Transcriptomic identification of genes expressed in invasive S. aureus diabetic foot ulcer infection
- 1Department of Orthopedics and Rehabilitation, Yale School of Medicine, New Haven, CT, United States
- 2Division of Life Science, Gyeongsang National University, Jinju, Republic of Korea
- 3Yale Center for Genome Analysis, Department of Genetics, Yale School of Medicine, New Haven, CT, United States
Introduction: Infection in diabetic foot ulcers (DFUs) is one of the major complications associated with patients with diabetes. Staphylococcus aureus is the most common offending pathogen in patients with infected DFU. Previous studies have suggested the application of species-specific antibodies against S. aureus for diagnosis and monitoring treatment response. Early and accurate identification of the main pathogen is critical for management of DFU infection. Understanding the host immune response against species-specific infection may facilitate diagnosis and may suggest potential intervention options to promote healing infected DFUs. We sought to investigate evolving host transcriptome associated with surgical treatment of S. aureus– infected DFU.
Methods: This study compared the transcriptome profile of 21 patients with S. aureus– infected DFU who underwent initial foot salvage therapy with irrigation and debridement followed by intravenous antibiotic therapy. Blood samples were collected at the recruitment (0 weeks) and 8 weeks after therapy to isolate peripheral blood mononuclear cells (PBMCs). We analyzed the PBMC expression of transcriptomes at two different time points (0 versus 8 weeks). Subjects were further divided into two groups at 8 weeks: healed (n = 17, 80.95%) versus non-healed (n = 4, 19.05%) based on the wound healing status. DESeq2 differential gene analysis was performed.
Results and discussion: An increased expression of IGHG1, IGHG2, IGHG3, IGLV3-21, and IGLV6-57 was noted during active infection at 0 weeks compared with that at 8 weeks. Lysine- and arginine-rich histones (HIST1H2AJ, HIST1H2AL, HIST1H2BM, HIST1H3B, and HIST1H3G) were upregulated at the initial phase of active infection at 0 weeks. CD177 and RRM2 were also upregulated at the initial phase of active infection (0 weeks) compared with that at 8 weeks of follow-up. Genes of heat shock protein members (HSPA1A, HSPE1, and HSP90B1) were high in not healed patients compared with that in healed patients 8 weeks after therapy. The outcome of our study suggests that the identification of genes evolution based on a transcriptomic profiling could be a useful tool for diagnosing infection and assessing severity and host immune response to therapies.
1 Introduction
Infection of diabetic foot ulcer (DFU) presents a significant social and economic burden to our society. Diabetes and its associated infection affect an estimated 9.3% of people in the United States, accounting for more than 20% of all hospitalization (Kim and Boye, 2009; Skrepnek et al., 2017; Schmidt, 2022; McDermott et al., 2023). They frequently result in prolonged hospitalization, poor clinical outcomes, readmission, and, in extreme cases, amputation (Herber et al., 2007; Phillips et al., 2016; Armstrong et al., 2017; Skrepnek et al., 2017; Smith-Strom et al., 2017; Polikandrioti et al., 2020; Raffetto et al., 2020; Zhao et al., 2020; Tai et al., 2021; Guo et al., 2023). Infection or re-current infection of DFU can have an impact on quality of life and present social challenges and a declined in psychological wellbeing (Phillips et al., 2016; Egan et al., 2020; Guo et al., 2023). Staphylococcus aureus is the most common pathogen (70%–80%) found in patients with DFU infection (Macdonald et al., 2020). The main challenge in treating musculoskeletal infection (MSKI) is the identification of main pathogen (Lipsky et al., 2012; Heravi et al., 2020; Lipof et al., 2021). False-negative or false-positive cultures in polymicrobial MSKI may result in inappropriate antibiotic coverage plan, subsequent recurrent infection, and loss of the limb. Although inflammatory markers, such as white blood cell count (WBC), erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP), have been utilized to diagnose and monitor treatment response, they are non-specific (Masters et al., 2019; Grondman et al., 2020; Lipof et al., 2021).
Previous investigations have demonstrated an increased host production of S. aureus– specific antibodies in patients with DFU. Specifically, antibodies against iron- regulated surface determinant (IsdB and IsdH) protein, recombinant staphylococcal complement inhibitor, and the recombinant clumping factors (ClfA and ClfB) were suggested as biomarker candidates to diagnose and monitor treatment response (Masters et al., 2019; Heravi et al., 2020; Choi et al., 2021; Hao et al., 2021; Lipof et al., 2021; Wang et al., 2021; Fritz et al., 2022). These endeavors to characterize humoral responses to S. aureus infection with the use of immunoassay techniques have shown some potential for clinical application, but there are certain limitations (Masters et al., 2019; Choi et al., 2021; Hao et al., 2021; Fritz et al., 2022). Because of the historical abundance of these antibodies produced by host’s previous exposure to infection, the elevated serum titers may not accurately reflect the state of the current infection.
Advancements in bioinformatics have suggested using RNA- seq technology to accelerate the knowledge on transcriptomic analysis. By profiling the transcriptome landscape of cells in heterogeneous tissues, it provides a deep insight into cell function and disease pathophysiology (Jiang et al., 2015). Transcriptome research focuses on mRNA and non-coding RNA acting as a regulator to numerous proteins and gene function/structure and reveals specific molecular mechanisms of disease occurrence (Jiang et al., 2015; Stubbington et al., 2017). The new technology paradigm provides a greater specificity for studying the immune response. The shift from the use of convectional culture-dependent methodologies and serum antibody levels to transcriptome analysis may help us better understand DFU infection, patient outcomes, and wound progression. Few research studies have recently elucidated the role of RNA-seq/transcriptomes in DFU (Theocharidis et al., 2020; Wang et al., 2020; Li Y, et al., 2022; Theocharidis et al., 2022; Sandoval-Schaefer et al., 2023). Transcriptome data analysis, for example, has been utilized to identify gene expression profiles, biological signal pathways, and biomarkers in various diseases such as cancer, tuberculosis, neuronal dysfunction, autoimmune diseases, and COVID-19 prognosis (Baine et al., 2011; Islam et al., 2017; Bellaver et al., 2019; Wang et al., 2019; Xiong et al., 2020; Zheng et al., 2020; Fang et al., 2021; Han et al., 2021; Ascoli et al., 2022; Ghosh et al., 2022; Maleknia et al., 2022; Mendelsohn et al., 2022; Zimmer et al., 2022). Theocharidis et al. recently reported the use of RNA-seq RNA-Seq to elucidate gene expression associated with wound healing in the single-cell transcriptome landscape of DFU in human and mice. The above study focuses on wound healing in DFU. However, in our study, we sought to investigate whether the transcriptome could be used to diagnose infected DFU. Our objective is to develop a more accurate transcriptome-based test specific for infection and its application for diagnosis and prognostication potential. Probing how host immune system responds to infection in DFU through the variability in gene expression will increase our knowledge in diagnosis and aid clinical decision in management of MSKI.
We aimed to analyze evolving human peripheral blood mononuclear cell (PBMC) transcriptome associated with treatment of clinically infected DFU to identify the genes involved in immunologic response to invasive S. aureus DFU infection. We sought to identify up- and downregulated genes among healed patients at the time of active infection (0 weeks) vs. 8 weeks after initiation of therapy. Secondary outcome was to investigate whether certain gene expression (singly or in conjunction with others) could serve as a prognostic indicator for resolution of infection and eventual wound healing.
2 Materials and methods
2.1 Ethics statement
This study was approved by the Yale Human Research Protection and Ethics Committee (Reference No. 2000030908). Informed consents were obtained from all patients enrolled.
2.2 Patient population and antibiotics regimen
Twenty-one patients with infected DFU whose bone culture obtained from the base of infected wound showed a growth of S. aureus formed the basis of the study. At the start of therapy, blood samples were taken to measure hemoglobin A1C, WBC, CRP, and ESR. All patients underwent initial foot salvage therapy with irrigation and debridement. Prior to surgery, intravenous (IV) antibiotics were withheld for all patients until intraoperative specimens were obtained for culture. Once obtained, a broad-spectrum IV antibiotic, such as piperacillin/tazobactam (Zosyn), was administered and continued until the culture results were available or until the regimen was changed by a consulting infectious disease (ID) specialist. For patients with a history of methicillin-resistant S. aureus infection, vancomycin was initiated instead. Histologic evaluation of resection margin was performed for all patients who underwent amputation or bony resection procedure. If a clear margin was confirmed, then patients were kept on oral antibiotics for additional 2 weeks after surgery to eliminate any residual infection in the soft tissue. However, if the resection margin was determined to be contaminated, then a culture-based targeted long-term (6 weeks) IV or oral antibiotics was administered as determined by the ID specialist. DFU infection and wound healing status were classified as healed vs. not healed based on wound status (size), clinical sign, and symptoms of infection (Wagner, 1981).
2.3 Sample collection
During wound debridement procedure, two pea-sized grossly infected bone samples were aseptically collected at the base of DFU (Oh et al., 2018; Choi et al., 2021). The samples were then separated into two sterile vials: One was transferred to the microbiology laboratory for standard culture, and the other vial was immediately stored in a −80°C. The DNA was extracted, and a quantitative polymerase chain reaction (qPCR) was performed with specific primers to determine the bacterial burden and antibiotic resistance genes.
