Rapid Detection and Differentiating of the Predominant Salmonella Serovars in Chicken Farm by TaqMan Multiplex Real-Time PCR Assay
- Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
Salmonella has been known as an important zoonotic pathogen that can cause a variety of diseases in both animals and humans. Poultry are the main reservoir for the Salmonella serovars Salmonella Pullorum (S. Pullorum), Salmonella Gallinarum (S. Gallinarum), Salmonella Enteritidis (S. Enteritidis), and Salmonella Typhimurium (S. Typhimurium). The conventional serotyping methods for differentiating Salmonella serovars are complicated, time-consuming, laborious, and expensive; therefore, rapid and accurate molecular diagnostic methods are needed for effective detection and prevention of contamination. This study developed and evaluated a TaqMan multiplex real-time PCR assay for simultaneous detection and differentiation of the S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium. In results, the optimized multiplex real-time PCR assay was highly specific and reliable for all four target genes. The analytical sensitivity corresponded to three colony-forming units (CFUs) for these four Salmonella serovars, respectively. The detection limit for the multiplex real-time PCR assay in artificially contaminated samples was 500 CFU/g without enrichment, while 10 CFU/g after pre-enrichment. Moreover, the multiplex real-time PCR was applied to the poultry clinical samples, which achieved comparable results to the traditional bacteriological examination. Taken together, these results indicated that the optimized TaqMan multiplex real-time PCR assay will be a promising tool for clinical diagnostics and epidemiologic study of Salmonella in chicken farm and poultry products.
Introduction
Salmonella is an important zoonotic pathogen that can cause a variety of diseases in both animals and humans (Majowicz et al., 2010). Salmonella is prevalent in domestic animals such as poultry, pigs, and cattle. Poultry are a main reservoir for Salmonella, including the most prevalent Salmonella serovars Salmonella Pullorum (S. Pullorum), Salmonella Gallinarum (S. Gallinarum), Salmonella Enteritidis (S. Enteritidis), and Salmonella Typhimurium (S. Typhimurium) (Medalla et al., 2016; Wang et al., 2020; Xu et al., 2020; Zhao et al., 2020; Yu et al., 2021). These Salmonella serovars can lead to serious avian salmonellosis, which causes economic losses in the poultry industry. In addition, Salmonella can be transmitted to humans by contaminated poultry products and cause acute gastroenteritis or diarrhea, being a threat to public health (Balasubramanian et al., 2019). Currently, animals, in particular poultry, are considered to be the primary cause for salmonellosis and numerous other foodborne outbreaks (Keerthirathne et al., 2017; Biswas et al., 2019; Liu et al., 2021). Thus, detection and differentiation of these Salmonella serovars in poultry farms are required to prevent, control, and eliminate the spread of Salmonella.
Rapid and accurate diagnosis is curial for effective prevention and control of the disease. Currently, more than 2,600 Salmonella serovars have been identified based on the O, H, and Vi antigens (Issenhuth-Jeanjean et al., 2014). Although bacteriological culture and serum agglutination test were considered to be the gold standard for differentiating Salmonella serovars, there were many disadvantages for this routine diagnosis in practice. The conventional culture method tends to be complex, time-consuming, and laborious. Moreover, false-negative result for O and H antigens agglutination test occurs occasionally due to the loss of surface antigens in non-culturable state (Schrader et al., 2008). In efforts to avoid such disadvantages, several nucleic acid amplification methods have been developed to detect and differentiate the Salmonella serovars (Shi et al., 2015). Although there is extensive sequence conservation in Salmonella genome, comparative genomic analysis is effective to validate novel serovar-specific genes. The unique genes had been identified among the different Salmonella serovars (Liu et al., 2011; Zhang et al., 2019). The gene lygD in Sdf locus has been found specific in S. Enteritidis. Serovar-specific gene STM4495 for identifying S. Typhimurium was obtained by comparative genomics (Agron et al., 2001; Akiba et al., 2011). In addition, comparative analysis of the glgC gene sequence identified an 11 bp (GATCGATCACG) deletion presented only in S. Gallinarum but not other Salmonella serovars (Kang et al., 2011). Based on these specific gene, PCR assays were applied for detecting different Salmonella serovars (Shah et al., 2005; Kim et al., 2006; Hong et al., 2008; Kang et al., 2011; Xu et al., 2018). Compared to conventional PCR, real-time PCR assay offers advantages in rapidity, quantitative measurement, and avoiding of cross-contamination. More importantly, real-time PCR assay enables to obtain both qualitative and quantitative measurement of the pathogen presented in samples. Thus, real-time PCR has been widely utilized to detect different pathogens (Ding et al., 2017; Liu et al., 2019). Recently, increasing studies developed single and multiplex real-time PCR for the specific detection of major Salmonella serovars in food products (Lee et al., 2009; Munoz et al., 2010; Prendergast et al., 2013; Kasturi and Drgon, 2017; Kim et al., 2017; Nair et al., 2019). The rapid detection and differentiation of Salmonella serovars are required for the epidemiologic investigation of Salmonella in chicken farms.
This study attempted to develop a rapid multiplex RT-PCR assay for the simultaneous detection and differentiation of the prevalent S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium. The specificity and sensitivity evaluations indicated that the developed multiplex real-time PCR assay appears to be a promising tool for clinical diagnostics and epidemiology studies for Salmonella in chicken farm and poultry products.
