Molecular Epidemiology of Hypervirulent Carbapenemase-Producing Klebsiella pneumoniae
- 1Department of Laboratory Medicine, Huashan Hospital, Fudan University, Shanghai, China
- 2Department of Intensive Care Unit, Huashan Hospital, Fudan University, Shanghai, China
- 3Zhejiang Provincial Demonstration Center of Laboratory Medicine Experimental Teaching, Wenzhou Medical University, Wenzhou, China
- 4Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
- 5Department of Microbiology, Children’s Hospital of Fudan University, Shanghai, China
- 6Zhuhai Precision Medical Center, Zhuhai People’s Hospital (Zhuhai Hospital Affiliated With Jinan University), Zhuhai, China
Objective: To investigate the overall distributions of key virulence genes in Klebsiella pneumoniae, especially the hypervirulent blaKPC-positive K. pneumoniae (Hv-blaKPC(+)-KP).
Methods: A total of 521 complete genomes of K. pneumoniae from GenBank were collected and analyzed. Multilocus sequence typing, molecular serotyping, antibiotic-resistance, virulence genes and plasmid replicon typing were investigated.
Results: Positive rates of virulence genes highly varied, ranging from 2.9 (c-rmpA/A2) to 99.6% (entB). Totally 207 strains presented positive fimH, mrkD, entB and wzi and 190 showed positive fimH, mrkD, entB, irp2 and wzi, which were the two primary modes. A total of 94, 165 and 29 strains were denoted as hypervirulent K. pneumoniae (HvKP), blaKPC(+)-KP and Hv-blaKPC(+)-KP. ST11 accounted for 17 among the 29 Hv-blaKPC(+)-KP strains; Genes iucA, p-rmpA2 and p-rmpA were positive in 28, 26 and 18 Hv-blaKPC(+)-KP strains respectively. Among the 29 Hv-blaKPC(+)-KP strains exhibiting four super clusters from GenBank, IncHI1B plasmids carrying virulence genes and IncFII ones with blaKPC were responsible for both 23 strains respectively.
Conclusions: Positive rates of virulence genes vary remarkably in K. pneumoniae. Genes iucA, p-rmpA2 and p-rmpA were primary ones inducing Hv-blaKPC(+)-KP. IncHI1B plasmids carrying virulence genes and IncFII ones with blaKPC constitute the primary combination responsible for Hv-blaKPC(+)-KP. The making of Hv-blaKPC(+)-KP is mostly via blaKPC(+)-KP acquiring another plasmid harboring virulence genes.
Introduction
Klebsiella pneumoniae, a ubiquitous and an opportunistic pathogen, can induce both nosocomial and community-acquired infections (Russo and Marr, 2019; Choby et al., 2020). The former consist of pneumonia, bacteremia, urinary tract infections, etc. The latter include pyogenic liver abscess, endophthalmitis, meningitis, necrotizing fasciitis, etc. K. pneumoniae inducing such “invasive syndrome” is termed as hypervirulent K. pneumoniae (HvKP), which is more virulent than “classical” K. pneumoniae (cKP) typically responsible for nosocomial infections (Russo and Marr, 2019). Many virulence factors are involved in such pathogenesis, e.g. capsule, lipopolysaccharide, Types 1 and 3 fimbriae, siderophores, allantoin metabolism, etc. (Paczosa and Mecsas, 2016). Further, numerous genes are determinants of those factors. Genes p-rmpA, p-rmpA2 and c-rmpA/A2 all could induce hypercapsule (Paczosa and Mecsas, 2016). Traditionally, HvKP was usually susceptible to most antibiotics except inherently resistant ampicillin (Fang et al., 2007).
With years passing, K. pneumoniae, regardless of cKP or HvKP, becomes more and more drug-resistant, among which carbapenem-resistance is of great concern. Carbapenem-resistance is mostly conferred by carbapenemase gene (blaKPC), New Delhi metallo-β-lactamase gene (blaNDM), and oxacillinases-48 gene (blaOXA-48), which are predominantly carried on the mobile genetic elements (Zhang et al., 2015; Lee et al., 2016). Among them, blaKPC, particularly blaKPC-2/3 is predominant (Kopotsa et al., 2019). Carbapenem-resistant K. pneumoniae (CRKP) has now become a great public health threat worldwide (Lee et al., 2016; Niu and Li, 2019), due to its causing high mortality and medical burden.
In the past decades, hypervirulence and drug-resistance advance separately in K. pneumoniae. CRKP was not usually considered hypervirulent (Zhang et al., 2017a). However, their convergence was found in recent years worldwide (Zhang et al., 2015; Lam et al., 2019; Wozniak et al., 2019). Not surprisingly, such K. pneumoniae strains could induce an overwhelming mortality (Gu et al., 2018). Due to the mobility of elements carrying virulence and drug-resistance genes, hypervirulent carbapenem-resistant K. pneumoniae (Hv-CRKP) gained more and more prevalence with its positive rate reaching 7.4–15.0% among CRKP in recent years (Lee et al., 2017). To date, the overall distribution of key virulence genes in K. pneumoniae strains, in particular hypervirulent blaKPC-positive K. pneumoniae (Hv-blaKPC(+)-KP), was rarely reported. Here, we collected 521 K. pneumoniae strains from GenBank. Upon the yielded data, we could get insight into the distributions of key virulence genes in K. pneumoniae, particularly Hv-blaKPC(+)-KP.
Materials and Methods
K. pneumoniae Strains
A total of 521 complete whole genomes (Table S1) of K. pneumoniae from the GenBank Database (https://www.ncbi.nlm.nih.gov/genome/815; download date: May 13th, 2020) were analyzed in this study. Those draft genomes (contigs and scaffolds) were not included. The 521 strains included 28.4% (148 strains) from Mainland China, 4.4% (23 strains) from Taiwan of China, 1.5% (eight strains) from Hong Kong of China, 25.7% (134 strains) from USA, 9.6% (50 strains) from Australia, 6.7% (35 strains) from UK, 3.8% (20 strains) from Germany, 2.7% (14 strains) from Korea, 2.3% (12 strains) from India, 2.1% (11 strains) from France, 1.5% (eight strains) from Japan and 11.1% (58 strains) from other countries.
Multilocus Sequence Typing (MLST)
The DNA fasta sequences of the 521 genomes were compared with the K. pneumoniae MLST database (Larsen et al., 2012) containing the seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB) and the STs were yielded.
Determination of Serotypes, Antibiotic-Resistance and Virulence Genes
For the genomes of K. pneumoniae from GenBank, the accession numbers were directly used to determine the capsular types via the database of Institute Pasteur (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). The potential beta-lactamase genes were determined using the Resfinder software version 3.2 (https://cge.cbs.dtu.dk/services/ResFinder/) (Zankari et al., 2012) with the minimum coverage of 60% and minimum identity of 90%, and the virulence genes were predicted using NCBI_BLAST (megablast) searches against the virulence genes of K. pneumoniae with experimental supports (Table S2) with the cut-off coverage of 80% and cut-off identity of 80%.
For virulence genes in this study, they could be classified as the following categories: metabolism (peg-344), colonization (allS), assembling channel protein for capsular polysaccharides or macromolecular exopolysaccharides (EPS, wzy-K1), regulator of mucoid phenotype (p-rmpA2, c-rmpA/A2, p-rmpA), Type 1 fimbriae (fimH), Type 3 fimbriae (mrkD), enterobactin (entB), yersiniabactin (irp2), salmochelin (iroN), and aerobactin (iucA) and capsular polysaccharide-anchor (wzi).
