Inflammatory Signatures of Pathogenic and Non-Pathogenic Leptospira Infection in Susceptible C3H-HeJ Mice
- 1Department of Pharmaceutical Sciences, The University of Tennessee Health Science Center, Memphis, TN, United States
- 2Department of Microbiology, Immunology and Biochemistry, The University of Tennessee Health Science Center, Memphis, TN, United States
The exact global impact of leptospirosis is unknown due to inadequate surveillance systems in place in most low-income countries. In this study, we analyzed the differences in mouse inflammatory signatures involved in pathogenic versus non-pathogenic Leptospira recognition at 24h and 72h post infection. Injection of C3H-HeJ mice with non-pathogenic L. biflexa increased circulation of a few chemokines (5/21, 24%) without secretion of cytokines in blood that resulted in engagement of resident macrophages, dendritic cells, neutrophils and NK cells without engagement of T cells. In contrast, pathogenic L. interrogans induced circulation of a much higher panel of chemokines (18/21, 86%) and pro- and anti-inflammatory cytokines (11/19, 58%) in blood with a resulting signaling cascade leading to engagement of macrophages, dendritic cells, monocytes, NK cells and T cells without engagement of neutrophils. Although neutrophils do not appear to be engaged, a considerable number of chemokines that recruit other granulocytes such as eosinophils and basophils were also increased at 72h post infection with L. interrogans. Overall, the data suggest that prevention of dissemination of L. biflexa is associated with an early engagement of the innate immune response characterized by upregulation of a few chemokines that results in an efficacious phagocytic response without an overwhelming increase of pro-inflammatory cytokines. However, when macrophages fail to clear a pathogenic serovar such as L. interrogans, the adaptive response (T cells) is engaged to help out, but the resulting chemo-cytokine storm mediates a robust but non-resolving inflammatory response to pathogenic Leptospira that results in dissemination, kidney colonization, pathology and disease.
Introduction
Leptospirosis is a neglected tropical disease with an incidence of a million annual human cases in temperate countries and more than 10 million during epidemic outbreaks in tropical areas (Costa et al., 2015; Roqueplo et al., 2019). About 10-15% patients develop severe leptospirosis with multi organ failure and high mortality rate (Cagliero et al., 2018). However, the exact global impact is unknown due to inadequate surveillance systems in most low-income countries (Guerra, 2013) and to under-reporting in developed countries (Fontes et al., 2015). Furthermore, the ubiquitous presence of pathogenic Leptospira in the environment and in different host species increases the zoonotic transmission of Leptospira (Cordonin et al., 2020). Thus, the disease has a severe socioeconomic impact due to infection of animals of agricultural interest (Schafbauer et al., 2019).
Leptospira pathogenesis depends on the virulence of the Leptospira serovar and how the immune system engages with the spirochete in the host (Cagliero et al., 2018). Despite extensive research to understand the nuances of host-pathogen interactions, several questions remain unanswered regarding the host recognition process. Prior reports have dissected differences in induction of proinflammatory cytokines and chemokines after infection of hamster and guinea pigs (Vernel-Pauillac and Merien, 2006; Lourdault et al., 2009; Fujita et al., 2015). However, studies on how the cellular arm of the immune system distinguishes between pathogenic and non-pathogenic Leptospira during early infection are very limited in mouse models (da Silva et al., 2009; Domingos et al., 2017). The first line of defense to pathogenic versus non-pathogenic Leptospira should be characterized by production of different signatures of attractant chemokines and cytokines that recruit specialized immune cells and drive inflammation (Santecchia et al., 2020) which ultimately leads to early elimination of the pathogen from the host and further modulate pathogenesis and disease progression.
Mice are reservoir hosts of pathogenic Leptospira. As such, this species is generally associated with resistance to infection (Adler and Faine, 1977). A possible factor underlying susceptibility to infection in humans may be a lack of recognition of leptospiral-LPS by human TLR4, whereas resistance in mice has been linked to production of antibody within 48-72h post infection (Adler and Faine, 1976) and the murine TLR4 ability to engage leptospiral-LPS (Werts et al., 2001; Nahori et al., 2005; Santecchia et al., 2020). However, mouse strains that express an impaired TLR4 in their immune cells (C3H-HeJ) are susceptible to lethal and sublethal leptospirosis (Pereira et al., 1998; Nally et al., 2005; Viriyakosol et al., 2006; Richer et al., 2015) and those that have been engineered to not express TLR4 (C57BL/6J-TLR4ko) succumb to infection (Chassin et al., 2009). We found recently that rather than leptospiral-LPS sensing, the presence of a fully functional TLR4 receptor in mice is necessary to control acute leptospirosis (Nair et al., 2021). In this study, we used the C3H-HeJ mouse model, which recapitulates susceptibility to leptospirosis, to analyze differences in cell-mediated immune markers engaged by pathogenic disseminators as well as those engaged by saprophytic non-disseminators during the earliest phase of infection with Leptospira, at 24h and 72h.
Materials and Methods
Animals
Male, 10-weeks old, C3H-HeJ mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Male mice were chosen as it is reasonable to expect they should be more susceptible to Leptospira infection as we observed previously for male hamsters (Gomes et al., 2018). Animals were maintained and used in a pathogen-free environment in compliance with the University of Tennessee Health Science Center Institutional Animal Care and Use Committee Protocol no. 19-0062.
Bacterial Strains and Infection
Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130 (LIC), culture passage 2 after hamster infection, and cultured Leptospira biflexa serovar Patoc were used to infect mice. Pathogenic L. interrogans was originally isolated from a patient in Brazil. Non-pathogenic L. biflexa serovar Patoc was obtained from ATCC. Leptospira was cultured as previously described (Sullivan et al., 2017) and enumerated by dark-field microscopy (Zeiss USA, Hawthorne, NY) using a Petroff- Hausser counting chamber. About 108 spirochetes were used to do intraperitoneal infections.
q-PCR and RT-PCR
Quantification of Leptospira was performed using TAMRA probe and primers (Eurofins) to leptospiral 16s rRNA (L. interrogans) and 23s rRNA (L. biflexa) by qPCR. Primers and probes are described in Table S1. Isolation of DNA from blood was carried out using a NucleoSpin tissue kit (Clontech, Mountain View, CA) according to the manufacturer’s instructions and qPCR was run against a standard curve of 106 to 1 L. interrogans. Extraction of total cellular mRNA from whole blood was done using Nucleospin RNA blood kit (Macherey Nagel) respectively. RNA purity was measured at wavelength A260/280 ratio using a Nanodrop instrument (Thermo Scientific). A high-capacity cDNA reverse transcription kit (Applied Biosystems) was used for cDNA preparation. TAMRA specific probes and primers for a panel of chemokines and cytokines as well as β-actin (Eurofins Genomics) were used (Table S1). Data were analyzed using the comparative CT method. Ct values over 38 were considered undetermined.
Flow Cytometry
Single cell suspension from spleen were prepared after RBC lysis using a previously described protocol with minor modifications (Richer et al., 2015). Dead cells were eliminated using Live/dead cell stain during cell counting using a Luna cell counter (Logos Biosystems, South Korea). Approximately 106 cells were seeded per well in a 96 well microtiter plate and blocked with anti-mouse CD16/32 antibody (1:50) for 15-20 min on ice in staining buffer. Surface staining was performed using appropriate primary conjugated antibody against different cell surface markers and incubated in the dark for 30 min at 4°C. Cells were washed twice with phosphate buffered saline (PBS, 1X; pH 7.4) and fixed with 4% Paraformaldehyde for 10 mins followed by a single PBS wash. Cells were resuspended in staining buffer, acquired on BioRad ZE5 Cell analyzer and data were analyzed using FlowJo software. The panel of fluorochrome conjugated antibodies (Table S2) and gating strategy (Figure S1) are described in Supplementary Material.
Proteome Profile Array
Proteome Profiler Mouse Cytokine Array kit (Panel A, R&D Systems Inc., Minneapolis, MN, USA) was used for the detection of cytokines and chemokines (Table S3) in serum (Chondrou et al., 2020), following the manufacturer’s instructions. Nitrocellulose membrane contains spotted capture antibodies in duplicate. In brief, serum samples were mixed with a cocktail of 40 different biotinylated detection antibodies and then incubated with the Mouse Cytokine Array membrane. A complex is formed between the above mixture and the immobilized capture antibody in the membrane. A washing step is performed to remove unbound conjugate. Lastly, chemiluminescent and Streptavidin-HRP were used for detection of these conjugates, which is proportional to the bound cytokine quantity. Images were acquired using a Chemi-Doc image analyzer; the mean pixel density was measured using imageJ software and plotted using GraphPad Prism software. The mean pixel density values were normalized by subtracting the background of 3 membranes for each exposure (total of 3 exposures). Specifics on the mouse proteome profile array are presented in Supplementary Material, Figures S2 and S3.
Statistical Analysis
Statistical analysis was performed using unpaired t-test with Welch’s correction to analyze differences between non-infected and infected groups, and between infected L. interrogans and L. biflexa groups, α = 0.05.
