
In the current study, ST2Pas, ST1Pas, ST422Pas, ST622Pas, and ST85Pas were the most prevalent sequence types among the A. baumannii isolates. This finding is consistent with the finding of Khuntayaporn et al., study indicated that ST2 is the predominant ST in Thailand (Khuntayaporn et al., 2021). Several studies have consistently shown that the majority of CRAB isolates are associated with the international clone ST2, which has been reported in various studies in Mediterranean countries According to reports from various countries, the clonal diversity of A. baumannii, according to the Pasteur scheme of MLST, shows that ST2pas, ST1pas, and ST3pas are the predominant carbapenem-resistant clones worldwide. In ST studies, ST2 was the most dominant (Abhari et al., 2019; Khuntayaporn et al., 2021). Between 1999 and 2009, a study conducted in four Mediterranean countries (Greece, Italy, Lebanon, and Turkey) revealed that A. baumannii outbreaks were predominantly driven by the dissemination of ST2, with fewer contributions to ST1, ST25, ST78, and ST20 (Cherubini et al., 2022). These clones were found to carry blaOXA-58, blaOXA-23, and blaOXA-72 (Nawfal Dagher et al., 2019; Li et al., 2023). In Greece, ST2 is the most common clone circulating in hospitals (Pournaras et al., 2017). In addition, the international clone ST2 is widely distributed in Lebanon (Nawfal Dagher et al., 2019). In a study by Thadtapong et al., ST2 was most frequently identified among colistin- and carbapenem resistant A. baumannii isolates. This observation aligns with the findings of the current study, indicating a consistent prevalence of ST2pas in these isolates (Thadtapong et al., 2021). Khorshid et al. investigated the prevalence of various STs associated with genes encoding aminoglycoside-modifying enzymes. Their findings revealed that ST2 was the most prevalent among these STs. Remarkably, ST2, identified as an international clone, exhibits substantial genetic capacity for acquiring antimicrobial resistance genes within its genome (Khurshid et al., 2020).
However, an increased resistance to carbapenems has been reported worldwide. MBLs and CHDL-producing A. baumannii isolates, which are responsible for outbreaks, have been reported in different regions worldwide (Reyes et al., 2023). Although OXA-like enzymes weakly hydrolyze carbapenems, they can confer high resistance to carbapenems when associated with ISAba1 and ISAba125 (Li et al., 2019). MBLs and CHDLs located on MGEs spread rapidly to clonal lineages of A. baumannii (Shropshire et al., 2022).
Conjugative plasmids are involved in the rapid spread of CRAB (Chen et al., 2017). Therefore, these strains carrying plasmids and chromosomes harboring different carbapenemases and other antimicrobial resistant genes could pose a major threat to the healthcare system. Therefore, genetic characterization of these plasmids and chromosomes plays an important role in the control of bacteria carrying carbapenemases.
This study provides insights into the genetic structure of plasmids and chromosomes harboring major carbapenemase genes in CRAB. Nevertheless, the study has several limitations. First, the initial dataset and completeness relied on GenBank submissions and annotations. Furthermore, sampling bias exists because the plasmids and isolates studied may not encompass the full spectrum of CRAB strains and carbapenemase gene variations worldwide. In summary, the limitations of this study stem from data source dependence and sampling bias, highlighting the need for caution when interpreting the findings.
5 Conclusion
Characterization of the genetic structures revealed that carbapenemase genes appear not only in plasmids but also in the chromosomes of CRAB. The co-existence of plasmids encoding carbapenemases with other antibiotic resistance genes, co-occurrence, and gene repetition of carbapenemases in plasmids and chromosomes were notable findings. Conjugative plasmids containing blaNDM-1 and blaOXA-58 pose a threat to the expansion of carbapenem resistance. On the other hand, plasmids harboring blaNDM are widespread. A. baumannii employ different genetic strategies such as gene repetition and various genetic elements (transposons, integrons, and insertion sequences) to develop efficient resistance against carbapenems. Gene repetition, association of resistance gene cassettes with mobile genetic elements, acquisition of conjugative plasmids, high capacity to acquire carbapenemase genes on plasmids and chromosomes, and expansion of carbapenemases through successful international clones (ST2Pas, ST1Pas, ST422Pas, ST622Pas, and ST85Pas) play major roles in the development of resistance to carbapenems in A. baumannii worldwide. The high consumption of antibiotics in clinical settings exacerbates antimicrobial resistance worldwide. Therefore, a global campaign is necessary to combat against CRAB infection.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/genbank/.