2.4 Microbiological analysis to identify bacterial species
A method for microbial identification previously described (Harmsen et al., 2003; Oh et al., 2018; Choi et al., 2021) was adopted. Briefly, we did microbial culture as a standard practice and bacterial abundance was determined by real-time qPCR using bacterial 16S primers, probe, and cloned plasmid standards. The region of bacterial 16S rRNA from V1 to V3 was amplified from extracted DNA sample via dual-indexed coded primers and Phusion high-fidelity polymerase (Thermo Fisher, Inc.). With the use of SequalPrep normalization plates (Life Technologies), all PCR amplicons were purified, normalized, pooled, and validated on an Agilent bioanalyzer (2200 TapeStation, D1000 tape or HSD1000 tape). The final library was paired end and sequenced on an Illumina MiSeq (2 × 300 bp). In each amplification and sequencing run, a blank sample was run through the extraction, amplification, and sequencing steps as a negative control, whereas positive controls included bacterial genomic DNA from a variety of genera. We identified different bacteria species from the polymicrobial infection of the DFU such as S. aureus, Enterococcus faecalis, Proteus mirabilis, Streptococcus agalactiae, and Serratia marcescens from different patient’s samples. Because S. aureus alone accounted for about 70% of MSKI, we are unsure that only samples that were S. aureus positive were included in this research. To further confirm that S. aureus is present in all the selected infected samples, we genotyped bacterial cultures for the staphylococcal virulence gene spa, infected samples were classified on the basis of the outcome of the result as PCR + or PCR− for S. aureus.
2.5 PBMC isolation
Blood samples from patients with DFU infection were collected at the enrollment (0 weeks) and at 8 weeks after initiation of treatment. PBMCs were isolated by density gradient centrifugation. Blood was gentle mixed by inversion several times to ensure a homogeneous suspension. The whole blood was mixed with equal volume of Phosphate-buffered saline (PBS) in ratio 1:1, gently layered the mixed blood-PBS on top of the density gradient medium (lyphoprep), and centrifuged at 1,500 g for 25 min at room temperature. PBMCs were harvest by inserting the pipette directly through the upper plasma layer to the mononuclear cells at the interface. The PBMC aliquot was stored in at −80°C for further analysis.
2.6 RNA-seq and differential gene expression analysis of the PBMCs
Methods for RNA-seq and differential gene expression analysis described were adopted with slight modification (Pertea et al., 2015). Briefly, gene expression levels were quantified using StringTie v1.3.3b with gene models (v27) from the GENCODE project. PBMCs were lysed on ice for 5 min in 10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 0.2 mM Ethylene-diamine-tetra acetic acid (EDTA), and 0.05% NP-40, and nuclei were spun at 2,500g for 3 min before being resuspended in QIAzol for RNA isolation using a miRNeasy kit, as directed by the manufacturer (Qiagen). The RNA-seq library was constructed using the Illumina TruSeq RNA Sample Preparation Kit v2 and sequenced on one lane of the HiSeq 2000 platform (100 bp, paired-end). Low- quality reads were trimmed, and adaptor contamination was removed using Trim Galore (v0.5.0). Trimmed reads were mapped to the human reference genome (hg38) using HISAT2 v2.1.0 and differentially expressed genes were identified using DESeq2 v 1.22.1 (Love et al., 2014; Pertea et al., 2015; Kim et al., 2019).
2.7 Statistical and ingenuity pathway analysis
Human PBMC Differentially Expressed (DE) gene lists were filtered using an Log2 Fold Change (LFC) cutoff to measure the statistical dependence between two variables (0 and 8 weeks) for which a value of ≥ 1 indicates a positive correlation and ≤ −1 indicates a negative correlation. Positively and negatively correlated genes that were statistically significant with reference to ≤ 0.05 False Discovery Rate (FDR) cut off value were uploaded into Ingenuity Pathway Analysis (IPA) (Qiagen), (https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-andvisualization/qiagen-ipa/) (Kramer et al., 2014). IPA software used Fisher’s exact binomial confidence interval test to detect statistically significant up- or downregulated genes for healed samples at 0 weeks before initiation of antibiotic vs. 8 weeks of follow-up after therapy and in healed vs. not healed at 8 weeks after antibiotic therapy.
2.8 Data availability
Transcriptomics of the RNA-seq data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) and can be accessed using GEO accession number GSE230426.
3 Results
3.1 Correlation of clinical variables
We analyzed 42 samples from 21 (n = 21) patients with S. aureus– infected DFU who underwent surgical debridement/treatment. Infection status was clinically determined on the basis of the assessment of 12 signs and symptoms (Gardner et al., 2013). Wound healing status (size), presence of infection such as purulent exudate, erythema, pain, or offensive odor was documented at 0 and 8 weeks after initiation of therapy. In this study, n = 17 (80.95%) patients clinically healed the infection at 8 weeks of follow-up, whereas n = 4 (19.05%) patients failed to heal. Those patients who failed to heal underwent repeat debridement or amputation (Figure 1). Analysis of our results was based on the comparison between 0 and 8 weeks among patients that were healed (n = 17) and between healers (n = 17) and non-healers (n = 3) at 8 weeks after antibiotic therapy. One sample (n = 1) was contaminated and was excluded from further analysis.
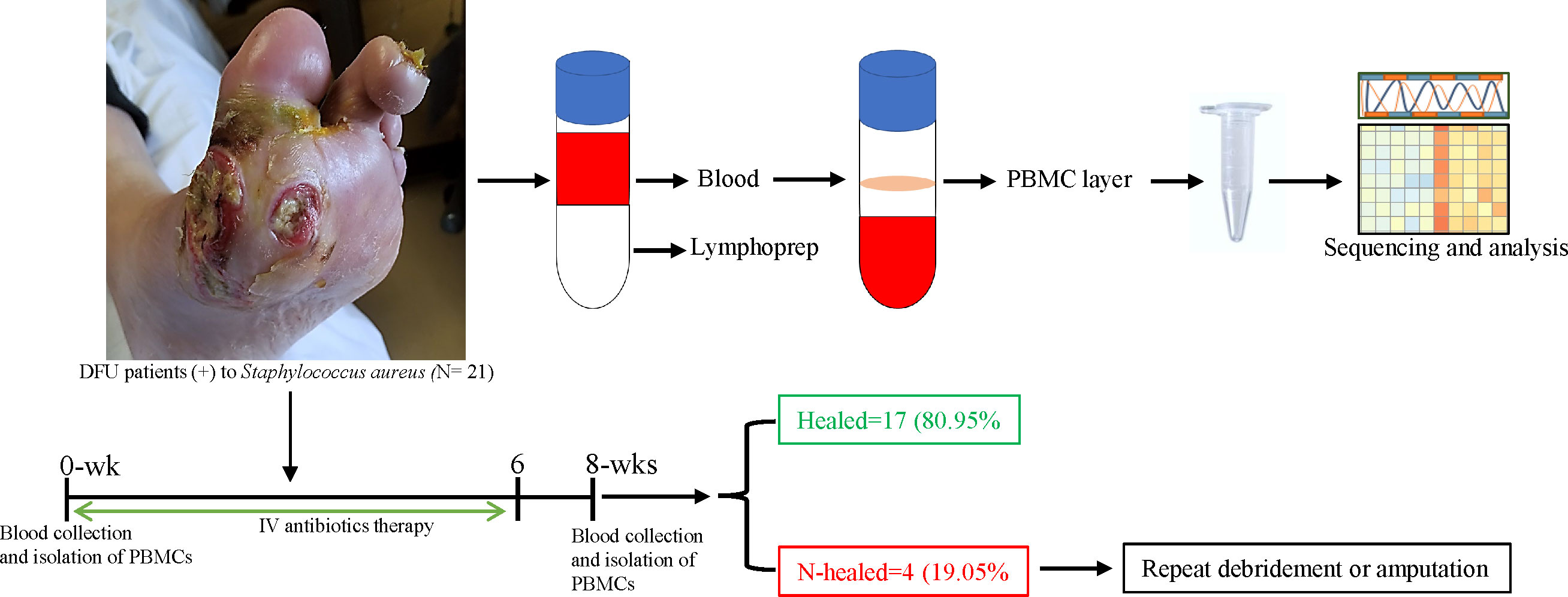
Figure 1 Schematic overview of the study design. Whole -blood sample was collected from patients with S. aureus– infected DFU before initiation of antibiotics (0 weeks) and after therapy (8 weeks) to isolate PBMC. Blood samples were mix with PBS (1:1), lymphoprep density gradients was used to separate PBMC at the interphase, and PBMC samples were harvested and used for RNA and transcriptome analysis. After 8 weeks of follow -up, subjects were divided into two groups: healed (n = 17) versus non-healed (n = 3) based on infection symptoms and the wound healing status; non-healers were either debrided again or undergo amputation.
