Profiling of miRNAs in Mouse Peritoneal Macrophages Responding to Echinococcus multilocularis Infection
- 1State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Gansu, China
- 2Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University College of Veterinary Medicine, Yangzhou, China
Alveolar echinococcosis (AE) is a zoonotic helminthic disease caused by infection with the larval of Echinococcus multilocularis in human and animals. Here, we compared miRNA profiles of the peritoneal macrophages of E. multilocularis-infected and un-infected female BALB/c mice using high-throughput sequencing. A total of 87 known miRNAs were differentially expressed (fold change ≥ 2, p < 0.05) in peritoneal macrophages in mice 30- and 90-day post infection compared with ones in un-infected mice. An increase of mmu-miR-155-5p expression was observed in peritoneal macrophages in E. multilocularis-infected mice. Compared with the control group, the production of nitric oxide (NO) was increased in peritoneal macrophages transfected with mmu-miR-155-5p mimics at 12 h after transfection (p < 0.001). Two key genes (CD14 and NF-κB) in the LPS/TLR4 signaling pathway were also markedly altered in mmu-miR-155-5p mimics transfected cells (p < 0.05). Moreover, mmu-miR-155-5p mimics suppressed IL6 mRNA expression and promoted IL12a and IL12b mRNA expression. Luciferase assays showed that mmu-miR-155-5p was able to bind to the 3′ UTR of the IKBKE gene and decreased luciferase activity. Finally, we found the expression of IKBKE was significantly downregulated in both macrophages transfected with mmu-miR-155-5p and macrophages isolated from E. multilocularis-infected mice. These results demonstrate an immunoregulatory effect of mmu-miR-155 on macrophages, suggesting a role in regulation of host immune responses against E. multilocularis infection.
Introduction
Alveolar echinococcosis (AE) is a serious life-threatening disease caused by Echinococcus multilocularis metacestodes in humans. AE is more prevalent in the northern hemisphere, including parts of central Asia and China (Weiss et al., 2010). In China, this disease is particularly high in pasturing regions of Northwest China, such as Xinjiang, Qinghai, and Gansu (Craig, 2006). Echinococcus multilocularis completes its entire life cycle in two different hosts: an intermediate host (rodents) and a definitive host (foxes or wild canids) (Kamiya and Sato, 1990). The adult E. multilocularis inhabits in the small intestine of domestic and wild carnivorous canids and disperses a large number of eggs into environment with host's feces (Mackenstedt et al., 2015). Grazing farm animals or humans were infected by peroral ingestion of the food or water contaminated with E. multilocularis eggs (Federer et al., 2015). AE is usually diagnosed and treated in a late stage and surgery is only effective treatment (Lointier et al., 1990). Up to now, our understandings of the interactions between E. multilocularis and its hosts are limited, and researches in this respect are urgently needed. As key post-transcriptional regulators, miRNAs play a regulatory roles in various physiologic and pathologic processes, including host-pathogen interactions (O'Connell et al., 2010; Britton, 2017). Using high-throughput sequencing and RNA microarray, host (tissue or circulating) miRNA profiles in response to helminth infection have been widely characterized in recent years (Han et al., 2013; Hansen et al., 2016; Hong et al., 2017). For example, Cryptosporidium parvum infection induces the expression of mir-30b, mir-125b-1, mir-21, and mir-23b-27b-24-1 in epithelial cells (Zhou et al., 2009). It was also indicated that miR-223 was significantly up-regulated in livers and sera of schistosome-infected mice (He et al., 2013). Our previous study found 46 miRNAs were differentially expressed in the livers of mice infected with E. multilocularis (Jin et al., 2017). In another study, we found circulating miRNAs in sera from E. multilocularis-infected mice were significantly dysregulated (Guo and Zheng, 2017a). Our recent study also showed that miRNA expression was disturbed in E. multilocularis- treated RAW264.7 macrophages (Guo and Zheng, 2017b). Moreover, some of altered miRNAs (miR-146a-5p and miR-155-5p) were regulators of immune and inflammatory response genes (Guo and Zheng, 2017b).