Materials and Methods
Bacterial Strains and Growth Conditions
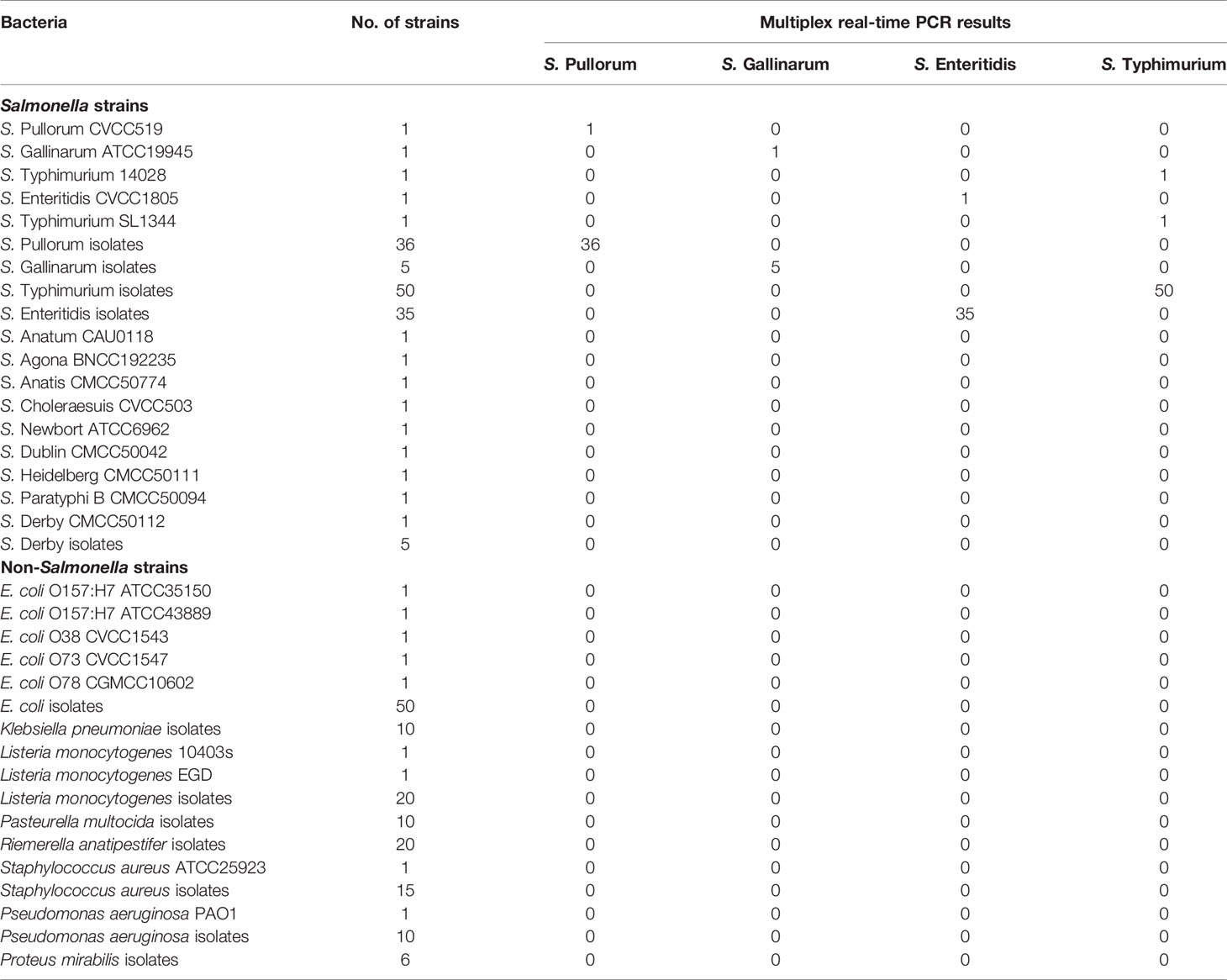
The Salmonella and non-Salmonella bacterial strains used to establish and verify the multiplex real-time PCR assay are listed in Table 1. The Salmonella, Escherichia coli, and Pseudomonas aeruginosa strains were cultured at 37°C in Luria-Bertani (LB) medium (BD, Detroit, MI, USA) with aeration. Other bacterial strains were grown in appropriate medium under recommended culture conditions.
DNA Extraction
Bacterial genomic DNA was extracted from fresh bacterial culture using TIANamp Bacteria DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions. Whereas, the total DNA from clinical samples was prepared using DNA Isolation Reagent for meat Products (Tiangen Biotech, Beijing, China). The concentration and purity of the DNA were measured with spectrophotometer.
Primers and Probes Designing
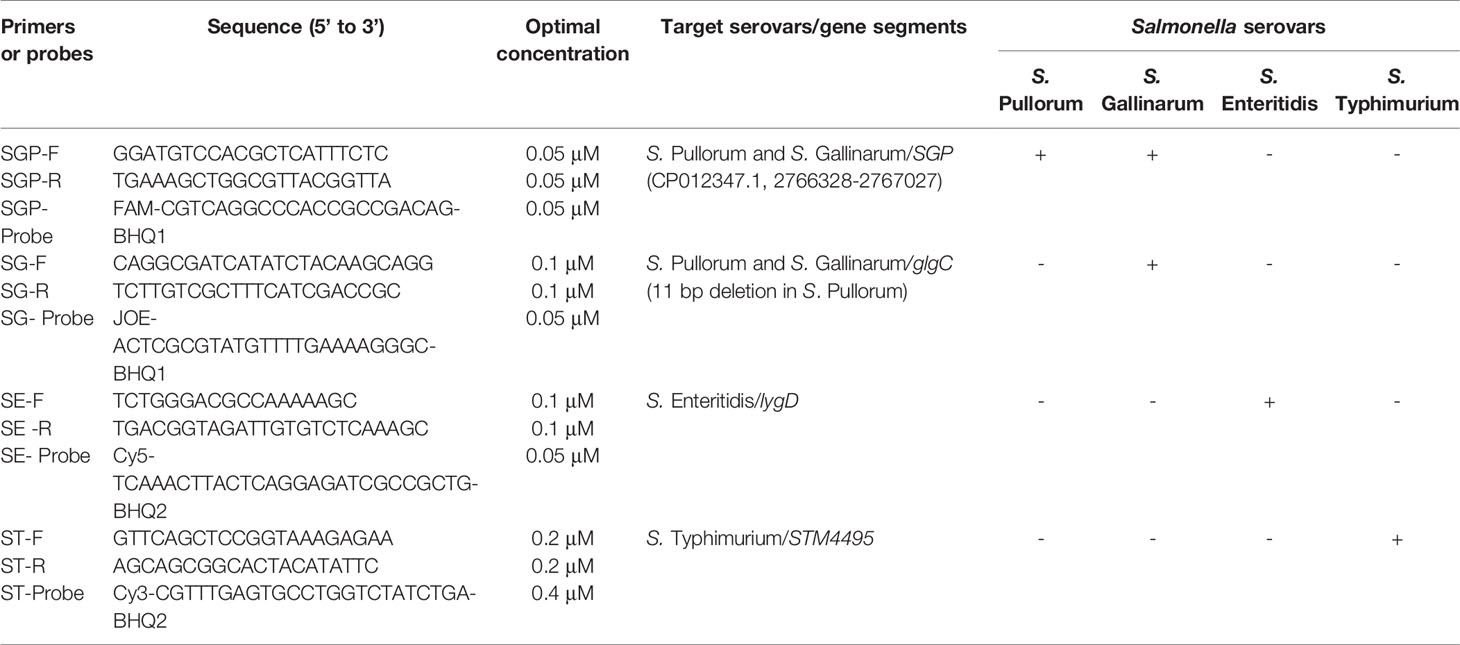
To design the suitable primers, the specific gene sequences of these Salmonella serovars were analyzed. By bioinformatics analysis, we found a specific gene segment (699 bp) SGP (GenBank No. CP012347.1 segment 2766328 to 2767027) was presented and generally conserved in S. Pullorum and S. Gallinarum. In addition, a previous study identified an 11 bp (GATCGATCACG) sequence deletion in glgC gene presented only in S. Gallinarum, but not other Salmonella serovars (Kang et al., 2011). Thus, the SGP gene segment and truncated sequence of glgC gene could be exploited to differentiate S. Pullorum and S. Gallinarum from other Salmonella serovars. The serovar-specific genes lygD and STM4495 for specifically identifying and differentiating S. Enteritidis and S. Typhimurium were selected as targets according to previous studies (Agron et al., 2001; Akiba et al., 2011). Then, the specific primers and probes were designed based on the specific gene sequences (Table 2 and Figure 1). Furthermore, the specificity of the primer sequences was tested by in silico analysis using BLAST at the National Center for Biotechnology Information (NCBI). All the primers and probes were synthesized by Sangon Biotech (Shanghai) Co., Ltd, China.