3.2 Immunoglobulin heavy G chain upregulated during active infection
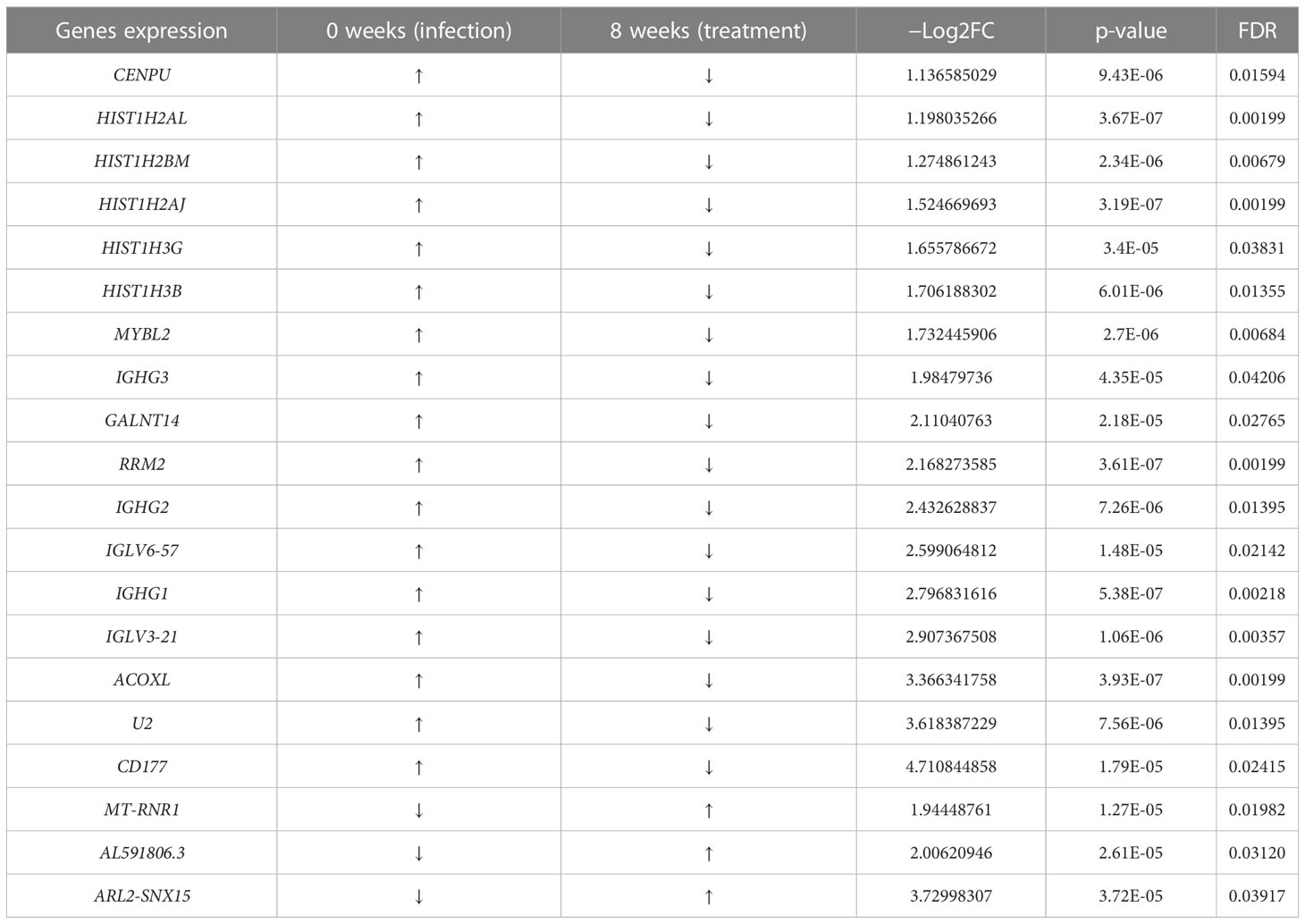
Immunoglobulins are glycoproteins that are exclusively produced by activated B lymphocytes and plasma cells and mediate the humoral response to pathogens. We identified differences in PBMC mRNA gene expression/pattern as demonstrated in the RNA sequence heatmap and volcano plot (Figure 2). The analysis showed a significant variation in the transcriptome profile observed among the members of immunoglobulin gamma heavy chain (IGHG1, IGHG2, and IGHG3). Among the groups that were healed of the infection after antibiotic therapy (n = 17), we observed a higher expression of IGHG1, IGHG2, and IGHG3 in the active phase of infection at 0 week, and the gene expression of these immunoglobulins was downregulated at 8 weeks of follow-up (p = 5.38 × 10−7, 7.26 × 10−6, and 4.35 × 10−5 with corresponding FDR values of 0.002183, 0.013948, and 0.042061, respectively). These findings indicate a significant association between evolving expression of these immunoglobulins and resolution of infection (Figure 3A; Table 1).
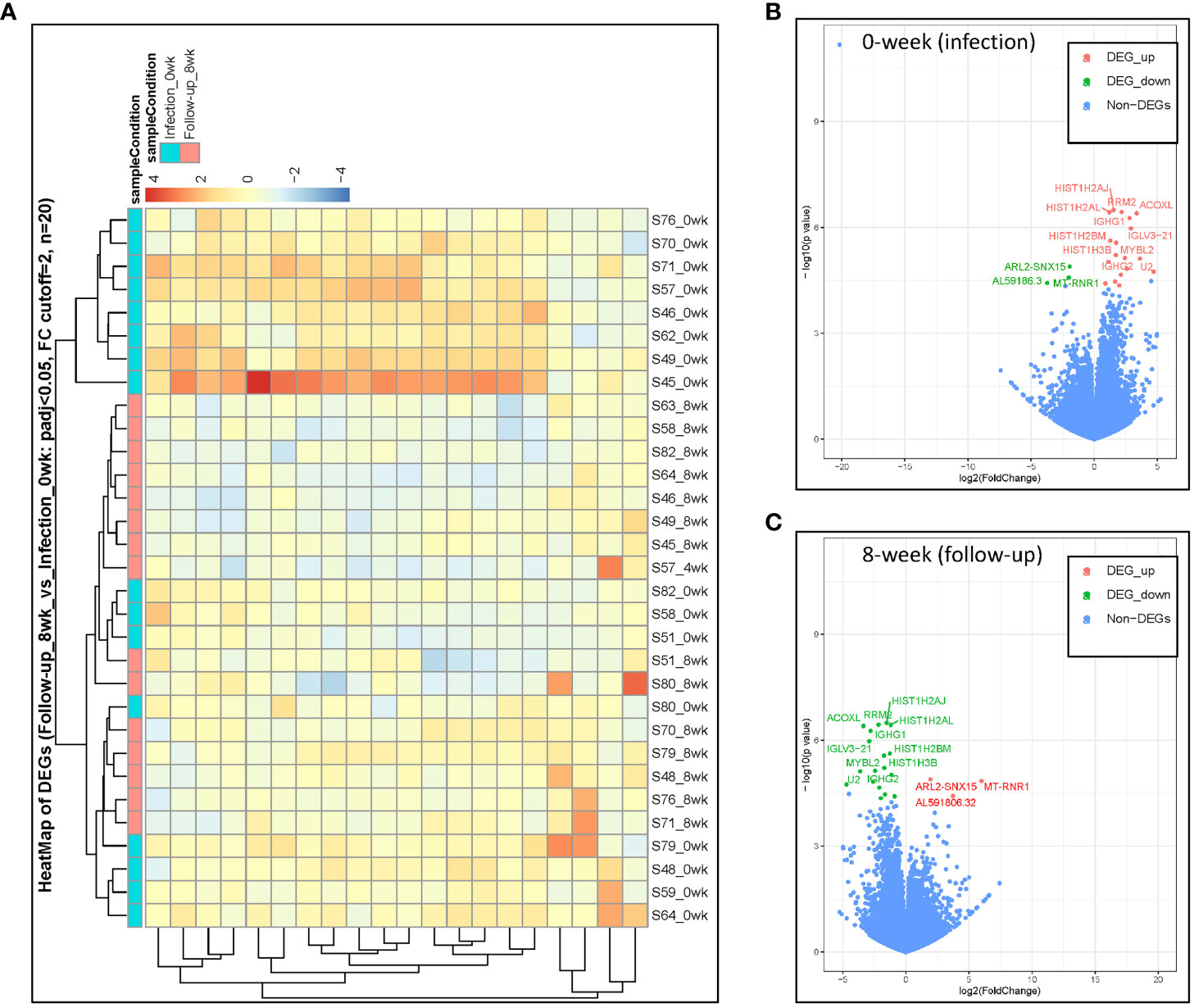
Figure 2 DESeq2 analysis was used to characterize the transcriptomes of host PBMC mRNA gene expression among healed patients (n = 17). (A) Heatmap identification of gene pattern of expression. (B, C) DE volcano plot of gene expression at 0 weeks during active infection compared with that at 8 weeks of follow-up among healed patients. Transcripts of selected upregulated genes colored in red, whereas downregulated genes are colored in green.