Macrophages are important for modulating immune system in host defense helminth infection (Reyes and Terrazas, 2010). However, the dynamic miRNA profiles in peritoneal macrophages against E. multilocularis infection remain unclear. Considering the important role of miRNAs in immune responses, we characterized miRNA profiles of peritoneal macrophages of E. multilocularis-infected mice and found some of altered miRNAs were related to the regulation of immune responses. These findings will provide rich resources for further studies on the functions of host miRNAs against E. multilocularis, which will help us to better understand the host defense mechanisms.
Materials and Methods
Ethics Approval and Consent to Participate
Animal experiments in the study were performed at Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences and handled in accordance with good animal practice according to the Animal Ethics Procedures.
Parasite Infection
Echinococcus multilocularis protoscoleces were obtained from the hydatid cysts of infected mouse in our lab. Sixty female BALB/c mice (4–6 weeks old) were purchased from Experimental Animal Center, Lanzhou Veterinary Research Institute, and were randomly divided into two groups. One group (n = 30) was intraperitoneally injected with 1,000 E. multilocularis protoscoleces as previously described (Guo and Zheng, 2017a). The other (n = 30) was inoculated with 0.85% NaCl solution as an uninfected group.
Peritoneal Macrophages Isolation, Culture, and Transfection
Peritoneal macrophages were recovered from female BALB/c mice sacrificed under sterile conditions at 30-, 60-, and 90-day post infection, respectively. Cells were centrifuged at 500 g for 15 min. After removing the supernatant, the cell pellets were resuspended in RPMI-1640 medium (11875093-Gibco) supplemented with 10 % fetal bovine serum (10099141-Gibco). Isolated macrophages were seeded into 6-well plates (costar, corning incorporated) with 5 × 106 cells per well and cultured in a 5% CO2 humified incubator at 37°C. Mouse peritoneal macrophages were, respectively, transfected with mmu-miR-155-5p mimics and miRNA mimic negative-control (NC) (Ambion/Invitrogen) using Lipofectamine™ RNAiMAX transfection Reagent (InvitrogenTM) according to the manufacturer's protocol (Yahiro et al., 2012). The transfection medium was replaced with RPMI-1640 medium supplemented with 10 % FBS 10 h post-transfection.
Deep Sequencing and Data Analysis
Peritoneal macrophages from three mice in each group were randomly selected. Total RNA was isolated from mouse peritoneal macrophages using TRIzol reagent (Invitrogen) according to the guidelines (Hummon et al., 2007). High-throughput sequencing of small RNA was conducted on an illumina Hiseq 2500/2000 platform in Beijing Novogene (China) (Malone et al., 2012). Raw data were firstly processed through in-hours perl and python scripts. In this step, the clean reads were obtained by removing ploy-N, 5′ and 3′ adaptor sequences, and low-quality reads. The Small RNA tags perfectly mapped to the mouse genome (http://www.ncbi.nlm.nih.gov/genome/genomes/52) were used to identify known miRNA. Mouse known miRNAs were identified by BLAST searching against the MirGeneDB database (http://www.mirgenedb.org). Relative expression levels of miRNAs in three groups were analyzed using DESeq R package (1.8.3) (Likun et al., 2010) and differential expressed miRNAs were identified by using a fold change cut-off of ≥2 and significance p < 0.05. Volcano plots were used to visualize distinct expression profiles of miRNAs between two groups.
Quantitative RT-PCR Analysis
The miRNA expression levels were examined using All-in-OneTM miRNA detection mix (GeneCopoia) according to the manufacturer's protocol as previously described (Guo and Zheng, 2017b). The miRNA expression levels were quantified based on the threshold cycle (Ct) values. U6 snRNA was selected as an endogenous control.
For examining mRNA expression levels, the first-strand cDNA was synthesized using 1 μg of total RNA by a ThermoScript™ RT-PCR System (Invitrogen). The reaction mixtures (20 μL) were incubated at 42°C for 1 h and then stopped by heating 75°C for 5 min. The cDNA mixture was diluted by 7-fold with nuclease-free water. Quantitative RT-PCR was performed with SYBR® Premix Ex TagTM II (TaKaRa) using 7500 Real Time PCR System (Applied Biosystems) the under following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 1 min. Gapdh was selected as an endogenous reference gene. The qRT-PCR primers were purchased from GeneCopoeia. The relative expression levels of miRNA or mRNA were calculated using the 2−ΔΔCt formula (Manzanoromán and Sileslucas, 2012). Statistical analysis data were taken from three independent experiments.