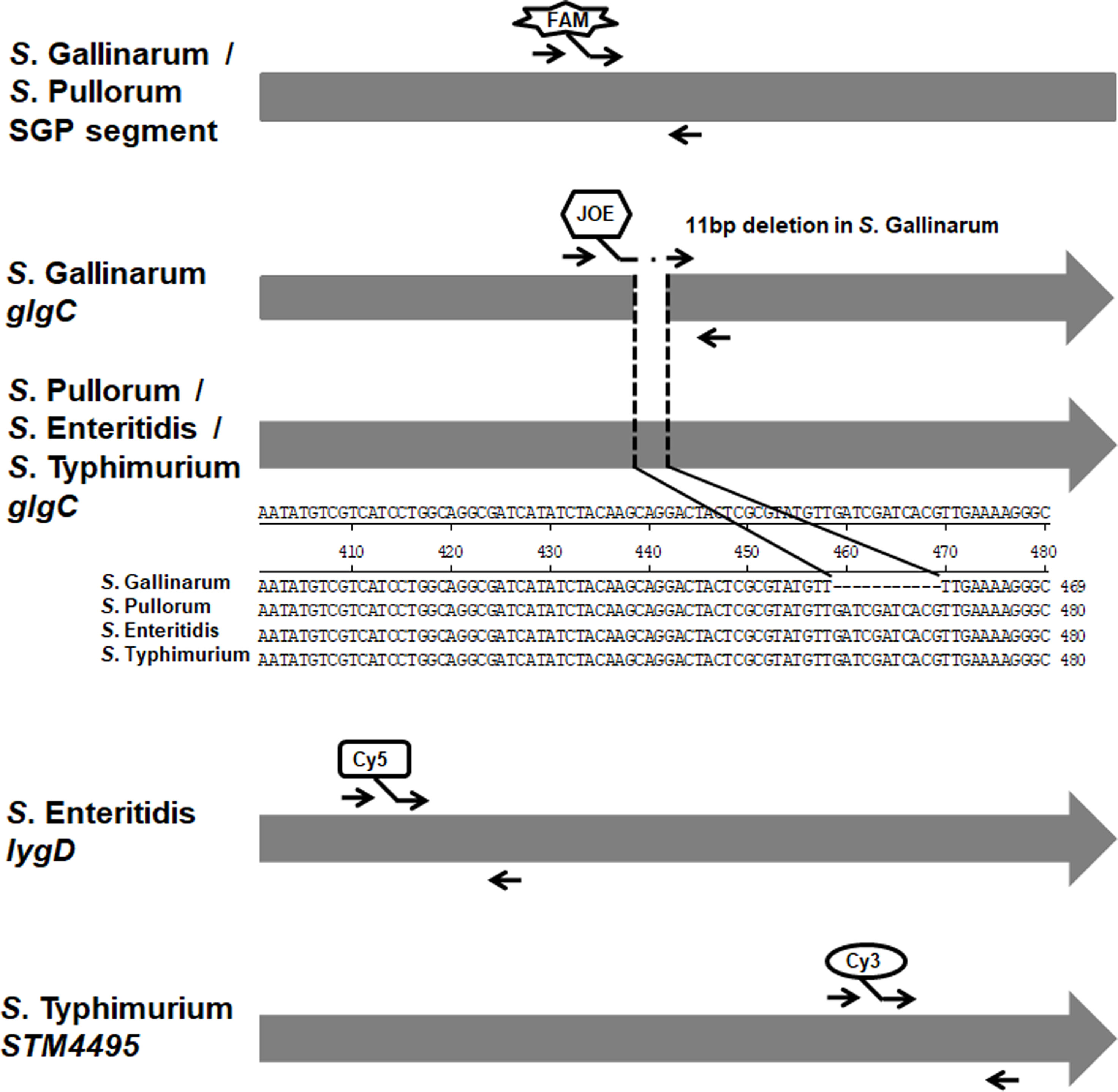
Figure 1 Diagram of the primers and probes designing for the multiplex real-time PCR. The specific gene or segment of these four Salmonella serovars was analyzed and exploited to design the primers and probes. The primers and fluorophore-labeled probes were indicated. The arrows indicated the positions of the designed primers. In addition, the alignments of glgC genes in S. Gallinarum, S. Pullorum, S. Enteritidis, and S. Typhimurium were shown.
Optimization and Development of Multiplex Real-Time PCR Assay
The multiplex real-time PCR was carried out in a final volume of 20.0 µl, and the concentrations of primers, probes, and reaction condition were optimized using the purified DNA of S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium reference strains (Table 1). Sterile distilled water was used as negative control. The multiplex real-time PCR was performed on the ABI 7500 Real-time PCR system (Applied Biosystems, CA, USA), and fluorescent signals were detected simultaneously during annealing/extension phase. All analyses were performed with ABI 7500 Software Version 1.4 (Applied Biosystems, CA, USA).
Specificity of the Multiplex Real-Time PCR Assay
A collection of bacterial strains, including various Salmonella serovars and non-Salmonella (Table 1), was used to evaluate the specificity of multiplex real-time PCR assay. All of the bacterial strains have been confirmed by biochemical identification, PCR, and serotyping with traditional agglutination assay. The bacterial genomic DNA was extracted and used as a template in the multiplex real-time PCR assay under optimized condition.