Results
Infection With L. interrogans Leads to Spirochetemia in Contrast to Infection With L. biflexa
We inoculated mice with 108 spirochetes and measured weight-loss as well as Leptospira load in blood by qPCR for 4 days. As expected, within the first 4 days of infection there is no significant change in body weight between the groups (Figure 1A). In addition, dissemination of pathogenic L. interrogans in blood was detected from day 1 to day 4 post infection (Figure 1B i) whereas no dissemination occurred for non-pathogenic L. biflexa (Figure 1B ii) infected mice.
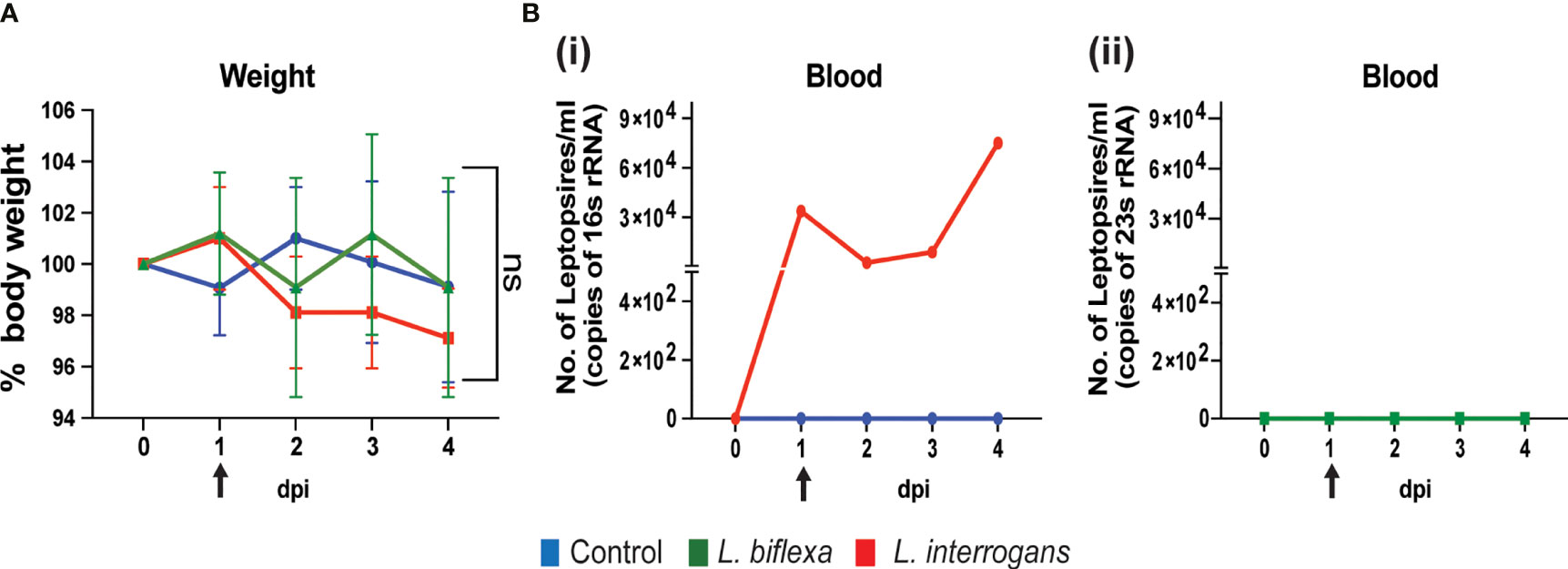
Figure 1 Body weight and dissemination of Leptospira in blood of infected mice. Male C3H-HeJ mice were infected with 108 L. interrogans serovar Copenhageni strain Fiocruz L1-130 and L. biflexa serovar Patoc whereas control mice received PBS. Panel (A) shows 4-day body weight measurements of mice injected with L. interrogans (red), L. biflexa (green), and PBS control (blue). Panel (B) depicts the total Leptospira burden in blood of L. interrogans (i) and L. biflexa (ii) infected mice over 4 days post infection by qPCR. n=4 mice per group. Arrow indicates the day of infection. Statistical analysis was performed by unpaired t-test with Welch’s correction between control and infected groups. ns, non-significance.
L. interrogans Infection Led to Increased Circulation of Chemokines and Cytokines in Serum Mostly at 72h, Whereas L. biflexa Infection Led to Increased Circulation of a Few Chemokines Mostly at 24h
We quantified circulating immune mediators in serum using a mouse Proteome Profile Array (Figures 2, 3 and Tables 1, 2, 4; Table S3) targeting the following 40 proteins: KC/CXCL1, MIP-2/CXCL2, MIG/CXCL9, IP-10/CXCL10, I-TAC/CXCL11, SDF-1/CXCL12, BLC/CXCL13, I-309/CCL1, JE/MCP-1/CCL2, MIP-1α/CCL3, MIP-1β/CCL4, RANTES/CCL5, Eotaxin/CCL11, MCP-5/CCL12, TARC/CCL17, sICAM-1, TIMP-1, TREM-1, G-CSF, M-CSF, GM-CSF, C5/C5a, IFN-γ, TNF-α, IL-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12p70, IL-13, IL-16, IL-17a, IL-23, IL-27. At 24h post infection, we found that L. biflexa increased secretion of 4/21 (19%) chemokines in serum as compared to control, whereas 17/21 (81%) were decreased; in addition, no cytokines were increased, whereas 16/18 (89%) were decreased. At 24h, L. interrogans infection increased secretion of 7/21 (33%) chemokines in serum, whereas 11/21 (52%) were decreased; furthermore, 2/18 (11%) cytokines were increased whereas 10/18 (56%) were decreased. Specifically, L. interrogans induced MIG, I-TAC, RANTES and the cytokines IL-1ra and IL-16 in addition to the 4 chemokines also induced by L. biflexa: IP-10, BLC, JE/MCP-1 and TIMP-1 (Figure 2A, Figure 3A; Tables 1, 2 and Summary in Table 4). At 72h post infection with L. biflexa, no chemo-cytokine activity was increased in serum as compared to control; furthermore, 12/21 (57%) chemokines as well as 12/18 (67%) cytokines were decreased. At 72h, L. interrogans infection increased 14/21 (67%) pro-inflammatory chemokines (KC, IP-10, BLC, I-309 JE/MCP-1, MIP-1β, Eotaxin, MCP-5, TARC, TIMP-1, TREM-1, G-CSF, M-CSF and GM-CSF) and 9/18 (50%) cytokines in serum as compared to control (TNF-α, IL-3, IL-4, IL-6, IL-10, IL-12p70, IL-13, IL-16 and IL-23); in addition, 3/21 (14%) chemokines (RANTES, SDF-1 and ICAM-1) and 2/18 (11%) cytokines (IFN-γ and IL-27) were decreased (Figure 2B, Figure 3B; Tables 1, 2 and Summary in Table 4). Both L. interrogans and L. biflexa induced increased concentration of complement factor C5/C5a in serum in comparison with the control.
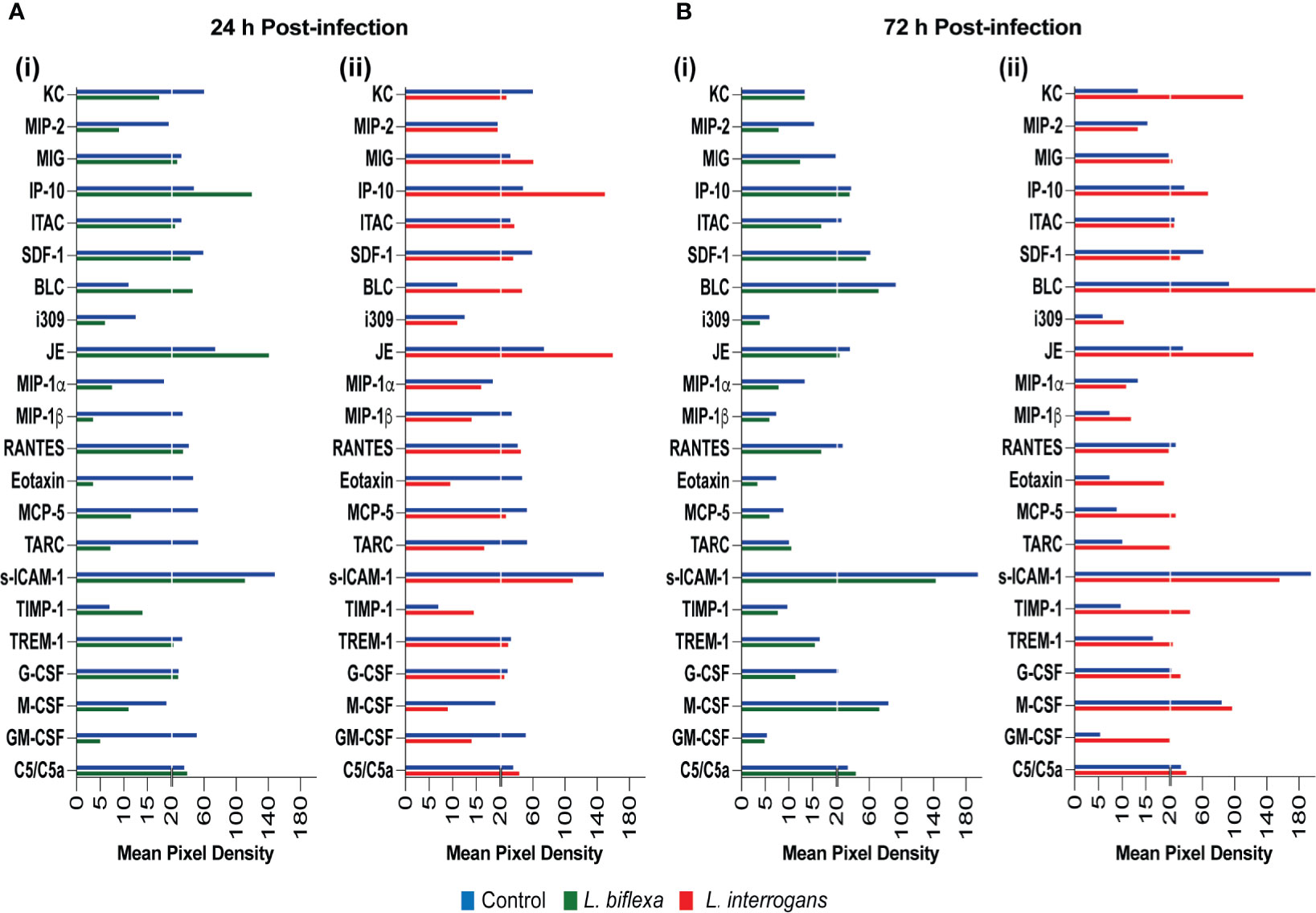
Figure 2 Proteome profiling of chemokines circulating in serum from mice infected with L. interrogans, L. biflexa and PBS controls. Detection of chemokine proteins in pooled serum (n=4 mice), represented as mean pixel density at 24h (A) and 72h (B) post infection. Sub-panel (i) depicts representative graphs of L. biflexa vs control, whereas (ii) depicts L. interrogans vs control. The chemiluminescence was measured and the mean pixel density value was determined using ImageJ software. The values were normalized by subtracting the background of 3 membranes for each exposure for a total of 3 exposures. Data represents the average pixel density between the two spots for each immune marker after subtraction of the normalized background.
