Revisiting ESKAPE Pathogens: virulence, resistance, and combating strategies focusing on quorum sensing
- 1Quorum Sensing Laboratory, Centre for Research in Infectious Diseases (CRID), School of Chemical and Biotechnology, SASTRA Deemed to be University, Thanjavur, India
- 2Division of Restorative Dental Sciences, Faculty of Dentistry, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
The human–bacterial association is long-known and well-established in terms of both augmentations of human health and attenuation. However, the growing incidents of nosocomial infections caused by the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp.) call for a much deeper understanding of these organisms. Adopting a holistic approach that includes the science of infection and the recent advancements in preventing and treating infections is imperative in designing novel intervention strategies against ESKAPE pathogens. In this regard, this review captures the ingenious strategies commissioned by these master players, which are teamed up against the defenses of the human team, that are equally, if not more, versatile and potent through an analogy. We have taken a basketball match as our analogy, dividing the human and bacterial species into two teams playing with the ball of health. Through this analogy, we make the concept of infectious biology more accessible.
Introduction
The incidence of bacterial players on the grounds of the human body is well-known (Ursell et al., 2012). The bacterial pathobionts play a significant role in assisting the human team in making them healthy by influencing stress levels, immune response, and cognition (Mohajeri et al., 2018). However, the opportunistic bacterial squad taking advantage of the immunocompromised state and the underlying dysbiosis in the human team are teamed up against the very human team, which they are an integral part of (Proença et al., 2017) (Figure 1). Studies show that antimicrobial resistance (AMR) causes more than 35,000 deaths annually and over 2.8 million recorded cases in the United States alone per year (Biggest Threats and Data | Antibiotic/Antimicrobial Resistance | CDC (Centers for Disease Control and Prevention); Martínez, 2014). Adding a feather to their cap, six prime players, namely, Enterococcus sp., Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp. (ESKAPE in short), have been shortlisted by the World Health Organization (WHO) owing to their mastery in the art of “escapism” (WHO, 2017). The human team is no less than the bacterial team, given its ability to defend itself by targeting diverse opponents consistently (Centers for Disease Control, 2019; Thakur et al., 2019). However, the bacterial team is versatile, wherein one species is reported to target multiple organs, just like an all-rounder including the lungs, kidneys, and skin (Bachman et al., 2011; Thomer et al., 2016; Okojie and Omorokpe, 2018). The highly coordinated human team is found to be constantly involved in keeping a check over any advances made by the bacterial team (Nicholson, 2016). Hence, this review aims to reinforce the human team by briefing about the strengths and strategies employed by the bacterial team and therefore augmenting the process of developing new strategies in preventing the bacterial team from scoring goals by infecting humans. It also attempts to capture this ingenious game between the bacterial team and the human team by recapitulating the various game plans and the substitutes employed by each team.
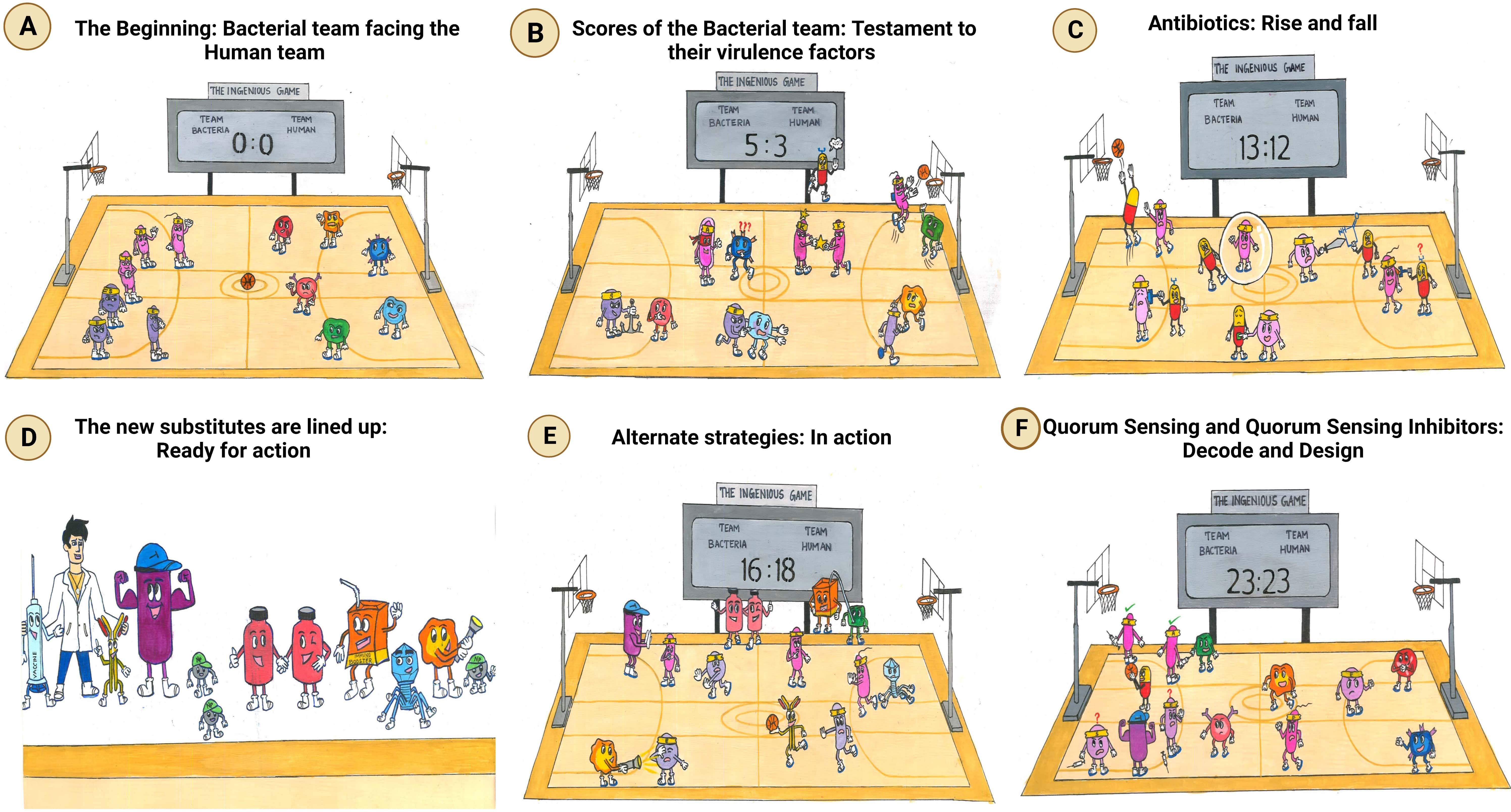
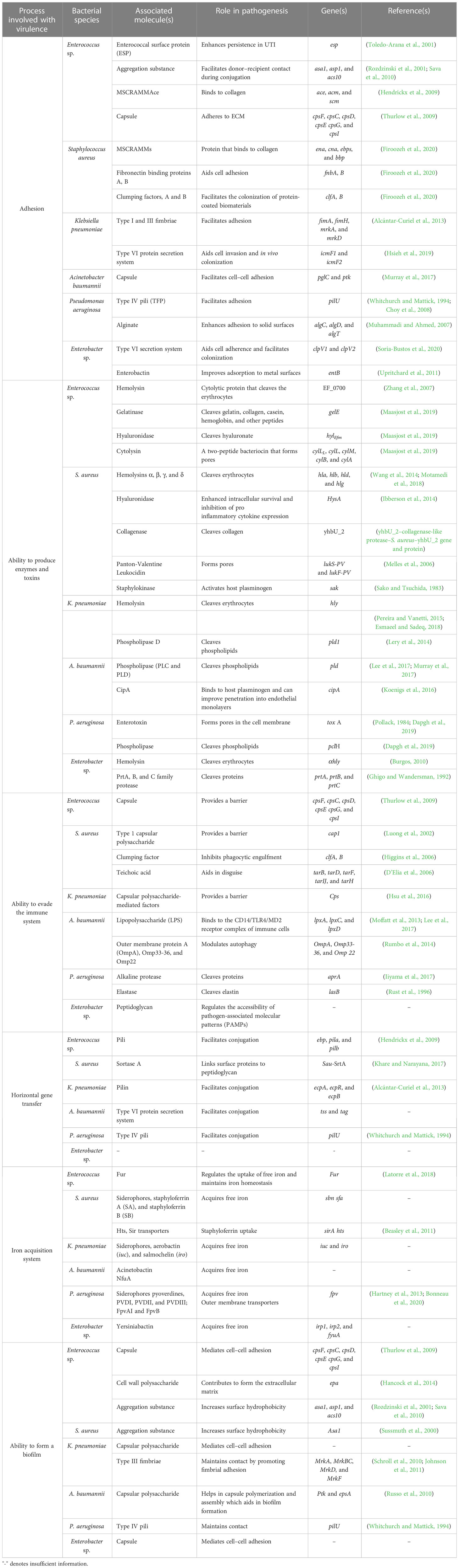
Figure 1 The ingenious game between team bacteria (ESKAPE) and team human. (A) The beginning: bacterial team facing the human team: bacterial team includes the Gram-positive Enterococcus sp., Staphylococcus aureus, and Enterobacter sp. and the Gram-negative Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa; the human team comprises macrophages, T-lymphocytes, B-lymphocytes, monocytes, eosinophils, and neutrophils. (B) Scores of the bacterial team: testament to their virulence factors. The bacterial players are rooted to the ground, closely adhering to the human body. The immune cells, however, cannot recognize them due to the masking effect of the bacterial capsule. To make things worse, another bacterium is spotted sharing its “special attribute” with their teammate. Ultimately, the bacterial team scores the goal, despite the efforts taken by the immune cell to block it. It is at this point that an antibiotic is spotted exclaiming its helplessness, being not recruited into the team. (C) Antibiotics: rise and fall. Although the antibiotics have achieved their goal, the bacteria have reduced their permeability, preventing the antibiotics from acting on them further. The bacterial players are also seen switching off the antibiotics by modifying them. Another bacterium is spotted in the act of slashing the functional antibiotic, rendering it inactive. Moreover, the antibiotic can no longer bind specifically to its target, as the bacterium has decoded the relentlessly used strategy of the human team and has modified the target. (D) The new substitutes are lined up: ready for action. The external coach, the researcher, is seen with a vaccine and monoclonal antibody on either side. Then comes the strong player representing various inhibitors—beta-lactamase inhibitor, efflux pump inhibitor, and conjugation inhibitor. Combinatorial drug molecules stand next to the highly versatile nanoparticles, winking and confirming their action plan. Next in the row is an immune booster. Adjacent to it, we see the grim-faced bacteriophage, which is waiting to take its toll! Lastly, we have the representative of antimicrobial light therapy holding a torch. (E) Alternate strategies: in action. The inhibitor is found to defend the antibiotic efficiently from the bacteria. Antimicrobial light therapy is affecting the bacteria. One bacterial player is alarmed at the entry of the combinatorial substitutes. Another bacterium is puzzled at the look of an immune cell drinking its energy potion! The monoclonal antibody has successfully recovered the ball of health from the bacterial team. Bacteriophage is doing its part by preventing bacterial players from entering human premises. (F) Quorum sensing and quorum-sensing inhibitors: decode and design. The bacterial players are spotted forming a protective shell (technically, biofilm) right below their goal post to defend their team. Among the four, two are caught communicating with each other, while the other pair is not, owing to the presence of a quorum-sensing inhibitor blocking their communication. On a closer look, the bacteria that cannot communicate with each other are equally unable to work with their injection (technically, express their virulence factor). This, in turn, has made them vulnerable to attack by the immune cell of the human team. Taking advantage of the current situation, the antibiotic has sneaked in and aims for the goal! Other players of the human team are seen guarding their goalpost against the entry of any bacterial player.
ESKAPE: players’ biology and characteristics
>Enterococcus sp.
Enterococcus sp. includes Enterococcus faecium and Enterococcus faecalis, ubiquitous pathogens with clinical relevance. They are Gram-positive and facultative anaerobes (Pendleton et al., 2013). As commensals, they are commonly found in the gut and modulate the immune system. They are opportunistic pathogens and translocate to different locations when there is an overgrowth in the gut due to antibiotic resistance or host inflammation (Krawczyk et al., 2021). Enterococci are associated with hospital-acquired infections, including catheter-associated urinary tract infections (CAUTIs), surgical site infections (SSIs), and bloodstream infections. Vancomycin-resistant enterococci (VREs) emerged in the 1980s and are still prevalent and estimated to cause 5,400 deaths in 2017 alone (Centers for Disease Control). Vancomycin-resistant E. faecium is on the WHO’s high-priority pathogen list (CDC, WHO).
Staphylococcus aureus
S. aureus is Gram-positive and is considered one of the major pathogens. S. aureus is a skin commensal and becomes a pathogen in susceptible patients (Guo et al., 2020). S. aureus is found in wound infections and can cause multiple infections from soft tissue infections to infective carditis to bacteremia to fatal pneumonia (Tong et al., 2015). Methicillin-resistant S. aureus (MRSA) was isolated in 1961 and evolved with only 2 years of treatment. The spread of MRSA infection is so alarming that the number of deaths by MRSA has surpassed deaths by acquired immune deficiency syndrome (AIDS) and Parkinson’s disease, as per the report in 2012 (Lessa et al., 2012). The prevalence of MRSA is alarmingly even today and is clinically relevant. MRSA is also on the WHO’s high-priority pathogen list (CDC, WHO).
Klebsiella pneumoniae
K. pneumoniae is a Gram-negative pathogen and belongs to the Enterobacteriaceae family. K. pneumoniae is most commonly associated with community-acquired pneumonia (Podschun and Ullmann, 1998; Piperaki et al., 2017). They are prominent extended-spectrum β-lactamase (ESBL) producers, making them a pathological threat in hospital settings. K. pneumoniae can infect multiple sites, including the lungs, urinary tract, blood stream, and brain. They are non-motile and encapsulated but present in both environments and on the surface of mammals. Hypervirulent strains of K. pneumoniae have also emerged (Russo and Marr, 2019), and carbapenem-resistant K. pneumoniae pose a significant threat. K. pneumoniae are intrinsically resistant to multiple antibiotics and found to cause sporadic cases worldwide (Lin et al., 2006).
Acinetobacter baumannii
Carbapenem-resistant A. baumannii is one of the WHO critical priority pathogens that need immediate action. A. baumannii is a Gram-negative, opportunistic pathogen that can adapt to various hostile conditions. It can survive in dry conditions, erratic temperatures, and pH ranges, making it stay in the dynamic host and environmental conditions. A. baumannii is intrinsically resistant to antibiotics and also possesses resistant islands to impart resistance not only to antibiotics but also to metals and ammonium-based disinfectants. It can easily acquire β-lactamases, and most OXA carbapenemases are isolated in different clinical isolates of A. baumannii. It infects critically ill patients who are severely immunocompromised. It can cause hospital-acquired respiratory infections and urinary tract infections and is also present in wound infections. Considering its versatility and adaptability, A. baumannii is a tough nut to crack.
Pseudomonas aeruginosa
Carbapenem-resistant P. aeruginosa is also one of the critical pathogens as defined by the WHO. P. aeruginosa is a Gram-negative, facultative anaerobe that infects immunocompromised patients and is often isolated from cystic fibrosis (CF) patients and burn patients (Moradali et al., 2017). P. aeruginosa can survive in harsh conditions and resist various antibiotics, mostly prominently fluoroquinolones (Livermore, 2002). It can cause infections at multiple sites, including the eye, skin, lungs, and urinary tract. Cystic fibrosis patients are most susceptible to P. aeruginosa infections from childhood (Malhotra et al., 2019), which is the prominent reason for mortality in CF adult patients (Doring et al., 2000). Nosocomial infections—ventilator-associated pneumonia, urinary tract infections, central line bloodstream infections, and surgical infections—are caused by P. aeruginosa and are considered the highest burden in healthcare settings (Lambert et al., 2011). Resistance to multiple classes of antibiotics combined with wide virulence factors to survive hostile conditions makes P. aeruginosa a mighty player to defeat.
Enterobacter sp.
Enterobacter sp. is a group of Gram-negative pathogens, usually rod-shaped and facultative anaerobes. Like other pathogens, it is also often found in bacteremia, urinary tract infections, surgical site infections, and device-related infections (Davin-Regli and Pagès, 2015). Enterobacter sp. usually cannot be distinguished since it causes similar infections to other Gram-negative rod bacteria. However, ESBL-producing, carbapenem-resistant Enterobacter sp. is also one of the three critical pathogens listed by the WHO. Enterobacter cloacae, Enterobacter asburiae, and Enterobacter hormaechei are some of the clinically relevant species that have caused nosocomial outbreaks [(7) Clinical and pathogenesis overview of Enterobacter infections | Request PDF].
The commonality between the bacterial team players is their prominence in multidrug resistance, targeting immunocompromised patients causing nosocomial outbreaks, ability to adapt and survive in harsh environments, and translocating from one site to another. Understanding the virulence mechanism and resistance pathways is the need of the hour to devise strategies to tackle them effectively.
Virulence factors: strengths of the bacterial team
The bacterial team has attained ascendancy in the game through a detailed pathogenesis process. The pathogenesis process is a multilevel and complex process involving various factors to establish a successful infection of the host (Wilson et al., 2002). Even though the elements and approach of pathogens vary, a similar pattern is followed. To mark their territory in the host, the bacterial members team up through a strong adhesion between them and the host team (Ribet and Cossart, 2015). Thus, the first step is the adhesion of the bacteria among themselves: auto-aggregation, microcolony formation, and ultimately biofilm formation, followed by solid adhesion to the host through mucosal surfaces. The adhesion step is crucial for bringing dysbiosis to the host microbiota and colonizing and invading the host cells (Pizarro-Cerdá and Cossart, 2006). Once they have adhered, the bacterial cells invade the host cells and release different toxins—proteins, enzymes, and siderophores—to affect the healthy host cells and evade the immune system (Siegel and Weiser, 2015). Table 1 elaborates the reported key genes involved in every step of the virulence process of ESKAPE pathogens. A part of the invaded bacteria goes to a quiescent state, termed “persisters”, to invoke recalcitrant infections later (Vasudevan et al., 2022). Understanding the underlying mechanisms dictating such survival mechanisms has been of utmost importance in recent days (Kaushik et al., 2022). Pathogens use the host environmental factors to drive this process and resist antibiotics (Hakansson et al., 2018). Several pathways and dedicated regulatory networks are involved in the pathogenesis (de Macedo et al., 2021). Figure 2 captures the virulence factors of each of the ESKAPE pathogens briefed below.