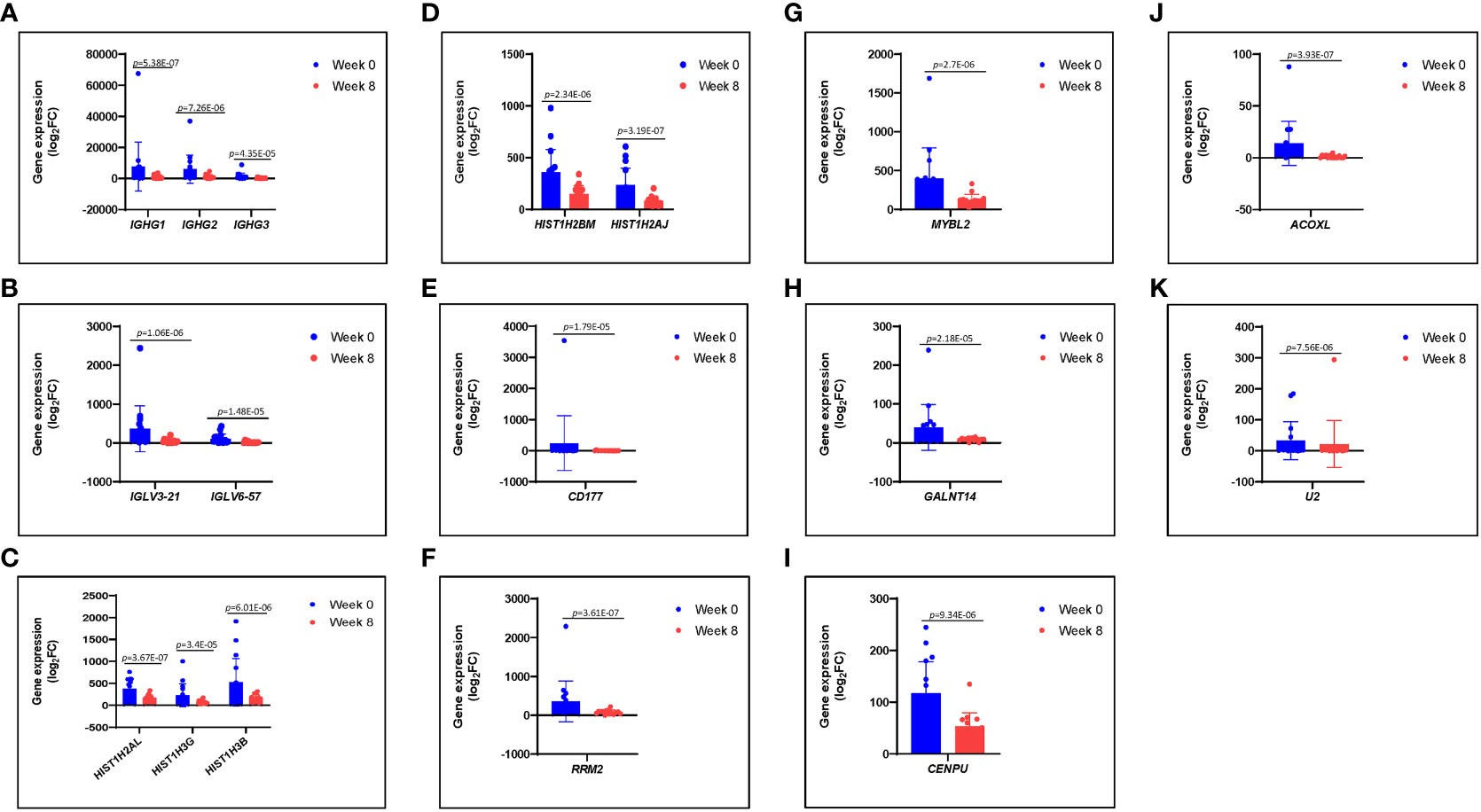
Figure 3 (A–K) Differences in gene expression patterns among healed patients (n = 17) during active infection at 0 (blue) and 8 (red) weeks. These genes were significantly expressed during active infection, but they were downregulated at 8 weeks following antibiotic therapy.
3.3 IGLV3-21 and IGLV6-57 downregulated after antibiotic therapy
Recently, studies have found significant variation in immunoglobulin loci, as well as links between germline immunoglobulin families and humoral response through B- cell receptors and antibodies expressed by B cells (Pennell et al., 2023). We found a significant increase in expression of IGLV3-21 and IGLV6-57 genes during active S. aureus DFU infection at 0 weeks; these genes and other immunoglobulin families of IGHG remain higher as observed among the patients (n = 3) with unresolved infection (data not shown) but became downregulated at 8 weeks associated with resolution of infection in healed patients (n = 17) after antibiotic therapy (p = 1.06E-06 and 1.48-E05) with corresponding FDR values of 0.003573 and 0.02142, respectively (Figure 3B; Table 1).
3.4 Core histone genes downregulated after antibiotic therapy
Histones are classified as lysine-rich (H1, H2A, and H2B) or arginine-rich (H3 and H4), and they play a significant role in DNA condensation (Li X, et al., 2022). They accumulate in the cytoplasm and on the plasma membrane, where they perform other functions such as antimicrobial activity. Quantitative transcriptome analyses with mRNA samples isolated from the PBMCs revealed that core histone genes such as HIST1H2AJ, HIST1H2AL, HIST1H2BM, HIST1H3B, and HIST1H3G were upregulated during active infection at 0 weeks and downregulated at 8 weeks following antibiotic therapy. Among the patients that were healed (n = 17), our data identified statistically significant downregulation of the aforementioned histone genes at 8 weeks (p = 3.19E-07, 3.67E-07, 2.34E-06, 6.01E-06, and 3.4E-05 with FDR values of 0.001994, 0.001994, 0.006794, 0.01354, and 0.038314, respectively) when compared with the active infection at 0 weeks (Figures 3C, D; Table 1). Despite the limitation of small sample size and variation in the severity of illness among the three unhealed patients compared with the healed patients, expression of histone genes remained relatively high in unresolved infection after 8 weeks (data not shown).
3.5 Cluster of differentiation 177 upregulated during active infection at 0 weeks
CD177 is a well-known neutrophil antigen/cell surface glycoprotein involved in neutrophil activation and migration. In response to inflammatory stimuli, neutrophils migrate from the circulating blood to infected tissues, where they protect the host by phagocytosing, killing, and digesting bacterial and fungal pathogens. In the healed group (n = 17), CD177 was highly expressed in response to active infection at 0 weeks and became downregulated at 8 weeks following antibiotic therapy. CD177 had the highest fold change among other genes at 8 weeks when compared with that at 0 weeks with −Log2FC of 4.7108 (p = 1.79E-05) with FDR value of 0.024148 (Figure 3E; Table 1).
3.6 RRM2 and MYBL2 upregulated during active infection at 0 weeks
Ribonucleotide reductase M2 (RRM2) is a subunit of ribonucleotide reductase, which is required for DNA replication and damage repair, whereas MYBL2 is a transcription factor in the Myb-related protein B (MYB) family that regulates cell cycle progression, survival, and differentiation. Our results also identified RRM2 and MYBL2 as part of differentially expressed mRNAs gene associated with PBMCs from patients with infected DFU. In the healed group (n = 17), RRM2 and MYBL2 were significantly upregulated during active infection at 0 weeks but became downregulated at 8 weeks following therapy (p = 3.61E-07 and 2.7E-06 with corresponding FDR values of 0.001994 and 0.006835, respectively) (Figures 3F, G; Table 1). In addition, the expression of RRM2 gene was also higher among the unhealed patients (n = 3) obtaining a significant p-value of 9.177E-05 with log2FC of 2.403 but failed to obtain a significant p-adjusted value that we taught might be due to small sample size and variation in infection severity among the unhealed patients when compared with healed patients (data not shown). The expression of RRM2 was significantly upregulated in parallel with CD177 and MYBL2. Other genes that were significantly upregulated during active infection include GALNT14, CENPU, ACOXL, and U2 (Figures 3H–K).