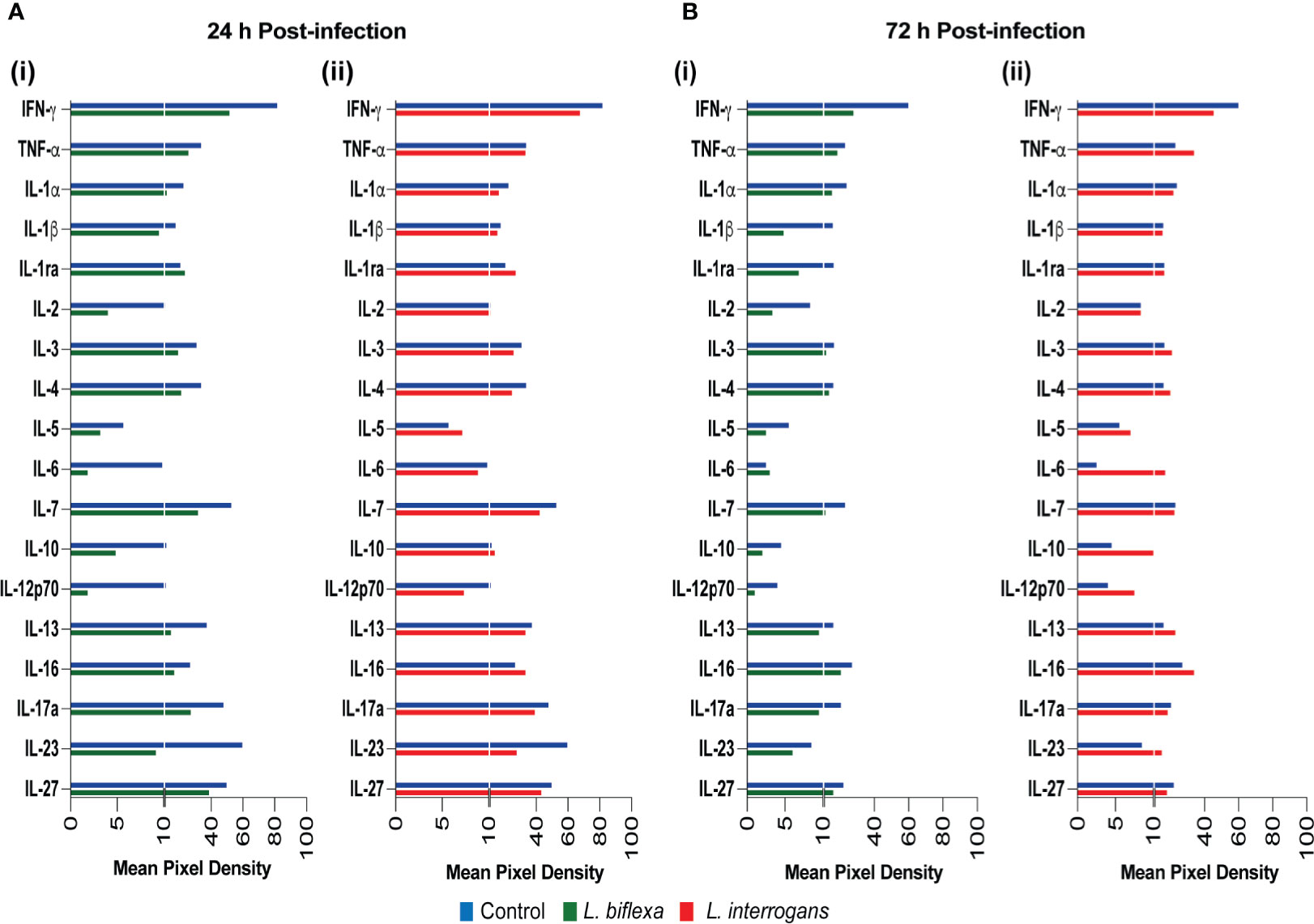
Figure 3 Proteome profiling of cytokines circulating in serum from mice infected with L. interrogans, L. biflexa and PBS controls. Detection of cytokine proteins in pooled serum (n=4 mice), represented as mean pixel density at 24h (A) and 72h (B) post infection. Sub-panel (i) depicts representative graphs of L. biflexa vs control, whereas (ii) depicts L. interrogans vs control. The chemiluminescence was measured and the mean pixel density value was determined using ImageJ software. The values were normalized by subtracting the background of 3 membranes for each exposure for a total of 3 exposures. Data represents the average pixel density between the two spots for each immune marker after subtraction of the normalized background.
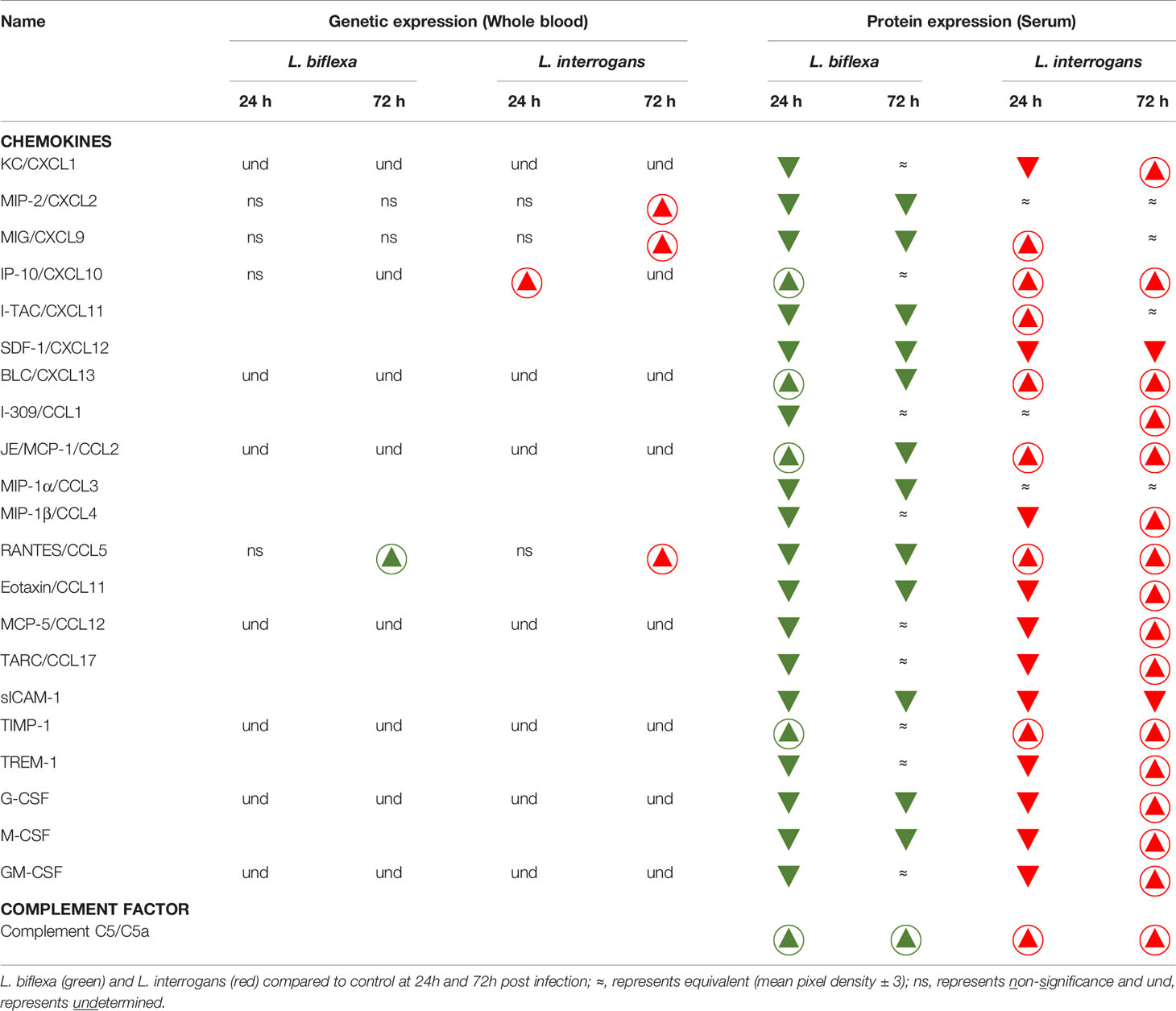
Table 1 Chemokines and complement factor affected during early infection with pathogenic and non-pathogenic Leptospira in comparison with control.