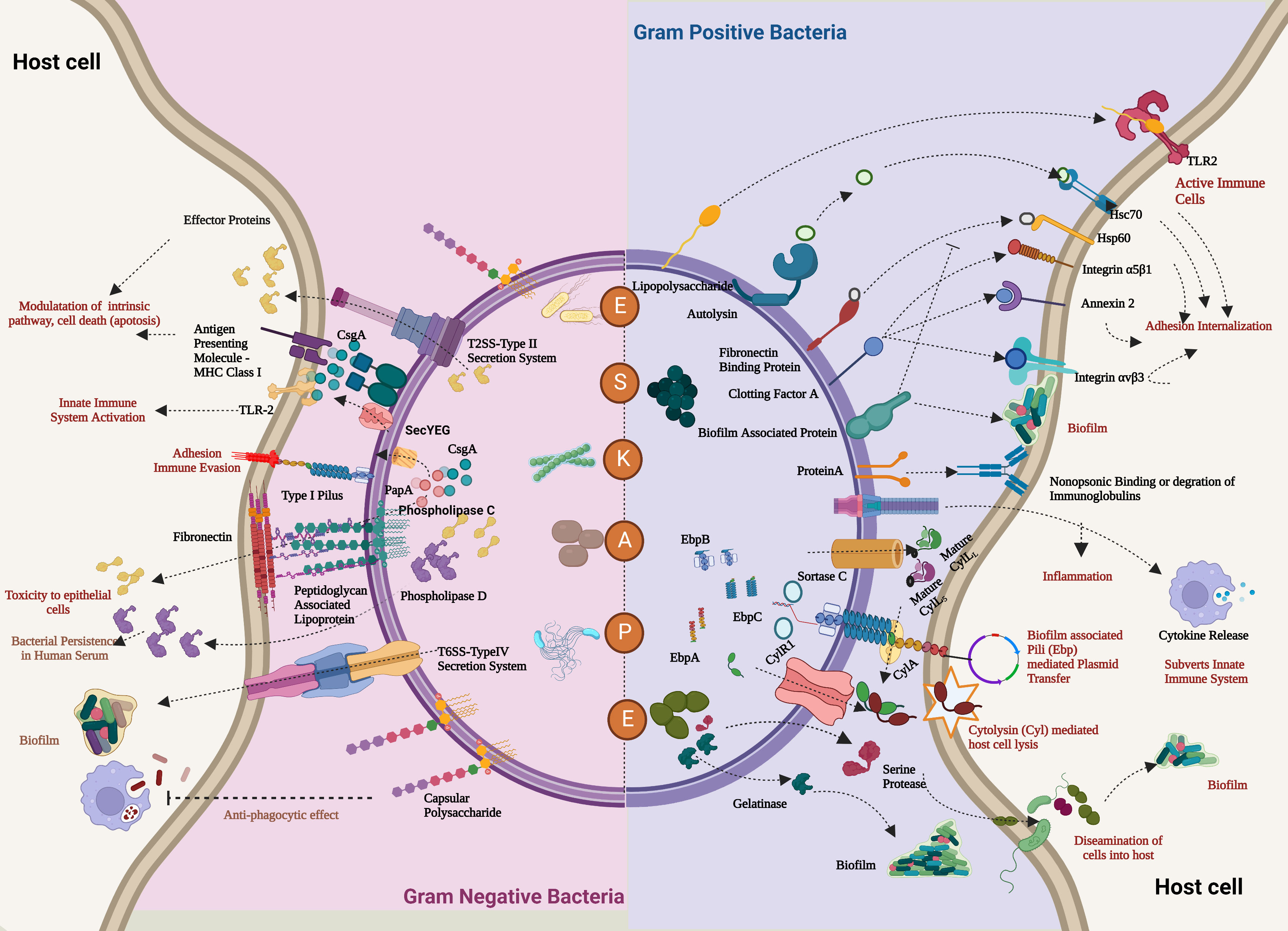
Figure 2 Comprehensive overview of the virulence factors of the ESKAPE pathogens. In the case of both Gram-positive bacteria (Enterococcus faecalis and Staphylococcus aureus) and Gram-negative bacteria (Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp.), the host evasion is orchestrated by the recurring events: adhesion to the host cells, Degradation by a range of degradative enzymes and toxins establishes biofilm to trigger the innate immune pathways and further deteriorates the cellular homeostasis of the host cell. In addition, these bacteria also transfer their virulence factors through horizontal gene transfer, which leads to persistent infections. Created with BioRender.com.
Biofilm formation
Biofilms are commonly associated with increasing antibiotic resistance due to their ability to protect pathogens from antibiotics and other environmental stress factors. Biofilms act as a physical barrier that prevents the diffusion of antimicrobials and upregulates specific biofilm-associated virulent genes contributing to antimicrobial resistance (Tuson and Weibel, 2013; Bowler et al., 2020). Understanding the course of biofilm formation and its regulation could be instrumental in preventing biofilm formation, re-structuring, and disintegrating existent biofilms (Dale et al., 2017). Biofilm formation predominantly involves four stages/moves by the bacterial team: 1) adhesion, 2) microcolony formation, 3) biofilm growth and maturation, and 4) dispersal.
Move 1: adhesion
The first and foremost step in forming a robust biofilm is surface adhesion. For instance, targeting this phase of biofilm formation, which depends on various factors, including surface charge, roughness, wettability, stiffness, topography, and bacterial motility, through different physical and chemical methods has been proven to be successful (Solanki et al., 2018; Zheng et al., 2021; Uneputty et al., 2022). Various adhesion-related genes, including the ones coding for aggregation substance agg1, collagen binding proteins ace, and enterococcal surface protein esp, were highly prevalent and were found to play a significant role in determining the virulence of E. faecalis clinically (Strateva et al., 2016). The deletion of ebp—the pilus-encoding gene—is reported to significantly impact the virulence and biofilm-forming ability of E. faecalis (Sillanpää et al., 2010). Another study by Soares et al. identified that genes that aid adhesion—esp and agg—are crucial for augmenting biofilm formation in the clinical isolates of Enterococcus sp. (Soares et al., 2014). However, a former study has observed enterococcal biofilms without esp, highlighting that this factor is not essentially indispensable for biofilm formation (Kristich et al., 2004). For a more detailed overview of enterococcal biofilm formation, the readers are directed to the review by Ch’ng et al. (Ch’ng et al., 2018). In the case of S. aureus, genes that encode fibronectin-binding protein (fib, fnbA, and B), clumping factors (clfA and B), elastin-binding protein (ebp), and serine-aspartate repeat family (sdr) are known to mediate surface adhesion (Chen Q et al., 2020). A recent study identified the presence of clfB, ebp, and sdrD in multidrug-resistant S. aureus strains isolated from periodontal lesions of patients and found an increased incidence of biofilms among these isolates (Uribe-García et al., 2021). Along these lines, it was reported that reduced expression of adhesion-related genes agr and sdr further diminished the ability of MRSA to form biofilms (Iwata et al., 2021).
Similarly, genes that promote adhesion, including the ones that encode type III fimbriae fim, a homolog of enterococcal ebp and protein secretion system icm, have proven to be attractive targets to reduce the biofilm formation ability of the opportunistic bacteria K. pneumoniae (Schroll et al., 2010; Alcántar-Curiel et al., 2013; Vuotto et al., 2014). Reiterating surface adhesion’s crucial role in biofilms’ structural organization, Raffaella Campana et al. proved that reduced bacterial adhesion impaired the biofilm-forming ability of K. pneumoniae in medical devices (Campana et al., 2017). Nevertheless, another study identified a direct correlation between the strength of adhesion and the biofilm-forming ability of K. pneumoniae, supporting the idea of targeting the first step in biofilm formation for attenuating virulence (Lenchenko et al., 2020). Taken together, it can be concluded that adhesion determines the strength of biofilms, and therefore, targeting this could prove to be a promising strategy for tackling ESKAPE-mediated infections. However, many factors influencing adhesion, including the surface that bacteria adhere upon, multi-species environment, and types of appendages employed for adhesion, should be considered while deciding upon the targets and designing novel strategies against these pathogens.
Move 2: microcolony formation
The bacterial cells adhered to the surface and then proliferate and form structurally organized micro-colonies embedded in a matrix of polysaccharides, proteins, lipids, and nucleic acids (Karygianni et al., 2020). The extracellular polysaccharides influence the architecture and the immediate surroundings of the bacterial cells by affecting the hydrophobicity, mechanical stability, charge, porosity, water content, and other essential nutrients. Interestingly, oxygen, hydrogen, and nutrient gradients also form during this stage, creating different microenvironmental conditions within the biofilm (Petrova et al., 2012). This phase, in which solitary bacterial cells come together to form a microcolony, is crucial in understanding biofilm formation and targeting novel preventative and therapeutic strategies. Recent studies identified the ability of Enterococcus faecalis to develop distinct microcolonies on the entire valvular regions. However, these colonies’ potential to advance and cause infection is still less explored (Barnes et al., 2022). In the case of S. aureus, the matrix is predominantly proteinaceous due to Bap protein. Bap protein has been identified to be a crucial player in promoting biofilm formation in S. aureus (Taglialegna et al., 2016). In addition, various other proteins, including FnBPA, FnBPB, and SdrC, have been shown to contribute to microcolony formation (Schilcher and Horswill, 2020). mifR is one of the significant factors contributing to microcolony formation in P. aeruginosa. Petrova et al. identified the importance of pyruvate and its utilization through fermentation to promote the development of microcolonies (Petrova et al., 2012). Although the specific genes and regulatory mechanisms dictating microcolony formation of ESKAPE pathogens are not fully understood, the evidence points to the importance of understanding and manipulating the same to better fight against these pathogens. Considering that this step is crucial in determining the structural organization of the biofilms, tampering with this phase could also help bring down the tower-like and mushroom-shaped biofilms (Dale et al., 2017).
Move 3: biofilm maturation and dispersal
Biofilm maturation is triggered by the accumulation of extracellular polymeric substances (EPS), eDNA, formation of channels for waste disposal and nutrient exchange, varying ionic concentrations, and most, importantly, quorum-sensing signals (Moormeier and Bayles, 2017; Wang T. et al., 2019). It has also been reported that it is at this phase that the genes responsible for flagellar development are downregulated, satisfying the need for building a stable biofilm architecture (de Kievit, 2011). To start with the case of E. faecalis, the crucial role played by eDNA in biofilm maturation has been re-iterated continuously. It has been reported that the reduction in eDNA levels, by either cleaving the eDNA by Dnase or by preventing its release by inhibiting AtlA, significantly disrupts the enterococcal biofilm and makes it susceptible to treatment (Yu et al., 2019). Staphylococcal biofilms, however, are identified to exist in two different microcolony structures based on the expression of cidABC and irgAB (Moormeier and Bayles, 2017). Various EPS components, including Psl, Pel, alginate, eDNA, and the proteinaceous components, have been reported to play specific roles in forming and maturing Pseudomonas biofilms (Wei and Ma, 2013). Overall, infectious biofilms often observed in clinical settings have been known to be highly matured, and targeting such structurally robust biofilms has been a difficult challenge. Various modern advancements in the field of therapeutics—CRISPR technology, quorum-sensing inhibition, and antimicrobial peptides, among others—have proven to be promising despite the need for extensive research in the respective domains (Jiang et al., 2020; Nadar et al., 2022). Inducing the dispersal of individual bacterial cells embedded in the EPS has also been instrumental in tackling the infection, considering the increased susceptibility of planktonic cells to antibiotics and other antimicrobial strategies. This strategy, however, also has an inherent risk of speeding up bacterial colonization by actively triggering biofilm dispersal. A deeper understanding of the dispersal mechanisms of the ESKAPE pathogens would be beneficial in translating various strategies to the bedside.
Colonization and invasion
The whole point of adhering to the host team is to infiltrate the human team and render them insufficient (Pizarro-Cerdá and Cossart, 2006). The pathogens must overcome the ever-dynamic physiological host environment—temperature, pH, and presence of other components—to colonize successfully. ESKAPE pathogens are mostly commensal-turned or hospital-acquired pathogens that affect the gut and cause bacteremia, oral infections, wound infections, and urinary tract infections. As can be seen, each host niche is unique, and to establish infection, host barriers are to be surpassed. The most prominent barrier is the acidic pH (2 to 5). Enterococcus sp. has adapted to tolerate acidic pH (Başaran et al., 1998). Also, commensals are reduced due to the antibiotic’s treatment, leaving the way for Enterococcus sp. to flourish. Adherence to the host site strongly supports the translocation to other sites, including blood, lymph nodes, blood, and spleen (Fiore et al., 2019). A similar trajectory is followed by S. aureus, where it has to overcome the host barriers to colonize the host (Liu, 2009). The breach of the intact microbiota, immune system evasion, and immune cell colonization support successful colonization. Both Enterococcus sp. and S. aureus, belonging to the Gram-positive group, teichoic acids, have primarily played a role in the successful colonization of the host. Once the propagation in the host site begins, the pathogens start to produce virulence factors—especially toxins and enzymes to disarm the host immune system and bring damage to the host. Taking an aggressive stance by making extracellular enzymes and toxins damage the host tissue has been customary in easing this process (Upadhyaya et al., 2009; Newman et al., 2017). Hemolysin encoded by EF_0700 gene is a potent toxin that cleaves the erythrocytes found in Enterococcus sp. (Zhang et al., 2007). Similarly, gelatinase, encoded by gelE, cleaves the host gelatin, collagen, casein, hemoglobin, and peptides. hylefm, which encodes hyaluronidase, cleaves hyaluronate present in the connective tissues (Maasjost et al., 2019). Enterococcus sp. produces cytolysins, which are two-peptide bacteriocins that form pores and damage the host tissue, encoded by gene cassettes cylLL, cylL, cylM, cylB, and cylA (10.2217/fmb-2021-0212). Hemolysins α, β, γ, and δ, which cleave erythrocytes encoded by hla, hlb, hld, and hlg, also present in S. aureus (Wang et al., 2014; Motamedi et al., 2018). hysA encodes hyaluronidase (Ibberson et al., 2014), ybhu_2 encodes collagenase, and lukS-PV and lukF-PV code Panton-Valentine Leukocidin, which forms pores (Melles et al., 2006) in the host system aid for S. aureus colonization process. Staphylokinase encoded by sak binds with the host plasminogen resulting in the plasmin enzyme, which essentially aids in the S. aureus penetration into the tissues (Sako and Tsuchida, 1983). In the case of Gram-negative pathogens, phospholipase D production, which cleaves phospholipase and hemolysin, is commonly used to damage the host. hly and pld1 genes in K. pneumoniae encode hemolysin (Pereira and Vanetti, 2015; Esmaeel and Sadeq, 2018) and phospholipase D (Lery et al., 2014), respectively. In A. baumannii, pld gene encodes phospholipases (PLC and PLD) (Lee et al., 2017; Murray et al., 2017), cipA gene encodes CipA, which has a similar function as staphylokinase, binds to plasminogen, and promotes penetration of A. baumannii in the endothelial monolayers (Koenigs et al., 2016). toxA encodes endotoxin in P. aeruginosa, which also forms pores in the cell membrane (Pollack, 1984; Dapgh et al., 2019) and also produces phospholipase encoded by pclH (Dapgh et al., 2019). In Enterobacter sp., hemolysin is encoded by hly (Burgos, 2010), whereas PtrA, B, and C families of proteases are encoded by prtA, prtB, and prtC, which cleave host proteins and promote colonization of the host (Ghigo and Wandersman, 1992).
Evading the immune system, the defending team is the next crucial step after getting hold of the ball (Finlay and McFadden, 2006). Different capsular serotypes, peptidoglycan, teichoic acid, and protein A have helped bacteria escape from the host humoral and cellular innate defenses by fooling them and turning them down (Leitão, 2020). Capsular polysaccharides have an evasion process to escape the immune system. These capsular polysaccharides surround the bacterial surface and evade complement activation, phagocytic killing, and opsonization (Merino and Tomás, 2010). cpsF, cpsC, cpsD, cpsE, cpsG, and cpsI in Enterococcus sp. encode the capsule (Thurlow et al., 2009). cap1 in S. aureus encodes type 1 capsular polysaccharide (Luong et al., 2002), and cps in K. pneumoniae (Hsu et al., 2016) and cps gene clusters in A. baumannii encode the capsule polysaccharide (Singh et al., 2019). In addition, clumping factors and teichoic acids encoded by clfA and B (Higgins et al., 2006) and tarB, tarD, tarF, tarIJ, and tarH (D’Elia et al., 2006) inhibit phagocytic engulfment in S. aureus. Cell membrane components play an essential role in the immune evasion process. In A. baumannii, lpxA, lpxC, and lpxD encode lipopolysaccharide, which effectively binds to the CD14/TLR4/MD2 receptor complex of immune cells and subverts its action (Moffatt et al., 2013; Lee et al., 2017). Outer membrane proteins modulate autophagy, which is mediated by ompA, omp33-36, and omp22 genes encoding for OmpA, Omp 33-36, and Omp-22, respectively. Alkaline protease encoded by aprA (Iiyama et al., 2017) and elastase encoded by lasB (Rust et al., 1996) evade the immune system by cleaving immunoglobulins, inactivating the complement system and several cytokines (TNF, IFN, IL1, and IL6).
Further, to improve the chances of winning, the bacterial team strengthens itself through horizontal gene transfer (Lerminiaux and Cameron, 2019). This trait has empowered the bacteria not primarily equipped with specific virulence factors and has posed an arduous challenge to the opponent team. For instance, a recent study reported the transfer of various virulence-related genes in Staphylococcus sp., which increased its pathogenicity (Smith and Andam, 2021). Bacterial cell wall appendages promote horizontal gene transfer to a large extent. Pili, hair-like appendages, primarily facilitate conjugation and transfer antibiotic resistance genes from one bacterium to another (Sun, 2018). ebp, pila, and pilb genes in Enterococcus sp. (Hendrickx et al., 2009); ecpA, ecpR, and ecpB in K. pneumoniae (Alcántar-Curiel et al., 2013); and pilU in P. aeruginosa (Whitchurch and Mattick, 1994) encode pili that facilitate conjugation. In addition, Sortase A enzyme of S. aureus, encoded by sau‐srtA that links the surface proteins to peptidoglycan (Khare and Narayana, 2017) and the type VI secretion system, also play a role in horizontal gene transfer.
The bacterial team also constantly competes with the human team for resources such as free iron (Kronstad and Caza, 2013). Iron is an essential metal that bacterial pathogens require for multiple processes like respiration, metabolism, and other iron-dependent cellular processes. The iron requirement is huge for bacteria, and iron acquisition is a prerequisite to sustaining them in the host environment. Similarly, iron is a co-factor for multiple enzymatic processes in the human system. They are also found in metalloprotein heme complexes: hemoglobin, myoglobin, catalases, cytochromes, and aconitase as Fe-S clusters. Immune cells, macrophages, and other cells are used as iron transporters during iron deficiency; thus, iron homeostasis is maintained. Hence, iron competition is fierce between the pathogens and the host. Bacteria have developed various mechanisms to sequester available iron from the environment. Ferric uptake regulatory proteins (Fur) are essential for maintaining iron homeostasis in most bacterial pathogens, especially Enterococcus sp. (Latorre et al., 2018). In S. aureus, sbn and sfa encode siderophores staphyloferrin A (SA) and staphyloferrin B (SB). K. pneumoniae has iuc and iro genes that encode siderophores aerobactin (iuc) and salmochelin (iro). Acinetobactin NfuA of A. baumannii and fpv in P. aeruginosa encode siderophores: pyoverdines (PVDI, PVDII, and PVDIII) and FpvAI and FpvB (Hartney et al., 2013; Bonneau et al., 2020). Yersiniabactins encoded by irp1, irp2, and fyuA are responsible for iron acquisition.