Determination of HvKP, cKP and Hv-blaKPC(+)-KP
The factors responsible for HvKP include hypercapsule (by p-rmpA2, c-rmpA/A2, p-rmpA), EPS (by wzy-K1) and excessive siderophores (Paczosa and Mecsas, 2016; Russo and Marr, 2019). In this study, HvKP could be defined as: positive wzy-K1, ≥3 positive siderophore genes (entB, irp2, iroN and iucA), or ≥1 positive capsule-regulating genes (p-rmpA2, c-rmpA/A2 and p-rmpA). Non-HvKP is termed as cKP. Hv-blaKPC(+)-KP is defined as HvKP carrying blaKPC.
Phylogenetic Analysis and Plasmid Replicon Analysis
The phylogenetic tree of K. pneumoniae strains was generated using kSNP3 (Gardner et al., 2015) software for K. pneumoniae chromosomes and displayed by iTOL (Letunic and Bork, 2016) with midpoint rooting. For the plasmids, the phylogenetic patterns were based on the presence/absence of orthologous gene families of all the plasmids under analysis. A binary gene presence/absence matrix was created using OrthoFinder (Emms and Kelly, 2019) with default settings and a hierarchical cluster result was shown by iTOL (Letunic and Bork, 2016).
Plasmid replicon typing was determined using the PlasmidFinder software version 2.0.1 with the minimum coverage of 60% and minimum identity of 95%.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software Inc., USA). Chi-square test was used to analyze comparisons between groups; p <0.05 was considered statistically significant.
Results
Distributions of Virulence Genes and Predicted Key Virulence Factors
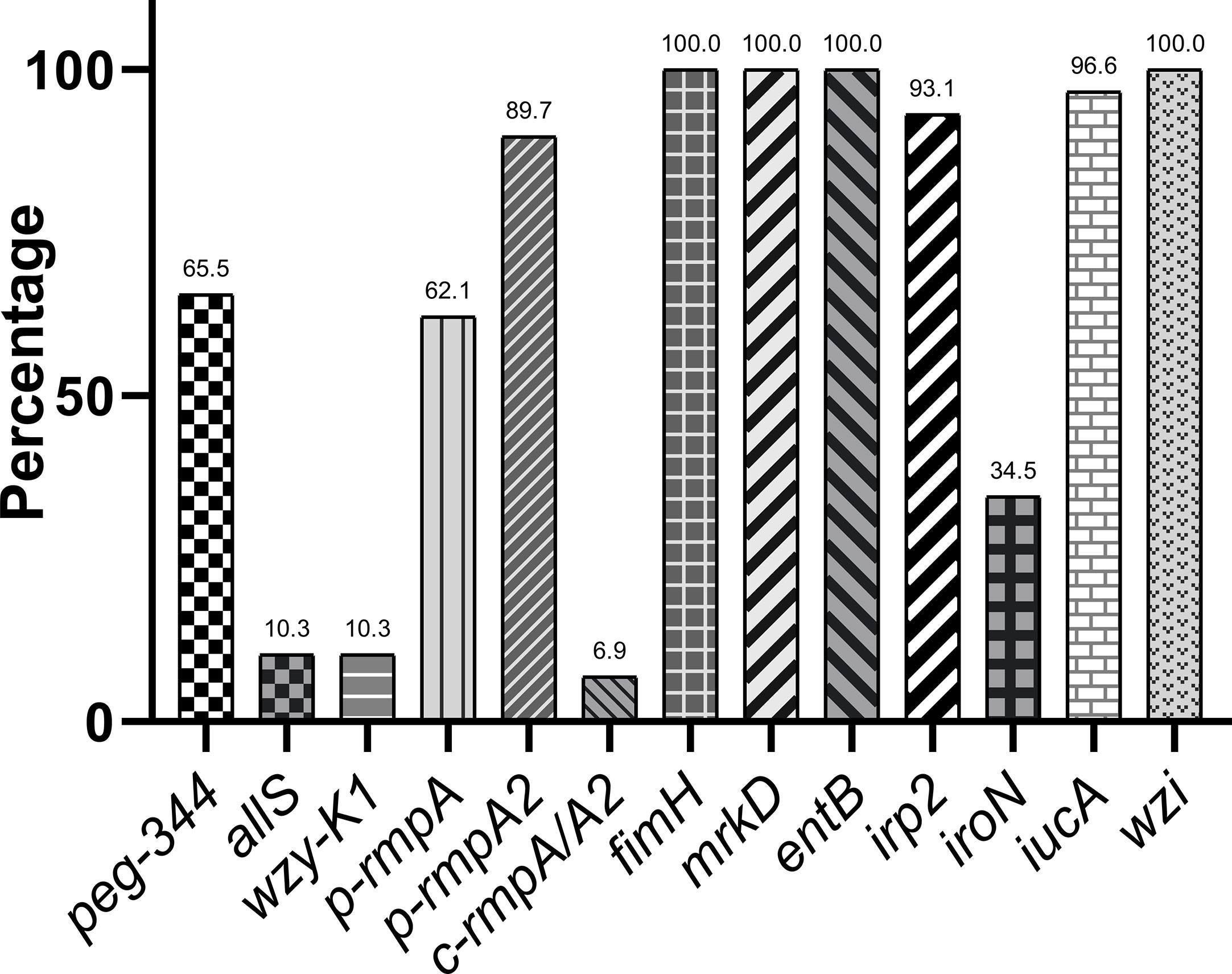
Figure 1A showed overwhelmingly different positive rates of virulence genes, ranging from 2.9 (c-rmpA/A2) to 99.6% (entB) among the 521 K. pneumoniae strains. Four genes (fimH, mrkD, entB and wzi) exhibited prevalence rates of > 90.0%, 1 (irp2) > 50.0% and the others < 25.0%. For the rmpAs, the order was: p-rmpA2 (12.5%), p-rmpA (10.6%) and c-rmpA/A2 (2.9%). For the four siderophore genes, the order was: entB (99.6%), irp2 (53.4%), iucA (15.7%) and iroN (9.2%). Positive rates of iroN and iucA were both lower than that of irp2 and entB (all p < 0.0001). Figure 1B presented different positive rates of predicted virulence factors, ranging from 0.2% (none siderophore) to 99.2% (Type 1 fimbriae). The factors (Types 1 and 3 fimbriae, regular capsule, one or two siderophores) were found more common; 436 (83.7%) strains were found possessing ≤ 2 siderophores. Figure 2 showed 23 modes of virulence genes in K. pneumoniae: each ≥2 strains. Totally 207 strains presented positive fimH, mrkD, entB and wzi and 190 showed positive fimH, mrkD, entB, irp2, and wzi simultaneously, which were the two primary modes and accounted for 39.7% and 36.5% respectively.