Author contributions
FB: Conceptualization, Data curation, Formal Analysis, Supervision, Writing – review & editing, Project administration, Software, Validation. MB: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. HS: Software, Validation, Writing – review & editing. VN: Methodology, Software, Writing – review & editing. MS: Methodology, Software, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank the personnel of the Bacteriology Department of the Pasteur Institute of Iran (IPI) for their assistance. This research is part of Ph.D. thesis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abhari, S. S., Badmasti, F., Modiri, L., Aslani, M. M., Asmar, M. (2019). Circulation of imipenem-resistant Acinetobacter baumannii ST10, ST2 and ST3 in a university teaching hospital from Tehran, Iran. J. Med. Microbiol. 68 (6), 860–865. doi: 10.1099/jmm.0.000987
Alrahmany, D., Omar, A. F., Alreesi, A., Harb, G., Ghazi, I. M. (2022). Acinetobacter baumannii infection-related mortality in hospitalized patients: risk factors and potential targets for clinical and antimicrobial stewardship interventions. Antibiotics (Basel) 11 (8), 1086–1096. doi: 10.3390/antibiotics11081086
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2017). GenBank. Nucleic Acids Res. 45 (D1), D37–D42. doi: 10.1093/nar/gkw1070
Chen, Y., Gao, J., Zhang, H., Ying, C. (2017). Spread of the bla(OXA-23)-Containing Tn2008 in Carbapenem-Resistant Acinetobacter baumannii Isolates Grouped in CC92 from China. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00163
Chen, Y., Guo, P., Huang, H., Huang, Y., Wu, Z., Liao, K. (2019). Detection of co-harboring OXA-58 and NDM-1 carbapenemase producing genes resided on a same plasmid from an Acinetobacter pittii clinical isolate in China. Iran J. Basic Med. Sci. 22 (1), 106–111. doi: 10.22038/ijbms.2018.28934.6994
Cherubini, S., Perilli, M., Segatore, B., Fazii, P., Parruti, G., Frattari, A., et al. (2022). Whole-genome sequencing of ST2 A. baumannii causing bloodstream infections in COVID-19 patients. Antibiotics (Basel) 11 (7), 955–965. doi: 10.3390/antibiotics11070955
Flores-Paredes, W., Luque, N., Albornoz, R., Rojas, N., Espinoza, M., Pons, M. J., et al. (2021). Evolution of antimicrobial resistance levels of ESKAPE microorganisms in a Peruvian IV-level hospital. Infect. Chemother. 53 (3), 449–462. doi: 10.3947/ic.2021.0015
Frenk, S., Temkin, E., Lurie-Weinberger, M. N., Keren-Paz, A., Rov, R., Rakovitsky, N., et al. (2022). Large-scale WGS of carbapenem-resistant Acinetobacter baumannii isolates reveals patterns of dissemination of ST clades associated with antibiotic resistance. J. Antimicrob. Chemother. 77 (4), 934–943. doi: 10.1093/jac/dkac010
Horne, T., Orr, V. T., Hall, J. P. (2023). How do interactions between mobile genetic elements affect horizontal gene transfer? Curr. Opin. Microbiol. 73, 102282. doi: 10.1016/j.mib.2023.102282
Hsieh, Y. C., Wang, S. H., Chen, Y. Y., Lin, T. L., Shie, S. S., Huang, C. T., et al. (2020). Association of capsular types with carbapenem resistance, disease severity, and mortality in Acinetobacter baumannii. Emerg. Microbes Infect. 9 (1), 2094–2104. doi: 10.1080/22221751.2020.1822757