Also, the constantly evolving host–bacterial interactions determine the extent of the underlying pathogenesis by influencing the process of adherence, invasion, and biofilm formation. For instance, Scherr TD et al. identified the differential expression of genes associated with biofilm formation in S. aureus when exposed to different subsets of immune cells, aiding in its persistence (Scherr et al., 2013). In addition to the immune factors, the host microenvironment in vivo influences the biofilm’s nature. Rahman MUA et al. identified the role of free collagen in determining the viscoelasticity of P. aeruginosa biofilms. Understanding biofilms’ stability and homogeneity and the way the host environment dictates it could prove instrumental in replicating in vivo conditions more accurately and in targeting biofilms more efficiently (Rahman et al., 2021). A recent study reported the role of interaction between host fibronectin and peptidoglycan-associated protein of A. baumannii in biofilm formation. It explored the possibility of therapeutic targeting of this bacterial protein to augment the immune response (Solanki et al., 2023).
One other key strategy is to form biofilms by aggregating with each other within and across species. A plethora of evidence suggests biofilm formation aggravates the infection by improving cell adhesion, colonization, and horizontal gene transfer. Significant factors, including the capsule, aggregation substance, pili, and fimbriae, are reported to be associated with assisting biofilm formation. In particular, the capsule contributes toward shielding the bacteria from various harsh conditions, including pH, temperature, ultraviolet (UV) radiation, antibiotics, and poor nutrients, by acting as physical barriers and by providing a confluent microenvironment, thereby sustaining survival and metabolism (Yin et al., 2019; Vor et al., 2020). Various stress conditions, including pH, temperature, and oxygen availability, are crucial in triggering biofilm formation in certain bacteria, such as S. aureus, P. aeruginosa, and Enterobacter sp. (Hoštacká et al., 2010; Gupta et al., 2016; Chu et al., 2018). It is essential to emphasize that more than one virulence factor generally acts in synergy to introduce the infection (Figure 1B) successfully.
Antibiotics: the substitutes
Time has arrived for the human team to employ innate and adaptive immune strategies to prove its competence against the bacterial squad, which has skillfully scored well in the first half of the match. Since relying only on the immune cells has proven inadequate, recruiting substitutes to strengthen the team has been hypothesized to be a good strategy (Figure 1C). Arsphenamine, a toxic dye, was one of the first substitutes that signed up for the match. Despite the effectiveness of this dye in treating syphilis, arsphenamine has not been employed widely owing to its toxicity to human cells, which ultimately kills the patients (FROM DYES TO PEPTIDES: THE EVOLUTION OF ANTIBIOTIC DRUGS | SCQ). Scrutinizing the target specificity and sensitivity of the drug is a crucial step in developing novel drug classes. Conscribing penicillin, the serendipitous drug, has manifested itself as one of the finest action plans until recently (Gaynes, 2017). Since then, an extensive range of antibiotics has been synthesized from various sources targeting Gram-positive and Gram-negative bacteria. Targeting the molecular mechanisms involved in cell growth (bacteriostatic) and bacterial survivability (bactericidal) has been authenticated to be an effective method (Pankey and Sabath, 2004). Antibiotics have proven to be a valuable addition to the human team by scoring goals (restoring “health”) and reducing the bacteria’s activity by binding with them.
Nonetheless, this effect was not persistent. The delimiting nature of monotherapy to tackle the infection has laid the foundation for recruiting more antibiotics against the skillful bacterial team. In this instance, the game started to change with a much-unexpected twist. Indiscriminate employment of players uninformed about the opponent team, such as the non-specific antibiotics, started turning down the strength of the human group (Om et al., 2016). To exacerbate the situation, the bacterial team has started unveiling their opponent’s strategies and devising new mechanisms to fool the combatants (Santajit and Indrawattana, 2016). Using the same class of antibiotics multiple times has been reported to be one major pitfall that alerted the bacterial team to decode our game plan. Nevertheless, modifying the scaffolds of the previously designed antibiotics has raised their potency and increased the chances of winning for the human team. Still, the bacterial team has formulated innovative plans, such as the utilization of efflux pumps and enzymes, chemical modification of drugs and the target, and alteration in membrane permeability, leading to the development of the pressing issue of antimicrobial resistance, the central feature that has raised the stature of the ESKAPE pathogens.
Antibiotic resistance mechanisms: the masterstroke
As mentioned, the bacterial team has emerging mechanisms to overcome antibiotic stress. ESKAPE pathogens have the gene(s) employed for each class of antibiotics for the resistance mechanism. The primary class of antibiotics is β-lactams, aminoglycosides, chloramphenicol, glycopeptides, tetracyclines, oxazolidinones, macrolides, ansamycins, streptogramins, and lipopeptides. Each class of antibiotic has a specific mechanism of action against bacteria and, hence, an exact resistance mechanism. The typical resistance mechanisms are antibiotic-inactivating enzymes, overexpression of efflux pumps, modifications in the target site, and the acquisition of resistance genes through horizontal gene transfer (Bhukta et al., 2022) (Figure 3). Table 2 elaborates on the specific set of genes essential for the resistance process of each antibiotic used.
Figure 3 Antibiotic resistance mechanism of ESKAPE pathogens. ESKAPE pathogens have developed various antibiotic resistance mechanisms against the different classes of antibiotics ranging from aminoglycosides to carbapenems. The exact ways each of these pathogens develops and disseminates resistance through biofilms vary widely. However, the most common mechanisms include the overexpression of efflux pumps, modification of cell wall composition and permeability, modification of the target, inactivation of the antibiotics, and reduction in antibiotic penetration through biofilm formation. Created with BioRender.com.
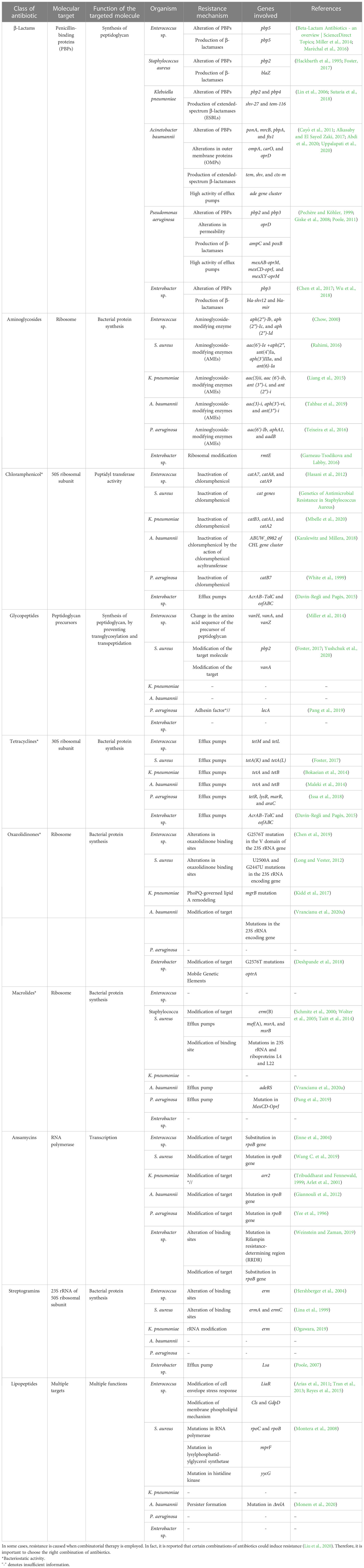
Table 2 Summary of the antibiotics employed and the resistance mechanisms evolved by the ESKAPE pathogens
β-Lactams
β-Lactams are one of the commonly administered drugs against bacterial infections. They target penicillin-binding proteins (PBPs) and carboxypeptidases involved in peptidoglycan synthesis. β-Lactams form a stable covalent complex with PBPs and stall the cell wall synthesis, leading to cell death. To overcome the survival pressures, bacteria have evolved to resist β-lactams by altering their proteins, producing β-lactam-degrading enzymes, and using excessive efflux pumps to efflux the antibiotics. In the case of Enterococcus sp., pbp5 is responsible for altering PBPs and β-lactamase production (Miller et al., 2014; Maréchal et al., 2016). In S. aureus, pbp2 gene is required for the protein alteration, whereas blaZ is responsible for β-lactamase (Hackbarth et al., 1995; Foster, 2017). K. pneumoniae utilizes pbp2 and pbp4 for altering PBPs and shv-27 and tem-116 for the production of ESBLs (Lin et al., 2006; Sutaria et al., 2018). A. baumannii alters PBPs using ponA, mrcB, pbpA, and fts1; tem, shv, and ctx-m for the production of ESBLs; ompA, carO, and oprD for the alteration of the outer membrane proteins; and ade gene cluster to accentuate the high efflux pump activity (Cayô et al., 2011; Alkasaby and El Sayed Zaki, 2017; Abdi et al., 2020; Uppalapati et al., 2020). P. aeruginosa and Enterobacter sp. rely on pbp3 for the alteration of PBPs. P. aeruginosa employs ampC and poxB for the production of β-lactamases; permeability modification and high efflux pump activity are brought about by oprD, mexAB-oprM, mexCD-oprJ, and mexXY-oprM (Pechère and Köhler, 1999; Giske et al., 2008; Poole, 2011). bla-shv12 and bla-mir of Enterobacter sp. are required to produce β-lactamases (Chen et al., 2017; Wu et al., 2018).
Aminoglycosides
Aminoglycosides are broad-spectrum antibiotics that can be used against Gram-negative and Gram-positive pathogens. They are known to bind to ribosomes and affect the translation of proteins. Structurally, aminoglycosides (AGs) are 2-deoxystreptamine (2-DOS) attached with amino-modified sugars. Owing to their structure, bacteria have developed intrinsic resistance by lowering the AGs’ permeability through the modified bacterial cell wall. They also employ modifying enzymes: Aminoglycoside modifying enzymes (AME) and RNA methyltransferases. AMEs are the most common AG resistance operated by the ESKAPE group. These are family enzymes that inactivate an aminoglycoside at a specific position; hence, the gene responsible carries the modification site number. These enzymes are further divided into three classes based on the modification of substrates: AG N-acetyltransferases (AACs), AG O-nucleotidyltransferases (ANTs), and AG O-phosphotransferases (APHs). aph(2″)-Ib, aph(2″)-Ic, and aph(2″)-Id of Enterococcus sp. encode AG O-phosphotransferases majorly (Chow, 2000). In S. aureus, genes such as aac(6′)-Ie +aph(2″), ant(4′)Ia, aph(3′)IIIa, and ant(6)-Ia are present and can target all three types of substrates (Rahimi, 2016). K. pneumoniae possesses genes aac(3)ii, aac(6′)-ib, ant(3″)-i, and ant(2″)-i, which focus on the AACs and ANTs (Liang et al., 2015). All three methyltransferases are present in A. baumannii encoded by aac(3)-i, aph(3′)-vi, and ant(3″)-i (Tahbaz et al., 2019). P. aeruginosa possesses genes aac(6′)-Ib, aphA1, and aadB, which are required for modifying AGs (Teixeira et al., 2016). In the case of Enterobacter sp., ribosomal modification is brought about by rmtE encoding ribosomal methyltransferase, which methylates the nucleotide G1405 at the N7 position and confers resistance to aminoglycosides (Garneau-Tsodikova and Labby, 2016).
Chloramphenicol
Chloramphenicol is a broad-spectrum antibiotic that is extracted from Streptomyces sp. Depending on the concentration, chloramphenicol can be bacteriostatic and bactericidal. It binds to the 50S subunit of the ribosome, blocking the peptide bond formation and, thus, the protein synthesis. Enzyme inactivation is the standard mechanism of resistance to chloramphenicol, especially by chloramphenicol acetyltransferase (CAT). CAT inactivates chloramphenicol by modifying the 3-hydroxyl group through acetyl-S-CoA-dependent acetylation. Another means is through the overexpression of efflux pumps. In Enterococcus sp., three prominent genes, catA7, catA8, and catA9, encode CAT (Hasani et al., 2012). S. aureus cat genes are also prevalent in the MRSA strains (Udo et al., 2021). In K. pneumoniae, catB3, catA1, and catA2 are expressed to inactivate chloramphenicol (Mbelle et al., 2020). In the case of A. baumannii, recent studies showed that mutations in ABUW_0982 of the CHL gene cluster encoding permease contribute to the intrinsic resistance and thereby reduce the permeability of the chloramphenicol into the cell (Karalewitz and Millera, 2018). catB7 gene in P. aeruginosa encodes CAT, leading to chloramphenicol resistance (White et al., 1999). In Enterobacter sp., efflux pumps are the primary cause of chloramphenicol resistance; mainly, AcrAB–TolC and eefABC encoded efflux pumps (Davin-Regli and Pagès, 2015).
Glycopeptides
Glycopeptide antibiotics (GPAs) are specifically administered against Gram-positive pathogens as a last-resort treatment. GPAs are glycosylated cyclic or polycyclic peptides (non-ribosomal) found naturally and synthetically. GPAs prevent the crosslinking of the peptidoglycan layer by specifically binding to the peptidoglycan precursors (d-Ala-d-Ala dipeptide), leading to incomplete transpeptidation and transglycosylation in Gram-positive pathogens. The perturbation in the peptidoglycan synthesis leads to defective cell walls, thereby leading to cell death. Gram-negative pathogens intrinsically resist GPAs based on their cell wall composition. The resistance to GPAs is brought about by modifying the target, unlike the shared mechanism of altering the antibiotic. Among the GPAs, vancomycin resistance is most common and reported widely (Yushchuk et al., 2020). The dipeptide sequence, d-Ala-d-Ala, is replaced by d-Ala-d-Lac or d-Ala-d-Ser, leading to the reduced affinity of the GPAs to the precursors. The genes bring about such replacements—vanH, vanA, and vanZ—in the case of Enterococcus sp. (Miller et al., 2014). It is shown that vancomycin resistance to S. aureus is through horizontal gene transfer from Enterococcus sp., and genes pbp2 and vanA are responsible for the modification of the target dipeptide (Foster, 2017; Yushchuk et al., 2020).
Tetracyclines
Tetracyclines are broad-spectrum antibiotics used to treat Gram-positive and Gram-negative pathogens and protozoan parasites in some cases. They are natural products obtained from Streptomyces sp. Tetracyclines bind to 30S ribosomal subunit and interact with 16S rRNA, interfering with the peptide elongation process (Grossman, 2016). They are generally bacteriostatic, but in some cases, bactericidal activity is also reported (Tessier and Nicolau, 2013). Both extrinsic and intrinsic resistance mechanisms bring about resistance to tetracycline. The critical resistance processes are overexpression of efflux pumps, mutations in the tetracycline binding site, inactivation of tetracycline, and expression of tetracycline-specific ribosomal protection proteins. The tetracycline-specific efflux pumps belong to the major facilitator superfamily (MFS), which excludes tetracycline at a proton’s expense. In Enterococcus sp., tetM and tetL encode the genes responsible for tetracycline exclusion, while tetK and tetL are required for S. aureus (Foster, 2017). Tet(K) and Tet(L) are expressed in Gram-positive pathogens, which are antiporters of monovalent H+ having 14 transmembrane segments of α and β domains. In both K. pneumoniae and A. baumannii, tetA and tetB are present and encode the H+ antiporters having 12 transmembrane segments of α and β domains (Bokaeian et al., 2014). Tet(A) and Tet(B) are present mainly in Gram-negative pathogens (Maleki et al., 2014). P. aeruginosa possesses tetR, lysR, marR, and araC genes that encode the efflux pumps (Issa et al., 2018). In contrast, acrAB–tolC and eefABC also play a role in tetracycline efflux in Enterobacter sp. (Davin-Regli and Pagès, 2015). Tetracycline-specific ribosomal protection proteins (RPPs), having significant similarity to elongation factors EF-G and EF-Tu, bring about conformational change in the ternary complex and enable translation even in the presence of tetracycline (Dönhöfer et al., 2012). Inactivation of tetracycline is facilitated by tet(X) gene that encodes Tet(X) monooxygenase enzyme that inactivates tetracycline by the addition of hydroxyl group in C11 position of the tetracycline core (Aminov, 2013). Such RPPs and Tet(X) enzymes are found in ESKAPE pathogens, leading to tetracycline resistance.
Oxazolidinones
Linezolid and tedizolid belong to oxazolidinones, synthetic drugs for treating Gram-positive pathogens resistant to other antibiotics. Gram-negative pathogens are also treated with these antibiotics in some cases. These bacteriostatic antibiotics inhibit protein synthesis by binding to the P site of the 50S ribosomal subunit (Bozdogan and Appelbaum, 2004). Development of resistance to oxazolidinones is rare, but reports show a common mechanism of resistance, unlike other antibiotics. Resistance is conferred by altering the oxazolidinone binding sites by mutations in 23S rRNA and acquiring mobile genetic elements (Brenciani et al., 2022). In Enterococcus sp., alterations in binding sites are through G2576T mutation in the V domain of the 23S rRNA gene (Chen et al., 2019), whereas in S. aureus, alterations in binding sites are through U2500A and G2447U mutations in the 23S rRNA encoding gene (Long and Vester, 2012). In K. pneumoniae, mgrB mutation leads to PhoPQ-mediated lipid A remodeling (Kidd et al., 2017). G2576T mutations that modify the target and optrA mobile genetic elements facilitate the resistance in Enterobacter sp. (Deshpande et al., 2018).
Macrolides and streptogramins
Macrolides are a class of antibiotics that primarily target Gram-positive pathogens but also have been shown to possess broad-spectrum activity. Structurally, they have 14-, 15-, or 16-membered lactone rings having sugar moieties and other substitutions in the lactone ring. Macrolide antibiotics target protein synthesis by binding to large subunits, leading to cell growth arrest (Nakajima, 1999). The primary resistance mechanisms are modification of the target site, 23S rRNA, mediated by erm gene, overexpression of efflux pumps, and inactivation of the antibiotics through esterase and macrolide phosphotransferase enzymes. erm gene encodes Erm methyltransferase, which catalyzes the demethylation of the macrolide binding site leading to the reduced affinity brought about by stearic hindrance (Gaynor and Mankin, 2003). S. aureus to overcome macrolide pressure—erm(B), mef(A), msrA, and msrB genes—to encode efflux pumps is present (Schmitz et al., 2000; Wolter et al., 2005; Taitt et al., 2014). A. baumannii overexpresses adeRS efflux pumps to reduce the accumulation of macrolides (Vrancianu et al., 2020b), whereas P. aeruginosa relies on the mutation in MexCD-OprJ efflux pumps (Pang et al., 2019). The other inactivating enzymes are not significantly reported in the clinical isolates.
A similar mechanism of action is followed by streptogramins, even though they are structurally diverse from macrolides. Streptogramins contain two subunits of distinct classes—type A and type B. They interfere with peptidyl transferase activity, inhibiting protein synthesis (Johnston et al., 2002). Individually, type A and type B are bacteriostatic, but they exhibit bactericidal activity when combined. Another commonality is the resistance mechanism against streptogramins—modification of target mediated by erm gene. Erm methyltransferase is present in Enterococcus sp. (Hershberger et al., 2004), S. aureus (Lina et al., 1999), and K. pneumoniae (Ogawara, 2019), leading to alteration of the target site and, thus, resistance. Enterobacter sp. uses lsa efflux pump to efflux out the streptogramins (Poole, 2007). Gram-negative pathogens are intrinsically resistant to streptogramins owing to the impermeability of their cell membrane.