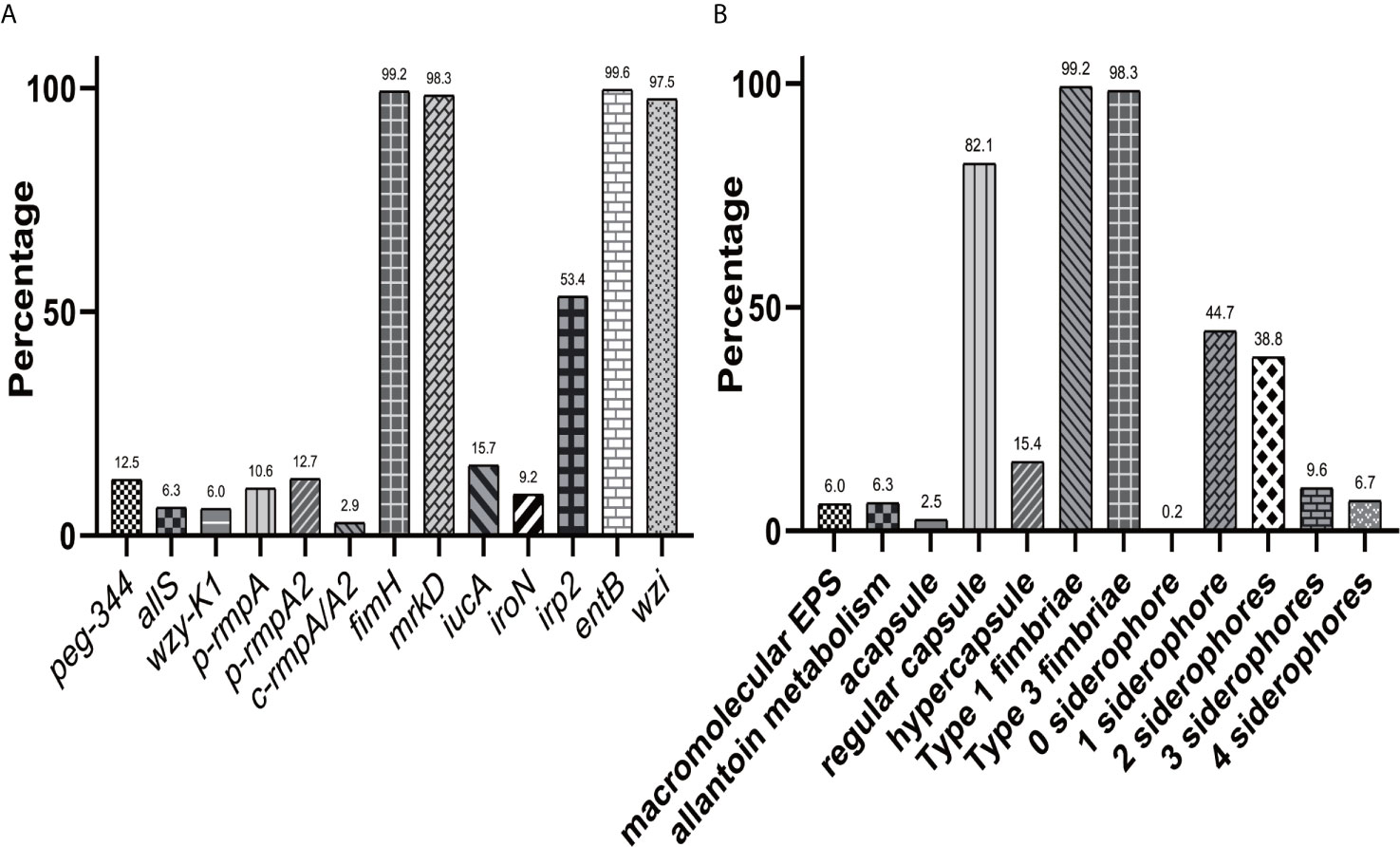
Figure 1 Distributions of virulence genes and factors in K. pneumoniae. (A) Distribution of 13 virulence genes in 521 K. pneumoniae strains. (B) Distribution of virulence factors in 521 K. pneumoniae strains.
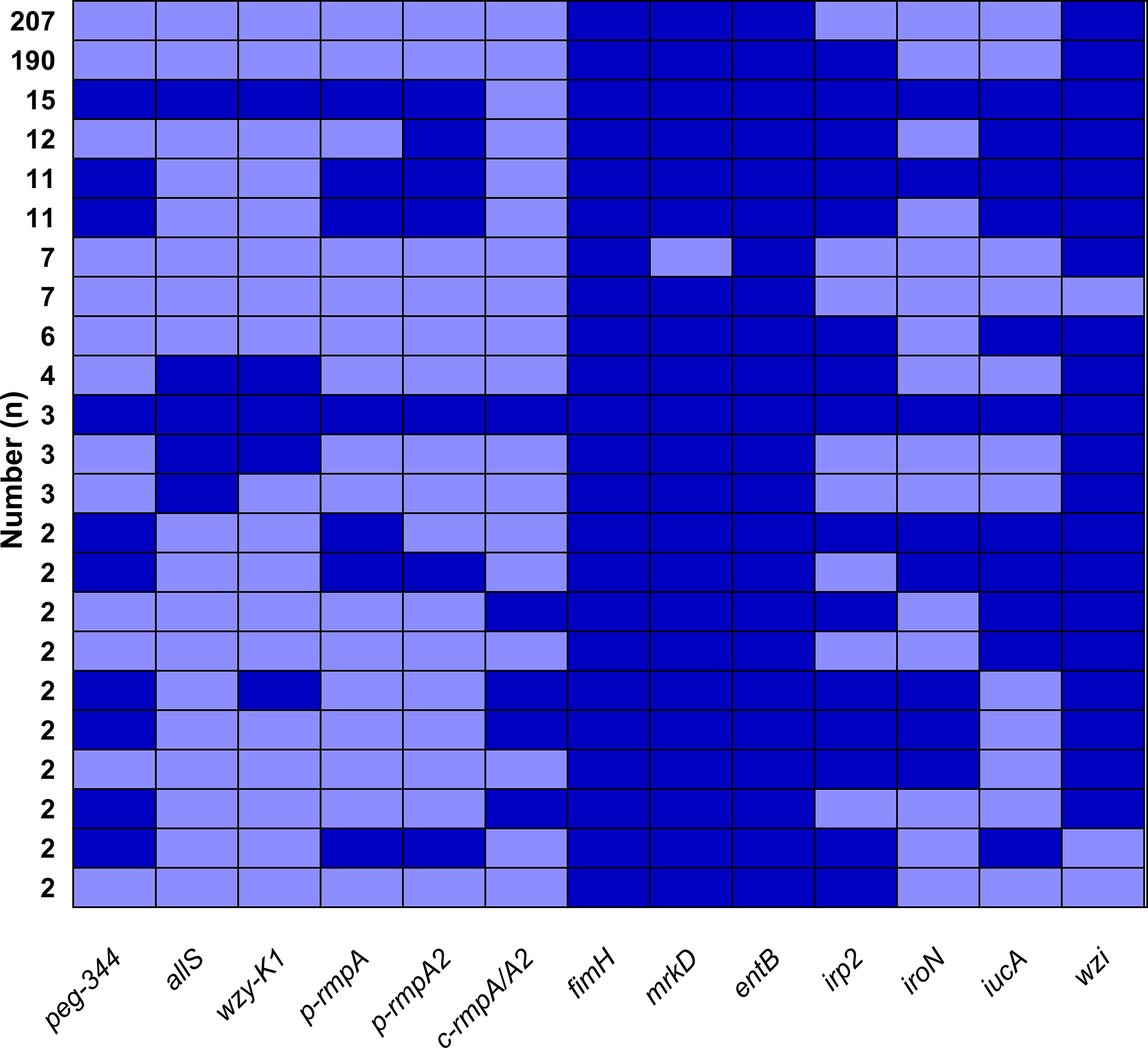
Figure 2 Modes of virulence genes in 521 K. pneumoniae strains. The presence of virulence genes is represented by a dark blue box and the absence of others is represented by a light blue box. Only those with ≥ 2 strains were included in Figure 2.