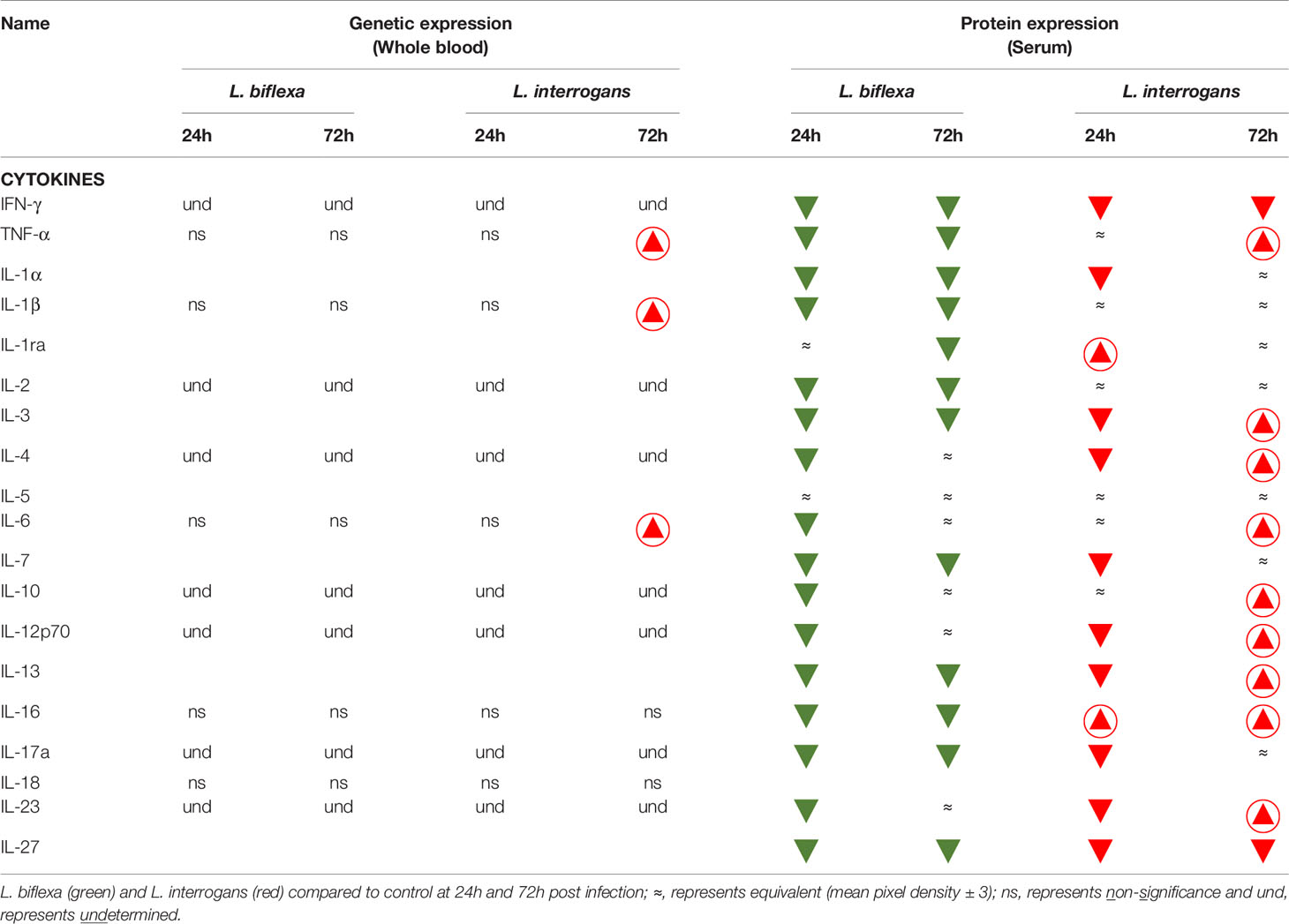
Table 2 Cytokines affected during early infection with pathogenic and non-pathogenic Leptospira in comparison with control.
L. interrogans Infection Increased Levels of Chemokine and Cytokine mRNA Mediators in Whole Blood at 72h Post Infection, Whereas L. biflexa Remained Mostly Unchanged
We quantified expression of mRNA of 23 transcripts KC/CXCL1, MIP-2/CXCL2, MIG/CXCL9, IP10/CXCL10, BLC/CXCL13, JE/MCP-1/CCL2, RANTES/CCL5, MCP-5/CCL12, TIMP-1, G-CSF, GM-CSF, IFN-γ, TNF-α, IL-1β, IL-2, IL-4, IL-6, IL-10, IL-12, IL-16, IL-17a, IL-18, IL-23 in cells from whole blood (Figure 4; Tables 1, 2, 4 and Table S3). Only immune markers with detectable levels of mRNA at 24h and 72h post infection were plotted in Figure 4. Significant differences between genetic expression of immune mediators were detected at 72h but not at 24h post infection with L. interrogans, except for an increase of IP-10 between L. interrogans and control (Table 1) at 24h, and a decrease in IL-1β between L. biflexa and L. interrogans infected mice (Figure 4E) at 24h. At 72h post-infection, RANTES was the only chemokine increased between L. biflexa and control groups (Figure 4A and Table 1). Between L. interrogans and control groups, the chemokines RANTES, MIP-2, and MIG, and the cytokines TNF-α, IL-1β, and IL-6 were increased (Figures 4A–F and Tables 1 and 2). IL-18 was increased in L. interrogans compared to L. biflexa but it was not different from the control (Figure 4G). The other 15 immune markers tested in mRNA purified from whole blood (KC, BLC, JE/MCP-1, MCP-5, TIMP-1, G-CSF, GM-CSF, IFN-γ, IL-2, IL-4, IL-10, IL-12, IL-16, IL-17a, IL- 23) were either not significant or not detected.
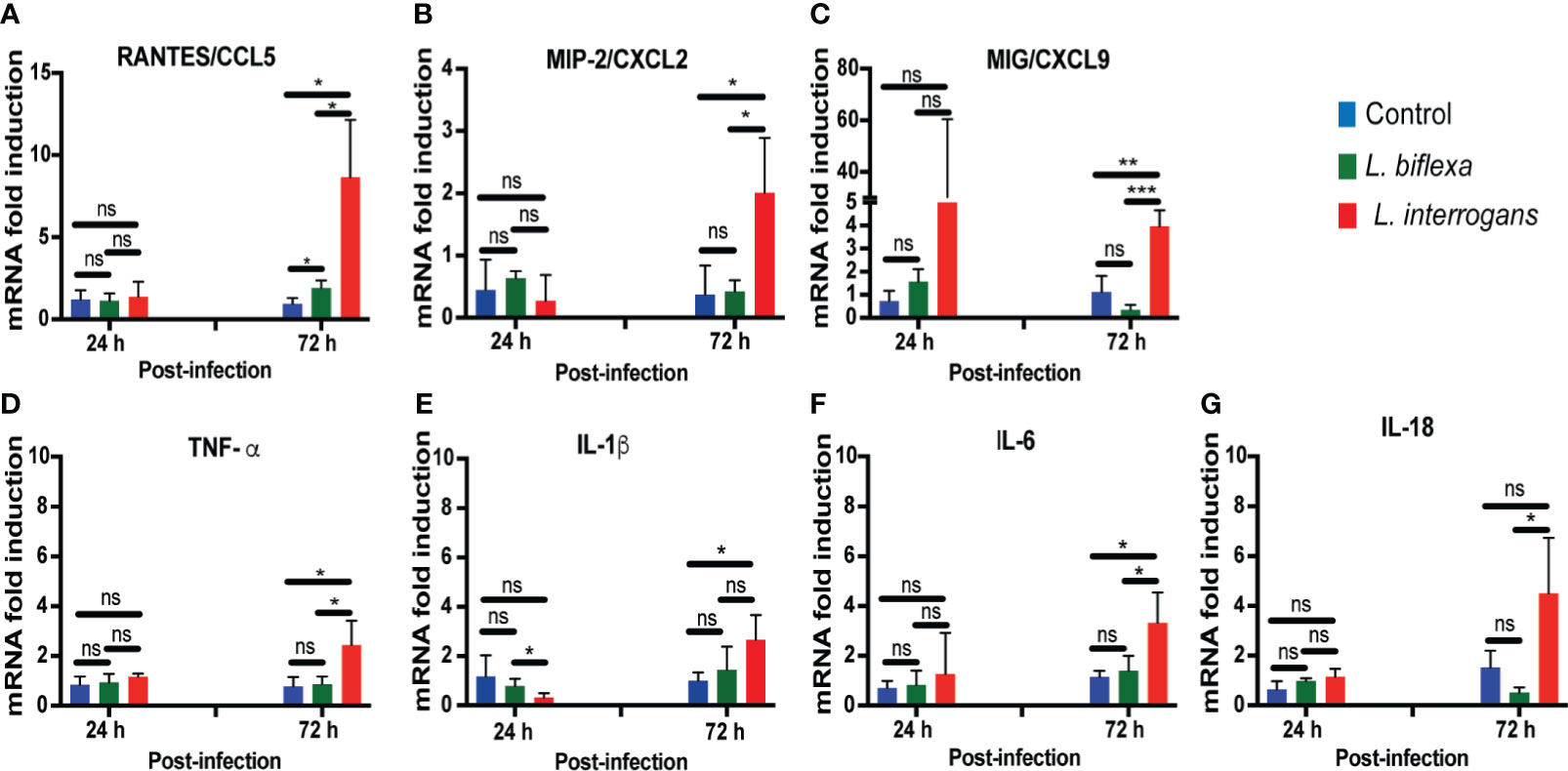
Figure 4 Genetic expression levels of chemo-cytokines in whole blood at 24h and 72h post infection with L. biflexa and L. interrogans. Infection was performed with pathogenic and non-pathogenic Leptospira in C3H-HeJ mice via intraperitoneal route with 108 bacteria for 24h and 72h. PBS was administered in control mice. We measured genetic expression of inflammatory signatures by RT-PCR (Panels A–G) from whole blood. Statistical significance was determined by unpaired t-test with Welch’s correction between control and infected groups, where *p<0.05, **p<0.01, ***p<0.001, ns, non-significance; n= 3-4 mice per group. Data represents one of two independent experiments.