Ansamycins
Ansamycins are rigid antibiotics because they have an aromatic nucleus and a long aliphatic bridge with a handle shape. This unique configuration confers unique biological properties. They target RNA polymerase (RNAP) in bacteria, which is essential but also structurally diverse from humans. Ansamycins bind to RNAP near the catalytic site, leading to abortive transcription. Thus, modification of the target site is the primary resistance mechanism and mainly maps to the ropB mutation. These mutations are single amino acid substitutions pointing to a few deletions or mutations in the case of Enterococcus sp. (Enne et al., 2004), S. aureus (Wang C. et al., 2019), A. baumannii (Giannouli et al., 2012), P. aeruginosa (Yee et al., 1996), and Enterobacter sp. (Weinstein and Zaman, 2019). Other resistance mechanisms include arr2 gene responsible for the inactivation of rifamycin through ribosylation (Tribuddharat and Fennewald, 1999; Arlet et al., 2001).
Lipopeptides
Lipopeptides are a class of antimicrobials derived naturally from Actinomyces, Bacillus, and Pseudomonas sp. Structurally, they are made of hydrophilic peptides and attached to a fatty acyl chain, which is hydrophobic. They exist in linear and cyclic forms, with up to 25 amino acid chains (Patel et al., 2015). The most prominent lipopeptides like polymyxins, daptomycin, surfactin, iturin, and pseudofactin take the cyclic form. Even though the exact mechanism of action of lipopeptides is yet to be elucidated, studies have shown interactions with the bacterial cell membrane calcium (Ho et al., 2008), and phospholipid phosphatidylglycerol has been shown to play a role in the antimicrobial action. These interactions improve the access to lipopeptide antibiotics in the bacterial cell membrane, thereby interfering with the integrity of the cell membrane, leading to cell death. Lipopeptide antibiotics insert in the cell membrane form pore, extract the lipid in the membrane, and translocate the membrane. Thus, resistance mechanism developed by bacteria is focused on modifications in the cell membrane protein. Through physical repulsion, bacteria evade the incoming antibiotic. In Enterococcus sp., liaR gene modifies the cell envelope stress response, and cls genes that encode cardiolipin synthase decrease the surface charge of the membrane and modify the phospholipid composition (Arias et al., 2011; Tran et al., 2013; Reyes et al., 2015). The resistance mechanism against lipopeptides are studied extensively in S. aureus. It was found that the changes in surface charge and modification or overexpression of lipopolysaccharide layer forming septa are the major mechanisms of resistance. mprF mutation encoding lysyl phosphatidyl glycerol synthetase leads to gain-of-function and thereby increases synthesis of positive charged lipopolysaccharide. Mutation in histidine kinase yycG leads to increased glycan chain length (Montera et al., 2008).
Alternate strategies: a way of escaping from ESKAPE pathogens
The bacterial team has seized the “ball of health” once again, despite recruiting new substitutes into the human team, which now cannot afford to increase the dosage of the recruited antibiotics due to the impending risk of toxicity. However, various alternate strategies are currently employed against the ESKAPE pathogens (Table 3; Figure 1D). Drug repurposing, where a drug used for another ailment, a previous-generation antibiotic currently in limited use or an orphan drug, is utilized as an antimicrobial agent, offers a new opportunity to invest in tuning up the strategies of the existing players. This is important, considering the significant time and money invested in identifying novel classes of antibiotics that are less prone to AMR (Silver, 2011). Modifying the functional groups helps build novel and effective antibiotics with the existing scaffold (Kamurai et al., 2020). Another quick-witted move along these lines is reinforcing combinatorial drugs with good chemistry in the team (Cheng et al., 2019). Adjuvants such as β-lactamase inhibitors prevent the degradation of the β-lactam antibiotics (Drawz and Bonomo, 2010; Ripoll et al., 2014), and efflux pump inhibitors inhibit the overexpressing efflux pumps, retaining the antibiotics to complete their course of action (Sharma et al., 2019; Verma et al., 2021), support the action of antibiotics by rendering a “double-attack defense”, and make it harder for the bacteria to shoot the target (González-Bello, 2017). Multiple strategies, such as monoclonal antibodies [which bind to the specific epitope of the bacterial cell targeting the conserved pathogenesis pathway and initiate immunological response leading to a second line of defense (Chames et al., 2009)], vaccines as a prophylactic tool to prevent the infection, and fecal microbiota transplant [one of the current trends where the stool from the healthy volunteer is transplanted into the patient helps in reversing the microbiome dysbiosis (Leshem et al., 2019)], are developed by tailoring specific drugs that target the rivals (Woodworth et al., 2019; Bekeredjian-Ding, 2020; Zurawski and McLendon, 2020). Consigning all-rounders like metal nanoparticles augments the team’s strength by targeting multiple mechanisms simultaneously (Borthagaray et al., 2018). There are multiple reports on the use of nanoparticles—metal, metal oxides, and polymeric—as a potential therapy to overcome the problem of resistance (Sharmin et al., 2021). Nanoparticles impart antibacterial activity at different levels: inhibit cell wall synthesis, inhibit biofilm, and target RNA and protein synthesis (Wang et al., 2017). The activity is achieved by increasing the reactive oxygen species that disintegrate the cell’s membrane potential (Slavin et al., 2017). These nanoparticles are also used as drug carriers for targeted action against pathogenic bacteria as against normal microbiota (Allahverdiyev et al., 2011). Silver, gold, zinc, copper, Cerium oxide, magnesium, chitosan, and cellulose-based nanoparticles are currently exploited as antimicrobials (Sánchez-López et al., 2020). Photo-antimicrobials are another interesting approach that combines the activity of dyes and light. Photo-antimicrobials absorb energy from the visible or infrared light and transfer it to molecular oxygen to generate reactive species—superoxide anions, singlet oxygen, and hydroxyl radicals—that can disrupt cells at multiple levels of proteins, lipids, and nucleic acids. Development of resistance is unlikely, as the target of action is not specific, and internalization of the drug is not mandatory in photodynamic therapy (Wainwright et al., 2017). To hold back the offending bacterial team, conjugation inhibitors and plasmid curing techniques are employed, which inhibit horizontal gene transfer and prevent the dissemination of the AMR genes into the bacterial community (Vrancianu et al., 2020b). Interestingly, taking inspiration from its opponents, the human team has been developing CRISPR-Cas-based systems to specifically compromise the antimicrobial resistant phenotype of the ESKAPE pathogens. Even though the guide-RNA based tool can be targeted against the virulent genes that contribute to antimicrobial resistance without affecting the natural microbiota, it comes with its own set of concerns including the possibility of off-target effects, reduced feasibility of the delivery system in vivo, and the involvement of the immune system (González de Aledo et al., 2021). Furthermore, various post-translational modifications (PTMs) of the ESKAPE pathogens could be targeted, considering their role in modulating the function of the proteins associated with bacterial virulence, motility, quorum sensing, and biofilm formation (Tiwari, 2019). Contrastingly, in the context of host–pathogen interactions, ESKAPE pathogens are reported to alter the PTMs of host proteins. Youssouf N et al. reported the ability of S. aureus to decrease the SUMOylation levels in the macrophages to enhance its chances of survival (Youssouf et al., 2021). One of the most elegant moves made by the human team is to recruit players with an excellent history of playing with the bacterial team. The involvement of bacteriophages in the game has proven to be a winning strategy because of its high specificity and efficiency (El Haddad et al., 2019). Phage therapy uses bacteriophages that infect pathogens as a treatment, which has been considered very potent in recent years. Precision medicine, i.e., phage preparations, can be performed for a specific set of clinical isolates that infect a patient. Phage cocktails and synergy with antibiotics are currently under consideration to prevent the development of resistance against phage therapy (Hatfull et al., 2022). In addition, lectin inhibition is considered promising, where naturally available lectins bind to the carbohydrates in the bacterial cell membrane. The interaction inhibits the invasion of the pathogen into the host and evokes the host’s immune response (Breitenbach Barroso Coelho et al., 2018). Along similar lines, essential oils have been shown to have antibacterial and anti-biofilm effects due to their ability to counter various virulence factors and quorum-sensing networks in ESKAPE pathogens. The ability to eradicate existing biofilms and their combinatorial effects on bacterial populations when employed with antimicrobials make them an attractive target (Panda et al., 2022). Iron chelation is also one of the promising approaches to overcoming antibiotic resistance. Iron is an important nutrient for pathogenic bacteria utilized for the essential growth and survival processes and in the host’s pathogenesis and invasion. Chelators (such as hydroxamates, catechols, and amino carboxylates) coordinate with Fe(III), reduce iron availability to the pathogens, and inhibit their growth (Vinuesa and McConnell, 2021). Several plant-based natural products are also exploited as antibacterial agents. Plants are a rich source of phytomolecules, which either alone or in combination impart antibacterial action against resistant pathogenic bacteria. They can act as efflux pump inhibitors, inhibit protein and nucleic acid synthesis, and disrupt cell membranes (Vaou et al., 2021). However, most of the strategies are at risk of inducing the onset of resistant phenotypes. However, immune boosters act as the energy drink for the human team and help build a strong defense, which complicates the process of scoring a goal by the bacterial team (Figure 1).
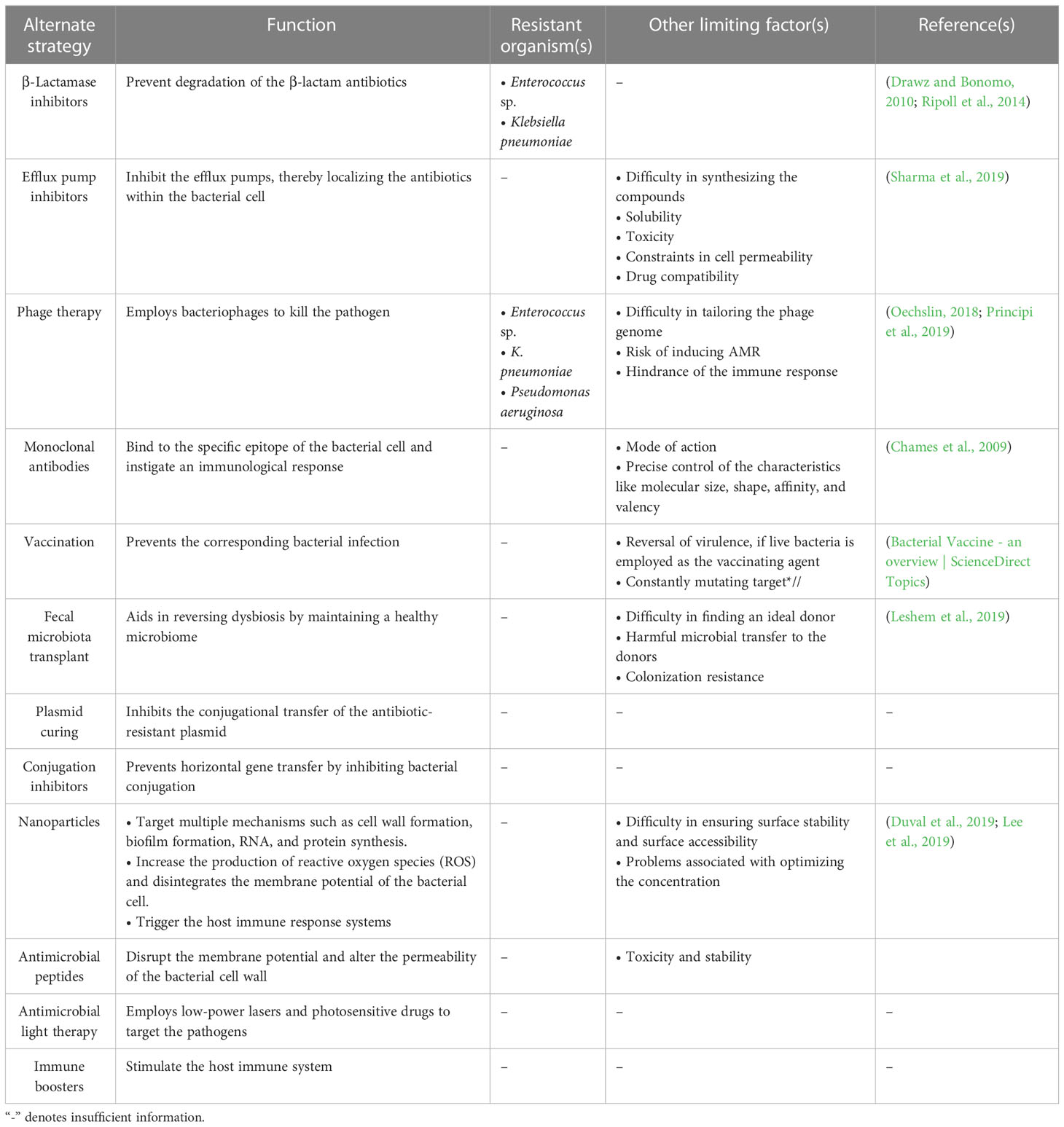
Table 3 Summary of the alternate strategies employed against the ESKAPE organisms and their limitations.
Quorum sensing: the game changer
One major obstacle preventing the human team from winning is the development of resistance by the bacterial team to the opponent’s strategies. The bacterial team is well-founded in two fundamental needs to succeed in the game: it maintains a strong defense by forming a nearly impassable biofilm and devising new tactics in scoring a goal by developing virulence against the opponents (de Macedo et al., 2021). Building a team that is proficient in both requires good communication and co-operation. In the bacterial squad, this is ensured by quorum sensing, a mechanism that aids the bacterial players to coordinate among themselves to infect the humans (Figure 4) (Santhakumari and Ravi, 2019). While teamwork depicted by the bacterial players is crucial in escalating the game, the competency of individual species is also a significant driver. It is important to recall that the virulence factors that elevate the proficiency of the bacterial players are controlled by “quorum-sensing circuits” (Table 4). Understanding the various systems involved in quorum sensing is, therefore, crucial to upgrade the plans of the human team.
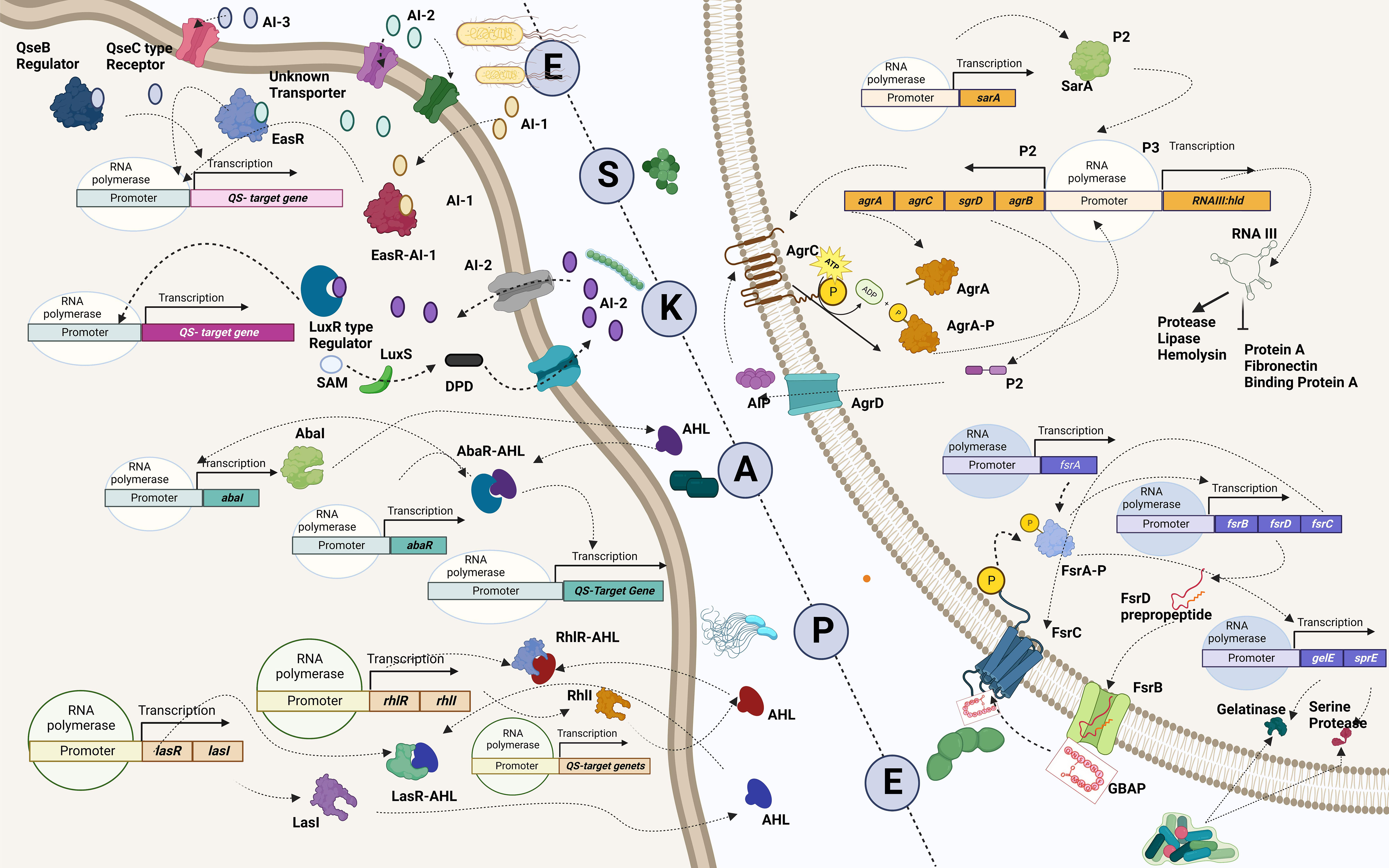
Figure 4 Quorum-sensing circuits of ESKAPE pathogens. All ESKAPE pathogens have been reported to have well-organized quorum-sensing circuits influencing their virulence and the ability to form biofilms. Four pathogens among the six, Enterococcus sp., Staphylococcus aureus, Klebsiella pneumoniae, and Enterobacter sp., involve LuxS system in altering antibiotic susceptibility and forming biofilms. More often than not, multiple quorum-sensing networks are involved in the biofilm formation process of these organisms. For instance, Pseudomonas aeruginosa is found to have a LasI–LasR system, RhII–RhIR system, and Quinolone and IQS systems in place to aid biofilm formation at various levels, including host tissue invasion and degradation. Similarly, the AbaI/AbaR system of Acinetobacter baumannii aids in its motility apart from contributing toward biofilm formation. Created with BioRender.com.