3.7 Heat shock protein upregulated in unresolved infection after antibiotic therapy
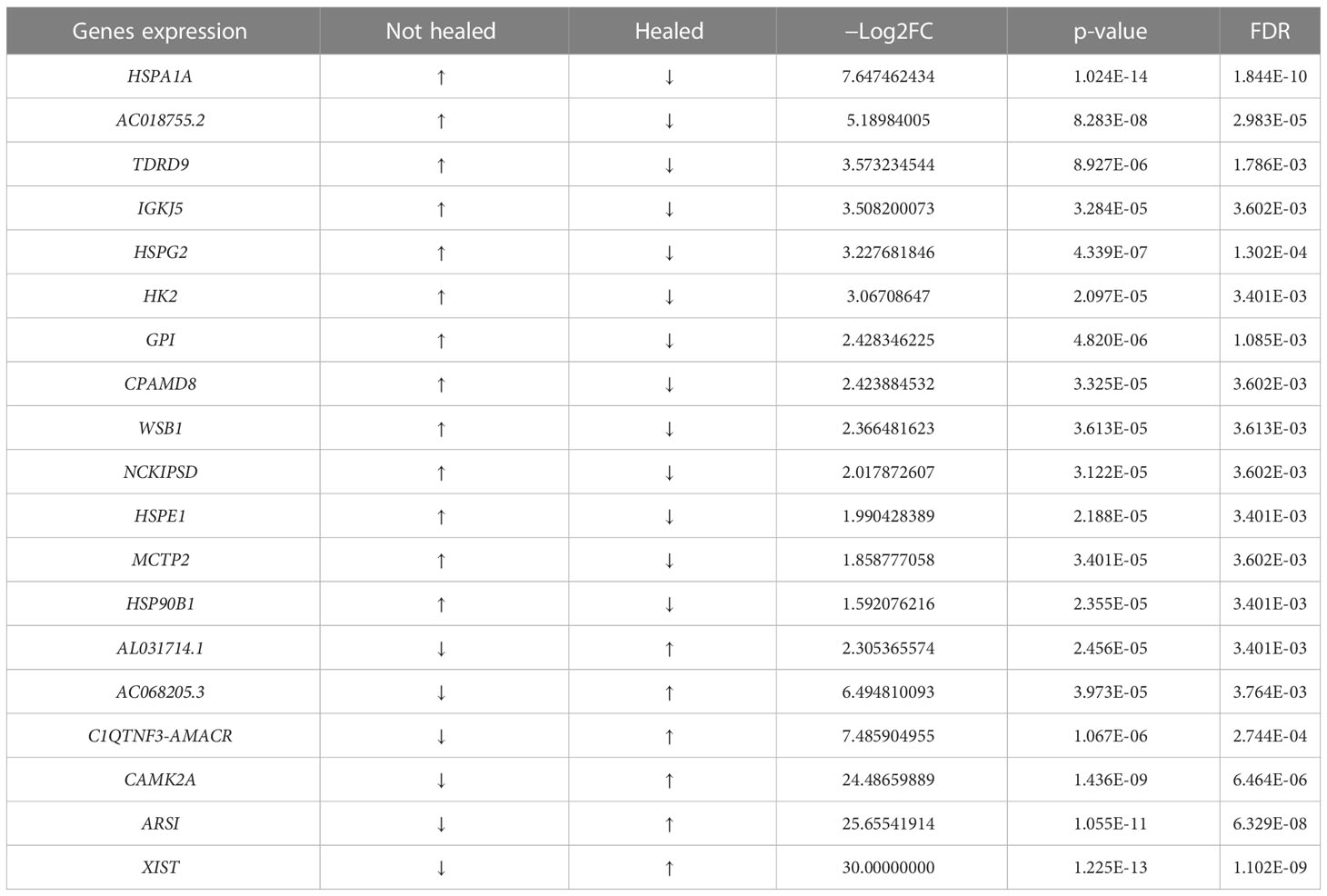
We reported that, following antibiotic therapy, subjects were divided into two groups: healed (n = 17) and not healed (n = 3) at 8 weeks. We compared the PBMC mRNA gene transcripts to check the difference in gene expression between these two groups (healed vs. not healed). Among the notable genes that were differentially expressed, we observed a significant increase in the gene expression of heat shock protein (HSP) members such as HSPA1A, HSPE1, and HSP90B1 in unhealed patients (n = 3) when compared with healed patients (n =1 7) at 8 weeks after antibiotic therapy (p = 3.284E-05, 2.188E-05, and 2.355E-05 with corresponding FDR values of 1.8444E-10, 3.401E-03, and 3.401E-03, respectively). Other differentially expressed genes that were either downregulated or upregulated in healed and not healed patients after therapy at 8 weeks are listed in Table 2.
4 Discussion
In this study, we performed RNA- seq transcriptome analysis of PBMCs to accurately and systematically profile gene expression in PBMCs of patients with S. aureus– infected DFU at the time of active infection (0 weeks) and after antibiotic therapy (8 weeks). Developing a robust system for earlier diagnosis of S. aureus infection is essential for diagnosis, treatment, and prognostication. Unlike the report by Theocharidis et al. and other authors on the genes profile of PBMC single-cell RNA that focused primarily on fibroblast- and keratinocyte-associated genes (wound healing) between patients with DFU whose ulcers heal (DFU healers) and those who do not heal (DFU non-healers), we focus primarily on profiling the genes that were differentially expressed in patients with infected DFU at different time points (0 vs. 8 weeks) to improve our understanding of earlier diagnosis, prognostication, and monitoring treatment response. We have identified various genes encoding different immunoglobulin and chemokines that are involved in the progression and healing of wounds in the PBMC samples of patients with S. aureus– infected DFU. Our findings suggest a complex interplay of host immune response via neutrophile and immunoglobulins.
We identified several members of histone genes that are highly regulated during S. aureus– infected DFU wound healing. Among the patients that were healed, we found that core histones such as HIST1H2AJ, HIST1H2AL, HIST1H2BM, HIST1H3B, and HIST1H3G were highly expressed during active infection at 0 weeks (and among the three unhealed patients) but became downregulated at 8 weeks after treatment follow-up, indicating that histone may play a significant role in host immune response to infection resolution. Histones are classified as lysine-rich (H1, H2A, and H2B) or arginine-rich (H3 and H4); among these two groups, H2A, H2B, H3, and H4 are known as “core histones”, whereas H1 is known as “linker histones” (Felsenfeld and Groudine, 2003; Li X, et al., 2022). Histones are the main components of nucleosomes, and they play a role in the packaging and arranging of DNA into functional units. However, there is growing evidence that histones can be found outside the nucleus and become extranuclear histones. These “extranuclear” histones have reported to enhance host defense functions and contribute to inflammatory responses (Urban et al., 2009; Venkatesh and Workman, 2015; Frydman et al., 2020).
Parallel to upregulation of core histones, the CD177 gene (a neutrophil-specific marker), MYBL2 and RRM2 were highly expressed during the active phase of infection (0 weeks) and the expressed genes were reduced at 8 weeks after therapy. These data are consistent with reported positive correlation of RRM2 with most chemokines and chemokine receptors (Zhou et al., 2022). Histones can act as antimicrobial peptides and directly kill bacteria and other pathogens in a variety of animal hosts. In addition, histones can trigger inflammatory responses, in some cases, acting through Toll-like receptors (Morita et al., 2013; Hoeksema et al., 2016). In addition, phagocytes are essential components of innate immune defense against infectious pathogens, and there is mounting evidence that histones and phagocytes have interactions (Li X, et al., 2022). Once phagocytosed, S. aureus is exposed to a variety of toxic products (NO, H2O2, superoxide and hydroxyl radicals, hydrolases, and proteolytic enzymes) that kill and degrade the engulfed bacteria.
Neutrophil extracellular traps (NETs) are fibrous structures with decondensed chromatin networks released from activated neutrophils during infection, which can capture and eliminate pathogens (Papayannopoulos, 2018; Song et al., 2019; Tan et al., 2021). NETs contain all core histones (H2A, H2B, H3, and H4), accounting for 65%–70% of all NET-related proteins; although their functions have not been fully elucidated, the presence of histones in NET structure suggested that histones could act as antimicrobial agents in NETs. Histones and their fragments play a vital role in host defense by binding to bacterial nucleic acid and lipopolysaccharide and by changing the permeability of bacterial cell membrane, and the antibacterial role of histones in different species is becoming more widely recognized (Howell et al., 2003; Patat et al., 2004; Lemaire et al., 2008; Urban et al., 2009; Mehta and Jeffrey, 2015; Shrestha et al., 2019; Doolin et al., 2020; Duong et al., 2020). High expression of CD177 and histones at 0 weeks may suggest a strong association with the severity of S. aureus infection.
We noted a high level expression of RRM2, MYBL2, and some other genes such as GALNT14, CENPU, ACOXL, and U2 at 0 weeks during active phase of infection, which were downregulated at 8 weeks after therapy (Table 1). We also observed a low level expression of ARL2-SNX15, MT-RNR1, and AL591806.3 at 0 weeks during active infection, which were upregulated at 8 weeks after therapy (Figures 4A–C; Table 1). The role of these genes in S. aureus– infected DFU wound healing and resolving infection is not completely understood. Among others, RRM2 has been reported as a member of the ferric iron-binding ferritin superfamily and that RRM2 engaged in the iron metabolism in hepatocellular carcinoma (Key et al., 2020; Shen et al., 2021). Although we did not know whether RRM2 is important for S. aureus pathogenicity, we observed a high expression of this seemly important gene among the three unhealed patients. Given that iron (heme) acquisition is a pathognomonic finding of invasive S. aureus infection, it would be prudent to further investigate factors associated with iron metabolism, which may potentially involve RRM2.