Among the 91 strains harboring wzy-K1, p-rmpA, p-rmpA2 or c-rmpA/A2, 49 (53.8%) possessed p-rmpA and p-rmpA2, 18 (19.8%) possessing wzy-K1, p-rmpA and p-rmpA2, 15 (16.5%) possessing merely p-rmpA2. Figure 3A showed strong relationships among wzy-K1/p-rmpA or p-rmpA/p-rmpA2. In the 520 strains positive in entB, irp2, iroN or iucA, 278 (53.5%) harbored entB and irp2, 241 (46.3%) harboring only entB, 35 (6.7%) harboring all the four genes. Figure 3B showed strong relationships between iucA/iroN and irp2. Other relationships were also shown in: Figure 3C (K1, peg-344, allS and ST23), Figure 3D (K2, p-rmpA, p-rmpA2 and c-rmpA/A2), Figure 3E (K2, peg-344, allS and ST14), Figure 3F (K2, irp2, iroN and iucA) and Figure 3G (K1, irp2, iroN and iucA). Gene wzy-K1 was completely restricted to K1 serotype (31/31), vice versa. High prevalence of peg-344 and allS was found in K1 strains (22/31, 28/31), but rarely in K2 ones (10/38, 0/38). Gene allS was mainly found in K1 strains (28/33), contrary to peg-344 (22/65). K1 strains mostly belonged to ST23 (23/31) while less than a half (17/38) of K2 ones belonged to ST14. K1 strains showed higher rates of rmpAs (p-rmpA/p-rmpA2/c-rmpA/A2) and siderophore genes (iroN/iucA) than K2 ones: 23/31 vs 10/38 (p < 0.0001), 23/31 vs 9/38 (p < 0.0001), which “confirmed” hypervirulence in K1 strains.
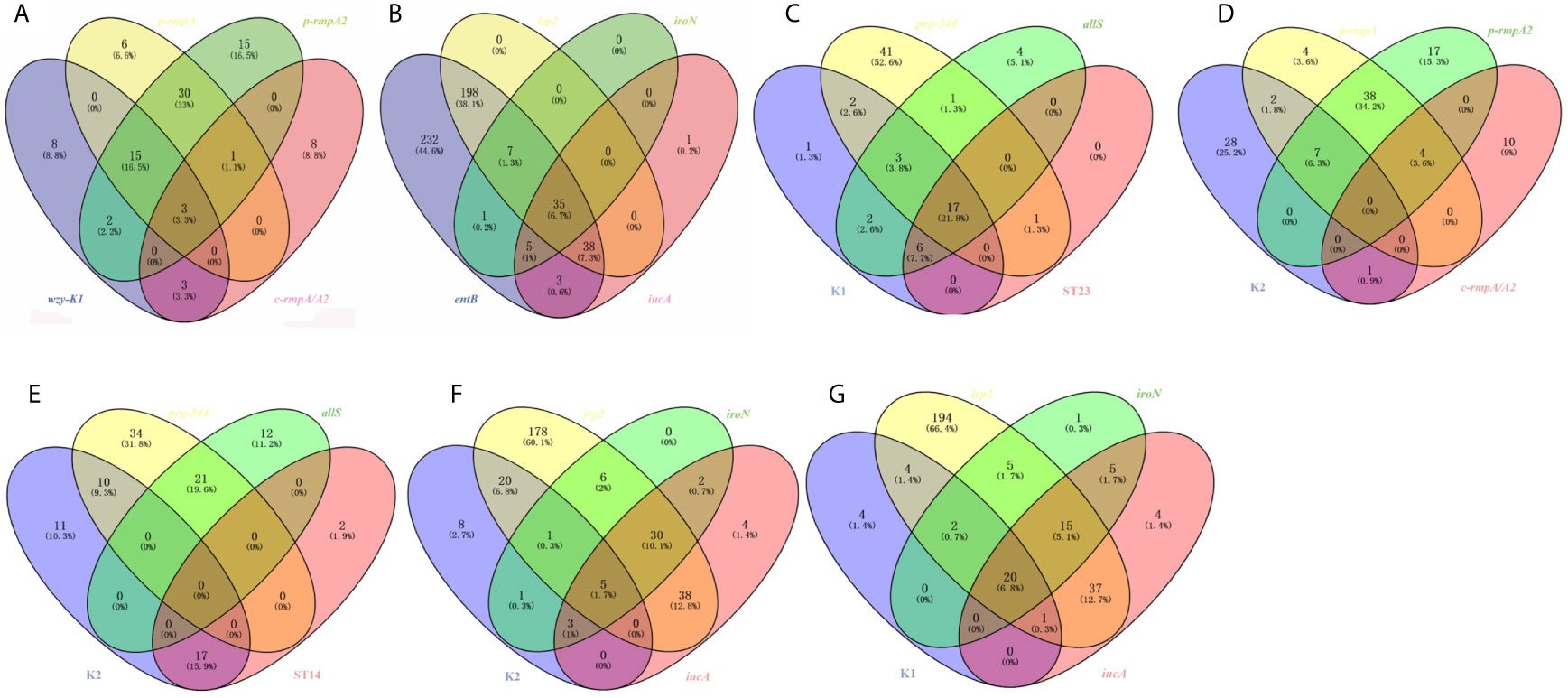
Figure 3 Venn diagrams of various relationships among virulence genes, serotypes and ST types. (A) Venn diagram of wzy-K1, p-rmpA, p-rmpA2 and c-rmpA/A2. (B) Venn diagram of entB, irp2, iroN and icuA. (C) Venn diagram of K1, peg-344, allS and ST23. (D) Venn diagram of K2, p-rmpA, p-rmpA2 and c-rmpA/A2. (E) Venn diagram of K2, peg-344, allS and ST14. (F) Venn diagram of K2, irp2, iroN and iucA. (G) Venn diagram of K1, irp2, iroN and iucA. Such relationships were shown in 521 K. pneumoniae strains.
According to the aforementioned criteria, 94 (18.0%), 165 (31.7%) and 29 (5.6%) strains were denoted as hypervirulent K. pneumoniae (HvKP), blaKPC(+)-KP and Hv-blaKPC(+)-KP, as shown in Figure S1. Consequently, 427 (82.0%) strains were cKP. Hv-blaKPC(+)-KP shared 17.6% (29/165) among blaKPC(+)-KP. For the blaKPC(+)-KP, ST11 accounted for 34.5% (57/165) while clonal group 258, including ST11, ST258, ST340 and ST437, was positive for 65.5% (108/165), indicating the focus of blaKPC(+)-KP.