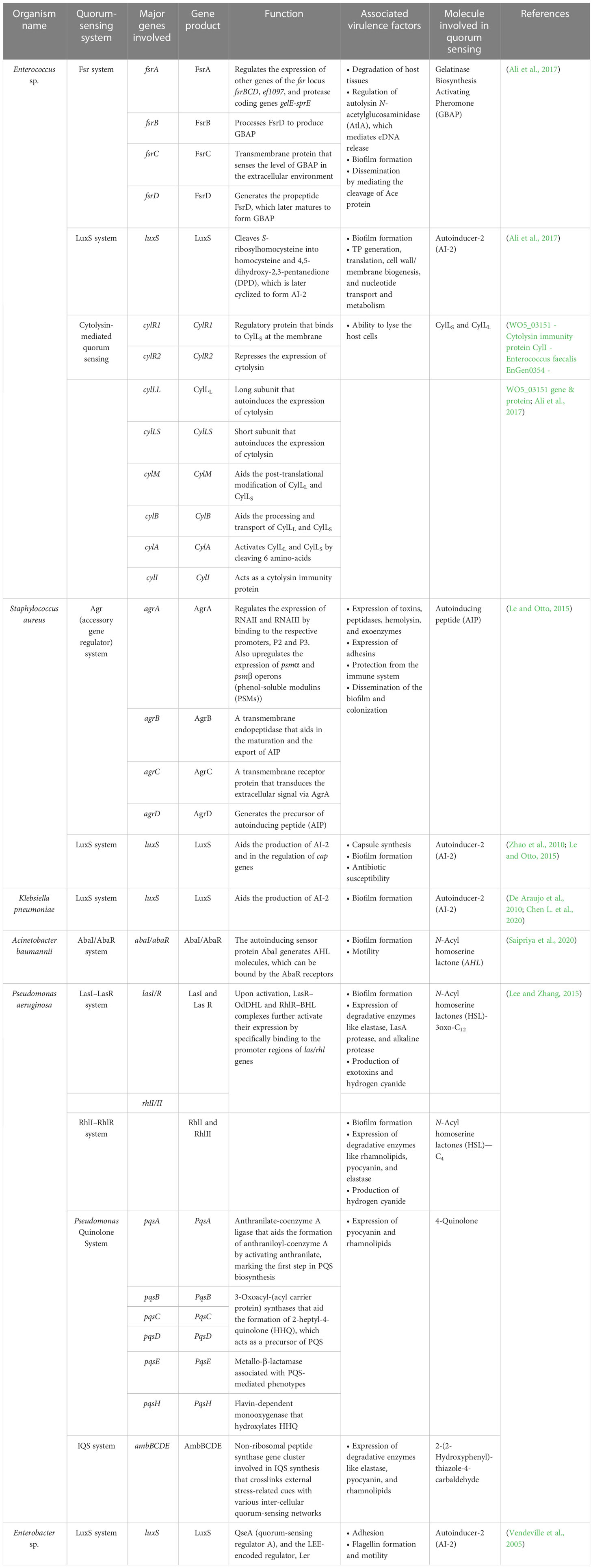
Table 4 Summary of the quorum-sensing systems employed by the ESKAPE organisms and the associated virulence factors.
Enterococcus sp. is reported to have three quorum-sensing circuits: Fsr, LuxS, and cytolysis-mediated systems. The fsr system senses the presence of gelatinase biosynthesis-activating pheromone (GBAP), the matured form of the pro-peptide FsrD, through the transmembrane protein FsrC. FsrB aids the processing of FsrD. It also involves the FsrA protein, which regulates the expression of other genes of the fsr locus (fsrBCD and ef1097) and protease coding genes (gelE-sprE). Fsr system is implicated in degrading the host tissues, regulating the autolysin N-acetylglucosaminidase (AtlA) and, thereby, the release of eDNA, biofilm formation, and the cleavage of Ace protein and subsequent dissemination (Ali et al., 2017). The LuxS system, however, regulates cell wall biogenesis, nucleotide transport, and metabolism. It cleaves S-ribosyl homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD), which is later cyclized to form AI-2 (Ali et al., 2017). Finally, the ability to lyse the host cells is conferred by the cytolysin system (WO5_03151–Cytolysin immunity protein CylI–E. faecalis EnGen0354–WO5_03151 gene and protein; Ali et al., 2017). On the contrary, the Agr and LuxS systems are known to be employed by S. aureus. The accessory regulatory system (Agr in short) involves AgrD, which generates the autoinducing peptide (AIP) precursor, which acts as the quorum-sensing molecule. AgrB, a transmembrane endopeptidase, aids in the AIP’s maturation and export. At the same time, AgrC transduces the extracellular signal via AgrA, which is also implicated in the regulation of the expression of RNAII and RNAIII and the upregulation of psmα and psmβ operons (phenol-soluble modulins (PSMs)). Signals associated with the Agr system influence the expression of toxins, peptidases, hemolysin, exoenzymes, and adhesins, in addition, to aiding in the protection from the immune system and the dissemination of the biofilm and colonization (Le and Otto, 2015). Furthermore, the LuxS system aids the production of AI-2 and regulates cap genes involved in capsule formation. It also affects biofilm formation and antibiotic susceptibility (Zhao et al., 2010; Le and Otto, 2015). A similar kind of LuxS networking is observed in K. pneumoniae, which aids in the production of AI-2 and enables biofilm formation (De Araujo et al., 2010; Chen L. et al., 2020). Biofilm-forming ability in A. baumannii, however, is reported to be influenced by the AbaI/AbaR system where the auto-inducing sensor protein, AbaI, generates N-acyl homoserine lactone (AHL) molecules, which can be bound by the AbaR receptors (Saipriya et al., 2020). Different quorum-sensing systems, including the LasI–LasR system, RhlI–RhlR system, Pseudomonas Quinolone System, and the IQS system, are reported in P. aeruginosa. Among these, the LasI–LasR system involves activated LasR–OdDHL and RhlR–BHL complexes, further activating their expression by specifically binding to the promoter regions of las/rhl genes, thereby regulating biofilm formation, production of exotoxins, and hydrogen cyanide. It is also reported to influence the expression of degradative enzymes like elastase, LasA protease, and alkaline protease. The RhII–RhIR system, however, is associated with the expression of degradative enzymes like rhamnolipids, pyocyanin, and elastase. It is also involved in the generation of hydrogen cyanide and biofilm formation. Alternatively, the Pseudomonas Quinolone System regulates the expression of pyocyanin and rhamnolipids. Finally, the IQS system is reported to be involved with a non-ribosomal peptide synthase gene cluster, which plays a role in IQS synthesis that crosslinks external stress-related cues with various inter-cellular quorum-sensing networks, thereby regulating the expression of degradative enzymes like elastase, pyocyanin, and rhamnolipids (Lee and Zhang, 2015). Finally, in Enterobacter sp., the LuxS system regulates adhesion, flagellin formation, and motility (Vendeville et al., 2005). In addition to facilitating bacterial virulence and biofilm formation, quorum-sensing molecules influence host–pathogen interactions. A recent study by Chakraborty et al. reported the hijacking role of 2-aminoacetophenone in altering the host autophagic and lipid biosynthesis mechanism in P. aeruginosa. Increased persistence of P. aeruginosa is attributed to the reduced expression of autophagy-mediating genes (Unc-51-like autophagy activating kinase 1 (ULK1) and Beclin1) and lipogenic gene [stearoyl-CoA desaturase 1 (Scd1)] (Chakraborty et al., 2023).
Insights on the quorum-sensing circuits have assisted the human team in advocating using quorum-sensing inhibitors (QSIs) as adjuvants to support the existing players—antibiotics and the immune cells (Table 5). Targeting one master player that supports and regulates other players is reported to be a successful strategy (Zhao et al., 2020). A gene knockout study involving LuxS/AI-2 deletion mutants observed reduced biofilm-forming ability in mutants compared to controls, thus proving the significant role played by the LuxS system in biofilm formation. This study, however, did not report any significant correlation between the proliferation ability of Enterococcus sp. and the absence of a functioning LuxS system (Yang et al., 2018). Another study involving a chemical inhibitor—siamycin I to block the fsr system of Enterococcus sp.—identified reduced growth, gelatinase activity, GBAP production, and biofilm-forming ability in the treated population in contrast to the control (Nakayama et al., 2007). Similarly, Balaban et al. reported reduced biofilm ability among the S. aureus population whose agr system was compromised (Balaban et al., 2007). Another study on K. pneumoniae reported decreased adherence and biofilm-forming ability of the chemically treated bacterial population as opposed to the controls with an effective C6-AHL system (Cadavid and Echeverri, 2019).

Table 5 Summary of the quorum-sensing inhibition methods employed against the ESKAPE organisms and their impact on pathogenicity.
Furthermore, a knockout gene study on this bacterial species revealed the decreased ability to form biofilm and to synthesize lipopolysaccharide with almost no significant influence over the ability to synthesize type 3 fimbriae in deletion mutants (Chen L. et al., 2020). A similar observation of decreased ability to form biofilms and to produce proteolytic enzymes, resistance to oxidative stress, twitching, and swarming motilities occurred when A. baumannii was treated with a chemical inhibitor that influences the Aba1/AbaR system (Seleem et al., 2020). The decreasing trends in the biofilm-forming ability and the surface-associated motility were reported in the corresponding gene knockout models (Mayer et al., 2020). Along these lines, inhibition of the LasR system in P. aeruginosa decreased the ability to form biofilm and generate pyocyanin, rhamnolipids, and elastin (Zhong et al., 2020). Gene knockout analyses revealed the decreased biofilm-forming ability, adhesion, and swarming motility in LasI mutants (Ouyang et al., 2020). It can be concluded that quorum sensing is quintessential in regulating virulence factors. Therefore, targeting the quorum-sensing networks can help counter the virulent traits of the ESKAPE pathogens.
QSIs have proven instrumental in cheating bacterial players by obstructing communication. Interfering with communication has aided in reducing the team’s strength by compromising its ability to form biofilms and to express the associated virulence factors (Munir et al., 2020). This, in turn, has boosted the chances of antibiotics and the immune cells in tackling the individual bacterial players (Brackman et al., 2011) (Figure 1F).
It is important to note that most of the substitutes in the human team resorted to conferring selective pressure against the bacterial squad, which is not the case with QSIs (Rasmussen and Givskov, 2006). A competition study by Gerdt et al. showed that the inadequacy of quorum-sensing signals by QSI-sensitive bacteria and their cheating mechanisms against the rare QSI-resistant bacteria would inherently reduce the spread of resistance against QSIs targeting QS receptor function (Gerdt and Blackwell, 2014). It is therefore perceived to be a safer move by the human team, as it does not come with an inherent risk of development of resistance by the bacterial players (Zhou et al., 2020).
Conclusion
The profound strategies employed by both teams make it equally hard for the opponent to win this never-ending “game of health”. However, understanding the opponent’s action plans would benefit the human team in devising holistic game plans. Employing quorum-sensing inhibitors along with specific antibiotics could prove to be an excellent combinatorial therapy in improving the chances of the human team winning by aiding the immune cells. However, the question of the efficacy of such combinations in treating well-established infections is yet to be addressed. Understanding the quorum-sensing signals might help us unravel the relationship between pathogens and normal microbiota of the host in disease progression in addition to answering the questions: i) do quorum-sensing signals of the pathogens aid in building a confluent microenvironment within the host? ii) Do the pathogens’ quorum-sensing signals influence the host’s natural microbiota? iii) Quorum-sensing signals ensure communication among a wide range of bacterial and fungal species. How can the pathogens be targeted with high specificity? Does the non-specific nature of QSIs disrupt the communication of normal microbiota, thereby exacerbating the condition? Recent studies report the development of resistance against quorum-sensing inhibitors. Therefore, the human team should constantly be vigilant to detect traces of resistance or “escaping” mechanisms that the bacterial players might develop.
Author contributions
APS and PN conceived the idea. PV, SV, and HD designed and drafted the manuscript. APS, AS, and KS provided the illustrations for the figures. APS and PN proofread the manuscript and suggested critical changes. All authors contributed to the article and approved the submitted version.
Acknowledgments
Our sincere admiration to all the players of the bacterial team and the human team, which passionate researchers immensely support. Authors acknowledge SASTRA Deemed to be University, Thanjavur for the encouragement and extending infrastructure support. APS acknowledges Prof. T.R. Rajagopalan Fund Scheme for the financial support. Thanks to Ms. Rajalakshmi, a real-time basketball player, for helping us frame the analogies. We also appreciate Ms. Aniritha for recommending focusing on the ESKAPE pathogens. Thanks to you, the readers, for turning up for the match.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdi, S. N., Ghotaslou, R., Ganbarov, K., Mobed, A., Tanomand, A., Yousefi, M., et al. (2020). Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect. Drug Resist. 13, 423–434. doi: 10.2147/IDR.S228089
Alcántar-Curiel, M. D., Blackburn, D., Saldaña, Z., Gayosso-Vázquez, C., Iovine, N., Cruz, M. A. D., et al. (2013). Multi-functional analysis of klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 4, 129. doi: 10.4161/VIRU.22974
Ali, L., Goraya, M. U., Arafat, Y., Ajmal, M., Chen, J. L., Yu, D. (2017). Molecular mechanism of quorum-sensing in enterococcus faecalis: its role in virulence and therapeutic approaches. Int. J. Mol. Sci. 18 (5), 960. doi: 10.3390/ijms18050960
Alkasaby, N. M., El Sayed Zaki, M. (2017). Molecular study of acinetobacter baumannii isolates for metallo- β -lactamases and extended-spectrum- β -lactamases genes in intensive care unit, mansoura university hospital, Egypt. Int. J. Microbiol. 2017. doi: 10.1155/2017/3925868
Allahverdiyev, A. M., Kon, K. V., Abamor, E. S., Bagirova, M., Rafailovich, M. (2011). Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents. Expert Rev. Anti Infect. Ther. 9, 1035–1052. doi: 10.1586/ERI.11.121
Aminov, R. I. (2013). Evolution in action: dissemination of tet(X) into pathogenic microbiota. Front. Microbiol. 4. doi: 10.3389/FMICB.2013.00192/BIBTEX
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for In vivo daptomycin resistance in enterococci. N Engl. J. Med. 365, 892–900. doi: 10.1056/nejmoa1011138
Arlet, G., Nadjar, D., Herrmann, J., Donay, J., Rouveau, M., Lagrange, P., et al. (2001). Plasmid-mediated rifampin resistance encoded by an arr-2-like gene cassette in klebsiella pneumoniae producing an aCC-1 class c β-lactamase [2]. Antimicrob. Agents Chemother. 45, 2971–2972. doi: 10.1128/AAC.45.10.2971-2972.2001
Bacterial vaccine - an overview | ScienceDirect topics. Available at: https://www.sciencedirect.com/topics/medicine-and-dentistry/bacterial-vaccine (Accessed February 18, 2021).
Bachman, M. A., Oyler, J. E., Burns, S. H., Caza, M., Lépine, F., Dozois, C. M., et al. (2011). Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79, 3309–3316. doi: 10.1128/IAI.05114-11
Balaban, N., Cirioni, O., Giacometti, A., Ghiselli, R., Braunstein, J. B., Silvestri, C., et al. (2007). Treatment of staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob. Agents Chemother. 51, 2226–2229. doi: 10.1128/AAC.01097-06
Barnes, A. M. T., Frank, K. L., Dale, J. L., Manias, D. A., Powers, J. L., Dunny, G. M. (2022). Enterococcus faecalis colonizes and forms persistent biofilm microcolonies on undamaged endothelial surfaces in a rabbit endovascular infection model. FEMS Microbes 2, xtab014. doi: 10.1093/FEMSMC/XTAB014
Başaran, Ü.N., Celayir, S., Eray, N., Öztürk, R., Şenyüz, O. F. (1998). The effect of an H2-receptor antagonist on small-bowel colonization and bacterial translocation in newborn rats. Pediatr. Surg. Int. 13, 118–120. doi: 10.1007/S003830050263/METRICS
Beasley, F. C., Marolda, C. L., Cheung, J., Buac, S., Heinrichs, D. E. (2011). Staphylococcus aureus transporters hts, sir, and sst capture iron liberated from human transferrin by staphyloferrin a, staphyloferrin b, and catecholamine stress hormones, respectively, and contribute to virulence. Infect. Immun. 79, 2345–2355. doi: 10.1128/IAI.00117-11
Bekeredjian-Ding, I. (2020). Challenges for clinical development of vaccines for prevention of hospital-acquired bacterial infections. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01755
Beta-lactam antibiotics - an overview | ScienceDirect topics. Available at: https://www.sciencedirect.com/topics/neuroscience/beta-lactam-antibiotics (Accessed February 11, 2021).
Bhukta, S., Samal, S. K., Vasudevan, S., Sarveswari, H. B., Shanmugam, K., Princy, S. A., et al. (2022). A prospective diversity of antibacterial small peptidomimetic and quorum sensing mediated drug: a review. ChemistrySelect 7, e202102743. doi: 10.1002/SLCT.202102743
Biggest threats and data | Antibiotic/Antimicrobial resistance | CDC. Available at: https://www.cdc.gov/drugresistance/biggest-threats.html (Accessed December 11, 2020).