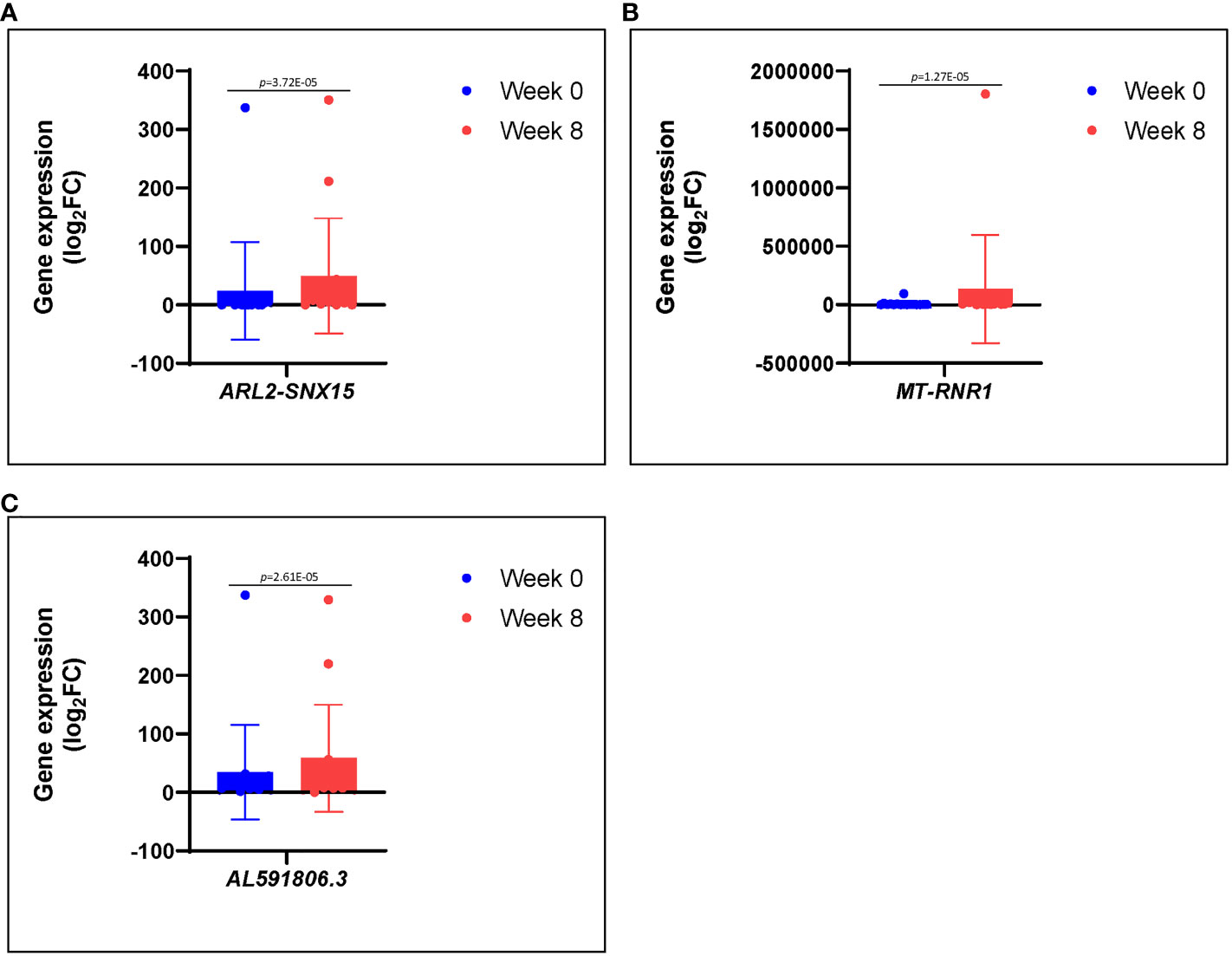
Figure 4 (A–C) Differences in gene expression patterns among healed patients (n = 17) during active infection at 0 (blue) and 8 (red) weeks. These genes were significantly repressed during active infection, but they were upregulated at 8 weeks following antibiotic therapy.
We reported a high expression of IGHG family (IGHG1, IGHG2, and IGHG3) and IGLV family (IGLV3-21 and IGLV6-57) at 0 weeks, which are clinically correlate with active infection. Conversely, IGHG and IGLV expression was reduced at 8 weeks after therapy but remain higher in unresolved infection. Because neutrophil- associated histones and CD117 were also reduced at 8 weeks, these findings reflect a reduction of S. aureus infection burden at 8 weeks after therapy. Our finding suggests that diverse IGHG and IGLV family may contribute to a protective host immune response against S. aureus infection in DFU and may serve as a potential biomarker for diagnosis and prognostication of infection and wound healing.
In addition, we noted a significant elevation in HSP levels in unresolved infection (not healed n = 3) compared with healed patients (n = 17) at 8 week after antibiotic therapy (Table 2). The presence of high levels of inducible HSPs in unhealed patients may be due to the continuous release of virulence machineries by S. aureus and the host immune response to unresolved infection. In line with our observation, HSPs have been reported in cellular response to heat shock/fever and oxidative stress signals and has been found significantly higher during bacterial infection (Borges et al., 2012; Varano Della Vergiliana et al., 2013; Tulapurkar et al., 2015). Further study is needed to investigate clinical utilization of HSPs as a potential transcriptome biomarker for early detection of recurrent or persistent infection after antibiotic therapy.
5 Conclusion
We found upregulated IGHG1, IGHG2, IGHG3, GLV3-21, IGLV6-57, CD177, HIST1H2AJ, HIST1H2AL, HIST1H2BM, HIST1H3B, and HIST1H3G genes from PBMC during active phase of S. aureus infection in patients with DFU at 0 weeks; these genes became downregulated at 8 weeks among healed patients, and these transcriptomes may reflect infectivity before and after antibiotic therapy. In line with the report by Sandoval-Schaefer et al. (2023) on transcriptional heterogeneity in human diabetic foot wounds, it was reported that profiling chronic foot ulcers from non-diabetic and diabetics patients using single-cell RNA sequencing display transcription changes in gene expression that may provide therapeutic baseline for treatment of DFU. Our findings further add evidence to the existing knowledge that transcriptome profiling of RNA-seq can delineate genes signatures and could be a potential valuable technique for diagnosing infection, determining severity, and measuring host immune response to therapies.
5.1 Clinical relevance
PBMC transcriptomes may serve as a tool for early diagnosis, monitoring and prognostication of infection, and wound healing in MSKI.
5.2 Limitation
This study is limited due to a small sample size, especially the group of patients who failed to heal at 8 weeks of follow-up. In addition, severity of illness, inflammatory, and immune response to infection differ from patients to patients.
Data availability statement
The data presented in the study are deposited in the Gene Expression Omnibus (GEO) repository, accession number “GSE230426”.
Ethics statement
The studies involving human participants were reviewed and approved by Human Research Protection and Ethics Committee (Reference No. 2000030908), Yale university. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization and supervision: IO; Methodology: IO, JK, DZ; Validation and interpretation of results: IO, FL, TA, H-KK, JK and DZ; writing of original draft: TA; review and editing: TA and IO, funding acquisition: IO. All authors contributed to the article and approved the submitted version.
Funding
The study was aided by a grant from the Orthopaedic Research and Education Foundation funded, in part, by the Musculoskeletal Transplant Foundation (MTF) Biological and The Dr. Dane and Mrs Mary Louise Miller Endowment Fund.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Armstrong, D. G., Boulton, A. J. M., Bus, S. A. (2017). Diabetic foot ulcers and their recurrence. N Engl. J. Med. 376 (24), 2367–2375. doi: 10.1056/NEJMra1615439
Ascoli, C., Schott, C. A., Huang, Y., Turturice, B. A., Wang, W., Ecanow, N., et al. (2022). Altered transcription factor targeting is associated with differential peripheral blood mononuclear cell proportions in sarcoidosis. Front. Immunol. 13, 848759. doi: 10.3389/fimmu.2022.848759
Baine, M. J., Menning, M., Smith, L. M., Mallya, K., Kaur, S., Rachagani, S., et al. (2011). Differential gene expression analysis of peripheral blood mononuclear cells reveals novel test for early detection of pancreatic cancer. Cancer biomark. 11 (1), 1–14. doi: 10.3233/CBM-2012-0260
Bellaver, B., Rocha, A. S., Souza, D. G., Leffa, D. T., De Bastiani, M. A., Schu, G., et al. (2019). Activated peripheral blood mononuclear cell mediators trigger astrocyte reactivity. Brain Behav. Immun. 80, 879–888. doi: 10.1016/j.bbi.2019.05.041
Borges, T. J., Wieten, L., van Herwijnen, M. J., Broere, F., van der Zee, R., Bonorino, C., et al. (2012). The anti-inflammatory mechanisms of Hsp70. Front. Immunol. 3, 95. doi: 10.3389/fimmu.2012.00095
Choi, Y., Oda, E., Waldman, O., Sajda, T., Beck, C., Oh, I. (2021). Next-generation sequencing for pathogen identification in infected foot ulcers. Foot Ankle Orthop 6 (3), 24730114211026933. doi: 10.1177/24730114211026933
Doolin, T., Amir, H. M., Duong, L., Rosenzweig, R., Urban, L. A., Bosch, M., et al. (2020). Mammalian histones facilitate antimicrobial synergy by disrupting the bacterial proton gradient and chromosome organization. Nat. Commun. 11 (1), 3888. doi: 10.1038/s41467-020-17699-z
Duong, L., Gross, S. P., Siryaporn, A. (2020). A novel antibacterial strategy: histone and antimicrobial peptide synergy. Microb. Cell 7 (11), 309–311. doi: 10.15698/mic2020.11.736
Egan, A. M., Dow, M. L., Vella, A. (2020). A review of the pathophysiology and management of diabetes in pregnancy. Mayo Clin. Proc. 95 (12), 2734–2746. doi: 10.1016/j.mayocp.2020.02.019
Fang, S., Zhang, L., Liu, Y., Xu, W., Wu, W., Huang, Z., et al. (2021). Lysosome activation in peripheral blood mononuclear cells and prognostic significance of circulating LC3B in COVID-19. Brief Bioinform. 22 (2), 1466–1475. doi: 10.1093/bib/bbab043
Fritz, B. G., Kirkegaard, J. B., Nielsen, C. H., Kirketerp-Moller, K., Malone, M., Bjarnsholt, T. (2022). Transcriptomic fingerprint of bacterial infection in lower extremity ulcers. APMIS 130 (8), 524–534. doi: 10.1111/apm.13234
Frydman, G. H., Tessier, S. N., Wong, K. H. K., Vanderburg, C. R., Fox, J. G., Toner, M., et al. (2020). Megakaryocytes contain extranuclear histones and may be a source of platelet-associated histones during sepsis. Sci. Rep. 10 (1), 4621. doi: 10.1038/s41598-020-61309-3
Gardner, S. E., Hillis, S. L., Heilmann, K., Segre, J. A., Grice, E. A. (2013). The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 62 (3), 923–930. doi: 10.2337/db12-0771