Plasmid Construction and Luciferase Assay
For luciferase plasmid construction, the 3′ UTR fragment of IKBKE with restriction enzyme sites was amplified by RT-PCR using a pair of primers: 5′-GAGCTCGCCATTGGCCATTGGCC-3′ (restriction enzyme sites were underlined) and 5′-CTCGAGCAGCCAGTTTAGGTAATAAAC-3′ with the following steps: 94°C for 5 min, 35 cycles of 94°C for 30 s, 54°C for 30 s and 72°C for 1 min, and then 72°C for 10 min. The PCR product was subcloned into PmirGLO Dual-Luciferase vector (Promega, USA) and sequenced. The full length of IKBKE 3′UTR with mutations in a binding site was artificially synthesized (BGI Genomics, China), and was also subcloned into PmirGLO Dual-Luciferase vector and sequenced.
HEK293T cells were plated into 24-well plates and transfected with 1 μg WT-IKBKE or Mut-IKBKE in combination with 30 pmol mmu-miR-155-5p mimic or negative control (NC) mimic (ThermoFisher Scientific) using Lipofectamine 2000 (ThermoFisher Scientific). The luciferase activity was measured 24 h post transfection using a Dual-Glo Luciferase Assay System (Promega). Fluorescent intensity was recorded using GloMax96 (Promega). Each transfection was independently repeated three times.
Determination of NO Secretion
Mouse peritoneal macrophages were transfected with mmu-miR-155-5p mimics or NC mimic for 12 h, followed by stimulation with 10 ng/mL IFN-γ and 100 ng/mL LPS for 12 h. The culture supernatants were harvested and nitrite levels were measured using Griess reagent (Invitrogen) (Zheng et al., 2016). Each supernatant sample was detected in triplicate and the data for final statistical analysis were taken from three independent experiments.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 5 (La Jolla, USA), and a one-tailed unpaired t-test or one-way ANOVA was used for comparing differences among two or more independent groups. A p < 0.05 was statistically significant.
Results
Identification of Differentially Expressed miRNAs in Mouse Peritoneal Macrophages Against E. multilocularis Infection
To understand the dynamic of miRNA expression at early (30 days) and late (90 days) time points post infection, we compared global miRNA abundance of peritoneal macrophages in E. multilocularis-infected and un-infected mice. A total of 995 known miRNAs were identified from three libraries. Compared with the uninfected group, 125 and 139 known mature miRNAs were differentially expressed in peritoneal macrophages in mice 30- and 90-day post infection of E. multilocularis (Tables S1, S2), respectively. Of them, 87 differentially expressed miRNAs were commonly shared in peritoneal macrophages in mice 30- and 90-day post infection (p < 0.05 and fold change ≥ 2 or≤ -2), with 51 miRNAs being upregulated and 36 downregulated (Figure 1).