L. interrogans Infection Led to Engagement of All Myeloid Cell Populations in Spleen Except Neutrophils, Whereas L. biflexa Infection Led to Engagement of Resident Macrophages, Dendritic Cells, NK Cells and Neutrophils
Mice infected with L. interrogans had enlarged spleens both at 24h and 72h post infection as compared to controls, whereas differences in spleens harvested from L. biflexa infected mice were less apparently enlarged as observed visually (Figure 5A), and by weight measurement (Figure 5B). We phenotyped the immune cells contributing with the elevated expression of inflammatory signatures during the earliest phase of infection by flow cytometry (Figures 5C–K and Table 3). We found that at 24h post infection, myeloid cells (Figure 5C), monocyte-macrophages (Figure 5G), resident macrophages (Figure 5H) and dendritic cells (Figure 5I) were significantly increased in L. interrogans infected groups compared to controls; at 72h post infection, myeloid cells (Figure 5C), monocytes (Figure 5D), NK cells (Figure 5E) were significantly increased and, dendritic cells (Figure 5I) and T cells (Figure 5J) were significantly decreased whereas neutrophils (Figure 5F) and macrophages (Figure 5G, H) were not engaged in L. interrogans infected groups compared to controls. Between L. biflexa and control groups, at 24h post-infection, resident-macrophages (Figure 5H) and dendritic cells (Figure 5I) were increased, and at 72h post infection, myeloid cells (Figure 5C) and NK cells (Figure 5E) where increased, whereas neutrophils (Figure 5F) were decreased. A comparison between L. interrogans and L. biflexa at 72h post infection shows that myeloid cells (Figure 5C), monocytes (Figure 5D), NK cells (Figure 5E) and neutrophils (Figure 5F) were significantly increased in L. interrogans, whereas dendritic cells (Figure 5I) and T cells (Figure 5J) are significantly decreased. No differences in B cells were observed between L. interrogans, L. biflexa and controls at any time points (Figure 5K).
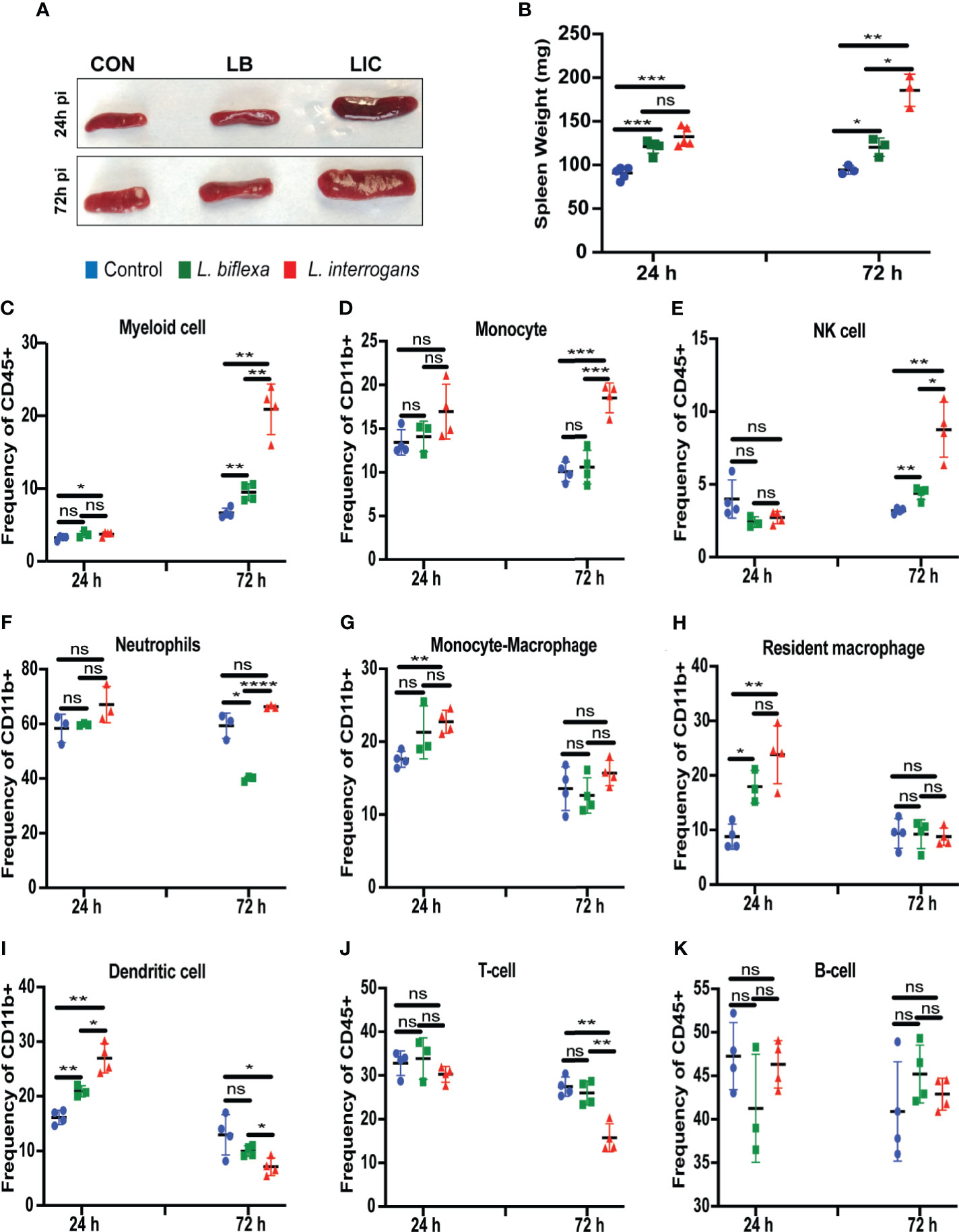
Figure 5 Spleen morphometry and flow cytometric analysis of immune cells after infection. (A) depicts clear L. interrogans induced splenomegaly, while L. biflexa spleen appears to retain normal splenic architecture as do controls at 24h and 72h pi. (B) is the representation of respective splenic weight in milligrams, differences in weight were measured both for L. biflexa and L. interrogans. Statistical analysis between groups was done by unpaired t-test with Welch’s correction, *p<0.05, **p<0.01, ***p<0.001, n= 3-5 mice per group. (C–K) represent different immune cell population in spleen at 24h and 72h pi. Myeloid cells like monocytes (D), neutrophils (F), monocyte-macrophage (G), resident macrophage (H), and dendritic cells (I) were determined based on CD11b+ frequency. Myeloid cells (C), NK-cell (E), T-cell (J) and B-cell (K) were measured in terms of CD45+ frequency. Statistical analysis was done by unpaired t-test with Welch’s correction between control and infected groups, where *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns, non-significance; n=4 mice per group. Data represents one of three independent experiments.