Standard Curve and Sensitivity Analysis
The standard curves and sensitivity of multiplex real-time PCR were determined using genomic DNA of various bacterial concentrations as described previously (Lee et al., 2009; Ding et al., 2017). The pure cultures of the S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium strains were 10-fold serially diluted to appropriate dilutions (ranging from 3 to 3×107 CFU/ml), which were counted by plating. The genomic DNA extracted from bacterial culture dilutions was used as templates for multiplex real-time PCR. Negative control includes sterile distilled water in place of DNA. The standard curves were calculated automatically based on the Cycle threshold (Ct) values using the ABI 7500 Software. The amplification efficiencies (E) were determined by using the slope of the standard curve and applying the equation: E = (10−1/slope)-1 (Kawasaki et al., 2010).
Evaluation of the Limit of Detection of Multiplex Real-Time PCR in Artificial Contamination Samples
The LOD of multiplex real-time PCR assay was evaluated for the artificial contamination samples with or without enrichment as previously described (Lee et al., 2009), with some modifications. Briefly, 100 μl of each bacterial dilutions (1 to 108 CFU/ml) were individually added to 1 g of chicken meat samples. Then, these contaminated samples were thoroughly homogenized with 9 ml of buffered peptone water (BPW). The pre-enriched homogenized samples were used for DNA extraction using DNA Isolation Reagent for meat Products (Tiangen Biotech, Beijing, China). In addition, the homogenized samples were incubated at 37°C for 6 h. After primary enrichment, DNA extraction was performed. These DNA were used as templates for multiplex real-time PCR. Non-inoculated meat was subjected to the same procedure and used as a negative control.
Detection of Clinical Samples
The multiplex real-time PCR assay was applied to evaluate the presence of Salmonella in 60 sick or dead chicken with clinical signs collected from five farms. All animal experiments were conducted in strict accordance with the guidelines of the Humane Treatment of Laboratory Animals and were approved by the Animal Care and Use Committee at the Shanghai Veterinary Research Institute, China. The liver samples were collected aseptically from the chickens. The samples were homogenized for primary enrichment or DNA isolation as described above. The DNA extracted from these samples was analyzed by multiplex real-time PCR method. Meanwhile, each sample was also subjected to standard bacterial culture methods and traditional serum agglutination assay.
Results
Development of the Multiplex Real-Time PCR Assay
Based on the bioinformatics analysis, the specific primers and probes were designed based on the target genes of these four Salmonella serovars (Figure 1). Then, multiplex real-time PCR was optimized by adjustment of different parameters. The optimal amplification reaction contained 10.0 µl Premix Ex Taq™ (Takara, Dalian, China), 0.2 μl ROX Reference Dye II (Takara, Dalian, China), each primer and probe at final concentrations of 0.05 to 0.4 μM (Table 2), 2.0 μl template and nuclease-free water. The reaction profile consisting of initial denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s and 40 s annealing/extension at 57°C. Accordingly, the reference strains of these four Salmonella serovars were specifically differentiated by the multiplex real-time PCR assay.
Analytical Specificity
The specificity of the multiplex real-time PCR was evaluated using the different bacterial templates listed in Table 1. All the S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium strains produced the corresponding amplified signals (Figure 2). Whereas, the non-target bacteria, including other Salmonella serovars and non-Salmonella strains, yielded negative results in the multiplex real-time PCR (Table 1). No false positive or negative results were found, indicating the multiplex real-time PCR was specific.
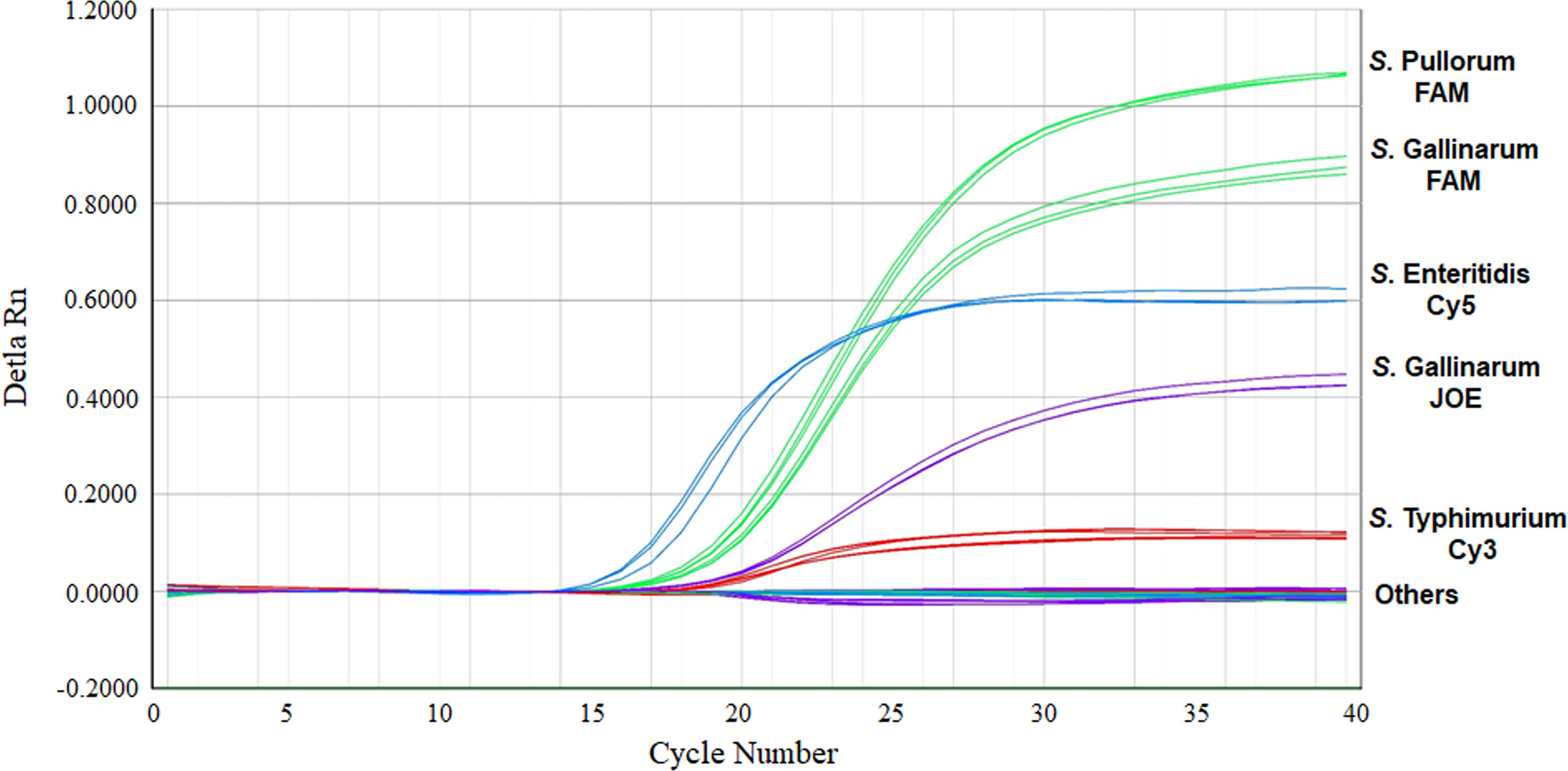
Figure 2 Specificity of the multiplex real-time PCR for the detection and differentiation of S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium. Others: None of these four Salmonella serovars bacteria.