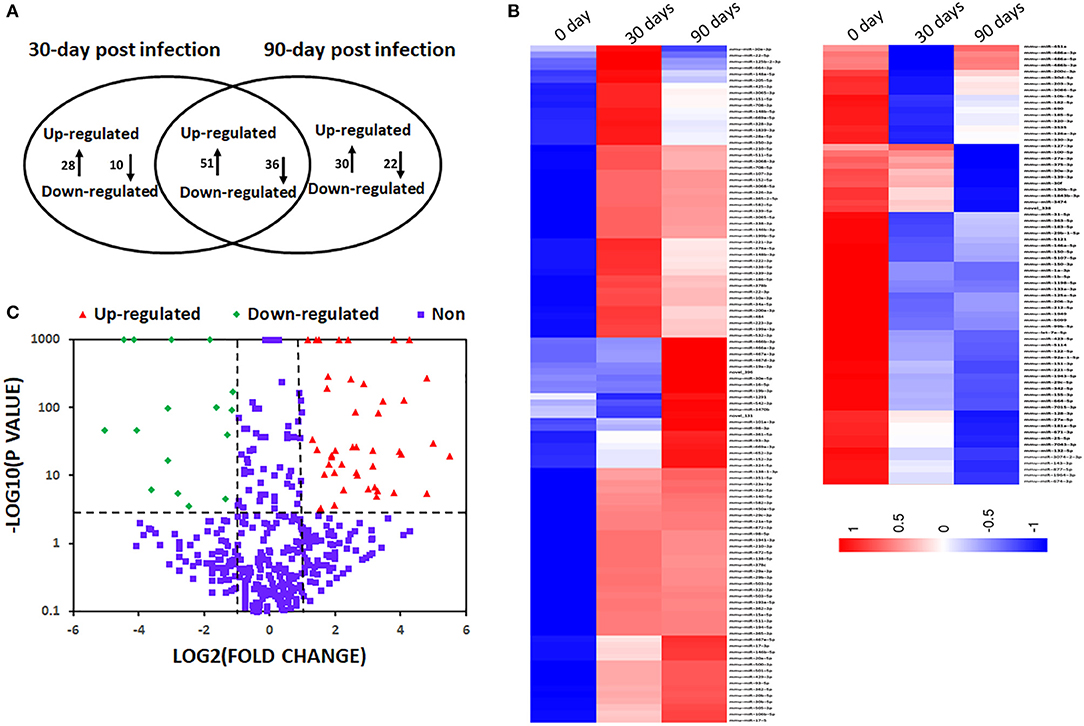
Figure 1. Differentially expressed miRNAs in mouse peritoneal macrophages during Echinococcus multilocularis infection. (A) Commonly shared and specific miRNAs between two E. multilocularis-infected libraries. Compared with the normal group, 125 and 139 miRNAs were differentially expressed in peritoneal macrophages in mice 30 and 90-day post infection of E. multilocularis, respectively. A total of 87 differentially expressed miRNAs were commonly shared between peritoneal macrophages in mice 30 and 90-day post infection. (B) A heat map of differentially expressed miRNAs between peritoneal macrophages in mice 30 and 90-day post infection. (C) A volcano plot of the differentially expressed miRNAs between peritoneal macrophages in mice 30 and 90-day post infection, where the red and green indicate significantly upregulated miRNAs and downregulated miRNAs, respectively.
The predicted target genes of 87 differentially expressed miRNAs were classified according to GO functional annotations (Table 1 and Figure S1). Go analysis revealed that they were highly enriched in metabolic process, regulation of signaling, protein binding, and intracellular organelle (Figure S1). The KEGG pathway analysis indicated that top 20 pathways were highly represented, including MAPK signaling pathway, pathways in cancer, regulating pluripotency of stem cells, and Rap1 signaling pathway (Figure S2).