Islam, M. M., Kim, J., Kim, K., Kim, D., Akter, S., Bang, S., et al. (2023). Complete genome sequence of the multidrug-resistant clinical isolate Acinetobacter baumannii KBN10P05679: Insights into antimicrobial resistance genotype and potential virulence traits. J. Glob Antimicrob. Resist. 33, 78–82. doi: 10.1016/j.jgar.2023.02.016
Katip, W., Oberdorfer, P., Kasatpibal, N. (2022). Effectiveness and Nephrotoxicity of Loading Dose Colistin-Meropenem versus Loading Dose Colistin-Imipenem in the Treatment of Carbapenem-Resistant Acinetobacter baumannii Infection. Pharmaceutics. 14 (6), 1266. doi: 10.3390/pharmaceutics14061266
Khuntayaporn, P., Kanathum, P., Houngsaitong, J., Montakantikul, P., Thirapanmethee, K., Chomnawang, M. T. (2021). Predominance of international clone 2 multidrug-resistant Acinetobacter baumannii clinical isolates in Thailand: a nationwide study. Ann. Clin. Microbiol. Antimicrob. 20 (1), 19. doi: 10.1186/s12941-021-00424-z
Khurshid, M., Rasool, M. H., Ashfaq, U. A., Aslam, B., Waseem, M., Ali, M. A., et al. (2020). Acinetobacter baumannii sequence types harboring genes encoding aminoglycoside modifying enzymes and 16SrRNA methylase; a multicenter study from Pakistan. Infect. Drug Resist. 13, 2855–2862. doi: 10.2147/IDR.S260643
Kyriakidis, I., Vasileiou, E., Pana, Z. D., Tragiannidis, A. (2021). Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10 (3), 373–404. doi: 10.3390/pathogens10030373
Lam, M. M. C., Koong, J., Holt, K. E., Hall, R. M., Hamidian, M. (2023). Detection and Typing of Plasmids in Acinetobacter baumannii Using rep Genes Encoding Replication Initiation Proteins. Microbiol. Spectr. 11 (1), e0247822. doi: 10.1128/spectrum.02478-22
Lasarte-Monterrubio, C., Guijarro-Sanchez, P., Belles, A., Vazquez-Ucha, J. C., Arca-Suarez, J., Fernandez-Lozano, C., et al. (2022). Carbapenem Resistance in Acinetobacter nosocomialis and Acinetobacter junii Conferred by Acquisition of bla(OXA-24/40) and Genetic Characterization of the Transmission Mechanism between Acinetobacter Genomic Species. Microbiol. Spectr. 10 (1), e0273421. doi: 10.1128/spectrum.02734-21
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W2W6. doi: 10.1093/nar/gkab301
Li, S., Duan, X., Peng, Y., Rui, Y. (2019). Molecular characteristics of carbapenem-resistant Acinetobacter spp. from clinical infection samples and fecal survey samples in Southern China. BMC Infect. Dis. 19 (1), 900. doi: 10.1186/s12879-019-4423-3
Li, J., Li, Y., Cao, X., Zheng, J., Zhang, Y., Xie, H., et al. (2023). Genome-wide identification and oxacillinase OXA distribution characteristics of Acinetobacter spp. based on a global database. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1174200
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46 (W1), W229–WW34. doi: 10.1093/nar/gky352
Mirzaei, B., Bazgir, Z. N., Goli, H. R., Iranpour, F., Mohammadi, F., Babaei, R. (2020). Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res. Notes. 13 (1), 380. doi: 10.1186/s13104-020-05224-w
Monnheimer, M., Cooper, P., Amegbletor, H. K., Pellio, T., Gross, U., Pfeifer, Y., et al. (2021). High prevalence of carbapenemase-producing acinetobacter baumannii in wound infections, Ghana, 2017/2018. Microorganisms. 9 (3), 537. doi: 10.3390/microorganisms9030537