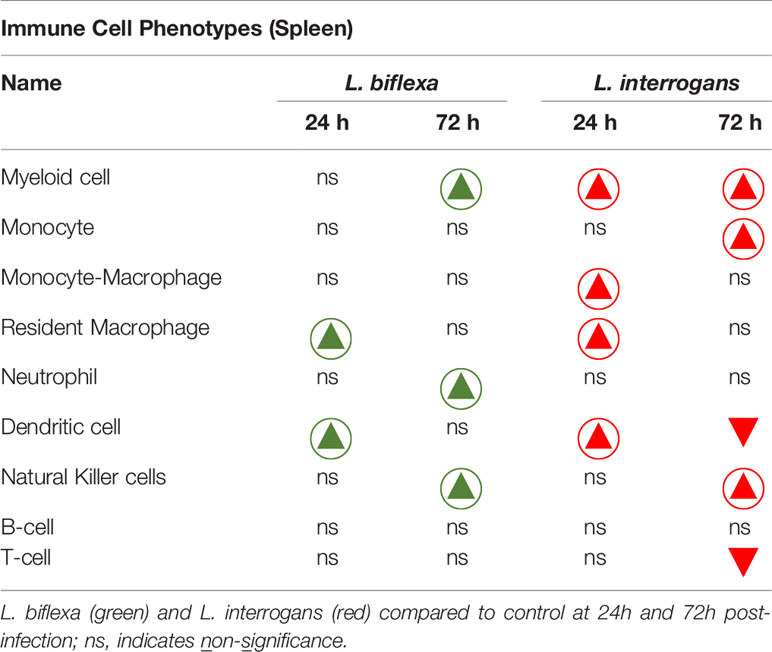
Table 3 Immune cells engaged during infection with pathogenic and non-pathogenic Leptospira at 24h and 72h post-infection in comparison with control.
A summary of all the factors measured such as protein circulating in serum, cellular mRNA from whole blood and immune cells homing to the secondary lymphoid organ - spleen - that orchestrate the inflammatory immune response to pathogenic L. interrogans and saprophytic L. biflexa is presented in Table 1 (21 chemokines and Complement component 5 - C5/C5a), Table 2 (19 cytokines), Table 3 (9 immune cell types), Table 4 (overall summary) and Table S3 (immune markers and cell function). Over the two time points, L. biflexa infection increased secretion of Complement component 5 (C5/C5a), 5/21 (24%) chemokines and 0/19 (0%) cytokines in serum and blood cells, and engaged 5/9 (56%) immune cell types in spleen. In contrast, L. interrogans increased secretion of C5/C5a, 18/21 (86%) chemokines and 11/19 (58%) inflammatory cytokines in serum and blood cells, and engaged 6/9 (67%) immune cell types in spleen.
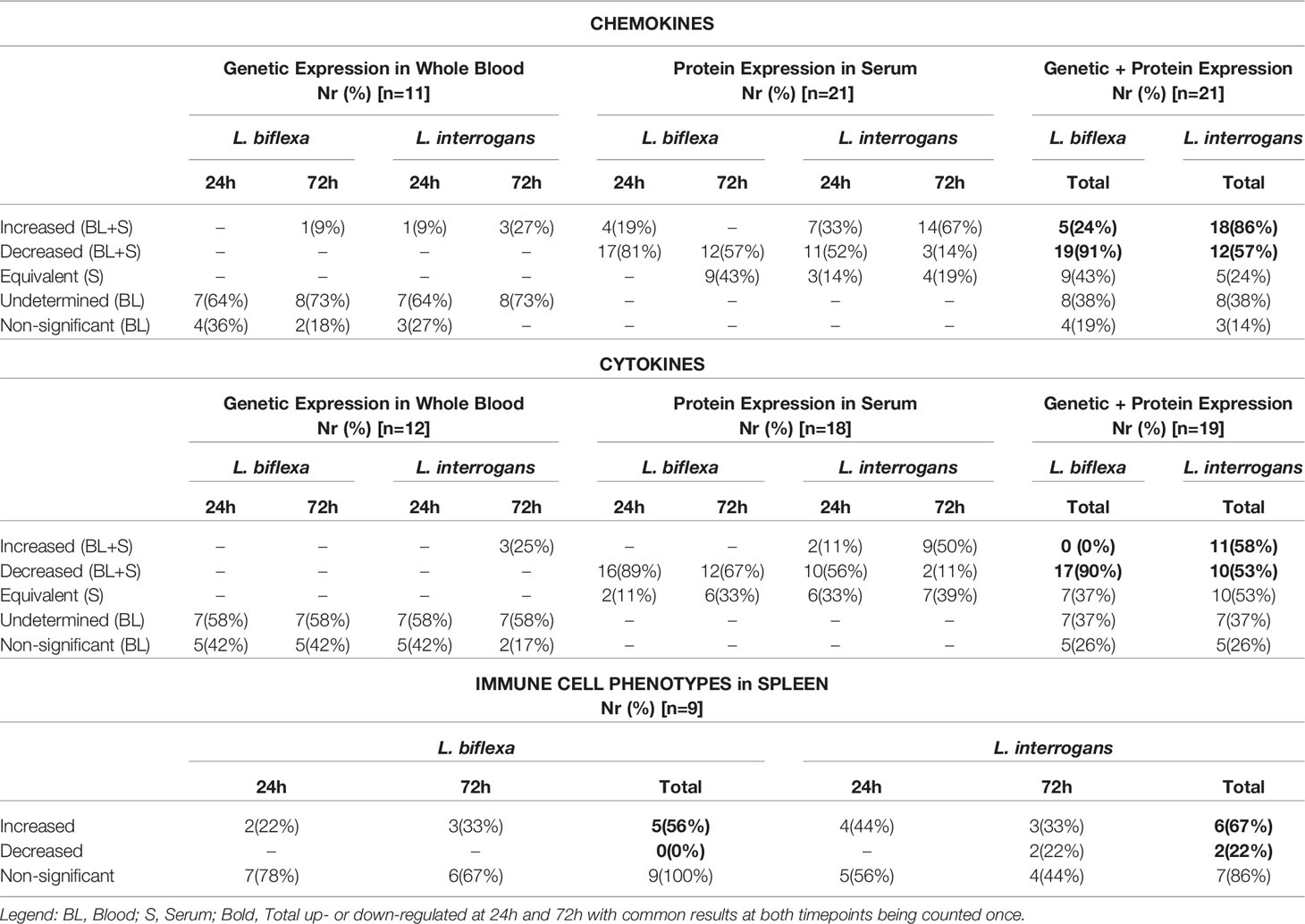
Table 4 Summary of immune markers and cells involved in responses to L. biflexa and L. interrogans infection.
Discussion
The engagement of the host immune system with pathogenic and non-pathogenic serovars of Leptospira at the point of entry defines the outcome of infection. In this study, we quantified immune cell mediators circulating in serum and genetic expression of chemo/cytokines in whole blood of C3H-HeJ mice infected with pathogenic and saprophytic Leptospira (L. interrogans and L. biflexa) and correlated these circulating signatures with the immune cell populations enriched in the secondary lymphoid organ (spleen) during the earliest phase of infection. We analyzed two timepoints (24h and 72h) within the first week of infection when Leptospira interrogans is known to circulate in blood and there are no weight differences - a clinical score of disease progression - in infected C3H-HeJ mice [Figure 1 (Richer et al., 2015; Nair et al., 2020)].
We found that infection with L. biflexa was generally characterized by induction of chemoattractant chemokines (IP-10, JE/MCP-1) that engage resident macrophages and dendritic cells in spleen at 24h post infection which was replaced by an increase of Natural Killer (NK) cells and a decrease of neutrophils at 72h post infection (Figures 2 and 5). Although RANTES and BLC chemoattractants were increased there was no evidence of engagement of T cells and B cells, respectively. The marked decrease in neutrophils frequency in spleen from mice infected with L. biflexa could be explained by its rapid recruitment to the spirochete point of entry as it is known that neutrophils phagocytize and destroy saprophytic Leptospira (Isogai et al., 1989; Scharrig et al., 2015; Charo et al., 2019). An efficacious phagocytic response engaged by resident macrophages (Toma et al., 2011), and neutrophils (Scharrig et al., 2015) without an overwhelming increase of pro-inflammatory cytokines contributes to elimination of the spirochete at the point of entry preventing Leptospira dissemination and absence of pathology and disease. However, it is not clear which immune mediators recruited neutrophils given that the neutrophil chemoattractants tested (KC, MIP-2 and IL-1α) were downregulated at 24h and 72h post infection with L. biflexa. Regarding the other two cell types (NK cells and dendritic cells) involved in clearance of L. biflexa, both are induced by JE/MCP-1 which was increased in serum at 24h post-infection. The protective effects of NK cells from vaccinated bovines (Zuerner et al., 2011) and from serum-thymic factor treated gerbils (Yukawa et al., 1994) have been described. Thus, we speculate that NK cells may directly eliminate cells that have phagocytized L. biflexa. Both pathogenic and non-pathogenic Leptospira induced maturation of human dendritic cells and production of pro-inflammatory cytokines (Gaudart et al., 2008; Santecchia et al., 2020). In another study, liposomes prepared from L. biflexa lipids activated bone marrow derived dendritic cells that released pro-inflammatory cytokines and induced strong humoral and cytotoxic T cell responses (Faisal et al., 2011). In our study, we did not detect any pro-inflammatory cytokines in serum or blood cells of C3H-HeJ mice infected with L. biflexa. Thus, the role played by these professional antigen presenting cells is unclear, but it may be related to the recruitment of cytotoxic T cells.