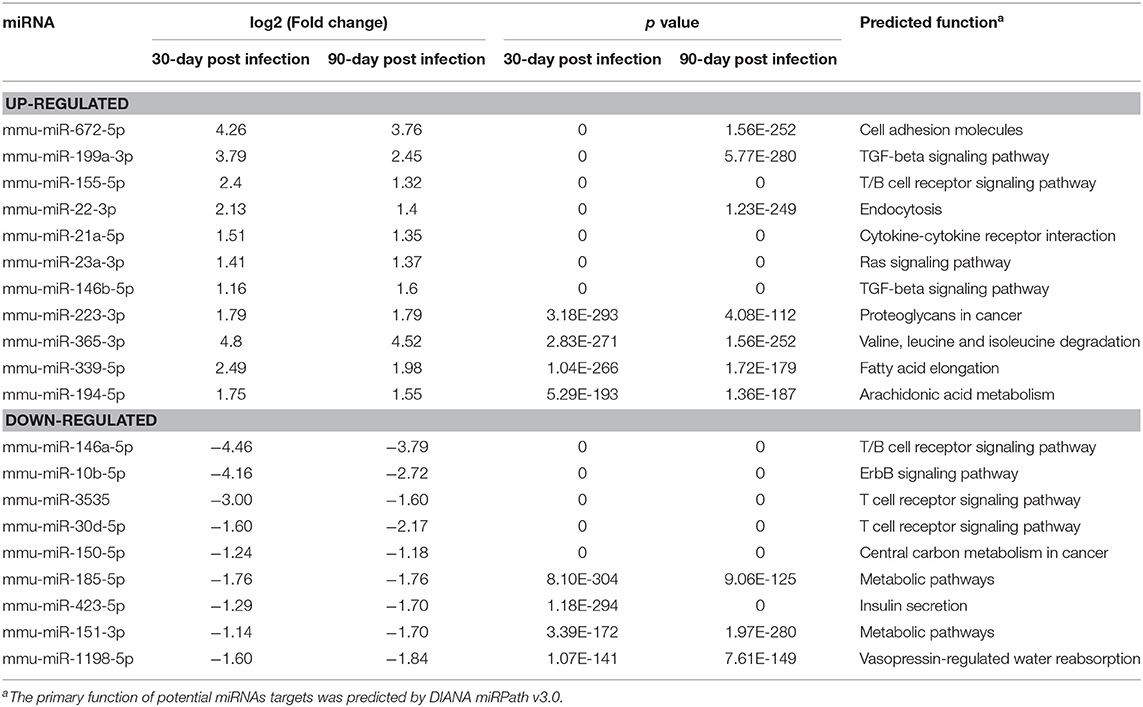
Table 1. Summary of the top 20 differentially expressed miRNAs with high abundance and small p value.
Dynamic Relative Expression of Differentially Expressed miRNAs in Peritoneal Macrophages of E. multilocularis-Infected Mice
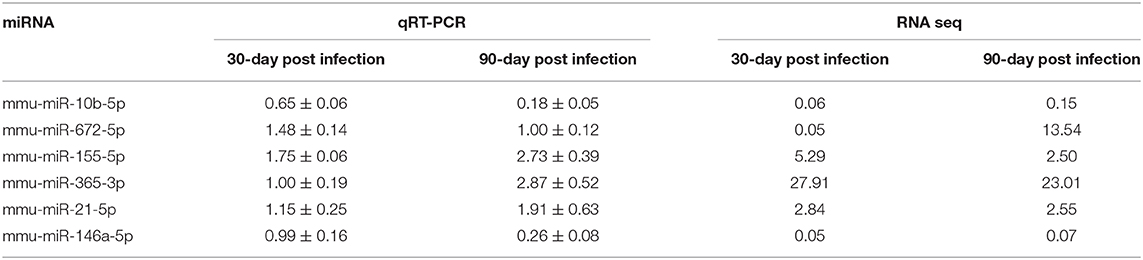
To verify the high-throughput sequencing data, the expression levels of six selected miRNAs with high fold change and small p value (miR-10b-5p, miR-672-5p, miR-155-5p, miR-365-3p, miR-21-5p, and miR-146a-5p) were analyzed in peritoneal macrophages in mice 30-, 60-, and 90-day post inoculation. With the extension of E. multilocularis infection time, the expression levels of miR-10b-5p and miR-146a-5p exhibited a gradual reduction (Figure 2 and Table 2; p < 0.01), whereas the expression levels of miR-155-5p and miR-365-3p showed a gradual increase (Figure 2 and Table 2). The expression level of miR-672-5p was remarkedly upregulated 30 days post infection (Figure 2 and Table 2; p < 0.05) but no difference was observed between 60- and 90-day post inoculation (Figure 2 and Table 2; p > 0.05). The level of miR-21-5p expression was significantly upregulated 60-day post infection compared with uninfected group (Figure 2 and Table 2; p < 0.01).