Naas, T., Oueslati, S., Bonnin, R. A., Dabos, M. L., Zavala, A., Dortet, L., et al. (2017). Beta-lactamase database (BLDB) - structure and function. J. Enzyme Inhib Med. Chem. 32 (1), 917–919. doi: 10.1080/14756366.2017.1344235
Nadella, R. K., Panda, S. K., Badireddy, M. R., Kurcheti, P. P., Raman, R. P., Mothadaka, M. P. (2022). Multi-drug resistance, integron and transposon-mediated gene transfer in heterotrophic bacteria from Penaeus vannamei and its culture environment. Environ. Sci. pollut. Res. Int. 29 (25), 37527–37542. doi: 10.1007/s11356-021-18163-1
Nascimento, M., Sousa, A., Ramirez, M., Francisco, A. P., Carrico, J. A., Vaz, C. (2017). PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33 (1), 128–129. doi: 10.1093/bioinformatics/btw582
Nawfal Dagher, T., Al-Bayssari, C., Chabou, S., Antar, N., Diene, S. M., Azar, E., et al. (2019). Investigation of multidrug-resistant ST2 Acinetobacter baumannii isolated from Saint George hospital in Lebanon. BMC Microbiol. 19 (1), 29. doi: 10.1186/s12866-019-1401-2
Nguyen, A. T., Pham, S. C., Ly, A. K., Nguyen, C. V. V., Vu, T. T., Ha, T. M. (2020). Overexpression of bla (OXA-58) Gene Driven by ISAba3 Is Associated with Imipenem Resistance in a Clinical Acinetobacter baumannii Isolate from Vietnam. BioMed. Res. Int. 2020, 7213429. doi: 10.1155/2020/7213429
Nocera, F. P., Attili, A. R., De Martino, L. (2021). Acinetobacter baumannii: its clinical significance in human and veterinary medicine. Pathogens 10 (2). doi: 10.3390/pathogens10020127
Nowak, P., Paluchowska, P. M., Budak, A. (2014). Co-occurrence of carbapenem and aminoglycoside resistance genes among multidrug-resistant clinical isolates of Acinetobacter baumannii from Cracow, Poland. Med. Sci. Monit Basic Res. 20, 9–14. doi: 10.12659/msmbr.889811
Ozer, E. A. (2018). ClustAGE: a tool for clustering and distribution analysis of bacterial accessory genomic elements. BMC Bioinf. 19 (1), 150. doi: 10.1186/s12859-018-2154-x
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 31 (22), 3691–3693. doi: 10.1093/bioinformatics/btv421
Pournaras, S., Dafopoulou, K., Del Franco, M., Zarkotou, O., Dimitroulia, E., Protonotariou, E., et al. (2017). Predominance of international clone 2 OXA-23-producing-Acinetobacter baumannii clinical isolates in Greece, 2015: results of a nationwide study. Int. J. Antimicrob. Agents. 49 (6), 749–753. doi: 10.1016/j.ijantimicag.2017.01.028
Ramoul, A., Loucif, L., Bakour, S., Amiri, S., Dekhil, M., Rolain, J. M. (2016). Co-occurrence of blaNDM-1 with blaOXA-23 or blaOXA-58 in clinical multidrug-resistant Acinetobacter baumannii isolates in Algeria. J. Glob Antimicrob. Resist. 6, 136–141. doi: 10.1016/j.jgar.2016.05.003
Reyes, J., Komarow, L., Chen, L., Ge, L., Hanson, B. M., Cober, E., et al. (2023). Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 4 (3), e159–ee70. doi: 10.1016/S2666-5247(22)00329-9
Reyes, J. A., Melano, R., Cárdenas, P. A., Trueba, G. (2020). Mobile genetic elements associated with carbapenemase genes in South American Enterobacterales. Braz. J. Infect. Dis. 24 (3), 231–238. doi: 10.1016/j.bjid.2020.03.002