Standard Curve and Sensitivity of the Multiplex Real-Time PCR Assay
The standard curve of the multiplex real-time PCR assay was constructed using the mean Ct values for various Salmonella concentrations corresponding to the genomic DNA. As shown in Figure 3, the slopes of the standard curves for S. Pullorum, S. Gallinarum, S. Enteritidis, S. Typhimurium were −3.567, −3.464, −3.448, and −3.360, respectively. The correlation coefficients (R2) were above 0.99, and the amplification efficiencies ranged from 90 to 110%, indicating high linearity for the multiplex real-time PCR assay. The sensitivity analysis showed that the bacterial DNA corresponding to 3 CFU of these four Salmonella serovars could be detected for the multiplex real-time PCR assay.
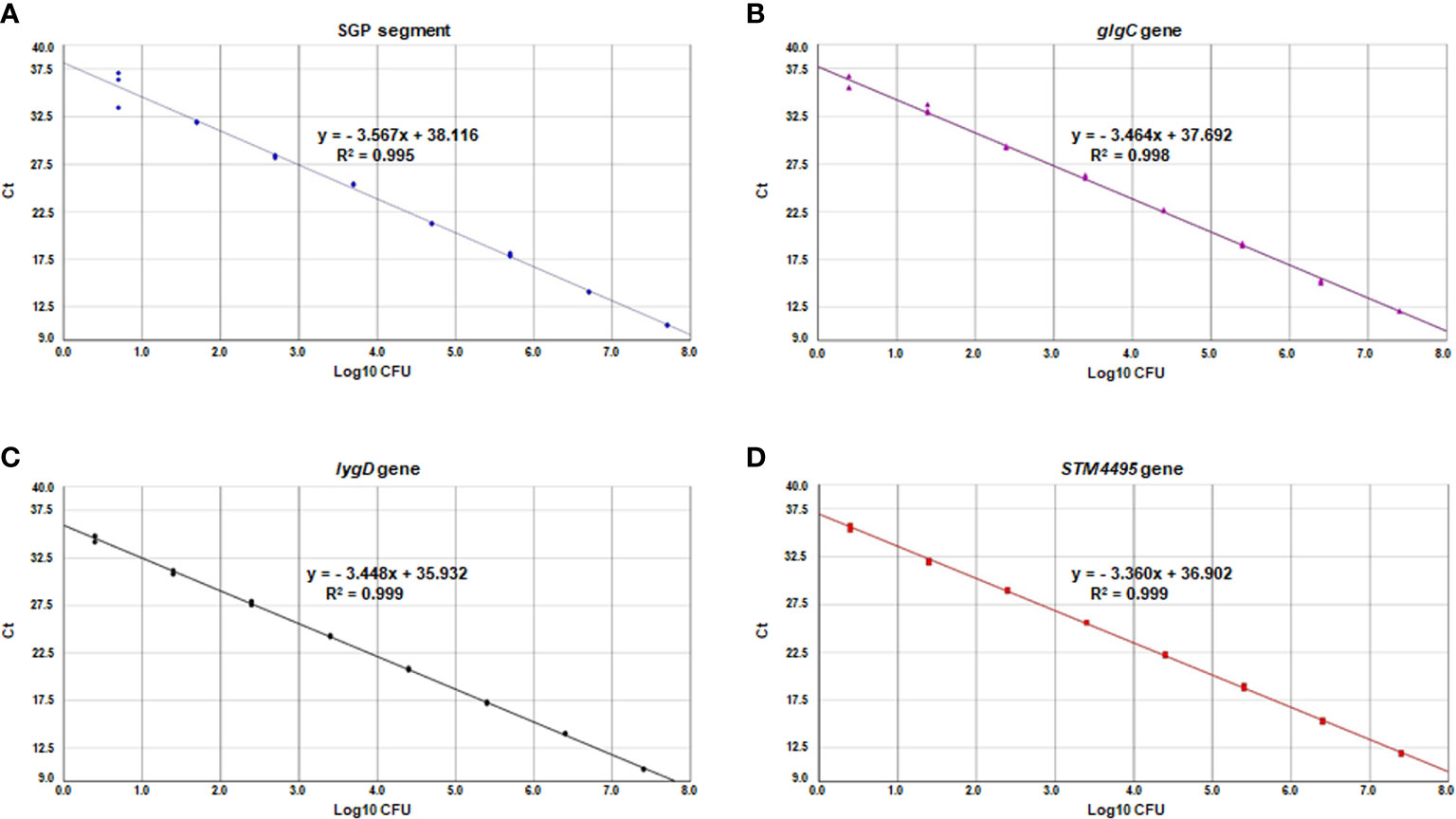
Figure 3 Standard curves of the multiplex real-time PCR for SGP segment (A), glgC gene (B), lygD gene (C), and STM4495 gene (D) using serially diluted S. Pullorum (A), S. Gallinarum (B), S. Enteritidis (C), and S. Typhimurium (D) bacterial DNA, respectively.
Limit of Detection in Artificially Contaminated Chicken Samples
The artificially contaminated samples with serial dilutions of each Salmonella were tested for the LOD of multiplex real-time PCR assay. For the samples without enrichment, each Salmonella of 500 CFU/g could be detected by the multiplex real-time PCR assay. However, when incubated for 6 h for the enrichment, the multiplex real-time PCR assay could successfully detect as low as 10 CFU of each Salmonella in 1 g chicken samples.
Clinical Samples Validation
To evaluate the discernibility and applicability of established method for clinical samples, a total of 60 suspected samples were collected and detected using our multiplex real-time PCR and conventional bacteriological tests. After enrichment, 35 of the 60 clinical samples were Salmonella positive by multiplex real-time PCR, whereas other samples had no Salmonella. Among these positive samples, 21 samples were identified as S. Pullorum, one for S. Gallinarum, nine for S. Enteritidis, and four for S. Typhimurium. Same samples were also examined by traditional bacteriological serotyping tests, which was in accordance with the multiplex real-time PCR results with enrichment. Whereas, two Salmonella positive samples gave negative results without the additional enrichment step (Table 3). This might be due to the limited bacterial amounts in the samples. Thus, the results indicated that the multiplex real-time PCR assay with enrichment is more sensitive than pre-enriched condition to detect the limited amounts of bacteria in samples.