In contrast to L. biflexa, infection with L. interrogans was characterized by a substantial induction of attractant chemokines whose function is to recruit myeloid phagocytic cells and pro- and anti-inflammatory cytokines whose main function is to engage lymphoid cells (Figures 2–5). The frequency of macrophages and dendritic cells was likely driven by MIG, IP-10 and JE/MCP-1 chemokines increased in blood at 24h post-infection. These cells were replaced by a subsequent increase in monocytes and Natural Killer cells and a decrease of dendritic cells driven by IP-10, I-309, JE/MCP-1, MIP-1β and MCP-5 at 72h post-infection. T cell recruitment may have been driven by IP-10, I-309, RANTES, MCP-5 and TARC at 72h post infection. As observed for L. biflexa, BLC which is involved in B cell recruitment was increased in serum of L. interrogans infected mice but differences in B cell frequency in spleen were not significant at this timepoint. This suggests engagement of the adaptive arm of the immune system as early as 24h-72h post infection with L. interrogans as described by others (Adler and Faine, 1976). Chemokine proteome arrays in L. interrogans infected mice have been done by other groups (Domingos et al., 2017; Silva et al., 2020). Differences observed in the panel of chemokines detected can be explained by differences in methodology in that we quantified circulating chemokines (serum and blood cells) and they quantified chemokines in spleen/lung (Domingos et al., 2017) and in liver/kidney (Silva et al., 2020). Nevertheless, MIG/CXCL9, IP-10/CXCL10, I-TAC/CXCL11, BLC/CXCL13 and JE/MCP-5/CCL2 detected in kidney of C3H-HeJ mice at 24h post infection (Silva et al., 2020) was also detected in serum/blood cells in our study.
Neutrophil chemoattractants, KC and MIP-2, were increased in the serum proteome and in whole blood gene transcripts of L. interrogans infected mice. However, neutrophils were not increased or decreased in spleen. Earlier studies have shown that neutrophils do not phagocytize pathogenic Leptospira (Isogai et al., 1989; Scharrig et al., 2015; Charo et al., 2019; Santecchia et al., 2020). Others have shown that macrophages, but not neutrophils, infiltrate organs of C3H-HeJ mice infected with L. interrogans (Chen et al., 2017). This may explain the lack of engagement of neutrophils in L. interrogans infected C3H-HeJ mice in our study.
Discrepant results between serum protein and genetic expression in blood may reflect the limited number of cell types circulating in blood at the time of blood collection, whereas serum receives proteins from all immune cell types from different organs and tissues. This may explain why we detected a more comprehensive panel of immune markers in serum than in transcripts from whole blood. Nevertheless, it is interesting to note that the immune markers detected by genetic expression represent the overall engagement of all arms of the immune cell response: MIP-2 (neutrophils), MIG and TNF-α (macrophages/NK cells), IP-10 (monocytes, macrophages, dendritic cells, NK cells and T cells), RANTES (T cells, eosinophils, basophils), IL-1β (immune cell proliferation, differentiation and apoptosis) and IL-6 (pro-inflammatory and regulatory-T cell suppression).
As expected, and in contrast to L. biflexa infection, L. interrogans induced a strong adaptive immune response with production of proinflammatory cytokines TNF-α (innate immune alarm), IL-1ra (inhibitor of IL-1), IL-3, IL-4 (Th2), IL-6 (pro- and anti-inflammatory, Treg suppression), IL-10 (anti-inflammatory), IL-12p70 (NK and Th1 cell function), IL-13 (Th2, IgE antibody), IL-16 (activated T cells, dendritic cells and eosinophils), and IL-23 (Th17 expansion, induces IL-6). Furthermore, a considerable number of chemokines that recruit eosinophils and basophils (RANTES, Eotaxin, MCP-5, G-CSF, GM-CSF) were also increased at 72h post infection with L. interrogans. Although neutrophils do not appear to be engaged by L. interrogans, these other granulocytes may contribute to inflammation.
Conclusion
The stark differences in expression of chemoattractant chemokines and pro- and anti-inflammatory cytokines can be correlated with pathogenesis. These data suggest that prevention of dissemination of L. biflexa is associated with an early engagement of the innate immune system characterized by secretion of a few chemoattractants that results in an efficacious phagocytic response by resident macrophages and neutrophils, which are helped by dendritic and Natural Killer cells without an overwhelming increase of pro-inflammatory cytokines. However, when macrophages fail to clear a pathogenic serovar such as L. interrogans, monocytes, NK cells, T cells and possibly other granulocytes (eosinophils and basophils) may be recruited to help out. The resulting chemo-cytokine storm mediates a robust but non-protective inflammatory response to pathogenic Leptospira (Cagliero et al., 2018) which results in dissemination, kidney colonization, pathology and disease.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by University of Tennessee Health Science Center Institutional Animal Care and Use Committee.
Author Contributions
AS and SK contributed equally. MGS is the corresponding author who conceptualized the project, procured funding, designed experiments, data analysis, prepared figures and wrote the manuscript. AS and SK designed and performed the experiments, assisted in writing the manuscript, data analysis and figure preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Public Health Service grants R43AI136551, R21AI142129 and R43AI155211 to MGS from the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Diedre Daria and Dr. Tony Marion of the Flow Cytometry and Cell Sorting Core facility at UTHSC. We also would like to acknowledge Dr. Ae-Kyung Yi’s lab for her support with the Chemi-Doc system for imaging the proteome profile array.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.677999/full#supplementary-material
References
Adler, B., Faine, S. (1976). Susceptibility of Mice Treated With Cyclophosphamide to Lethal Infection With Leptospira Interrogans Serovar Pomona. Infect. Immun. 14 (3), 703–708. doi: 10.1128/IAI.14.3.703-708.1976
Adler, B., Faine, S. (1977). Host Immunological Mechanisms in the Resistance of Mice to Leptospiral Infections. Infect. Immun. 17 (1), 67–72. doi: 10.1128/iai.17.1.67-72.1977
Cagliero, J., Villanueva, S., Matsui, M. (2018). Leptospirosis Pathophysiology: Into the Storm of Cytokines. Front. Cell Infect. Microbiol. 8, 204. doi: 10.3389/fcimb.2018.00204
Charo, N., Scharrig, E., Ferrer, M. F., Sanjuan, N., Carrera Silva, E. A., Schattner, M., et al. (2019). Leptospira Species Promote a Pro-Inflammatory Phenotype in Human Neutrophils. Cell Microbiol. 21 (2), e12990. doi: 10.1111/cmi.12990
Chassin, C., Picardeau, M., Goujon, J. M., Bourhy, P., Quellard, N., Darche, S., et al. (2009). TLR4- and TLR2-Mediated B Cell Responses Control the Clearance of the Bacterial Pathogen, Leptospira Interrogans. J. Immunol. 183 (4), 2669–2677. doi: 10.4049/jimmunol.0900506
Chen, X., Li, S. J., Ojcius, D. M., Sun, A. H., Hu, W. L., Lin, X., et al. (2017). Mononuclear-Macrophages But Not Neutrophils Act as Major Infiltrating Anti-Leptospiral Phagocytes During Leptospirosis. PLoS One 12 (7), e0181014. doi: 10.1371/journal.pone.0181014
Chondrou, P., Karapetsas, A., Kiousi, D. E., Vasileiadis, S., Ypsilantis, P., Botaitis, S., et al. (2020). Assessment of the Immunomodulatory Properties of the Probiotic Strain Lactobacillus Paracasei K5 In Vitro and In Vivo. Microorganisms 8 (5), p709. doi: 10.3390/microorganisms8050709
Cordonin, C., Turpin, M., Bringart, M., Bascands, J. L., Flores, O., Dellagi, K., et al. (2020). Pathogenic Leptospira and Their Animal Reservoirs: Testing Host Specificity Through Experimental Infection. Sci. Rep. 10 (1), 1–8. doi: 10.1038/s41598-020-64172-4
Costa, F., Hagan, J. E., Calcagno, J., Kane, M., Torgerson, P., Martinez-Silveira, M. S., et al. (2015). Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 9 (9), e0003898. doi: 10.1371/journal.pntd.0003898