Bokaeian, M., Saeidi, S., Shahi, Z., Kadaei, V. (2014). tetA and tetB genes in klebsiella pneumoniae isolated from clinical samples. Gene Cell Tissue 1 (2), e18152. doi: 10.17795/gct-18152
Bonneau, A., Roche, B., Schalk, J. (2020). Iron acquisition in pseudomonas aeruginosa by the siderophore pyoverdine: an intricate interacting network including periplasmic and membrane proteins. Sci. Rep. 10, 120. doi: 10.1038/s41598-019-56913-x
Borthagaray, G., Mondelli, M., Facchin, G., Torre, M. H. (2018). “Silver-containing nanoparticles in the research of new antimicrobial agents against ESKAPE pathogens,” in Inorganic frameworks as smart nanomedicines (Frameworks as Smart Nanomedicines, William Andrew Publishing), 317–386. doi: 10.1016/B978-0-12-813661-4.00008-0
Bowler, P., Murphy, C., Wolcott, R. (2020). Biofilm exacerbates antibiotic resistance: is this a current oversight in antimicrobial stewardship? Antimicrob. Resist. Infect. Control 9, 1–5. doi: 10.1186/S13756-020-00830-6/METRICS
Bozdogan, B., Appelbaum, P. C. (2004). Oxazolidinones: activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 23, 113–119. doi: 10.1016/J.IJANTIMICAG.2003.11.003
Brackman, G., Cos, P., Maes, L., Nelis, H. J., Coenye, T. (2011). Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55, 2655–2661. doi: 10.1128/AAC.00045-11
Breitenbach Barroso Coelho, L. C., Marcelino dos Santos Silva, P., Felix de Oliveira, W., de Moura, M. C., Viana Pontual, E., Soares Gomes, F., et al. (2018). Lectins as antimicrobial agents. J. Appl. Microbiol. 125, 1238–1252. doi: 10.1111/JAM.14055
Brenciani, A., Morroni, G., Schwarz, S., Giovanetti, E. (2022). Oxazolidinones: mechanisms of resistance and mobile genetic elements involved. J. Antimicrob. Chemother. 77, 2596–2621. doi: 10.1093/JAC/DKAC263
Burgos, Y. (2010). Common origin of plasmid encoded alpha-hemolysin genes in escherichia coli. BMC Microbiol. 10, 193. doi: 10.1186/1471-2180-10-193
Cadavid, E., Echeverri, F. (2019). The search for natural inhibitors of biofilm formation and the activity of the autoinductor C6-AHL in klebsiella pneumoniae ATCC 13884. Biomolecules 9(2), 49. doi: 10.3390/biom9020049
Campana, R., Casettari, L., Ciandrini, E., Illum, L., Baffone, W. (2017). Chitosans inhibit the growth and the adhesion of klebsiella pneumoniae and escherichia coli clinical isolates on urinary catheters. Int. J. Antimicrob. Agents 50, 135–141. doi: 10.1016/J.IJANTIMICAG.2017.03.031
Cayô, R., Rodríguez, M. C., Espinal, P., Fernández-Cuenca, F., Ocampo-Sosa, A. A., Pascual, Á., et al. (2011). Analysis of genes encoding penicillin-binding proteins in clinical isolates of acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 5907–5913. doi: 10.1128/AAC.00459-11
Centers for Disease Control. (2019). U. antibiotic resistance threats in the united states (Centers for Disease Control and Prevention (.gov)). doi: 10.15620/cdc:82532
Chakraborty, A., Kabashi, A., Wilk, S., Rahme, L. G. (2023). Quorum-sensing signaling molecule 2-aminoacetophenone mediates the persistence of pseudomonas aeruginosa in macrophages by interference with autophagy through epigenetic regulation of lipid biosynthesis. MBio 14, e0015923. doi: 10.1128/MBIO.00159-23
Chames, P., Van Regenmortel, M., Weiss, E., Baty, D. (2009). Therapeutic antibodies: successes, limitations and hopes for the future. Br. J. Pharmacol. 157, 220–233. doi: 10.1111/j.1476-5381.2009.00190.x
Chen, H., Wang, X., Yin, Y., Li, S., Zhang, Y., Wang, Q., et al. (2019). Molecular characteristics of oxazolidinone resistance in enterococci from a multicenter study in China. BMC Microbiol. 19, 162. doi: 10.1186/s12866-019-1537-0
Chen, L., Wilksch, J. J., Liu, H., Zhang, X., Torres, V. V. L., Bi, W., et al. (2020). Investigation of lux s-mediated quorum sensing in klebsiella pneumoniae. J. Med. Microbiol. 69, 402–413. doi: 10.1099/jmm.0.001148
Chen, Q., Xie, S., Lou, X., Cheng, S., Liu, X., Zheng, W., et al. (2020). Biofilm formation and prevalence of adhesion genes among staphylococcus aureus isolates from different food sources. Microbiologyopen 9 (1), e00946. doi: 10.1002/MBO3.946
Chen, W., Zhang, Y. M., Davies, C. (2017). Penicillin-binding protein 3 is essential for growth of pseudomonas aeruginosa. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.01651-16
Cheng, Y. S., Williamson, P. R., Zheng, W. (2019). Improving therapy of severe infections through drug repurposing of synergistic combinations. Curr. Opin. Pharmacol. 48, 92–98. doi: 10.1016/j.coph.2019.07.006
Ch’ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J., Kline, K. A. (2018). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2018 172 17, 82–94. doi: 10.1038/s41579-018-0107-z
Chow, J. W. (2000). Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31, 586–589. doi: 10.1086/313949
Choy, W.-K., Bajic, V. B., Heng, M.-W., Veronika, M., Swarup, S. (2008). Regulatory networks of genes affected by mora, a global regulator containing ggdef and eal domains in pseudomonas aeruginosa (World Scientific Pub Co Pte Lt), 123–129. doi: 10.1142/9781848162525_0022
Chu, E. K., Kilic, O., Cho, H., Groisman, A., Levchenko, A. (2018). Self-induced mechanical stress can trigger biofilm formation in uropathogenic escherichia coli. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-06552-z
Dale, J. L., Nilson, J. L., Barnes, A. M. T., Dunny, G. M. (2017). Restructuring of enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. NPJ Biofilms Microbiomes 3, 15. doi: 10.1038/S41522-017-0023-4
Dapgh, A. N., Hakim, A. S., Abouelhag, H. A., Abdou, A. M., Elgabry, E. A. (2019). Detection of virulence and multidrug resistance operons in pseudomonas aeruginosa isolated from Egyptian baladi sheep and goat. Vet. World 12, 1524–1528. doi: 10.14202/vetworld.2019.1524-1528
Davin-Regli, A., Pagès, J. M. (2015). Enterobacter aerogenes and enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00392
De Araujo, C., Balestrino, D., Roth, L., Charbonnel, N., Forestier, C. (2010). Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in klebsiella pneumoniae. Res. Microbiol. 161, 595–603. doi: 10.1016/j.resmic.2010.05.014
de Kievit, T. (2011). Biofilms. Compr. Biotechnol. Second Ed. 1, 547–558. doi: 10.1016/B978-0-08-088504-9.00064-7
D’Elia, M. A., Pereira, M. P., Chung, Y. S., Zhao, W., Chau, A., Kenney, T. J., et al. (2006). Lesions in teichoic acid biosynthesis in staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188, 4183–4189. doi: 10.1128/JB.00197-06
de Macedo, G. H. R. V., Costa, G. D. E., Oliveira, E. R., Damasceno, G. V., Mendonça, J. S. P., Silva, L. D. S., et al. (2021). Interplay between eskape pathogens and immunity in skin infections: an overview of the major determinants of virulence and antibiotic resistance. Pathogens 10, 1–34. doi: 10.3390/pathogens10020148
Deshpande, L. M., Castanheira, M., Flamm, R. K., Mendes, R. E. (2018). Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY antimicrobial surveillance program. J. Antimicrob. Chemother. 73, 2314–2322. doi: 10.1093/jac/dky188
Dönhöfer, A., Franckenberg, S., Wickles, S., Berninghausen, O., Beckmann, R., Wilson, D. N. (2012). Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. U. S. A. 109, 16900–16905. doi: 10.1073/PNAS.1208037109/SUPPL_FILE/SAPP.PDF
Doring, G., Conway, S. P., Heijerman, H. G. M., Hodson, M. E., Hoiby, N., Smyth, A., et al. (2000). Antibiotic therapy against pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur. Respir. J. 16, 749–767. doi: 10.1034/J.1399-3003.2000.16D30.X
Drawz, S. M., Bonomo, R. A. (2010). Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201. doi: 10.1128/CMR.00037-09
Duval, R. E., Gouyau, J., Lamouroux, E. (2019). Limitations of recent studies dealing with the antibacterial properties of silver nanoparticles: fact and opinion. Nanomaterials 9 (12), 1775. doi: 10.3390/nano9121775
El Haddad, L., Harb, C. P., Gebara, M. A., Stibich, M. A., Chemaly, R. F. (2019). A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin. Infect. Dis. 69, 167–178. doi: 10.1093/cid/ciy947
Enne, V. I., Delsol, A. A., Roe, J. M., Bennett, P. M. (2004). Rifampicin resistance and its fitness cost in enterococcus faecium. J. Antimicrob. Chemother. 53, 203–207. doi: 10.1093/jac/dkh044
Esmaeel, J. R., Sadeq, J. N. (2018). Hemolysin gene detection in some isolates of klebsiella pneumonia by PCR. Al-Qadisiyah J. Vet. Med. Sci. 17, 49–52. doi: 10.29079/vol17iss2art504
Finlay, B. B., McFadden, G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782. doi: 10.1016/j.cell.2006.01.034
Fiore, E., Van Tyne, D., Gilmore, M. S. (2019). Pathogenicity of enterococci. Microbiol. Spectr. 7 (4), 10.1128/microbiolspec.GPP3-0053-2018. doi: 10.1128/microbiolspec.gpp3-0053-2018
Firoozeh, F., Omidi, M., Saffari, M., Sedaghat, H., Zibaei, M. (2020). Molecular analysis of methicillin-resistant staphylococcus aureus isolates from four teaching hospitals in Iran: the emergence of novel MRSA clones. Antimicrob. Resist. Infect. Control 9 (1), 112. doi: 10.1186/s13756-020-00777-8
Foster, T. J. (2017). Antibiotic resistance in staphylococcus aureus. current status and future prospects. FEMS Microbiol. Rev. 41, 430–449. doi: 10.1093/femsre/fux007
FROM DYES TO PEPTIDES: THE EVOLUTION OF ANTIBIOTIC DRUGS | SCQ. Available at: https://www.scq.ubc.ca/from-dyes-to-peptides-the-evolution-of-antibiotic-drugs/ (Accessed December 29, 2020).
Garneau-Tsodikova, S., Labby, K. J. (2016). Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 7, 11–27. doi: 10.1039/c5md00344j
Gaynes, R. (2017). The discovery of penicillin–new insights after more than 75 years of clinical use. Emerg. Infect. Dis. 23, 849–853. doi: 10.3201/eid2305.161556
Gaynor, M., Mankin, A. (2003). Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 3, 949–960. doi: 10.2174/1568026033452159
Genetics of antimicrobial resistance in staphylococcus aureus. Available at: https://www.medscape.com/viewarticle/710150_6 (Accessed February 12, 2021).
Gerdt, J. P., Blackwell, H. E. (2014). Competition studies confirm two major barriers that can preclude the spread of resistance to quorum-sensing inhibitors in bacteria. ACS Chem. Biol. 9, 2291–2299. doi: 10.1021/CB5004288/SUPPL_FILE/CB5004288_SI_001.PDF
Ghigo, J. M., Wandersman, C. (1992). Cloning, nucleotide sequence and characterization of the gene encoding the erwinia chrysanthemi B374 PrtA metalloprotease: a third metalloprotease secreted via a c-terminal secretion signal. MGG Mol. Gen. Genet. 236, 135–144. doi: 10.1007/BF00279652
Giannouli, M., Di Popolo, A., Durante-Mangoni, E., Bernardo, M., Cuccurullo, S., Amato, G., et al. (2012). Molecular epidemiology and mechanisms of rifampicin resistance in acinetobacter baumannii isolates from Italy. Int. J. Antimicrob. Agents 39, 58–63. doi: 10.1016/j.ijantimicag.2011.09.016
Giske, C. G., Buarø, L., Sundsfjord, A., Wretlind, B. (2008). Alterations of porin, pumps, and penicillin-binding proteins in carbapenem resistant clinical isolates of pseudomonas aeruginosa. Microb. Drug Resist. 14, 23–30. doi: 10.1089/mdr.2008.0778
González-Bello, C. (2017). Antibiotic adjuvants – a strategy to unlock bacterial resistance to antibiotics. Bioorganic Med. Chem. Lett. 27, 4221–4228. doi: 10.1016/j.bmcl.2017.08.027
González de Aledo de Aledo, M., González-Bardanca, M., Blasco, L., Pacios, O., Bleriot, I., Fernández-García, L., et al. (2021). CRISPR-cas, a revolution in the treatment and study of ESKAPE infections: pre-clinical studies. Antibiotics 10. doi: 10.3390/ANTIBIOTICS10070756
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6 (4), a025387. doi: 10.1101/CSHPERSPECT.A025387
Guo, Y., Song, G., Sun, M., Wang, J., Wang, Y. (2020). Prevalence and therapies of antibiotic-resistance in staphylococcus aureus. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/FCIMB.2020.00107/BIBTEX
Gupta, S., Laskar, N., Kadouri, D. E. (2016). Evaluating the effect of oxygen concentrations on antibiotic sensitivity, growth, and biofilm formation of human pathogens. Microbiol. Insights 9, 37–46. doi: 10.4137/MBI.S40767
Hackbarth, C. J., Kocagoz, T., Kocagoz, S., Chambers, H. F. (1995) Point mutations in staphylococcus aureus PBP 2 gene affect penicillin-binding kinetics and are associated with resistance. Available at: http://aac.asm.org/ (Accessed February 11, 2021).
Hakansson, A. P., Orihuela, C. J., Bogaert, D. (2018). Bacterial-host interactions: physiology and pathophysiology of respiratory infection. Physiol. Rev. 98, 781–811. doi: 10.1152/PHYSREV.00040.2016/ASSET/IMAGES/LARGE/Z9J0021828410002.JPEG
Hancock, L. E., Murray, B. E., Sillanpää, J. (2014) Enterococcal cell wall components and structures (Massachusetts Eye and Ear Infirmary). (Accessed December 28, 2020).
Hartney, S. L., Mazurier, S., Girard, M. K., Mehnaz, S., Davis, E. W., Gross, H., et al. (2013). Ferric-pyoverdine recognition by fpv outer membrane proteins of pseudomonas protegens pf-5. J. Bacteriol. 195, 765–776. doi: 10.1128/JB.01639-12
Hasani, A., Sharifi, Y., Ghotaslou, R., Naghili, B., Hasani, A., Aghazadeh, M., et al. (2012). Molecular screening of virulence genes in high-level gentamicin-resistant enterococcus faecalis and enterococcus faecium isolated from clinical specimens in Northwest Iran. Indian J. Med. Microbiol. 30, 175–181. doi: 10.4103/0255-0857.96687
Hatfull, G. F., Dedrick, R. M., Schooley, R. T. (2022). Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211. doi: 10.1146/ANNUREV-MED-080219-122208
Hendrickx, A. P. A., Willems, R. J. L., Bonten, M. J. M., van Schaik, W. (2009). LPxTG surface proteins of enterococci. Trends Microbiol. 17, 423–430. doi: 10.1016/j.tim.2009.06.004
Hershberger, E., Donabedian, S., Konstantinou, K., Zervos, M. J. (2004). Quinupristin-dalfopristin resistance in gram-positive bacteria: mechanism of resistance and epidemiology. Clin. Infect. Dis. 38, 92–98. doi: 10.1086/380125
Higgins, J., Loughman, A., van Kessel, K. P. M., van Strijp, J. A. G., Foster, T. J. (2006). Clumping factor a of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol. Lett. 258, 290–296. doi: 10.1111/j.1574-6968.2006.00229.x
Ho, S. W., Jung, D., Calhoun, J. R., Lear, J. D., Okon, M., Scott, W. R. P., et al. (2008). Effect of divalent cations on the structure of the antibiotic daptomycin. Eur. Biophys. J. 37, 421–433. doi: 10.1007/S00249-007-0227-2/METRICS
Hoštacká, A., Čižnár, I., Štefkovičová, M. (2010). Temperature and pH affect the production of bacterial biofilm. Folia Microbiol. (Praha). 55, 75–78. doi: 10.1007/s12223-010-0012-y
Hsieh, P. F., Lu, Y. R., Lin, T. L., Lai, L. Y., Wang, J. T. (2019). Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647. doi: 10.1093/infdis/jiy534
Hsu, C. R., Liao, C. H., Lin, T. L., Yang, H. R., Yang, F. L., Hsieh, P. F., et al. (2016). Identification of a capsular variant and characterization of capsular acetylation in klebsiella pneumoniae PLA-associated type K57. Sci. Rep. 6, 1–13. doi: 10.1038/srep31946
Ibberson, C. B., Jones, C. L., Singh, S., Wise, M. C., Hart, M. E., Zurawski, D. V., et al. (2014). Staphylococcus aureus hyaluronidase is a CodY-regulated virulence factor. Infect. Immun. 82, 4253–4264. doi: 10.1128/IAI.01710-14
Iiyama, K., Takahashi, E., Lee, J. M., Mon, H., Morishita, M., Kusakabe, T., et al. (2017). Alkaline protease contributes to pyocyanin production in pseudomonas aeruginosa. FEMS Microbiol. Lett. 364, 51. doi: 10.1093/femsle/fnx051
Issa, K. H. B., Phan, G., Broutin, I. (2018). Functional mechanism of the efflux pumps transcription regulators from pseudomonas aeruginosa based on 3D structures. Front. Mol. Biosci. 5. doi: 10.3389/fmolb.2018.00057
Iwata, Y., Sakai, N., Yoneda, I., Senda, Y., Sakai-Takemori, Y., Oshima, M., et al. (2021). D-serine inhibits the attachment and biofilm formation of methicillin-resistant staphylococcus aureus. Biochem. Biophys. Res. Commun. 537, 50–56. doi: 10.1016/J.BBRC.2020.12.078
Jiang, Y., Geng, M., Bai, L. (2020). Targeting biofilms therapy: current research strategies and development hurdles. Microorganisms 8, 1–34. doi: 10.3390/MICROORGANISMS8081222
Johnson, J. G., Murphy, C. N., Sippy, J., Johnson, T. J., Clegg, S. (2011). Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in klebsiella pneumoniae. J. Bacteriol. 193, 3453–3460. doi: 10.1128/JB.00286-11
Johnston, N., Mukhtar, T., Wright, G. (2002). Streptogramin antibiotics: mode of action and resistance. Curr. Drug Targets 3, 335–344. doi: 10.2174/1389450023347678
Kamurai, B., Mombeshora, M., Mukanganyama, S. (2020). Repurposing of drugs for antibacterial activities on selected ESKAPE bacteria staphylococcus aureus and pseudomonas aeruginosa. Int. J. Microbiol. 2020. doi: 10.1155/2020/8885338
Karalewitz, A. P. A., Millera, S. I. (2018). Multidrug-resistant acinetobacter baumannii chloramphenicol resistance requires an inner membrane permease. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.00513-18
Karygianni, L., Ren, Z., Koo, H., Thurnheer, T. (2020). Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/J.TIM.2020.03.016
Kaushik, V., Tiwari, M., Tiwari, V. (2022). Interaction of RecA mediated SOS response with bacterial persistence, biofilm formation, and host response. Int. J. Biol. Macromol. 217, 931–943. doi: 10.1016/J.IJBIOMAC.2022.07.176
Khare, B., Narayana, V. L. (2017). Pilus biogenesis of gram-positive bacteria: roles of sortases and implications for assembly. Protein Sci. 26, 1458–1473. doi: 10.1002/pro.3191
Kidd, T. J., Mills, G., Sá-Pessoa, J., Dumigan, A., Frank, C. G., Insua, J. L., et al. (2017). A klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 9, 430–447. doi: 10.15252/emmm.201607336
Koenigs, A., Stahl, J., Averhoff, B., Göttig, S., Wichelhaus, T. A., Wallich, R., et al. (2016). CipA of acinetobacter baumannii is a novel plasminogen binding and complement inhibitory protein. J. Infect. Dis. 213, 1388–1399. doi: 10.1093/infdis/jiv601
Krawczyk, B., Wityk, P., Gałęcka, M., Michalik, M. (2021). The many faces of enterococcus spp.–commensal, probiotic and opportunistic pathogen. Microorg 9, 1900. doi: 10.3390/MICROORGANISMS9091900
Kristich, C. J., Li, Y. H., Cvitkovitch, D. G., Dunny, G. M. (2004). Esp-independent biofilm formation by enterococcus faecalis. J. Bacteriol. 186, 154. doi: 10.1128/JB.186.1.154-163.2004
Kronstad, J. W., Caza, M. (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 4. doi: 10.3389/fcimb.2013.00080
Lambert, M. L., Suetens, C., Savey, A., Palomar, M., Hiesmayr, M., Morales, I., et al. (2011). Clinical outcomes of health-care-associated infections and antimicrobial resistance in patients admitted to European intensive-care units: a cohort study. Lancet Infect. Dis. 11, 30–38. doi: 10.1016/S1473-3099(10)70258-9
Latorre, M., Quenti, D., Travisany, D., Singh, K. V., Murray, B. E., Maass, A., et al. (2018). The role of fur in the transcriptional and iron homeostatic response of enterococcus faecalis. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01580
Le, K. Y., Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6, 1174. doi: 10.3389/fmicb.2015.01174
Lee, N.-Y., Ko, W.-C., Hsueh, P.-R. (2019). Nanoparticles in the treatment of infections caused by multidrug-resistant organisms. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.01153
Lee, C. R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00055
Lee, J., Zhang, L. (2015). The hierarchy quorum sensing network in pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Leitão, J. H. (2020). Microbial virulence factors. Int. J. Mol. Sci. 21, 1–6. doi: 10.3390/ijms21155320
Lenchenko, E., Blumenkrants, D., Sachivkina, N., Shadrova, N., Ibragimova, A. (2020). Morphological and adhesive properties of klebsiella pneumoniae biofilms. Vet. World 13, 197. doi: 10.14202/VETWORLD.2020.197-200
Lerminiaux, N. A., Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi: 10.1139/cjm-2018-0275
Lery, L. M. S., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative analysis of klebsiella pneumoniae genomes identifies a phospholipase d family protein as a novel virulence factor. BMC Biol. 12, 41. doi: 10.1186/1741-7007-12-41
Leshem, A., Horesh, N., Elinav, E. (2019). Fecal microbial transplantation and its potential application in cardiometabolic syndrome. Front. Immunol. 10. doi: 10.3389/fimmu.2019.01341
Lessa, F. C., Mu, Y., Ray, S. M., Dumyati, G., Bulens, S., Gorwitz, R. J., et al. (2012). Impact of USA300 methicillin-resistant staphylococcus aureus on clinical outcomes of patients with pneumonia or central line–associated bloodstream infections. Clin. Infect. Dis. 55, 232–241. doi: 10.1093/CID/CIS408
Liang, C., Xing, B., Yang, X., Fu, Y., Feng, Y., Zhang, Y. (2015). Molecular epidemiology of aminoglycosides resistance on klebsiella pneumonia in a hospital in China. Int. J. Clin. Exp. Med. 8, 1381–1385.