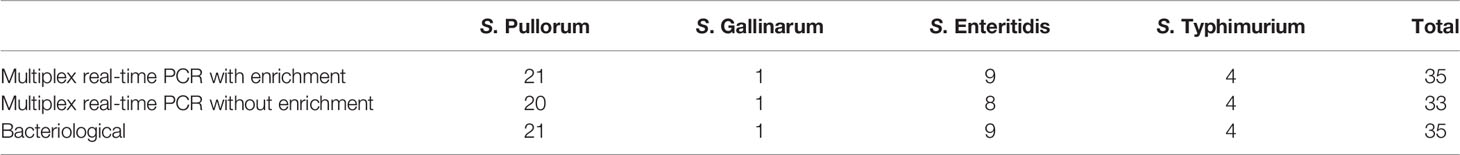
Table 3 Consistency evaluation of the multiplex real-time PCR and conventional bacteriological method for the clinical samples.
Discussion
It has been shown that S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium were the prevalent pathogens of salmonellosis in chicken farms (Medalla et al., 2016; Wang et al., 2020; Xu et al., 2020; Zhao et al., 2020; Yu et al., 2021). In addition, S. Enteritidis and S. Typhimurium could lead to serious zoonotic diseases via contaminated food, including poultry products (Coburn et al., 2007). Conventional, Salmonella serovars were identified according to the Kauffman-White scheme based on the specific cell-surface O and H antigens. Although serum agglutination assay offers a reliable method for differentiating Salmonella serovars, it is labor-intensive, complex, costly, and time-consuming (Schrader et al., 2008). Nowadays, reducing cost and time of experiment are critical for pathogen detection. Therefore, the rapid and accurate detection method has the potential to be of great significance for preventing the spread of salmonellosis. Several molecular methods, such as PCR and loop-mediated isothermal amplification, exist for identifying various Salmonella serovars with advantages in sensitivity, specificity, and speed (Shi et al., 2015; Yang et al., 2018). Among them, real-time PCR is more sensitive and suitable for high-throughput analysis (Kralik and Ricchi, 2017). Thus, this study developed a multiplex real-time PCR assay that simultaneously detected and differentiated the prevalent S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium, which exhibited efficiently identification in cultured bacteria and chicken samples.
Selection of specific target genes and design of compatible primers and probes are critical for the proper detection specificity of nucleic acid amplification. Although the homology of the genomes of various Salmonella serovars was very high, some genes were found to be related to specific serovars. Various genes, such as genes encoding the O, H, and Vi antigens (rfb, fliC, fliB, viaB, ipaJ), have been candidates suitable for the specific detection and serotyping of Salmonella in diverse clinical samples (Hirose et al., 2002; Hong et al., 2008; Xu et al., 2018). In the present study, analysis of genomic sequences identified a gene segment SGP (GenBank No. CP012347.1 segment 2766328 to 2767027) specifically existing in all S. Pullorum and S. Gallinarum. A previous study has revealed that the glgC gene is deemed to be the preferred target for differentiating S. Pullorum and S. Gallinarum from other Salmonella serovars (Kang et al., 2011). Based on literature (Agron et al., 2001; Akiba et al., 2011), the S. Enteritidis specific lygD gene and STM4495 gene specific for S. Typhimurium were chosen as targets in this study. As a result, the primers and probes were designed and optimized targeting these specific genes. Furthermore, no mismatch in the primers and probes with the available bacterial genome in GenBank was found by in silico analysis. The developed multiplex real-time PCR showed excellent specificity and exclusivity by the detection of Salmonella strains as well as other bacterial species. No cross-reactivity, false positives, or false negatives were observed. Previously, real-time PCR assays targeting various specific genes had been applied for Salmonella (Lee et al., 2009; Kasturi and Drgon, 2017; Kim et al., 2017; Nair et al., 2019). This study incorporated these four target genes into a unified multiplex real-time PCR assay for the detection and differentiation of multiple Salmonella serovars in chicken samples.
In the analytical sensitivity evaluation, the developed multiplex real-time PCR assay was shown to detect as low as the bacterial DNA corresponding to 3 CFU/ml bacterial cultures. Although the LOD of this multiplex real-time PCR was 500 CFU/g in artificially contaminated chicken samples without enrichment, it had more improved detection limits and yielded positive results even at the lowest contamination levels tested (10 CFU/g chicken samples) for the enriched samples. There was some uncertainty, such as PCR inhibitors, competitor organisms, which might result in the lower LOD without enrichment step. Indeed, the pre-enrichment step was effective to increase the number of bacterial cells and to dilute inhibitory substances that exist in the sample. Thus, an additional enrichment step was actually applied to increase the sensitivity of the multiplex real-time PCR (Lee et al., 2009; Ding et al., 2017). The LOD of this multiplex real-time PCR assay was similar to previous studies (Munoz et al., 2010; Kasturi and Drgon, 2017; Liu et al., 2019), indicating its sensitivity for diagnostic purpose. Our multiplex real-time PCR was applied to detect these four Salmonella serovars in poultry clinical samples, which achieved same results comparable to the traditional bacteriological examination. However, two of the positive samples were tested as negative by multiplex real-time PCR lack of enrichment step. The possibility remains the amplification inhibition factors or low bacterial concentration in the samples (Lee et al., 2009; Ding et al., 2017). In terms of shortening time for multiple bacterial detection, the entire process of the multiplex real-time PCR assay from sample enrichment to data analysis can be completed in 12 h. The effectiveness of multiplex real-time PCR was significantly improved compared to the traditional culture method.
In summary, this study developed a TaqMan multiplex real-time PCR assay for simultaneous detection and differentiation of prevalent S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium. Considering the specificity, sensitivity, and effectiveness, the multiplex real-time PCR assay developed herein appears to be a promising tool for clinical diagnostics and epidemiologic study of Salmonella in chicken farm and poultry products.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee at the Shanghai Veterinary Research Institute, China.