Distributions of Virulence Genes in Hv-blaKPC(+)-KP
Figure 4 presented greatly different prevalence of virulence genes in 29 Hv-blaKPC(+)-KP strains, ranging from fimH (100.0%), mrkD (100.0%), entB (100.0%), wzi (100.0%) to c-rmpA/A2 (6.9%). Genes iucA, p-rmpA2 and p-rmpA were positive in 28 (96.6%), 26 (89.7%) and 18 (62.1%) Hv-blaKPC(+)-KP strains respectively. A sum of 28 (96.6%) strains presented ≥ 3 siderophores and 29 (100.0%) carried p-rmpA/p-rmpA2 (p > 0.9999).
A total of nine modes of virulence genes were found among the 29 Hv-blaKPC(+)-KP strains, as shown in Figure 5. And the first four modes consisted of eight (27.6%), seven (24.1%), five (17.2%) and three (10.3%) strains, which constituted the majority.
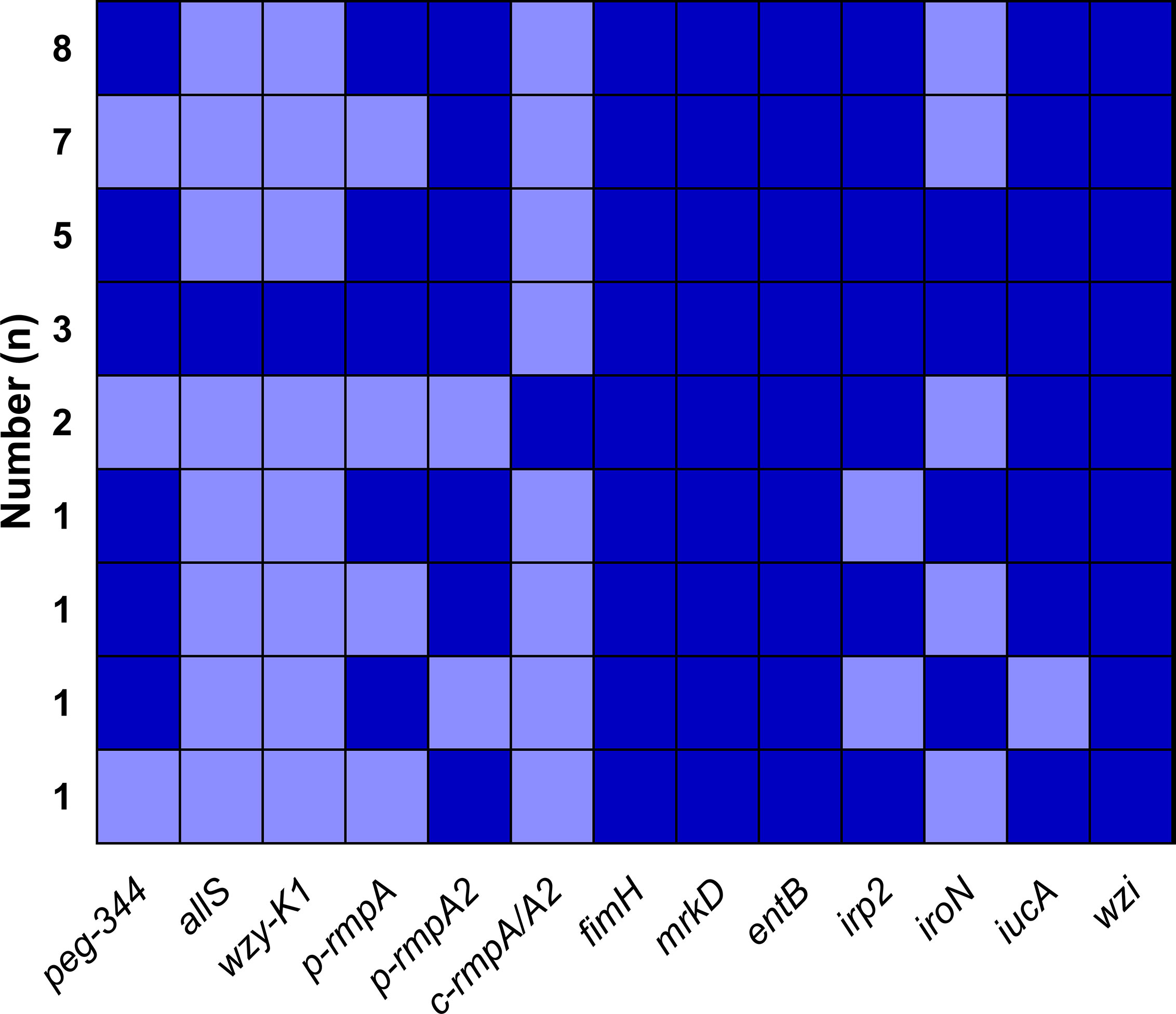
Figure 5 Modes of virulence genes in Hv-blaKPC(+)-KP. The presence of virulence genes is represented by a dark blue box and the absence of others is represented by a light blue box.
Distributions of STs and Serotypes in Hv-blaKPC(+)-KP
Among the 29 Hv-blaKPC(+)-KP strains, ST11 accounted for the majority (17, 58.6%) although more than 10 STs were found in total (Figure 6A). And five serotypes were found (Figure 6B), among which K64 (11, 37.9%) and K47 (10, 34.5%) made the majority.
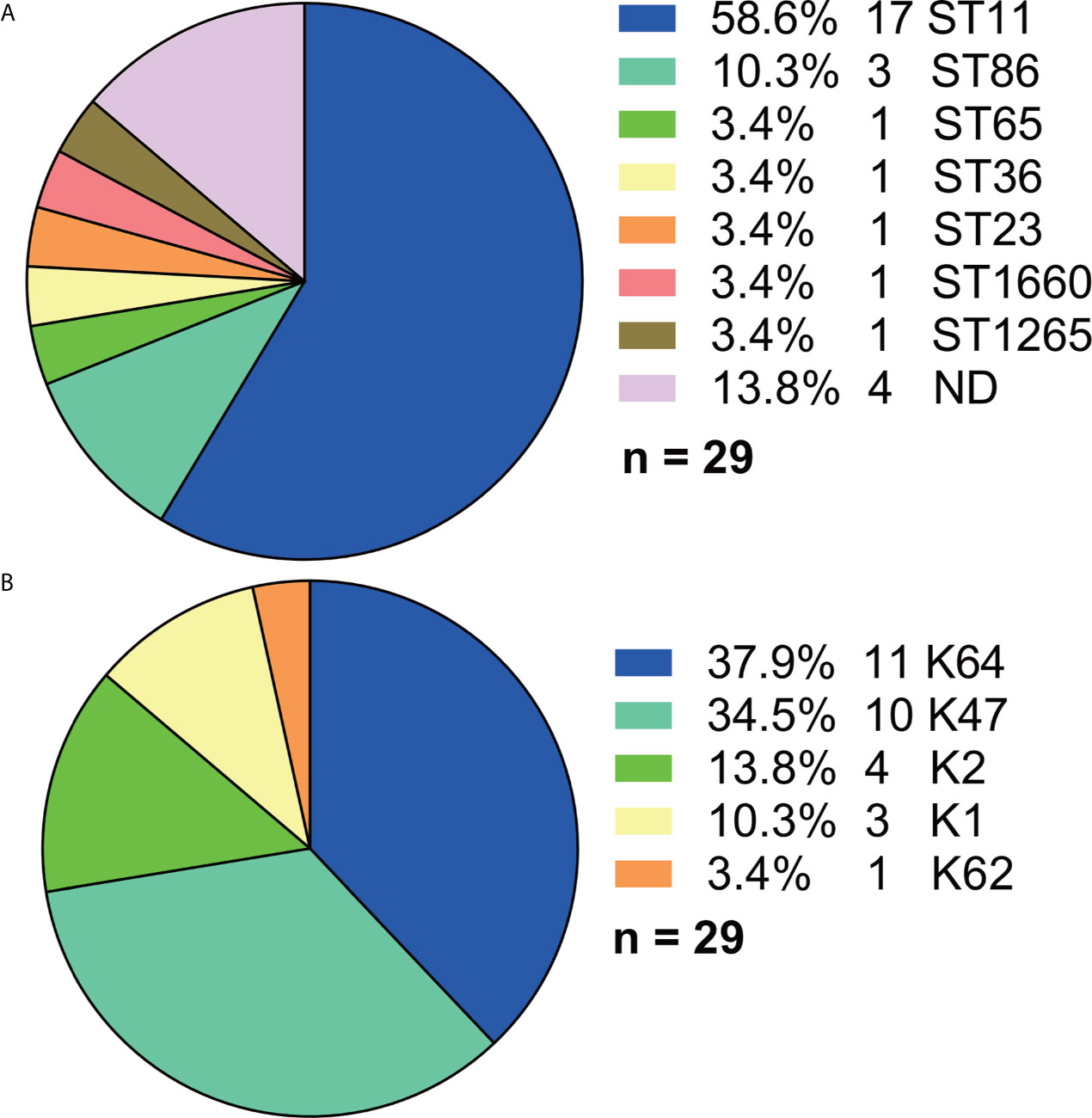
Figure 6 Distributions of STs and serotypes in Hv-blaKPC(+)-KP. (A) Distribution of STs in Hv-blaKPC(+)-KP. (B) Distribution of serotypes in Hv-blaKPC(+)-KP. Statistics were made among the 29 Hv-blaKPC(+)-KP strains. ND, not defined.