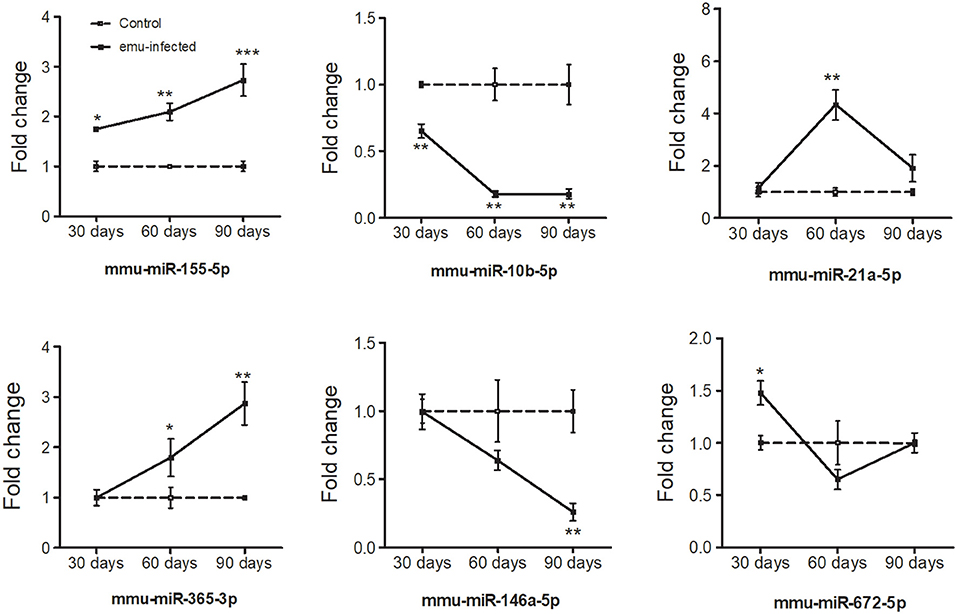
Figure 2. Dynamic expression of six differentially expressed miRNAs in the peritoneal macrophage cells in E. multilocularis-infected mice 30-, 60-, and 90-day post-infection. The expression of these miRNAs was detected by qRT-PCR and normalized to U6 snRNA. Data for final statistical analysis were taken from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Immunomodulation of mmu-miR-155-5p on Mouse Peritoneal Macrophages
To investigate the immunoregulatory capacity of mmu-miR-155-5p on peritoneal macrophages, mmu-miR-155-5p mimics was transfected into mouse peritoneal macrophages. Compared with NC-transfected macrophages, the expression level of mmu-miR-155-5p was significantly upregulated in the mmu-miR-155-5p mimics-transfected peritoneal macrophages (p < 0.05, Figure 3A). In these mmu-miR-155-5p-transfected cells, it was found that the iNOS mRNA expression was upregulated (p < 0.05, Figure 3B). In agreement with the above result, compared with NC-transfected macrophages, NO secretion was increased in mmu-miR-155-5p mimic-transfected peritoneal macrophages (p < 0.05, Figure 3C). Moreover, the mRNA expression levels of eight genes in the TLR4 signaling pathway were also detected in mmu-miR-155-5p-transfected macrophages. Compared with NC-transfected macrophages, NF-κB was significantly upregulated, while CD14 was significantly downregulated in mmu-miR-155-5p mimics-transfected macrophages (Figure 3D). Moreover, transfection of mmu-miR-155-5p mimics increased IL-12a and IL-12b mRNA expression and decreased IL-6 mRNA expression (Figure 3E).