Author Contributions
SW conceived the project. SX and HZ developed the TaqMan multiplex real-time PCR assay and wrote the original manuscript draft. CT, BZ, LY, YZ, and DA were responsible for sampling and sample test. TL, MT, JQ, CD, SY, and SW analyzed and discussed the experimental results. SW and SY directed the experiments, funded the research, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0500800, 2018YFD0500500), the National Natural Science Foundation of China (31972654), Shanghai Pujiang Program (2019PJD057), Scientific and Technical Innovation Project of the Chinese Academy of Agricultural Sciences, China (SHVRI-ASTIP-2014-8), and the National Basic Fund for Research Institutes, which is supported by the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (2021JB07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agron, P. G., Walker, R. L., Kinde, H., Sawyer, S. J., Hayes, D. C., Wollard, J., et al. (2001). Identification by Subtractive Hybridization of Sequences Specific for Salmonella Enterica Serovar Enteritidis. Appl. Environ. Microbiol. 67, 4984–4991. doi: 10.1128/AEM.67.11.4984-4991.2001
Akiba, M., Kusumoto, M., Iwata, T. (2011). Rapid Identification of Salmonella Enterica Serovars, Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by Multiplex PCR. J. Microbiol. Methods 85, 9–15. doi: 10.1016/j.mimet.2011.02.002
Balasubramanian, R., Im, J., Lee, J. S., Jeon, H. J., Mogeni, O. D., Kim, J. H., et al. (2019). The Global Burden and Epidemiology of Invasive non-Typhoidal Salmonella Infections. Hum. Vaccines Immunotherapeut. 15, 1421–1426. doi: 10.1080/21645515.2018.1504717
Biswas, S., Li, Y., Elbediwi, M., Yue, M. (2019). Emergence and Dissemination of Mcr-Carrying Clinically Relevant Salmonella Typhimurium Monophasic Clone St34. Microorganisms 7, 298. doi: 10.3390/microorganisms7090298
Coburn, B., Grassl, G. A., Finlay, B. B. (2007). Salmonella, the Host and Disease: A Brief Review. Immunol. Cell Biol. 85, 112–118. doi: 10.1038/sj.icb.7100007
Ding, T., Suo, Y., Zhang, Z., Liu, D., Ye, X., Chen, S., et al. (2017). A Multiplex RT-PCR Assay for S. Aureus, L. Monocytogenes, and Salmonella Spp. Detection in Raw Milk With Pre-Enrichment. Front. Microbiol. 8, 989. eCollection 2017. doi: 10.3389/fmicb.2017.00989.eCollection2017
Hirose, K., Itoh, K., Nakajima, H., Kurazono, T., Yamaguchi, M., Moriya, K., et al. (2002). Selective Amplification of Tyv (Rfbe), Prt (Rfbs), Viab, and fliC Genes by Multiplex PCR for Identification of Salmonella Enterica Serovars Typhi and Paratyphi A. J. Clin. Microbiol. 40, 633–636. doi: 10.1128/JCM.40.02.633-636.2002
Hong, Y., Liu, T., Lee, M. D., Hofacre, C. L., Maier, M., White, D. G., et al. (2008). Rapid Screening of Salmonella Enterica Serovars Enteritidis, Hadar, Heidelberg and Typhimurium Using a Serologically-Correlative Allelotyping PCR Targeting the O and H Antigen Alleles. BMC Microbiol. 8:178. doi: 10.1186/1471-2180-8-178
Issenhuth-Jeanjean, S., Roggentin, P., Mikoleit, M., Guibourdenche, M., De Pinna, E., Nair, S., et al. (2014). Supplement 2008-2010 (No. 48) to the White-Kauffmann-Le Minor Scheme. Res. Microbiol. 165, 526–530. doi: 10.1016/j.resmic.2014.07.004
Kang, M. S., Kwon, Y. K., Jung, B. Y., Kim, A., Lee, K. M., An, B. K., et al. (2011). Differential Identification of Salmonella Enterica Subsp. Enterica Serovar Gallinarum Biovars Gallinarum and Pullorum Based on Polymorphic Regions of glgC and speC Genes. Vet. Microbiol. 147, 181–185. doi: 10.1016/j.vetmic.2010.05.039
Kasturi, K. N., Drgon, T. (2017). Real-Time PCR Method for Detection of Salmonella Spp. In Environmental Samples. Appl. Environ. Microbiol. 83, e00644–17. doi: 10.1128/AEM.00644-17
Kawasaki, S., Fratamico, P. M., Horikoshi, N., Okada, Y., Takeshita, K., Sameshima, T., et al. (2010). Multiplex Real-Time Polymerase Chain Reaction Assay for Simultaneous Detection and Quantification of Salmonella Species, Listeria Monocytogenes, and Escherichia Coli O157:H7 in Ground Pork Samples. Foodborne. Pathog. Dis. 7, 549–554. doi: 10.1089/fpd.2009.0465
Keerthirathne, T. P., Ross, K., Fallowfield, H., Whiley, H. (2017). Reducing Risk of Salmonellosis Through Egg Decontamination Processes. Int. J. Environ. Res. Public Health 14, 335. doi: 10.3390/ijerph14030335
Kim, T. H., Hwang, H. J., Kim, J. H. (2017). Development of a Novel, Rapid Multiplex Polymerase Chain Reaction Assay for the Detection and Differentiation of Salmonella Enterica Serovars Enteritidis and Typhimurium Using Ultra-Fast Convection Polymerase Chain Reaction. Foodborne. Pathog. Dis. 14, 580–586. doi: 10.1089/fpd.2017.2290
Kim, H. J., Park, S. H., Kim, H. Y. (2006). Comparison of Salmonella Enterica Serovar Typhimurium LT2 and non-LT2 Salmonella Genomic Sequences, and Genotyping of Salmonellae by Using PCR. Appl. Environ. Microbiol. 72, 6142–6151. doi: 10.1128/AEM.00138-06
Kralik, P., Ricchi, M. (2017). A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 8, 108. doi: 10.3389/fmicb.2017.00108
Lee, S. H., Jung, B. Y., Rayamahji, N., Lee, H. S., Jeon, W. J., Choi, K. S., et al. (2009). A Multiplex Real-Time PCR for Differential Detection and Quantification of Salmonella Spp., Salmonella Enterica Serovar Typhimurium and Enteritidis in Meats. J. Vet. Sci. 10, 43–51. doi: 10.4142/jvs.2009.10.1.43