Locations of Virulence and blaKPC Genes in Hv-blaKPC(+)-KP
Trends in virulence among Hv-blaKPC(+)-KP infections revealed that the prevalence of Hv-blaKPC(+)-KP significantly increased between 2018 and 2020, mainly from China, especially Mainland China (Figure 7). We found that IncHI1B plasmids were predominantly responsible for the virulence genes (23 strains, 79.3%) and IncFII plasmids were the main contributors for the gene blaKPC (23 strains, 79.3%), suggesting that Hv-blaKPC(+)-KP strains were mainly induced by two different plasmids (Figure 8). IncHI1B and IncFII plasmids constituted the alarmingly successful combination among Hv-blaKPC(+)-KP strains. ST11 accounted for 17 (58.6%) among the 29 Hv-blaKPC(+)-KP strains. Those Hv-blaKPC(+)-KP strains with ST11 typically corresponded to K47 (9/17) and K64 (8/17) serotypes and were divided into four super subgroups. Those with ST86 were all K2 serotype (4/4).
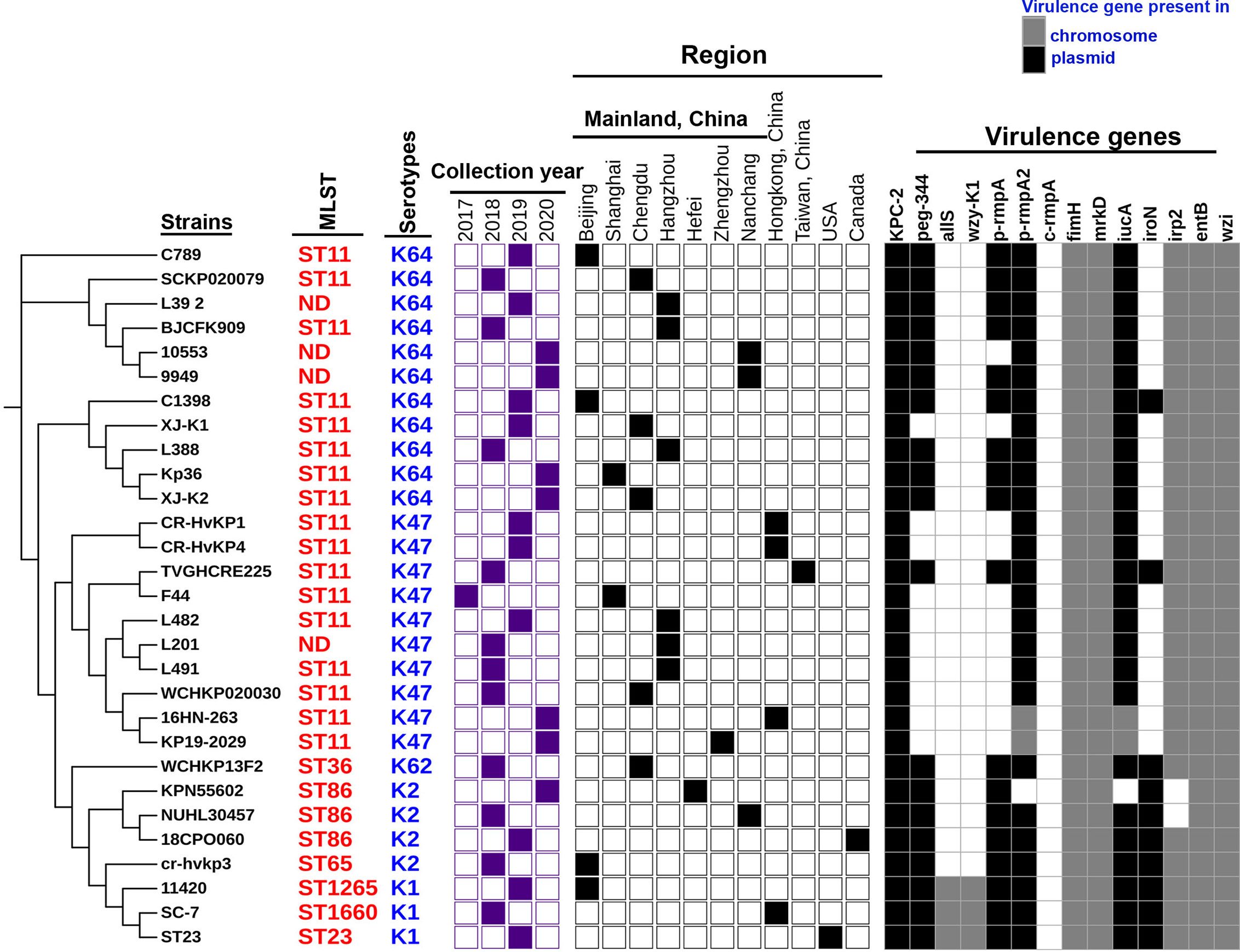
Figure 7 Characteristics of the 29 Hv-blaKPC(+)-KP strains. The phylogenetic relationship of the 29 Hv-blaKPC(+)-KP strains was analyzed by kSNP 3.0 Based on the predicted results, the binary gene presence/absence matrix was created reflecting the collection year, collection region, blaKPC-2 gene and core virulence genes. The STs of Hv-blaKPC(+)-KP strains were marked on the right of the phylogenetic tree. ND: not defined. The presence of genes, etc. is represented by a solid box. and the absence of others is represented by a white box.