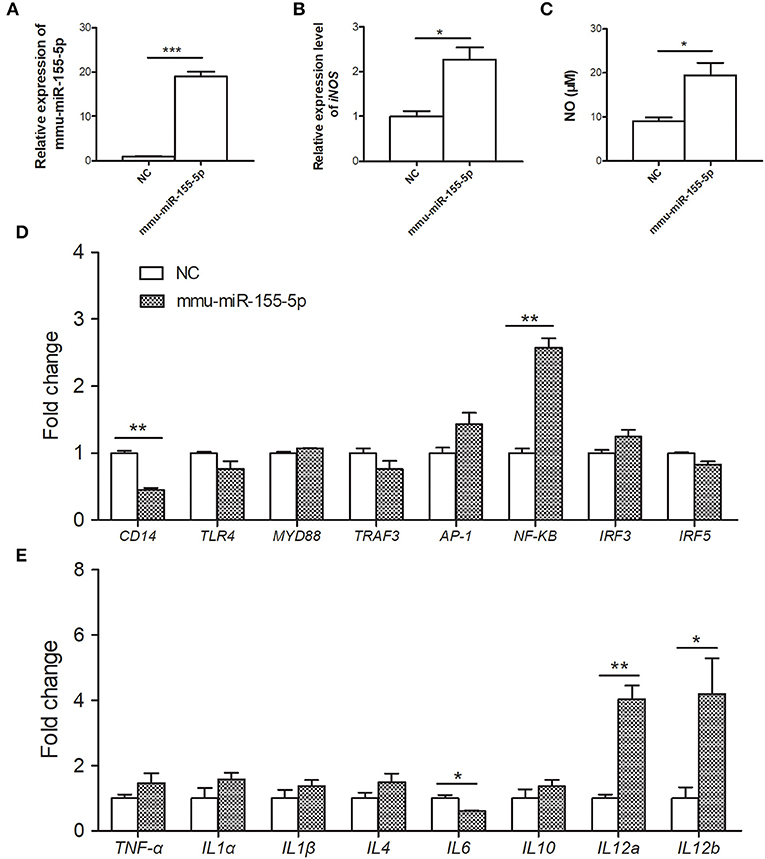
Figure 3. Immunoregulatory effects of mmu-miR-155-5p in peritoneal macrophage cells. (A) Validation of mmu-miR-155-5p expression in peritoneal macrophage cells transfected with mmu-miR-155-5p mimics. (B) The iNOS expression of peritoneal macrophages transfected with mmu-miR-155-5p mimics. (C) NO secretion was markedly increased in peritoneal macrophages transfected with mmu-miR-155-5p mimics. (D) The expression of eight genes of the TLR4 signaling pathway in mmu-miR-155-5p mimics-transfected macrophages. (E) The expression analysis of cytokines in macrophages over expressing mmu-miR-155-5p. Data for final statistical analysis were taken from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Validation of Predicted Target Genes of mmu-miR-155-5p
Six inflammation-related genes, including toll-like receptor 4/5 (TLR4/5), suppressor of cytokine signaling1 (SOCS1), inhibitor of kappaB kinase epsilon (IKBKE), and mitogen-activated protein kinase 8/10 (MAPK8/10), were selected as potential target genes of mmu-miR-155-5p. Among these candidates, only IKBKE gene was significantly down-regulated in mmu-miR-155-5p-transfected macrophages compared with NC-transfected macrophages (Figure 4A). IKBKE 3′UTR contained only one putative binding site that was located at the nucleotides 146-152 (Figure 4B). Luciferase reporting assay results showed mmu-miR-155-5p was able bind to the 3′ UTR of the WT-IKBKE gene and significantly decreased the luciferase activity in WT-IKBKE-transfected HEK293T cells compared to the NC-transfected cells (Figure 4C). The repression was drastically abolished in HEK293T cells co-transfected with mmu-miR-155-5p mimics plus Mut-IKBKE (Figure 4C), suggesting that mmu-miR-155-5p is able to bind to the IKBKE 3′-UTR.
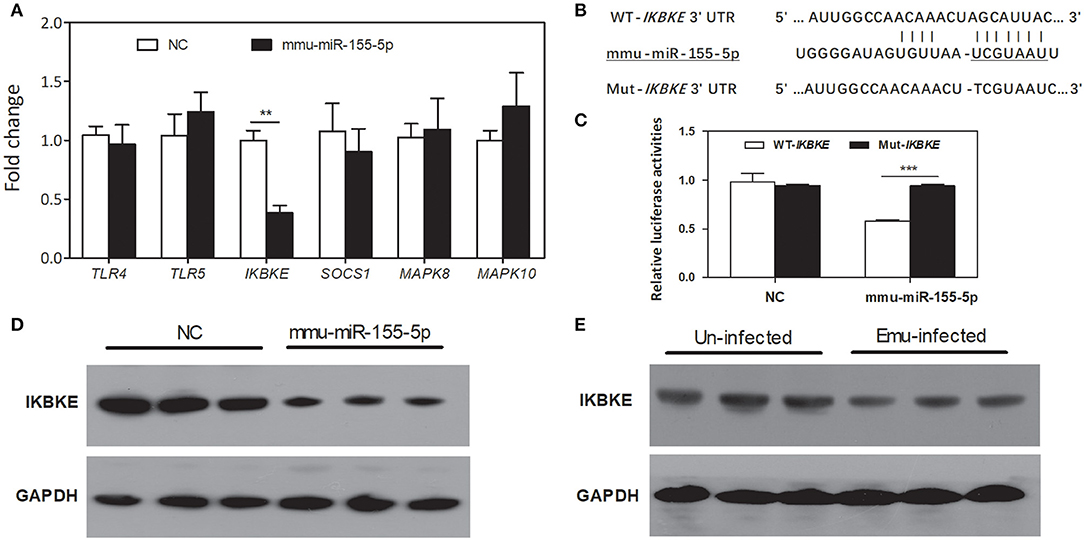
Figure 4. Prediction and validation of mmu-miR-155-5p target genes. (A) qPCR analysis of TLR4/5, SOCS1, IKBKE, and MAPK8/10 mRNA expression in mmu-miR-155-5p-transfected peritoneal macrophages. Data for the final analysis were obtained from 3 independent experiments. **p < 0.01. (B) Schematic representation of a putative binding site for mmu-miR-155-5p in the 3′UTR of IKBKE gene. (C) mmu-miR-155-5p binding to the IKBKE 3′UTR in vitro. WT-IKBKE or Mut- IKBKE (with mutations) construct was co-transfected into 293T cells with either negative control (NC) or mmu-miR-155-5p mimics. Luciferase activities were measured at 24 h after transfection. Data for the final analysis were obtained from 3 independent experiments. ***p < 0.001. (D) Western blotting analysis of IKBKE protein expression in mmu-miR-155-5p-transfected peritoneal macrophages. (E) Western blotting analysis of IKBKE protein expression in macrophages from E. multilocularis-infected mice.