Liu, Y., Cao, Y., Wang, T., Dong, Q., Li, J., Niu, C. (2019). Detection of 12 Common Food-Borne Bacterial Pathogens by TaqMan Real-Time PCR Using a Single Set of Reaction Conditions. Front. Microbiol. 10, 222. doi: 10.3389/fmicb.2019.00222
Liu, Y., Jiang, J., Ed-Dra, A., Li, X., Peng, X., Xia, L., et al. (2021). Prevalence and Genomic Investigation of Salmonella Isolates Recovered From Animal Food-Chain in Xinjiang, China. Food Res. Int. 142, 110198. doi: 10.1016/j.foodres.2021.110198
Liu, B., Zhang, L., Zhu, X., Shi, C., Chen, J., Liu, W., et al. (2011). PCR Identification of Salmonella Serogroups Based on Specific Targets Obtained by Comparative Genomics. Int. J. Food Microbiol. 144, 511–518. doi: 10.1016/j.ijfoodmicro.2010.11.010
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’brien, S. J., et al. (2010). The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Medalla, F., Gu, W., Mahon, B. E., Judd, M., Folster, J., Griffin, P. M., et al. (2016). Estimated Incidence of Antimicrobial Drug-Resistant Nontyphoidal Salmonella Infections, United States 2004-2012. Emerging. Infect. Dis. 23, 29–37. doi: 10.3201/eid2301.160771
Munoz, N., Diaz-Osorio, M., Moreno, J., Sanchez-Jimenez, M., Cardona-Castro, N. (2010). Development and Evaluation of a Multiplex Real-Time Polymerase Chain Reaction Procedure to Clinically Type Prevalent Salmonella Enterica Serovars. J. Mol. Diagn. 12, 220–225. doi: 10.2353/jmoldx.2010.090036
Nair, S., Patel, V., Hickey, T., Maguire, C., Greig, D. R., Lee, W., et al. (2019). Real-Time PCR Assay for Differentiation of Typhoidal and Nontyphoidal Salmonella. J. Clin. Microbiol. 57, e00167–19. doi: 10.1128/JCM.00167-19
Prendergast, D. M., Hand, D., Niota Ghallchoir, E., Mccabe, E., Fanning, S., Griffin, M., et al. (2013). A Multiplex Real-Time PCR Assay for the Identification and Differentiation of Salmonella Enterica Serovar Typhimurium and Monophasic Serovar 4,[5],12:I. Int. J. Food Microbiol. 166, 48–53. doi: 10.1016/j.ijfoodmicro.2013.05.031
Schrader, K. N., Fernandez-Castro, A., Cheung, W. K., Crandall, C. M., Abbott, S. L. (2008). Evaluation of Commercial Antisera for Salmonella Serotyping. J. Clin. Microbiol. 46, 685–688. doi: 10.1128/JCM.01808-07
Shah, D. H., Park, J. H., Cho, M. R., Kim, M. C., Chae, J. S. (2005). Allele-Specific PCR Method Based on rfbS Sequence for Distinguishing Salmonella Gallinarum From Salmonella Pullorum: Serotype-Specific rfbS Sequence Polymorphism. J. Microbiol. Methods 60, 169–177. doi: 10.1016/j.mimet.2004.09.005
Shi, C., Singh, P., Ranieri, M. L., Wiedmann, M., Moreno Switt, ,. A. I. (2015). Molecular Methods for Serovar Determination of Salmonella. Crit. Rev. Microbiol. 41, 309–325. doi: 10.3109/1040841X.2013.837862
Wang, J., Li, J., Liu, F., Cheng, Y., Su, J. (2020). Characterization of Salmonella Enterica Isolates From Diseased Poultry in Northern China Between 2014 and 2018. Pathogens 9, 95. doi: 10.3390/pathogens9020095
Xu, L., Liu, Z., Li, Y., Yin, C., Hu, Y., Xie, X., et al. (2018). A Rapid Method to Identify Salmonella Enterica Serovar Gallinarum Biovar Pullorum Using a Specific Target Gene ipaJ. Avian Pathol. 47, 238–244. doi: 10.1080/03079457.2017.1412084
Xu, Y., Zhou, X., Jiang, Z., Qi, Y., Ed-Dra, A., Yue, M. (2020). Epidemiological Investigation and Antimicrobial Resistance Profiles of Salmonella Isolated From Breeder Chicken Hatcheries in Henan, China. Front. Cell. Infect. Microbiol. 10, 497. doi: 10.3389/fcimb.2020.00497
Yang, Q., Domesle, K. J., Ge, B. (2018). Loop-Mediated Isothermal Amplification for Salmonella Detection in Food and Feed: Current Applications and Future Directions. Foodborne. Pathog. Dis. 15, 309–331. doi: 10.1089/fpd.2018.2445
Yu, X., Zhu, H., Bo, Y., Li, Y., Zhang, Y., Liu, Y., et al. (2021). Prevalence and Antimicrobial Resistance of Salmonella Enterica Subspecies Enterica Serovar Enteritidis Isolated From Broiler Chickens in Shandong Province, China 2013-2018. Poult. Sci. 100, 1016–1023. doi: 10.1016/j.psj.2020.09.079
Zhang, X., Payne, M., Lan, R. (2019). In Silico Identification of Serovar-Specific Genes for Salmonella Serotyping. Front. Microbiol. 10, 835. doi: 10.3389/fmicb.2019.00835
Keywords: multiplex real-time PCR, chicken, detection, differentiation, Salmonella serovars
Citation: Xin S, Zhu H, Tao C, Zhang B, Yao L, Zhang Y, Afayibo DJA, Li T, Tian M, Qi J, Ding C, Yu S and Wang S (2021) Rapid Detection and Differentiating of the Predominant Salmonella Serovars in Chicken Farm by TaqMan Multiplex Real-Time PCR Assay. Front. Cell. Infect. Microbiol. 11:759965. doi: 10.3389/fcimb.2021.759965
Received: 17 August 2021; Accepted: 10 September 2021;
Published: 29 September 2021.
Edited by:
Jianmin Zhang, South China Agricultural University, ChinaReviewed by:
Zhiming Pan, Yangzhou University, ChinaWenliang Li, Jiangsu Academy of Agricultural Sciences (JAAS), China
Zhe Ma, Nanjing Agricultural University, China
Copyright © 2021 Xin, Zhu, Tao, Zhang, Yao, Zhang, Afayibo, Li, Tian, Qi, Ding, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaohui Wang, shwang0827@126.com; Shengqing Yu, yus@shvri.ac.cn
†These authors have contributed equally to this work
 Suhua Xin†
Suhua Xin†  Hong Zhu
Hong Zhu Chenglin Tao
Chenglin Tao Yaodong Zhang
Yaodong Zhang Tao Li
Tao Li Mingxing Tian
Mingxing Tian Jingjing Qi
Jingjing Qi Chan Ding
Chan Ding Shengqing Yu
Shengqing Yu Shaohui Wang
Shaohui Wang