Figure 8 Details for the characteristics of the 57 antibiotic-resistance or virulence plasmids carried by the 29 Hv-blaKPC(+)-KP strains. The phylogenetic patterns were based on the presence/absence of orthologous gene families of 57 plasmids under analysis. Seven categories of information were presented in this figure, including the phylogenetic tree of 57 plasmids, STs of host strains, collection year, collection region, blaKPC-2 gene, core virulence genes and replicon types of plasmids. ND, not defined. The presence of genes, etc. is represented by a solid box and the absence of others is represented by a white box.
Discussion
This study investigated the general distributions of key virulence genes in K. pneumoniae, in particular Hv-blaKPC(+)-KP.
Among the 521 strains, 65 were positive for peg-344, of which 63 were denoted as HvKP. A sensitivity of 96.9% was therefore yielded, similar as the report (p = 0.5791) (Russo et al., 2018). Gene allS was not restricted to K1 and K2 strains, different from the document (Yu et al., 2008). The reason may lie in the different specimen types of analyzed strains. Gene wzy-K1 (formerly designated magA), corresponding to K1 serotype, vice versa, could help K. pneumoniae yield macromolecular EPS, which confers hypervirulence (Fang et al., 2004). Wzi is a protein riveting capsular polysaccharides, loss of which K. pneumoniae should be acapsular (Rahn et al., 2003). Acapsule was found in 13 (2.5%) strains, which means low virulence. A total of four kinds of siderophores were found in K. pneumoniae strains: enterobactin, salmochelin, yersiniabactin, and aerobactin (Russo and Marr, 2019). Intriguingly, one strain (strain AR_0096, accession number: CP027612.1) was found for none siderophore, indicating other ferric uptake systems than siderophores may also provide a certain amount of iron for growth and reproductivity (Hsieh et al., 2008).
Except for macromolecular EPS and excessive siderophores, hypercapsule could also contribute to hypervirulence (Russo and Marr, 2019), which is typically conferred by p-rmpA, p-rmpA2 or c-rmpA/A2 genes. Hypercapsule played an equal role with excessive siderophores (15.4 vs 16.3%, p = 0.6714) in hypervirulence of K. pneumoniae. The reason lies in the same pLVPK-like plasmids harboring rmpAs and siderophore genes concurrently.
Gene blaKPC was first reported from USA in 1996 (Yigit et al., 2001). Then, the first blaKPC-2(+)-KP strain was reported in mainland China in 2007 (Wei et al., 2007). CRKP has now shared 70 – 90% of carbapenem-resistant Enterobacteriaceae in the European Union and China (Grundmann et al., 2017; Zhang et al., 2017b). To date, blaKPC consists of more than 50 subtypes, among which blaKPC-2 is the most successful one and predominates CRKP worldwide. blaKPC-2 was positive in 132 (25.3%) strains while blaKPC-3 was found in 30 (5.8%) strains. Our study also showed clonal group 258 but not ST11 made up the majority of blaKPC(+)-KP (Wang et al., 2018; Fu et al., 2019); The reason comes from the global distribution of the 521 strains.
The first Hv-CRKP, belonging to K2 and ST65, was unveiled in mainland China in 2015, which was isolated from blood in Wuhan City in March 2013 (Zhang et al., 2015). Armed with hypervirulence and extreme drug-resistance, Hv-CRKP causes greater mortality and becomes notorious (Gu et al., 2018). Our study showed a positive rate of 5.6% for Hv-blaKPC(+)-KP worldwide. Different prevalence of iucA, p-rmpA2 and p-rmpA in Hv-blaKPC(+)-KP strains suggested their different roles in hypervirulence. The modes of virulence genes were rather diverse in Hv-blaKPC(+)-KP. Similar prevalence of ≥ 3 siderophores and p-rmpA/p-rmpA2 (p > 0.9999) indicated their equal roles in hypervirulence of Hv-blaKPC(+)-KP strains, which also originated from the same pLVPK-like plasmids harboring rmpAs and siderophore genes simultaneously. The proportion of K64 was (11, 37.9%), lower than another report (Zhang et al., 2020) (p < 0.0001). Further, IncHI1B plasmids carrying virulence genes and IncFII ones with blaKPC were responsible for both 23 strains, suggesting IncHI1B and IncFII plasmids jointly constitute the most successful combination. Furthermore, the phylogenetic trees revealed that the 29 Hv-blaKPC(+)-KP strains belonged to four super clusters although three clusters all possessed ST11 strains.
Hv-blaKPC(+)-KP evolution may occur through two mechanisms. The first pathway is via HvKP acquiring a plasmid carrying drug-resistance determinants (Wei et al., 2016; Feng et al., 2018) or by the insertion of resistance genes into virulence plasmid or chromosome harbored by HvKP (Zhang et al., 2016; Fu et al., 2018). The second pathway is via multidrug-resistant/extreme drug-resistant cKP acquiring a pK2044- or pLVPK-like virulence plasmid or integrated virulence genes into drug-resistance plasmids (Gu et al., 2018). Our data showed it was most likely that Hv-blaKPC(+)-KP mainly evolved through the second pathway, i.e. via blaKPC(+)-KP acquiring another plasmid harboring virulence genes. Zhou et al. (2020) and Tang et al. (2020) preached that CRISPR-Cas system deficiency in ST11 may play a vital role. However, the two papers elucidated only blaKPC entering ST11 strains; IncHI1B plasmids are different from IncFII ones: rare protospacers were found and they lacked Type IV secretion systems, e.g. traM gene. Therefore, the mechanisms behind IncHI1B plasmids entering ST11 strains would be sophisticated and intriguing.
This study has some limitations. First, the specimen types of 521 K. pneumoniae strains are not well known. Second, some positive virulence genes do not inevitably mean “exact” hypervirulence.
Taken together, positive rates of virulence genes vary overwhelmingly in K. pneumoniae. Hypercapsule plays an equal proportion with excessive siderophores in hypervirulence of K. pneumoniae. Virulence genes iucA, p-rmpA2 and p-rmpA are primary ones inducing Hv-blaKPC(+)-KP. IncHI1B plasmids carrying virulence genes and IncFII ones with blaKPC constitute the primary combination responsible for Hv-blaKPC(+)-KP. Hv-blaKPC(+)-KP urges more insightful investigations.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://pan.baidu.com/s/1sbsl_phsx8IRoQeeY87e-w (password: xf5l).