Ghosh, N., Saha, I., Plewczynski, D. (2022). Unveiling the biomarkers of cancer and COVID-19 and their regulations in different organs by integrating RNA-seq expression and protein-protein interactions. ACS Omega 7 (48), 43589–43602. doi: 10.1021/acsomega.2c04389
Grondman, I., Pirvu, A., Riza, A., Ioana, M., Netea, M. G. (2020). Biomarkers of inflammation and the etiology of sepsis. Biochem. Soc. Trans. 48 (1), 1–14. doi: 10.1042/BST20190029
Guo, Q., Ying, G., Jing, O., Zhang, Y., Liu, Y., Deng, M., et al. (2023). Influencing factors for the recurrence of diabetic foot ulcers: a meta-analysis. Int. Wound J. 20 (5), 1762–1775. doi: 10.1111/iwj.14017
Han, Z., Feng, W., Hu, R., Ge, Q., Ma, W., Zhang, W., et al. (2021). RNA-Seq profiling reveals PBMC RNA as a potential biomarker for hepatocellular carcinoma. Sci. Rep. 11 (1), 17797. doi: 10.1038/s41598-021-96952-x
Hao, S. P., Masters, E. A., Ninomiya, M. J., Beck, C. A., Schwarz, E. M., Daiss, J. L., et al. (2021). Species-specific immunoassay aids identification of pathogen and tracks infectivity in foot infection. Foot Ankle Int. 42 (3), 363–372. doi: 10.1177/1071100720965136
Harmsen, D., Claus, H., Witte, W., Rothganger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41 (12), 5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003
Heravi, F. S., Zakrzewski, M., Vickery, K., Malone, M., Hu, H. (2020). Metatranscriptomic analysis reveals active bacterial communities in diabetic foot infections. Front. Microbiol. 11, 1688. doi: 10.3389/fmicb.2020.01688
Herber, O. R., Schnepp, W., Rieger, M. A. (2007). A systematic review on the impact of leg ulceration on patients' quality of life. Health Qual Life Outcomes 5, 44. doi: 10.1186/1477-7525-5-44
Hoeksema, M., van Eijk, M., Haagsman, H. P., Hartshorn, K. L. (2016). Histones as mediators of host defense, inflammation and thrombosis. Future Microbiol. 11 (3), 441–453. doi: 10.2217/fmb.15.151
Howell, S. J., Wilk, D., Yadav, S. P., Bevins, C. L. (2003). Antimicrobial polypeptides of the human colonic epithelium. Peptides 24 (11), 1763–1770. doi: 10.1016/j.peptides.2003.07.028
Islam, M. A., Grosse-Brinkhaus, C., Proll, M. J., Uddin, M. J., Aqter Rony, S., Tesfaye, D., et al. (2017). PBMC transcriptome profiles identifies potential candidate genes and functional networks controlling the innate and the adaptive immune response to PRRSV vaccine in pietrain pig. PloS One 12 (3), e0171828. doi: 10.1371/journal.pone.0171828
Jiang, Z., Zhou, X., Li, R., Michal, J. J., Zhang, S., Dodson, M. V., et al. (2015). Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol. Life Sci. 72 (18), 3425–3439. doi: 10.1007/s00018-015-1934-y
Key, J., Sen, N. E., Arsovic, A., Kramer, S., Hulse, R., Khan, N. N., et al. (2020). Systematic surveys of iron homeostasis mechanisms reveal ferritin superfamily and nucleotide surveillance regulation to be modified by PINK1 absence. Cells 9 (10). doi: 10.3390/cells9102229
Kim, S., Boye, K. S. (2009). Excessive hospitalizations and its associated economic burden among people with diabetes in the united states. Value Health 12 (2), 267–272. doi: 10.1111/j.1524-4733.2008.00443.x
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi: 10.1038/s41587-019-0201-4
Kramer, A., Green, J., Pollard, J., Jr., Tugendreich, S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30 (4), 523–530. doi: 10.1093/bioinformatics/btt703
Lemaire, S., Trinh, T. T., Le, H. T., Tang, S. C., Hincke, M., Wellman-Labadie, O., et al. (2008). Antimicrobial effects of H4-(86-100), histogranin and related compounds–possible involvement of DNA gyrase. FEBS J. 275 (21), 5286–5297. doi: 10.1111/j.1742-4658.2008.06659.x
Li, Y., Ju, S., Li, X., Li, W., Zhou, S., Wang, G., et al. (2022b). Characterization of the microenvironment of diabetic foot ulcers and potential drug identification based on scRNA-seq. Front. Endocrinol. (Lausanne) 13, 997880. doi: 10.3389/fendo.2022.997880
Li, X., Ye, Y., Peng, K., Zeng, Z., Chen, L., Zeng, Y. (2022a). Histones: the critical players in innate immunity. Front. Immunol. 13, 1030610. doi: 10.3389/fimmu.2022.1030610
Lipof, J. S., Jones, C. M. C., Daiss, J., Oh, I. (2021). Comparative study of culture, next-generation sequencing, and immunoassay for identification of pathogen in diabetic foot ulcer. J. Orthop Res. 39 (12), 2638–2645. doi: 10.1002/jor.25001
Lipsky, B. A., Berendt, A. R., Cornia, P. B., Pile, J. C., Peters, E. J., Armstrong, D. G., et al. (2012). 2012 infectious diseases society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin. Infect. Dis. 54 (12), e132–e173. doi: 10.1093/cid/cis346
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Macdonald, K. E., Jordan, C. Y., Crichton, E., Barnes, J. E., Harkin, G. E., Hall, L. M. L., et al. (2020). A retrospective analysis of the microbiology of diabetic foot infections at a Scottish tertiary hospital. BMC Infect. Dis. 20 (1), 218. doi: 10.1186/s12879-020-4923-1
Maleknia, S., Tavassolifar, M. J., Mottaghitalab, F., Zali, M. R., Meyfour, A. (2022). Identifying novel host-based diagnostic biomarker panels for COVID-19: a whole-blood/nasopharyngeal transcriptome meta-analysis. Mol. Med. 28 (1), 86. doi: 10.1186/s10020-022-00513-5
Masters, E. A., Trombetta, R. P., de Mesy Bentley, K. L., Boyce, B. F., Gill, A. L., Gill, S. R., et al. (2019). Evolving concepts in bone infection: redefining "biofilm", "acute vs. chronic osteomyelitis", "the immune proteome" and "local antibiotic therapy". Bone Res. 7, 20. doi: 10.1038/s41413-019-0061-z
McDermott, K., Fang, M., Boulton, A. J. M., Selvin, E., Hicks, C. W. (2023). Etiology, epidemiology, and disparities in the burden of diabetic foot ulcers. Diabetes Care 46 (1), 209–221. doi: 10.2337/dci22-0043
Mehta, S., Jeffrey, K. L. (2015). Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity. Immunol. Cell Biol. 93 (3), 233–244. doi: 10.1038/icb.2014.101
Mendelsohn, S. C., Mbandi, S. K., Fiore-Gartland, A., Penn-Nicholson, A., Musvosvi, M., Mulenga, H., et al. (2022). Prospective multicentre head-to-head validation of host blood transcriptomic biomarkers for pulmonary tuberculosis by real-time PCR. Commun. Med. (Lond) 2 (1). doi: 10.1038/s43856-022-00086-8
Morita, S., Tagai, C., Shiraishi, T., Miyaji, K., Iwamuro, S. (2013). Differential mode of antimicrobial actions of arginine-rich and lysine-rich histones against gram-positive staphylococcus aureus. Peptides 48, 75–82. doi: 10.1016/j.peptides.2013.07.025
Oh, I., Muthukrishnan, G., Ninomiya, M. J., Brodell, J. D., Jr., Smith, B. L., Lee, C. C., et al. (2018). Tracking anti-staphylococcus aureus antibodies produced In vivo and ex vivo during foot salvage therapy for diabetic foot infections reveals prognostic insights and evidence of diversified humoral immunity. Infect. Immun. 86 (12). doi: 10.1128/IAI.00629-18
Papayannopoulos, V. (2018). Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18 (2), 134–147. doi: 10.1038/nri.2017.105
Patat, S. A., Carnegie, R. B., Kingsbury, C., Gross, P. S., Chapman, R., Schey, K. L. (2004). Antimicrobial activity of histones from hemocytes of the pacific white shrimp. Eur. J. Biochem. 271 (23-24), 4825–4833. doi: 10.1111/j.1432-1033.2004.04448.x
Pennell, M., Rodriguez, O. L., Watson, C. T., Greiff, V. (2023). The evolutionary and functional significance of germline immunoglobulin gene variation. Trends Immunol. 44 (1), 7–21. doi: 10.1016/j.it.2022.11.001