Lin, T. L., Tang, S. I., Fang, C. T., Hsueh, P. R., Chang, S. C., Wang, J. T. (2006). Extended-spectrum β-lactamase genes of klebsiella pneumoniae strains in Taiwan: recharacterization of shv-27, shv-41, and tem-116. Microb. Drug Resist. 12, 12–15. doi: 10.1089/mdr.2006.12.12
Lina, G., Quaglia, A., Reverdy, M. E., Leclercq, R., Vandenesch, F., Etienne, J. (1999). Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43, 1062–1066. doi: 10.1128/aac.43.5.1062
Liu, G. Y. (2009). Molecular pathogenesis of staphylococcus aureus infection. Pediatr. Res. 65, 71–77. doi: 10.1203/PDR.0b013e31819dc44d
Liu, J., Gefen, O., Ronin, I., Bar-Meir, M., Balaban, N. Q. (2020). Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204. doi: 10.1126/science.aay3041
Livermore, D. M. (2002). Multiple mechanisms of antimicrobial resistance in pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34, 634–640. doi: 10.1086/338782/2/34-5-634-FIG002.GIF
Long, K. S., Vester, B. (2012). Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56, 603–612. doi: 10.1128/AAC.05702-11
Luong, T. T., Ouyang, S., Bush, K., Lee, C. Y. (2002). Type 1 capsule genes of staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184, 3623–3629. doi: 10.1128/JB.184.13.3623-3629.2002
Maasjost, J., Lüschow, D., Kleine, A., Hafez, H. M., Mühldorfer, K., Bondi, M. (2019). Presence of virulence genes in enterococcus species isolated from meat turkeys in Germany does not correlate with chicken embryo lethality. BioMed. Res. Int. 2019, 6147695. doi: 10.1155/2019/6147695
Maleki, M. H., Sekawi, Z., Soroush, S., Azizi-Jalilian, F., Asadollahi, K., Mohammadi, S., et al. (2014). Phenotypic and genotypic characteristics of tetracycline resistant acinetobacter baumannii isolates from nosocomial infections at Tehran hospitals. Iran. J. Basic Med. Sci. 17, 21–26.
Malhotra, S., Hayes, D., Wozniak, D. J. (2019). Cystic fibrosis and pseudomonas aeruginosa: the host-microbe interface. Clin. Microbiol. Rev. 32. doi: 10.1128/CMR.00138-18
Maréchal, M., Amoroso, A., Morlot, C., Vernet, T., Coyette, J., Joris, B. (2016). Enterococcus hirae LcpA (Psr), a new peptidoglycan-binding protein localized at the division site. BMC Microbiol. 16, 1–13. doi: 10.1186/s12866-016-0844-y
Martínez, J. L. (2014). Short-sighted evolution of bacterial opportunistic pathogens with an environmental origin. Front. Microbiol. 5. doi: 10.3389/FMICB.2014.00239/BIBTEX
Mayer, C., Muras, A., Parga, A., Romero, M., Rumbo-Feal, S., Poza, M., et al. (2020). Quorum sensing as a target for controlling surface associated motility and biofilm formation in acinetobacter baumannii ATCC® 17978TM. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.565548
Mbelle, N. M., Feldman, C., Sekyere, J. O., Maningi, N. E., Modipane, L., Essack, S. Y. (2020). Pathogenomics and evolutionary epidemiology of multi-drug resistant clinical klebsiella pneumoniae isolated from Pretoria, south Africa. Sci. Rep. 10, 1–17. doi: 10.1038/s41598-020-58012-8
Melles, D. C., Van Leeuwen, W. B., Boelens, H. A. M., Peeters, J. K., Verbrugh, H. A., Van Belkum, A. (2006). Panton-valentine leukocidin genes in staphylococcus aureus [9]. Emerg. Infect. Dis. 12, 1174–1175. doi: 10.3201/eid1207.050865
Merino, S., Tomás, J. (2010). Bacterial capsules and evasion of immune responses. eLS. doi: 10.1002/9780470015902.A0000957.PUB3
Miller, W. R., Munita, J. M., Arias, C. A. (2014). Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 12, 1221–1236. doi: 10.1586/14787210.2014.956092
Moffatt, J. H., Harper, M., Mansell, A., Crane, B., Fitzsimons, T. C., Nation, R. L., et al. (2013). Lipopolysaccharide-deficient acinetobacter baumannii shows altered signaling through host toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect Immun. 81 (3), 684-689. doi: 10.1128/IAI.01362-12
Mohajeri, M. H., Brummer, R. J. M., Rastall, R. A., Weersma, R. K., Harmsen, H. J. M., Faas, M., et al. (2018). The role of the microbiome for human health: from basic science to clinical applications. Eur. J. Nutr. 57, 1. doi: 10.1007/s00394-018-1703-4
Monem, S., Furmanek-Blaszk, B., Łupkowska, A., Kuczyńska-Wiśnik, D., Stojowska-Swędrzyńska, K., Laskowska, E. (2020). Mechanisms protecting acinetobacter baumannii against multiple stresses triggered by the host immune response, antibiotics, and outside host environment. Int. J. Mol. Sci. 21, 1–30. doi: 10.3390/ijms21155498
Montera, C. I., Stock, F., Murray, P. R. (2008). Mechanisms of resistance to daptomycin in enterococcus faecium. Antimicrob. Agents Chemother. 52, 1167–1170. doi: 10.1128/AAC.00774-07
Moormeier, D. E., Bayles, K. W. (2017). Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 104, 365. doi: 10.1111/MMI.13634
Moradali, M. F., Ghods, S., Rehm, B. H. A. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/FCIMB.2017.00039/BIBTEX
Motamedi, H., Asghari, B., Tahmasebi, H., Arabestani, M. (2018). Identification of hemolysine genes and their association with antimicrobial resistance pattern among clinical isolates of staphylococcus aureus in West of Iran. Adv. Biomed. Res. 7, 153. doi: 10.4103/abr.abr_143_18
Muhammadi, Ahmed, N. (2007). Genetics of bacterial alginate: alginate genes distribution, organization and biosynthesis in bacteria. Curr. Genomics 8, 191–202. doi: 10.2174/138920207780833810
Munir, S., Shah, A. A., Shahid, M., Manzoor, I., Aslam, B., Rasool, M. H., et al. (2020). Quorum sensing interfering strategies and their implications in the management of biofilm-associated bacterial infections. Braz. Arch. Biol. Technol. 63, 2020. doi: 10.1590/1678-4324-2020190555
Murray, G. L., Tsyganov, K., Kostoulias, X. P., Bulach, D. M., Powell, D., Creek, D. J., et al. (2017). Global gene expression profile of acinetobacter baumannii during bacteremia. J. Infect. Dis. 215 (suppl_1), S52–S57. doi: 10.1093/infdis/jiw529
Nadar, S., Khan, T., Patching, S. G., Omri, A. (2022). Development of antibiofilm therapeutics strategies to overcome antimicrobial drug resistance. Microorganisms 10 (2), 303. doi: 10.3390/MICROORGANISMS10020303
Nakajima, Y. (1999). Mechanisms of bacterial resistance to macrolide antibiotics. J. Infect. Chemother. 5, 61–74. doi: 10.1007/S101560050011/METRICS
Nakayama, J., Tanaka, E., Kariyama, R., Nagata, K., Nishiguchi, K., Mitsuhata, R., et al. (2007). Siamycin attenuates fsr quorum sensing mediated by a gelatinase biosynthesis-activating pheromone in enterococcus faecalis. J. Bacteriology (American Soc. Microbiol. Journals) 189 (4), 1358–1365. doi: 10.1128/JB.00969-06
Newman, J. W., Floyd, R. V., Fothergill, J. L. (2017). The contribution of pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS Microbiol. Lett. 364, 124. doi: 10.1093/femsle/fnx124
Oechslin, F. (2018). Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10 (7), 351. doi: 10.3390/v10070351
Ogawara, H. (2019). Comparison of antibiotic resistance mechanisms in antibiotic-producing and pathogenic bacteria. Molecules 24, 1–55. doi: 10.3390/molecules24193430
Okojie, R. O., Omorokpe, V. O. (2018). A survey on urinary tract infection associated with two most common uropathogenic bacteria. Afr. J. Clin. Exp. Microbiol. 19, 111. doi: 10.4314/ajcem.v19i3.3
Om, C., Daily, F., Vlieghe, E., McLaughlin, J. C., McLaws, M. L. (2016). “If it’s a broad spectrum, it can shoot better”: inappropriate antibiotic prescribing in Cambodia. Antimicrob. Resist. Infect. Control 5, 58. doi: 10.1186/s13756-016-0159-7
Ouyang, J., Feng, W., Lai, X., Chen, Y., Zhang, X., Rong, L., et al. (2020). Quercetin inhibits pseudomonas aeruginosa biofilm formation via the vfr-mediated lasIR system. Microb. Pathog. 149, 104291. doi: 10.1016/j.micpath.2020.104291
Panda, S. K., Buroni, S., Swain, S. S., Bonacorsi, A., da Fonseca Amorim, E. A., Kulshrestha, M., et al. (2022). Recent advances to combat ESKAPE pathogens with special reference to essential oils. Front. Microbiol. 13. doi: 10.3389/FMICB.2022.1029098
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., Cheng, Z. (2019). Antibiotic resistance in pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Pankey, G. A., Sabath, L. D. (2004). Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of gram-positive bacterial infections. Clin. Infect. Dis. 38, 864–870. doi: 10.1086/381972
Patel, S., Ahmed, S., Eswari, J. S. (2015). Therapeutic cyclic lipopeptides mining from microbes: latest strides and hurdles. World J. Microbiol. Biotechnol. 31, 1177–1193. doi: 10.1007/S11274-015-1880-8/FIGURES/4
Pechère, J. C., Köhler, T. (1999). Patterns and modes of β-lactam resistance in pseudomonas aeruginosa. Clin. Microbiol. Infection 5 (Suppl 1), S15–S18. doi: 10.1111/j.1469-0691.1999.tb00719.x
Pendleton, J. N., Gorman, S. P., Gilmore, B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308. doi: 10.1586/ERI.13.12
Pereira, S. C. L., Vanetti, M. C. D. (2015). Potential virulence of klebsiella sp. isolates from enteral diets. Braz. J. Med. Biol. Res. 48, 782–789. doi: 10.1590/1414-431X20154316
Petrova, O. E., Schurr, J. R., Schurr, M. J., Sauer, K. (2012). Microcolony formation by the opportunistic pathogen pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 86, 819. doi: 10.1111/MMI.12018
Piperaki, E. T., Syrogiannopoulos, G. A., Tzouvelekis, L. S., Daikos, G. L. (2017). Klebsiella pneumoniae: virulence, biofilm and antimicrobial resistance. Pediatr. Infect. Dis. J. 36, 1002–1005. doi: 10.1097/INF.0000000000001675
Pizarro-Cerdá, J., Cossart, P. (2006). Bacterial adhesion and entry into host cells. Cell 124, 715–727. doi: 10.1016/J.CELL.2006.02.012
Podschun, R., Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Pollack, M. (1984). The virulence of pseudomonas aeruginosa. Rev. Infect. Dis. 6 (Suppl_3), S617–S626. doi: 10.1093/clinids/6.supplement_3.s617
Poole, K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162–176. doi: 10.1080/07853890701195262
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00065
Principi, N., Silvestri, E., Esposito, S. (2019). Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.00513
Proença, J. T., Barral, D. C., Gordo, I. (2017). Commensal-to-pathogen transition: one-single transposon insertion results in two pathoadaptive traits in escherichia coli -macrophage interaction. Sci. Rep. 2017 71 7, 1–12. doi: 10.1038/s41598-017-04081-
Rahimi, F. (2016). Characterization of resistance to aminoglycosides in methicillin-resistant staphylococcus aureus strains isolated from a tertiary care hospital in Tehran, Iran. Jundishapur J. Microbiol. 9, 29237. doi: 10.5812/jjm.29237
Rahman, M. U., Fleming, D. F., Sinha, I., Rumbaugh, K. P., Gordon, V. D., Christopher, G. F. (2021). Effect of collagen and EPS components on the viscoelasticity of pseudomonas aeruginosa biofilms. Soft Matter 17, 6225. doi: 10.1039/D1SM00463H
Rasmussen, T. B., Givskov, M. (2006). Quorum sensing inhibitors: a bargain of effects. Microbiology 152, 895–904. doi: 10.1099/mic.0.28601-0
Reyes, J., Panesso, D., Tran, T. T., Mishra, N. N., Cruz, M. R., Munita, J. M., et al. (2015). A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant enterococcus faecalis. J. Infect. Dis. 211, 1317–1325. doi: 10.1093/infdis/jiu602
Ribet, D., Cossart, P. (2015). How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 17, 173–183. doi: 10.1016/j.micinf.2015.01.004
Ripoll, A., Galán, J. C., Rodríguez, C., Tormo, N., Gimeno, C., Baquero, F., et al. (2014). Detection of resistance to beta-lactamase inhibitors in strains with CTX-m beta-lactamases: a multicenter external proficiency study using a well-defined collection of escherichia coli strains. J. Clin. Microbiol. 52, 122–129. doi: 10.1128/JCM.02340-13
Rozdzinski, E., Marre, R., Susa, M., Wirth, R., Muscholl-Silberhorn, A. (2001). Aggregation substance-mediated adherence of enterococcus faecalis to immobilized extracellular matrix proteins. Microb. Pathog. 30, 211–220. doi: 10.1006/mpat.2000.0429
Rumbo, C., Tomás, M., Moreira, E. F., Soares, N. C., Carvajal, M., Santillana, E., et al. (2014). The acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect. Immun. 82, 4666–4680. doi: 10.1128/IAI.02034-14
Russo, T. A., Luke, N. R., Beanan, J. M., Olson, R., Sauberan, S. L., MacDonald, U., et al. (2010). The K1 capsular polysaccharide of acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 78, 3993–4000. doi: 10.1128/IAI.00366-10
Russo, T. A., Marr, C. M. (2019). Hypervirulent klebsiella pneumoniae. Clin. Microbiol. Rev. 32 (3), e00001-19. doi: 10.1128/CMR.00001-19
Rust, L., Pesci, E. C., Iglewski, B. H. (1996). Analysis of the pseudomonas aeruginosa elastase (lasB) regulatory region. J. Bacteriol. 178, 1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996
Saipriya, K., Swathi, C. H., Ratnakar, K. S., Sritharan, V. (2020). Quorum-sensing system in Acinetobacter baumannii: a potential target for new drug development. J. Appl. Microbiol. 128, 15–27. doi: 10.1111/jam.14330
Sako, T., Tsuchida, N. (1983). Nucleotide sequence of the staphylokinase gene from staphylococcus aurens. Nucleic Acids Res. 11, 7679–7693. doi: 10.1093/nar/11.22.7679
Sánchez-López, E., Gomes, D., Esteruelas, G., Bonilla, L., Lopez-Machado, A. L., Galindo, R., et al. (2020). Metal-based nanoparticles as antimicrobial agents: an overview. Nanomater 10, 292. doi: 10.3390/NANO10020292
Santajit, S., Indrawattana, N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed. Res. Int. 2016, 2475067. doi: 10.1155/2016/2475067
Santhakumari, S., Ravi, A. V. (2019). Targeting quorum sensing mechanism: an alternative anti-virulent strategy for the treatment of bacterial infections. South Afr. J. Bot. 120, 81–86. doi: 10.1016/j.sajb.2018.09.028
Sava, I. G., Heikens, E., Huebner, J. (2010). Pathogenesis and immunity in enterococcal infections. Clin. Microbiol. Infect. 16, 533–540. doi: 10.1111/j.1469-0691.2010.03213.x
Scherr, T. D., Roux, C. M., Hanke, M. L., Angle, A., Dunman, P. M., Kielian, T. (2013). Global transcriptome analysis of staphylococcus aureus biofilms in response to innate immune cells. Infect. Immun. 81 (12), 4363–4376. doi: 10.1128/IAI.00819-13
Schilcher, K., Horswill, A. R. (2020). Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84 (3), e00001-19. doi: 10.1128/MMBR.00026-19
Schmitz, F. J., Sadurski, R., Kray, A., Boos, M., Geisel, R., Koöhrer, K., et al. (2000). Prevalence of macrolide-resistance genes in staphylococcus aureus and enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45, 891–894. doi: 10.1093/jac/45.6.891
Schroll, C., Barken, K. B., Krogfelt, K. A., Struve, C. (2010). Role of type 1 and type 3 fimbriae in klebsiella pneumoniae biofilm formation. BMC Microbiol. 10, 179. doi: 10.1186/1471-2180-10-179
Seleem, N. M., Abd El Latif, H. K., Shaldam, M. A., El-Ganiny, A. (2020). Drugs with new lease of life as quorum sensing inhibitors: for combating MDR acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1687–1702. doi: 10.1007/s10096-020-03882-z
Sharma, A., Gupta, V. K., Pathania, R. (2019). Efflux pump inhibitors for bacterial pathogens: from bench to bedside. Indian J. Med. Res. 149, 129–145. doi: 10.4103/ijmr.IJMR_2079_17
Sharmin, S., Rahaman, M. M., Sarkar, C., Atolani, O., Islam, M. T., Adeyemi, O. S. (2021). Nanoparticles as antimicrobial and antiviral agents: a literature-based perspective study. Heliyon 7, e06456. doi: 10.1016/J.HELIYON.2021.E06456
Siegel, S. J., Weiser, J. N. (2015). Mechanisms of bacterial colonization of the respiratory tract 69, 425–444. doi: 10.1146/ANNUREV-MICRO-091014-104209
Sillanpää, J., Nallapareddy, S. R., Singh, K. V., Prakash, V. P., Fothergill, T., Ton-That, H., et al. (2010). Characterization of the ebpfm pilus-encoding operon of enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1, 236. doi: 10.4161/viru.1.4.11966
Silver, L. L. (2011). Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24, 71. doi: 10.1128/CMR.00030-10
Singh, J. K., Adams, F. G., Brown, M. H. (2019). Diversity and function of capsular polysaccharide in acinetobacter baumannii. Front. Microbiol. 10. doi: 10.3389/FMICB.2018.03301/BIBTEX
Slavin, Y. N., Asnis, J., Häfeli, U. O., Bach, H. (2017). Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J. Nanobiotechnology 15, 1–20. doi: 10.1186/S12951-017-0308-Z/FIGURES/4
Smith, J. T., Andam, C. P. (2021). Extensive horizontal gene transfer within and between species of coagulase-negative staphylococcus. Genome Biol. Evol. 13 (9), evab206. doi: 10.1093/GBE/EVAB206
Soares, R. O., Fedi, A. C., Reiter, K. C., Caierão, J., D’Azevedo, P. A. (2014). Correlation between biofilm formation and gelE, esp, and agg genes in enterococcus spp. clinical isolates. Virulence 5, 634. doi: 10.4161/VIRU.28998
Solanki, V., Tiwari, M., Tiwari, V. (2018). Host-bacteria interaction and adhesin study for development of therapeutics. Int. J. Biol. Macromol. 112, 54–64. doi: 10.1016/J.IJBIOMAC.2018.01.151
Solanki, V., Tiwari, M., Tiwari, V. (2023). Investigation of peptidoglycan-associated lipoprotein of acinetobacter baumannii and its interaction with fibronectin to find its therapeutic potential. Infect. Immun. 91 (5), e0002323. doi: 10.1128/IAI.00023-23
Soria-Bustos, J., Ares, M. A., Gómez-Aldapa, C. A., González-y-Merchand, J. A., Girón, J. A., de la Cruz, M. A. (2020). Two type VI secretion systems of enterobacter cloacae are required for bacterial competition, cell adherence, and intestinal colonization. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.560488
Strateva, T., Atanasova, D., Savov, E., Petrova, G., Mitov, I. (2016). Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 20, 127–133. doi: 10.1016/J.BJID.2015.11.011
Sun, D. (2018). Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.02154/BIBTEX
Sussmuth, S. D., Muscholl-Silberhorn, A., Wirth, R., Susa, M., Marre, R., Rozdzinski, E. (2000). Aggregation substance promotes adherence, phagocytosis, and intracellular survival of enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immun. 68, 4900–4906. doi: 10.1128/IAI.68.9.4900-4906.2000
Sutaria, D. S., Moya, B., Green, K. B., Kim, T. H., Tao, X., Jiao, Y., et al. (2018). First penicillin-binding protein occupancy patterns of - lactams and -lactamase inhibitors in klebsiella pneumoniae. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.00282-18
Taglialegna, A., Navarro, S., Ventura, S., Garnett, J. A., Matthews, S., Penades, J. R., et al. (2016). Staphylococcal bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PloS Pathog. 12 (6), e1005711. doi: 10.1371/JOURNAL.PPAT.1005711
Tahbaz, S. V., Azimi, L., Lari, A. R. (2019). Characterization of aminoglycoside resistance mechanisms in acinetobacter baumannii isolates from burn wound colonization. Ann. Burns Fire Disasters 32, 115–121.