da Silva, J. B., Ramos, T. M., de Franco, M., Paiva, D., Ho, P. L., Martins, E. A., et al. (2009). Chemokines Expression During Leptospira Interrogans Serovar Copenhageni Infection in Resistant BALB/c and Susceptible C3H/HeJ Mice. Microb. Pathog. 47 (2), 87–93. doi: 10.1016/j.micpath.2009.05.002
Domingos, R. H., Pavanel, E. B., Nakajima, E., Schons-Fonseca, L., Da Costa, R. M. A., De Franco, M., et al. (2017). Resistance of Mice to Leptospira Infection and Correlation With Chemokine Response. Immunobiology 222 (11), 1004–1013. doi: 10.1016/j.imbio.2017.05.017
Faisal, S. M., Chen, J. W., McDonough, S. P., Chang, C. F., Teng, C. H., Chang, Y. F. (2011). Immunostimulatory and Antigen Delivery Properties of Liposomes Made Up of Total Polar Lipids From Non-Pathogenic Bacteria Leads to Efficient Induction of Both Innate and Adaptive Immune Responses. Vaccine 29 (13), 2381–2391. doi: 10.1016/j.vaccine.2011.01.110
Fontes, R. M., Cavalcanti, L. P., Oliveira, A. C., Bezerra, L. F., Gomes, A. M., Colares, J. K., et al. (2015). A New Possibility for Surveillance: Do We Identify All Cases of Leptospirosis? Rev. Inst Med. Trop. Sao Paulo 57 (5), 443–446. doi: 10.1590/S0036-46652015000500014
Fujita, R., Koizumi, N., Sugiyama, H., Tomizawa, R., Sato, R., Ohnishi, M. (2015). Comparison of Bacterial Burden and Cytokine Gene Expression in Golden Hamsters in Early Phase of Infection With Two Different Strains of Leptospira Interrogans. PLoS One 10 (7), e0132694. doi: 10.1371/journal.pone.0132694
Gaudart, N., Ekpo, P., Pattanapanyasat, K., van Kooyk, Y., Engering, A. (2008). Leptospira Interrogans Is Recognized Through DC-SIGN and Induces Maturation and Cytokine Production by Human Dendritic Cells. FEMS Immunol. Med. Microbiol. 53 (3), 359–367. doi: 10.1111/j.1574-695X.2008.00437.x
Gomes, C. K., Guedes, M., Potula, H. H., Dellagostin, O. A., Gomes-Solecki, M. (2018). Sex Matters: Male Hamsters Are More Susceptible to Lethal Infection With Lower Doses of Pathogenic Leptospira Than Female Hamsters. Infect. Immun. 86 (10), e00369–18 . doi: 10.1128/IAI.00369-18
Guerra, M. A. (2013). Leptospirosis: Public Health Perspectives. Biologicals 41 (5), 295–297. doi: 10.1016/j.biologicals.2013.06.010
Isogai, E., Isogai, H., Wakizaka, H., Miura, H., Kurebayashi, Y. (1989). Chemiluminescence and Phagocytic Responses of Rat Polymorphonuclear Neutrophils to Leptospires. Zentralbl Bakteriol. 272 (1), 36–46. doi: 10.1016/S0934-8840(89)80090-8
Lourdault, K., Aviat, F., Picardeau, M. (2009). Use of Quantitative Real-Time PCR for Studying the Dissemination of Leptospira Interrogans in the Guinea Pig Infection Model of Leptospirosis. J. Med. Microbiol. 58 (Pt 5), 648–655. doi: 10.1099/jmm.0.008169-0
Nahori, M. A., Fournie-Amazouz, E., Que-Gewirth, N. S., Balloy, V., Chignard, M., Raetz, C. R., et al. (2005). Differential TLR Recognition of Leptospiral Lipid A and Lipopolysaccharide in Murine and Human Cells. J. Immunol. 175 (9), 6022–6031. doi: 10.4049/jimmunol.175.9.6022
Nair, N., Guedes, M. S., Hajjar, A. M., Werts, C., Gomes-Solecki, M. (2021). Role of TLR4 in Persistent Leptospira Interrogans Infection: A Comparative In Vivo Study in Mice. Front. Immunol. 11, 572999. doi: 10.3389/fimmu.2020.572999
Nair, N., Guedes, M. S., Werts, C., Gomes-Solecki, M. (2020). The Route of Infection With Leptospira Interrogans Serovar Copenhageni Affects the Kinetics of Bacterial Dissemination and Kidney Colonization. PLoS Negl. Trop. Dis. 14 (1), e0007950. doi: 10.1371/journal.pntd.0007950
Nally, J. E., Fishbein, M. C., Blanco, D. R., Lovett, M. A. (2005). Lethal Infection of C3H/HeJ and C3H/SCID Mice With An Isolate of Leptospira Interrogans Serovar Copenhageni. Infect. Immun. 73 (10), 7014–7017. doi: 10.1128/IAI.73.10.7014-7017.2005
Pereira, M. M., Andrade, J., Marchevsky, R. S., Ribeiro dos Santos, R. (1998). Morphological Characterization of Lung and Kidney Lesions in C3H/HeJ Mice Infected With Leptospira Interrogans Serovar Icterohaemorrhagiae: Defect of CD4+ and CD8+ T-Cells Are Prognosticators of the Disease Progression. Exp. Toxicol. Pathol. 50 (3), 191–198. doi: 10.1016/S0940-2993(98)80083-3
Richer, L., Potula, H. H., Melo, R., Vieira, A., Gomes-Solecki, M. (2015). Mouse Model for Sublethal Leptospira Interrogans Infection. Infect. Immun. 83 (12), 4693–4700. doi: 10.1128/IAI.01115-15
Roqueplo, C., Kodjo, A., Demoncheaux, J. P., Scandola, P., Bassene, H., Diatta, G., et al. (2019). Leptospirosis, One Neglected Disease in Rural Senegal. Vet. Med. Sci. 5 (4), 536–544. doi: 10.1002/vms3.186
Santecchia, I., Ferrer, M. F., Vieira, M. L., Gomez, R. M., Werts, C. (2020). Phagocyte Escape of Leptospira: The Role of TLRs and Nlrs. Front. Immunol. 11, 571816. doi: 10.3389/fimmu.2020.571816
Schafbauer, T., Dreyfus, A., Hogan, B., Rakotozandrindrainy, R., Poppert, S., Straubinger, R. K. (2019). Seroprevalence of Leptospira Spp. Infection in Cattle From Central and Northern Madagascar. Int. J. Environ. Res. Public Health 16 (11), p–2014. doi: 10.3390/ijerph16112014
Scharrig, E., Carestia, A., Ferrer, M. F., Cedola, M., Pretre, G., Drut, R., et al. (2015). Neutrophil Extracellular Traps Are Involved in the Innate Immune Response to Infection With Leptospira. PLoS Negl. Trop. Dis. 9 (7), e0003927. doi: 10.1371/journal.pntd.0003927
Silva, P. L., Nakajima, E., Costa, R. M. D., Lee Ho, P., Martins, E. A., Carvalho, E., et al. (2020). Chemokine Expression Profiles in Liver and Kidney of Mice With Different Susceptibilities to Leptospirosis. Microb. Pathog. 149, 104580. doi: 10.1016/j.micpath.2020.104580
Sullivan, J. P., Nair, N., Potula, H. H., Gomes-Solecki, M. (2017). Eyedrop Inoculation Causes Sublethal Leptospirosis in Mice. Infect. Immun. 85 (4), e01050–16. doi: 10.1128/IAI.01050-16
Toma, C., Okura, N., Takayama, C., Suzuki, T. (2011). Characteristic Features of Intracellular Pathogenic Leptospira in Infected Murine Macrophages. Cell Microbiol. 13 (11), 1783–1792. doi: 10.1111/j.1462-5822.2011.01660.x
Vernel-Pauillac, F., Merien, F. (2006). Proinflammatory and Immunomodulatory Cytokine mRNA Time Course Profiles in Hamsters Infected With a Virulent Variant of Leptospira Interrogans. Infect. Immun. 74 (7), 4172–4179. doi: 10.1128/IAI.00447-06
Viriyakosol, S., Matthias, M. A., Swancutt, M. A., Kirkland, T. N., Vinetz, J. M. (2006). Toll-Like Receptor 4 Protects Against Lethal Leptospira Interrogans Serovar Icterohaemorrhagiae Infection and Contributes to In Vivo Control of Leptospiral Burden. Infect. Immun. 74 (2), 887–895. doi: 10.1128/IAI.74.2.887-895.2006
Werts, C., Tapping, R. I., Mathison, J. C., Chuang, T. H., Kravchenko, V., Saint Girons, I., et al. (2001). Leptospiral Lipopolysaccharide Activates Cells Through a TLR2-Dependent Mechanism. Nat. Immunol. 2 (4), 346–352. doi: 10.1038/86354
Yukawa, M., Mochizuki, K., Kosaka, T., Kamata, H., Awaya, A., Kobayashi, H., et al. (1994). Protective Effects of Serum Thymic Factor to Leptospira Interrogans Serovar Copenhageni Infection in Mongolian Gerbils. Vet. Microbiol. 41 (1-2), 99–106. doi: 10.1016/0378-1135(94)90139-2
Keywords: immune cells, flow cytometry, proteome array, cytokines, chemokines, L. biflexa, L. interrogans, Leptospirosis
Citation: Shetty A, Kundu S and Gomes-Solecki M (2021) Inflammatory Signatures of Pathogenic and Non-Pathogenic Leptospira Infection in Susceptible C3H-HeJ Mice. Front. Cell. Infect. Microbiol. 11:677999. doi: 10.3389/fcimb.2021.677999
Received: 08 March 2021; Accepted: 25 May 2021;
Published: 24 June 2021.
Edited by:
Charles Robert Brown, University of Missouri, United StatesReviewed by:
Ricardo Martin Gomez, CONICET Instituto de Biotecnologia y Biologia Molecular (IBBM), ArgentinaTatiana Rodrigues Fraga, University of São Paulo, Brazil
Copyright © 2021 Shetty, Kundu and Gomes-Solecki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Gomes-Solecki, mgomesso@uthsc.edu
†These authors have contributed equally to this work
 Advait Shetty
Advait Shetty Suman Kundu
Suman Kundu Maria Gomes-Solecki
Maria Gomes-Solecki