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Phillips, C. J., Humphreys, I., Fletcher, J., Harding, K., Chamberlain, G., Macey, S. (2016). Estimating the costs associated with the management of patients with chronic wounds using linked routine data. Int. Wound J. 13 (6), 1193–1197. doi: 10.1111/iwj.12443
Polikandrioti, M., Vasilopoulos, G., Koutelekos, I., Panoutsopoulos, G., Gerogianni, G., Babatsikou, F., et al. (2020). Quality of life in diabetic foot ulcer: associated factors and the impact of Anxiety/Depression and adherence to self-care. Int. J. Low Extrem Wounds 19 (2), 165–179. doi: 10.1177/1534734619900415
Raffetto, J. D., Ligi, D., Maniscalco, R., Khalil, R. A., Mannello, F. (2020). Why venous leg ulcers have difficulty healing: overview on pathophysiology, clinical consequences, and treatment. J. Clin. Med. 10 (1). doi: 10.3390/jcm10010029
Sandoval-Schaefer, T., Phan, Q., Dash, B. C., Prassinos, A. J., Duan, K., Gazes, M. I., et al. (2023). Transcriptional heterogeneity in human diabetic foot wounds. bioRxiv. doi: 10.1101/2023.02.16.528839
Schmidt, B. M. (2022). Emerging diabetic foot ulcer microbiome analysis using cutting edge technologies. J. Diabetes Sci. Technol. 16 (2), 353–363. doi: 10.1177/1932296821990097
Shen, H., Wu, H., Sun, F., Qi, J., Zhu, Q. (2021). A novel four-gene of iron metabolism-related and methylated for prognosis prediction of hepatocellular carcinoma. Bioengineered 12 (1), 240–251. doi: 10.1080/21655979.2020.1866303
Shrestha, B., Ito, T., Kakuuchi, M., Totoki, T., Nagasato, T., Yamamoto, M., et al. (2019). Recombinant thrombomodulin suppresses histone-induced neutrophil extracellular trap formation. Front. Immunol. 10, 2535. doi: 10.3389/fimmu.2019.02535
Skrepnek, G. H., Mills S.R, J. L., Lavery, L. A., Armstrong, D. G. (2017). Health care service and outcomes among an estimated 6.7 million ambulatory care diabetic foot cases in the U.S. Diabetes Care 40 (7), 936–942. doi: 10.2337/dc16-2189
Smith-Strom, H., Iversen, M. M., Igland, J., Ostbye, T., Graue, M., Skeie, S., et al. (2017). Severity and duration of diabetic foot ulcer (DFU) before seeking care as predictors of healing time: a retrospective cohort study. PloS One 12 (5), e0177176. doi: 10.1371/journal.pone.0177176
Song, Y., Kadiyala, U., Weerappuli, P., Valdez, J. J., Yalavarthi, S., Louttit, C., et al. (2019). Antimicrobial microwebs of DNA-histone inspired from neutrophil extracellular traps. Adv. Mater 31 (14), e1807436. doi: 10.1002/adma.201807436
Stubbington, M. J. T., Rozenblatt-Rosen, O., Regev, A., Teichmann, S. A. (2017). Single-cell transcriptomics to explore the immune system in health and disease. Science 358 (6359), 58–63. doi: 10.1126/science.aan6828
Tai, C. H., Hsieh, T. C., Lee, R. P., Lo, S. F. (2021). Prevalence and medical resource of patients with diabetic foot ulcer: a nationwide population-based retrospective cohort study for 2001-2015 in Taiwan. Int. J. Environ. Res. Public Health 18 (4). doi: 10.3390/ijerph18041891
Tan, C., Aziz, M., Wang, P. (2021). The vitals of NETs. J. Leukoc. Biol. 110 (4), 797–808. doi: 10.1002/JLB.3RU0620-375R
Theocharidis, G., Baltzis, D., Roustit, M., Tellechea, A., Dangwal, S., Khetani, R. S., et al. (2020). Integrated skin transcriptomics and serum multiplex assays reveal novel mechanisms of wound healing in diabetic foot ulcers. Diabetes 69 (10), 2157–2169. doi: 10.2337/db20-0188
Theocharidis, G., Thomas, B. E., Sarkar, D., Mumme, H. L., Pilcher, W. J. R., Dwivedi, B., et al. (2022). Single cell transcriptomic landscape of diabetic foot ulcers. Nat. Commun. 13 (1), 181. doi: 10.1038/s41467-021-27801-8
Tulapurkar, M. E., Ramarathnam, A., Hasday, J. D., Singh, I. S. (2015). Bacterial lipopolysaccharide augments febrile-range hyperthermia-induced heat shock protein 70 expression and extracellular release in human THP1 cells. PloS One 10 (2), e0118010. doi: 10.1371/journal.pone.0118010
Urban, C. F., Ermert, D., Schmid, M., Abu-Abed, U., Goosmann, C., Nacken, W., et al. (2009). Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against candida albicans. PloS Pathog. 5 (10), e1000639. doi: 10.1371/journal.ppat.1000639
Varano Della Vergiliana, J. F., Lansley, S. M., Porcel, J. M., Bielsa, S., Brown, J. S., Creaney, J., et al. (2013). Bacterial infection elicits heat shock protein 72 release from pleural mesothelial cells. PloS One 8 (5), e63873. doi: 10.1371/journal.pone.0063873
Venkatesh, S., Workman, J. L. (2015). Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16 (3), 178–189. doi: 10.1038/nrm3941
Wagner, F. W., Jr. (1981). The dysvascular foot: a system for diagnosis and treatment. Foot Ankle 2 (2), 64–122. doi: 10.1177/107110078100200202
Wang, S., He, L., Wu, J., Zhou, Z., Gao, Y., Chen, J., et al. (2019). Transcriptional profiling of human peripheral blood mononuclear cells identifies diagnostic biomarkers that distinguish active and latent tuberculosis. Front. Immunol. 10, 2948. doi: 10.3389/fimmu.2019.02948
Wang, Y., Shao, T., Wang, J., Huang, X., Deng, X., Cao, Y., et al. (2021). An update on potential biomarkers for diagnosing diabetic foot ulcer at early stage. BioMed. Pharmacother. 133, 110991. doi: 10.1016/j.biopha.2020.110991
Wang, A., Toma, M. A., Ma, J., Li, D., Vij, M., Chu, T., et al. (2020). Circular RNA hsa_circ_0084443 is upregulated in diabetic foot ulcer and modulates keratinocyte migration and proliferation. Adv. Wound Care (New Rochelle) 9 (4), 145–160. doi: 10.1089/wound.2019.0956
Xiong, Y., Liu, Y., Cao, L., Wang, D., Guo, M., Jiang, A., et al. (2020). Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 9 (1), 761–770. doi: 10.1080/22221751.2020.1747363
Zhao, H., McClure, N. S., Johnson, J. A., Soprovich, A., Al Sayah, F., Eurich, D. T. (2020). A longitudinal study on the association between diabetic foot disease and health-related quality of life in adults with type 2 diabetes. Can. J. Diabetes 44 (3), 280–286 e1. doi: 10.1016/j.jcjd.2019.08.008
Zheng, H. Y., Xu, M., Yang, C. X., Tian, R. R., Zhang, M., Li, J. J., et al. (2020). Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct Target Ther. 5 (1), 294. doi: 10.1038/s41392-020-00457-4
Zhou, Z., Song, Q., Yang, Y., Wang, L., Wu, Z. (2022). Comprehensive landscape of RRM2 with immune infiltration in pan-cancer. Cancers (Basel) 14 (12). doi: 10.3390/cancers14122938
Keywords: DFU, S. aureus-infection, transcriptome, PBMC, wound healing, host-immunity
Citation: Agidigbi TS, Kwon H-K, Knight JR, Zhao D, Lee FY and Oh I (2023) Transcriptomic identification of genes expressed in invasive S. aureus diabetic foot ulcer infection. Front. Cell. Infect. Microbiol. 13:1198115. doi: 10.3389/fcimb.2023.1198115
Received: 03 April 2023; Accepted: 25 May 2023;
Published: 26 June 2023.
Edited by:
Marat R Sadykov, University of Nebraska Medical Center, United StatesReviewed by:
Natasa Strbo, University of Miami, United StatesSrijon Kaushik Banerjee, University of Pittsburgh, United States
Copyright © 2023 Agidigbi, Kwon, Knight, Zhao, Lee and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irvin Oh, irvin.oh@yale.edu
 Taiwo Samuel Agidigbi
Taiwo Samuel Agidigbi Hyuk-Kwon Kwon
Hyuk-Kwon Kwon James R. Knight3
James R. Knight3  Dejian Zhao
Dejian Zhao Francis Y. Lee
Francis Y. Lee