Taitt, C. R., Leski, T. A., Stockelman, M. G., Craft, D. W., Zurawski, D. V., Kirkup, B. C., et al. (2014). Antimicrobial resistance determinants in acinetobacter baumannii isolates taken from military treatment facilities. Antimicrob. Agents Chemother. 58, 767–781. doi: 10.1128/AAC.01897-13
Teixeira, B., Rodulfo, H., Carreño, N., Guzmán, M., Salazar, E., Dedonato, M. (2016). Aminoglycoside resistance genes in pseudomonas aeruginosa isolates from cumana, Venezuela. Rev. Inst. Med. Trop. Sao Paulo 58, 13. doi: 10.1590/S1678-9946201658013
Tessier, P. R., Nicolau, D. P. (2013). Tigecycline displays In vivo bactericidal activity against extended-spectrum-β-lactamase-producing enterobacteriaceae after 72-hour exposure period. Antimicrob. Agents Chemother. 57, 640–642. doi: 10.1128/AAC.01824-12/ASSET/B5B1D12E-E93F-46C0-8A71-04DC5D210AD8/ASSETS/GRAPHIC/ZAC9991015090002.JPEG
Thakur, A., Mikkelsen, H., Jungersen, G. (2019). Intracellular pathogens: host immunity and microbial persistence strategies. J. Immunol. Res. 2019, 1356540. doi: 10.1155/2019/1356540
Thomer, L., Schneewind, O., Missiakas, D. (2016). Pathogenesis of staphylococcus aureus bloodstream infections. Annu. Rev. Pathol. Mech. Dis. 11, 343–364. doi: 10.1146/annurev-pathol-012615-044351
Thurlow, L. R., Thomas, V. C., Hancock, L. E. (2009). Capsular polysaccharide production in enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191, 6203–6210. doi: 10.1128/JB.00592-09
Tiwari, V. (2019). Post-translational modification of ESKAPE pathogens as a potential target in drug discovery. Drug Discovery Today 24, 814–822. doi: 10.1016/J.DRUDIS.2018.12.005
Toledo-Arana, A., Valle, J., Solano, C., Arrizubieta, M. J., Cucarella, C., Lamata, M., et al. (2001). The enterococcal surface protein, esp, is involved in enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67, 4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., Fowler, V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Tran, T. T., Panesso, D., Mishra, N. N., Mileykovskaya, E., Guan, Z., Munita, J. M., et al. (2013). Daptomycin-resistant enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 4 (4), e00281-13. doi: 10.1128/mBio.00281-13
Tribuddharat, C., Fennewald, M. (1999). Integron-mediated rifampin resistance in pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 960–962. doi: 10.1128/aac.43.4.960
Tuson, H. H., Weibel, D. B. (2013). Bacteria-surface interactions. Soft Matter 9, 4368. doi: 10.1039/C3SM27705D
Udo, E. E., Boswihi, S. S., Mathew, B., Noronha, B., Verghese, T. (2021). Resurgence of chloramphenicol resistance in methicillin-resistant staphylococcus aureus due to the acquisition of a variant florfenicol exporter (Fexav)-mediated chloramphenicol resistance in Kuwait hospitals. Antibiotics 10, 1250. doi: 10.3390/ANTIBIOTICS10101250/S1
Uneputty, A., Dávila-Lezama, A., Garibo, D., Oknianska, A., Bogdanchikova, N., Hernández-Sánchez, J. F., et al. (2022). Strategies applied to modify structured and smooth surfaces: a step closer to reduce bacterial adhesion and biofilm formation. Colloid Interface Sci. Commun. 46, 100560. doi: 10.1016/J.COLCOM.2021.100560
Upadhyaya, G. P. M., Ravikumar, K. L., Umapathy, B. L. (2009). Review of virulence factors of enterococcus: AAAn emerging nosocomial pathogen. Indian J. Med. Microbiol. 27, 301–305. doi: 10.4103/0255-0857.55437
Uppalapati, S. R., Sett, A., Pathania, R. (2020). The outer membrane proteins OmpA, CarO, and OprD of acinetobacter baumannii confer a two-pronged defense in facilitating its success as a potent human pathogen. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.589234
Upritchard, H. G., Yang, J., Bremer, P. J., Lamont, I. L., McQuillan, A. J. (2011). Adsorption of enterobactin to metal oxides and the role of siderophores in bacterial adhesion to metals. Langmuir 27, 10587–10596. doi: 10.1021/la202352j
Uribe-García, A., Paniagua-Contreras, G. L., Monroy-Pérez, E., Bustos-Martínez, J., Hamdan-Partida, A., Garzón, J., et al. (2021). Frequency and expression of genes involved in adhesion and biofilm formation in staphylococcus aureus strains isolated from periodontal lesions. J. Microbiol. Immunol. Infect. 54, 267–275. doi: 10.1016/J.JMII.2019.05.010
Ursell, L. K., Metcalf, J. L., Parfrey, L. W., Knight, R. (2012). Defining the human microbiome. Nutr. Rev. 70 Suppl 1. doi: 10.1111/J.1753-4887.2012.00493.X
Vaou, N., Stavropoulou, E., Voidarou, C., Tsigalou, C., Bezirtzoglou, E. (2021). Towards advances in medicinal plant antimicrobial activity: a review study on challenges and future perspectives. Microorg 9, 2041. doi: 10.3390/MICROORGANISMS9102041
Vasudevan, S., David, H., Chanemougam, L., Ramani, J., Ramesh Sangeetha, M., Solomon, A. P. (2022). Emergence of persister cells in staphylococcus aureus: calculated or fortuitous move? Crit Rev Microbiol. 22, 1–12. doi: 10.1080/1040841X.2022.2159319
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., Hardie, K. R. (2005). Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi: 10.1038/nrmicro1146
Verma, P., Tiwari, M., Tiwari, V. (2021). Strategies to combat bacterial antimicrobial resistance: a focus on mechanism of the efflux pumps inhibitors. SN Compr. Clin. Med. 3, 510–527. doi: 10.1007/S42399-021-00780-Z
Vinuesa, V., McConnell, M. J. (2021). Recent advances in iron chelation and gallium-based therapies for antibiotic resistant bacterial infections. Int. J. Mol. Sci. 22, 2876. doi: 10.3390/IJMS22062876
Vor, L., Rooijakkers, S. H. M., Strijp, J. A. G. (2020). Staphylococci evade the innate immune response by disarming neutrophils and forming biofilms. FEBS Lett. 594, 2556–2569. doi: 10.1002/1873-3468.13767
Vrancianu, C. O., Gheorghe, I., Czobor, I. B., Chifiriuc, M. C. (2020a). Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of acinetobacter baumannii. Microorganisms 8, 1–40. doi: 10.3390/microorganisms8060935
Vrancianu, C. O., Popa, L. I., Bleotu, C., Chifiriuc, M. C. (2020b). Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00761
Vuotto, C., Longo, F., Balice, M. P., Donelli, G., Varaldo, P. E. (2014). Antibiotic resistance related to biofilm formation in klebsiella pneumoniae. Pathogens 3, 743. doi: 10.3390/PATHOGENS3030743
Wainwright, M., Maisch, T., Nonell, S., Plaetzer, K., Almeida, A., Tegos, G. P., et al. (2017). Photoantimicrobials–are we afraid of the light? Lancet Infect. Dis. 17, e49–e55. doi: 10.1016/S1473-3099(16)30268-7
Wang, C., Fang, R., Zhou, B., Tian, X., Zhang, X., Zheng, X., et al. (2019). Evolution of resistance mechanisms and biological characteristics of rifampicin-resistant staphylococcus aureus strains selected in vitro. BMC Microbiol. 19, 220. doi: 10.1186/s12866-019-1573-9
Wang, T., Flint, S., Palmer, J. (2019). Magnesium and calcium ions: roles in bacterial cell attachment and biofilm structure maturation. Biofouling 35, 959–974. doi: 10.1080/08927014.2019.1674811
Wang, Z., Guo, C., Xu, Y., Liu, G., Lu, C., Liu, Y. (2014). Two novel functions of hyaluronidase from streptococcus agalactiae are enhanced intracellular survival and inhibition of proinflammatory cytokine expression. Infect. Immun. 82, 2615–2625. doi: 10.1128/IAI.00022-14
Wang, L., Hu, C., Shao, L. (2017). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomedicine 12, 1227–1249. doi: 10.2147/IJN.S121956
Wei, Q., Ma, L. Z. (2013). Biofilm matrix and its regulation in pseudomonas aeruginosa. Int. J. Mol. Sci. 14, 20983. doi: 10.3390/IJMS141020983
Weinstein, Z. B., Zaman, M. H. (2019). Evolution of rifampin resistance in escherichia coli and mycobacterium smegmatis due to substandard drugs. Antimicrob. Agents Chemother. 63 (1), e01243-18. doi: 10.1128/AAC.01243-18
Whitchurch, C. B., Mattick, J. S. (1994). Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in pseudomonas aeruginosa. Mol. Microbiol. 13, 1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x
White, P. A., Stokes, H., Bunny, K. L., Hall, R. M. (1999). Characterisation of a chloramphenicol acetyltransferase determinant found in the chromosome of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 175, 27–35. doi: 10.1111/j.1574-6968.1999.tb13598.x
WHO (2017). Global antimicrobial resistance surveillance system (GLASS) report (Geneva: World Health Organization). Available at: https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061-eng.pdf?ua=1.
Wilson, J. W., Schurr, M. J., LeBlanc, C. L., Ramamurthy, R., Buchanan, K. L., Nickerson, C. A. (2002). Mechanisms of bacterial pathogenicity. Postgrad. Med. J. 78, 216–224. doi: 10.1136/PMJ.78.918.216
Wolter, N., Smith, A. M., Farrell, D. J., Schaffner, W., Moore, M., Whitney, C. G., et al. (2005). Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49, 3554–3557. doi: 10.1128/AAC.49.8.3554-3557.2005
Woodworth, M. H., Hayden, M. K., Young, V. B., Kwon, J. H. (2019). The role of fecal microbiota transplantation in reducing intestinal colonization with antibiotic-resistant organisms: the current landscape and future directions. Open Forum Infect. Dis. 6 (10), ofz391. doi: 10.1093/ofid/ofz288
Wu, C., Lin, C., Zhu, X., Liu, H., Zhou, W., Lu, J., et al. (2018). The β-lactamase gene profile and a plasmid-carrying multiple heavy metal resistance genes of enterobacter cloacae. Int. J. Genomics 2018, 4989602. doi: 10.1155/2018/4989602
WO5_03151 - cytolysin immunity protein CylI - enterococcus faecalis EnGen0354 - WO5_03151 gene & protein. Available at: https://www.uniprot.org/uniprot/A0A0M2AP93#function (Accessed April 3, 2021).
Yang, Y., Li, W., Hou, B., Zhang, C. (2018). Quorum sensing LuxS/autoinducer-2 inhibits enterococcus faecalis biofilm formation ability. J. Appl. Oral. Sci. 26, e20170566. doi: 10.1590/1678-7757-2017-0566
Yee, Y. C., Kisslinger, B., Yu, V. L., Jin, D. J. (1996). A mechanism of rifamycin inhibition and resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 38, 133–137. doi: 10.1093/jac/38.1.133
Yin, W., Wang, Y., Liu, L., He, J. (2019). Biofilms: the microbial “Protective clothing” in extreme environments. Int. J. Mol. Sci. 20 (14), 3423. doi: 10.3390/IJMS20143423
Youssouf, N., Recasens-zorzo, C., Molle, V., Bossis, G., Soubeyran, P., Gannoun-zaki, L. (2021). Staphylococcus aureus decreases SUMOylation host response to promote intramacrophage survival. Int. J. Mol. Sci. 22 (15), 8108. doi: 10.3390/IJMS22158108
Yu, M. K., Kim, M. A., Rosa, V., Hwang, Y. C., Del Fabbro, M., Sohn, W. J., et al. (2019). Role of extracellular DNA in enterococcus faecalis biofilm formation and its susceptibility to sodium hypochlorite. J. Appl. Oral. Sci. 27 , e20180699. doi: 10.1590/1678-7757-2018-0699
Yushchuk, O., Binda, E., Marinelli, F. (2020). Glycopeptide antibiotic resistance genes: distribution and function in the producer actinomycetes. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01173
yhbU_2 - collagenase-like protease - staphylococcus aureus - yhbU_2 gene & protein. Available at: https://www.uniprot.org/uniprot/W8TVT7 (Accessed December 28, 2020).
Zhang, R., Tan, K., Zhou, M., Bargassa, M., Joachimiak, A. (2007). The crystal structure of a hemolysin domain from enterococcus faecalis V583.
Zhao, L., Xue, T., Shang, F., Sun, H., Sun, B. (2010). Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 78, 3506–3515. doi: 10.1128/IAI.00131-10
Zhao, X., Yu, Z., Ding, T. (2020). Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 8 (3), 425. doi: 10.3390/microorganisms8030425
Zheng, S., Bawazir, M., Dhall, A., Kim, H. E., He, L., Heo, J., et al. (2021). Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 9. doi: 10.3389/FBIOE.2021.643722/BIBTEX
Zhong, L., Ravichandran, V., Zhang, N., Wang, H., Bian, X., Zhang, Y., et al. (2020). Attenuation of pseudomonas aeruginosa quorum sensing by natural products: virtual screening, evaluation and biomolecular interactions. Int. J. Mol. Sci. 21 (6), 2190. doi: 10.3390/ijms21062190
Zhou, L., Zhang, Y., Ge, Y., Zhu, X., Pan, J. (2020). Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 11, 3925868. doi: 10.3389/fmicb.2020.589640
Keywords: ESKAPE, virulence, antimicrobial resistance, biofilm, quorum sensing
Citation: Venkateswaran P, Vasudevan S, David H, Shaktivel A, Shanmugam K, Neelakantan P and Solomon AP (2023) Revisiting ESKAPE Pathogens: virulence, resistance, and combating strategies focusing on quorum sensing. Front. Cell. Infect. Microbiol. 13:1159798. doi: 10.3389/fcimb.2023.1159798
Received: 06 February 2023; Accepted: 08 June 2023;
Published: 29 June 2023.
Edited by:
Vishvanath Tiwari, Central University of Rajasthan, IndiaReviewed by:
Guangshun Wang, University of Nebraska Medical Center, United StatesSagar Kiran Khadke, School of Medicine Wonkwang University, Republic of Korea
Copyright © 2023 Venkateswaran, Vasudevan, David, Shaktivel, Shanmugam, Neelakantan and Solomon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adline Princy Solomon, adlineprinzy@sastra.ac.in
†These authors have contributed equally to this work
 Parvathy Venkateswaran
Parvathy Venkateswaran Sahana Vasudevan
Sahana Vasudevan Helma David
Helma David Adityan Shaktivel
Adityan Shaktivel Karthik Shanmugam
Karthik Shanmugam Prasanna Neelakantan
Prasanna Neelakantan Adline Princy Solomon
Adline Princy Solomon