Sánchez-Urtaza, S., Ocampo-Sosa, A., Molins-Bengoetxea, A., Rodríguez-Grande, J., El-Kholy, M. A., Hernandez, M., et al. (2023). Co-Existence of blaNDM-1, blaOXA-23, blaOXA-64, blaPER-7 and blaADC-57 in a Clinical Isolate of Acinetobacter baumannii from Alexandria, Egypt. Int. J. Mol. Sci. 24 (15), 12515. doi: 10.3390/ijms241512515
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30 (14), 2068–2069. doi: 10.1093/bioinformatics/btu153
Shropshire, W. C., Konovalova, A., McDaneld, P., Gohel, M., Strope, B., Sahasrabhojane, P., et al. (2022). Systematic analysis of mobile genetic elements mediating beta-lactamase gene amplification in noncarbapenemase-producing carbapenem-resistant enterobacterales bloodstream infections. mSystems. 7 (5), e0047622. doi: 10.1128/msystems.00476-22
Thadtapong, N., Chaturongakul, S., Soodvilai, S., Dubbs, P. (2021). Colistin and carbapenem-resistant acinetobacter baumannii aci46 in Thailand: genome analysis and antibiotic resistance profiling. Antibiotics (Basel). 10 (9), 1054. doi: 10.3390/antibiotics10091054
Wang, T., Zhu, Y., Zhu, W., Cao, M., Wei, Q. (2023). Molecular characterization of class 1 integrons in carbapenem-resistant Enterobacterales isolates. Microb. Pathog. 177, 106051. doi: 10.1016/j.micpath.2023.106051
Wasfi, R., Rasslan, F., Hassan, S. S., Ashour, H. M., Abd El-Rahman, O. A. (2021). Co-existence of carbapenemase-encoding genes in acinetobacter baumannii from cancer patients. Infect. Dis. Ther. 10 (1), 291–305. doi: 10.1007/s40121-020-00369-4
Xu, J., Guo, H., Li, L., He, F. (2023). Molecular epidemiology and genomic insights into the transmission of carbapenem-resistant NDM-producing Escherichia coli. Comput. Struct. Biotechnol. J. 21, 847–855. doi: 10.1016/j.csbj.2023.01.004
Yao, M., Zhu, Q., Zou, J., Shenkutie, A. M., Hu, S., Qu, J., et al. (2022). Genomic Characterization of a Uropathogenic Escherichia coli ST405 Isolate Harboring bla (CTX-M-15)-Encoding IncFIA-FIB Plasmid, bla (CTX-M-24)-Encoding IncI1 Plasmid, and Phage-Like Plasmid. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.845045
Zalegh, I., Chaoui, L., Maaloum, F., Zerouali, K., Mhand, R. A. (2023). Prevalence of multidrug-resistant and extensively drug-resistant phenotypes of Gram-negative bacilli isolated in clinical specimens at Centre Hospitalo-Universitaire Ibn Rochd. Morocco. Pan Afr Med. J. 45, 41. doi: 10.11604/pamj.2023.45.41.34457
Keywords: Acinetobacter baumannii, carbapenem resistant, carbapenemase, gene repetition, sequence type
Citation: Beig M, Badmasti F, Solgi H, Nikbin VS and Sholeh M (2023) Carbapenemase genes distribution in clonal lineages of Acinetobacter baumannii: a comprehensive study on plasmids and chromosomes. Front. Cell. Infect. Microbiol. 13:1283583. doi: 10.3389/fcimb.2023.1283583
Received: 26 August 2023; Accepted: 14 November 2023;
Published: 01 December 2023.
Edited by:
Haiyang Liu, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyMatej Medvecky, University of Warwick, United Kingdom
Copyright © 2023 Beig, Badmasti, Solgi, Nikbin and Sholeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Farzad Badmasti, fbadmasti2008@gmail.com
 Masoumeh Beig
Masoumeh Beig Farzad Badmasti
Farzad Badmasti Hamid Solgi
Hamid Solgi Vajihe Sadat Nikbin
Vajihe Sadat Nikbin Mohammad Sholeh
Mohammad Sholeh