Author Contributions
DH, YL and PR conceived the study. DT, WC, PF, WW and XJ collected the 521 genomes. DH, YL, PR and XL did bioinformation analysis. DH and YL wrote the manuscript, which was revised by XL and XJ. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by research grants from the National Natural Science Foundation of China (grants 81871692, 81572031, and 82002170) and the Shanghai Municipal Science and Technology Commission (grant number 19JC1413002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.661218/full#supplementary-material
References
Choby J. E., Howard-Anderson J., Weiss D. S. (2020). Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 287 (3), 283–300. doi: 10.1111/joim.13007
Emms D. M., Kelly S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Gen. Biol. 20, 1, 238. doi: 10.1186/s13059-019-1832-y
Fang C. T., Chuang Y. P., Shun C. T., Chang S. C., Wang J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199 (5), 697–705. doi: 10.1084/jem.20030857
Fang C. T., Lai S. Y., Yi W. C., Hsueh P. R., Liu K. L., Chang S. C. (2007). Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45 (3), 284–293. doi: 10.1086/519262
Feng Y., Lu Y., Yao Z., Zong Z. (2018). Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae of Sequence Type 36. Antimicrob. Agents Chemother. 62 (7), e02644–17. doi: 10.1128/AAC.02644-17
Fu L., Tang L., Wang S., Liu Q., Liu Y., Zhang Z., et al. (2018). Co-location of the blaKPC-2, blaCTX-M-65, rmtB and virulence relevant factors in an IncFII plasmid from a hypermucoviscous Klebsiella pneumoniae isolate. Microb. Pathog. 124, 301–304. doi: 10.1016/j.micpath.2018.08.055
Fu P., Tang Y., Li G., Yu L., Wang Y., Jiang X. (2019). Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int. J. Antimicrob. Agents 54 (2), 117–124. doi: 10.1016/j.ijantimicag.2019.03.014
Gardner S. N., Slezak T., Hall B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31 (17), 2877–2878. doi: 10.1093/bioinformatics/btv271
Grundmann H., Glasner C., Albiger B., Aanensen D. M., Tomlinson C. T., Andrasevic A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17 (2), 153–163. doi: 10.1016/S1473-3099(16)30257-2
Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18 (1), 37–46. doi: 10.1016/S1473-3099(17)30489-9
Hsieh P. F., Lin T. L., Lee C. Z., Tsai S. F., Wang J. T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197 (12), 1717–1727. doi: 10.1086/588383
Kopotsa K., Osei Sekyere J., Mbelle N. M. (2019). Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann. N Y Acad. Sci. 1457 (1), 61–91. doi: 10.1111/nyas.14223
Lam M. M. C., Wyres K. L., Wick R. R., Judd L. M., Fostervold A., Holt K. E., et al. (2019). Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 74 (5), 1218–1222. doi: 10.1093/jac/dkz028
Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 (4), 1355–1361. doi: 10.1128/jcm.06094-11
Lee C. R., Lee J. H., Park K. S., Kim Y. B., Jeong B. C., Lee S. H. (2016). Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 7:895:895. doi: 10.3389/fmicb.2016.00895
Lee C. R., Lee J. H., Park K. S., Jeon J. H., Kim Y. B., Cha C. J., et al. (2017). Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell Infect. Microbiol. 7:483. doi: 10.3389/fcimb.2017.00483
Letunic I., Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44 (W1), W242–W245. doi: 10.1093/nar/gkw290
Niu G., Li W. (2019). Next-Generation Drug Discovery to Combat Antimicrobial Resistance. Trends Biochem. Sci. 44 (11), 961–972. doi: 10.1016/j.tibs.2019.05.005
Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 80 (3), 629–661. doi: 10.1128/MMBR.00078-15
Rahn A., Beis K., Naismith J. H., Whitfield C. (2003). A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J. Bacteriol 185 (19), 5882–5890. doi: 10.1128/jb.185.19.5882-5890.2003
Russo T. A., Olson R., Fang C. T., Stoesser N., Miller M., MacDonald U., et al. (2018). Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 56 (9), e00776. doi: 10.1128/JCM.00776-18
Russo T. A., Marr C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32 (3), e00001–19. doi: 10.1128/CMR.00001-19
Tang Y., Fu P., Zhou Y., Xie Y., Jin J., Wang B., et al. (2020). Absence of the type I-E CRISPR-Cas system in Klebsiella pneumoniae clonal complex 258 is associated with dissemination of IncF epidemic resistance plasmids in this clonal complex. J. Antimicrob. Chemother. 75 (4), 890–895. doi: 10.1093/jac/dkz538
Wang Q., Wang X., Wang J., Ouyang P., Jin C., Wang R., et al. (2018). Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: Data From a Longitudinal Large-scale CRE Study in Chin-2016). Clin. Infect. Dis. 67 (suppl_2), S196–S205. doi: 10.1093/cid/ciy660
Wei Z. Q., Du X. X., Yu Y. S., Shen P., Chen Y. G., Li L. J. (2007). Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob. Agents Chemother. 51 (2), 763–765. doi: 10.1128/AAC.01053-06
Wei D. D., Wan L. G., Deng Q., Liu Y. (2016). Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn. Microbiol. Infect. Dis. 85 (2), 192–194. doi: 10.1016/j.diagmicrobio.2015.03.012
Wozniak J. E., Band V. I., Conley A. B., Rishishwar L., Burd E. M., Satola S. W., et al. (2019). A Nationwide Screen of Carbapenem-Resistant Klebsiella pneumoniae Reveals an Isolate with Enhanced Virulence and Clinically Undetected Colistin Heteroresistance. Antimicrob. Agents Chemother. 63 (5), e00107–19. doi: 10.1128/AAC.00107-19
Yigit H., Queenan A. M., Anderson G. J., Domenech-Sanchez A., Biddle J. W., Steward C. D., et al. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45 (4), 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Yu W. L., Ko W. C., Cheng K. C., Lee C. C., Lai C. C., Chuang Y. C. (2008). Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62 (1), 1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 (11), 2640–2644. doi: 10.1093/jac/dks261
Zhang Y., Zeng J., Liu W., Zhao F., Hu Z., Zhao C., et al. (2015). Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Infect. 71 (5), 553–560. doi: 10.1016/j.jinf.2015.07.010
Zhang R., Lin D., Chan E. W., Gu D., Chen G. X., Chen S. (2016). Emergence of Carbapenem-Resistant Serotype K1 Hypervirulent Klebsiella pneumoniae Strains in China. Antimicrob. Agents Chemother. 60 (1), 709–711. doi: 10.1128/AAC.02173-15
Zhang R., Chan E. W., Zhou H., Chen S. (2017a). Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet Infect. Dis. 17 (3), 256–257. doi: 10.1016/S1473-3099(17)30072-5
Zhang R., Liu L., Zhou H., Chan E. W., Li J., Fang Y., et al. (2017b). Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang Y., Jin L., Ouyang P., Wang Q., Wang R., Wang J., et al. (2020). Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 75 (2), 327–336. doi: 10.1093/jac/dkz446
Keywords: Klebsiella pneumoniae, virulence, plasmid, blaKPC, epidemiology
Citation: Hu D, Li Y, Ren P, Tian D, Chen W, Fu P, Wang W, Li X and Jiang X (2021) Molecular Epidemiology of Hypervirulent Carbapenemase-Producing Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 11:661218. doi: 10.3389/fcimb.2021.661218
Received: 30 January 2021; Accepted: 16 March 2021;
Published: 07 April 2021.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Luchang Zhu, Houston Methodist Research Institute, United StatesJosé Alejandro Di Conza, University of Buenos Aires, Argentina
Copyright © 2021 Hu, Li, Ren, Tian, Chen, Fu, Wang, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofei Jiang, jiangxi2154@sina.com; Xiaobin Li, xiaobinli@sjtu.edu.cn
†These authors have contributed equally to this work
 Dakang Hu
Dakang Hu Yuming Li2†
Yuming Li2†  Pan Fu
Pan Fu Xiaobin Li
Xiaobin Li Xiaofei Jiang
Xiaofei Jiang