To further test the effect of mmu-miR-155-5p on IKBKE protein expression in vitro, peritoneal macrophages were transfected by mmu-miR-155-5p mimics. Western blotting results showed that mmu-miR-155-5p mimics significantly downregulated the level of IKBKE protein compared to the negative control (Figure 4D). Furthermore, we found the expression of IKBKE was significantly downregulated in macrophages from E. multilocularis-infected mice (Figure 4E). Taken together, these results indicate that mmu-miR-155-5p can repress IKBKE expression by directly targeting its 3′-UTR.
Discussion
miRNAs are widely deemed as an important regulator of gene expression that plays a critical role in response to parasite infections (Arora et al., 2017). Increasing evidence has shown that the dysregulation of hosts miRNA expression is associated with the development of parasitic diseases, reflecting their key roles in host–pathogen interaction and immune regulation against infections (Manzanoromán and Sileslucas, 2012). miRNA profiling was mainly characterized in the liver or serum of E. multilocularis-infected mice (Jin et al., 2017), while the dynamics of miRNAs in peritoneal macrophages in E. multilocularis-infected mice remain unknown. In this study, we identified a larger member of miRNAs that were aberrantly expressed in mouse peritoneal macrophages in response to E. multilocularis infection. As expected, some of altered miRNAs (including miR-146a-5p, miR-155-5p, miR-21-5p, and miR-10b-5p) might be associated with immune responses (Sonkoly and Pivarcsi, 2009).
To date, increasing evidence has shown that miR-155 act as a regulator of inflammation and immune responses (Testa et al., 2017). For example, miR-155-5p is preferentially expressed in activated immune cells, such as monocytes and macrophages, and its expression is induced by type I interferons or LPS (Elton et al., 2013). Previous study showed that miR-155-5p was dramatically induced in bone marrow-derived macrophages in response to H. pylori infection (Elton et al., 2013). In addition, miR-155 was significantly upregulated in brains of Toxoplasma-infected mice during persistence infection (Cannella et al., 2014). We previously reported that the expression level of mmu-miR-155-5p was significantly upregulated in E. multilocularis-treated RAW264.7 macrophage cells (Guo and Zheng, 2017a). Similarly, we herein observed an increased expression of mmu-miR-155-5p in peritoneal macrophages during E. multilocularis infection. With the infection time prolonged, the expression of mmu-miR-155-5p was gradually increased.
miR-155-5p can regulate cytokine expression and NF-κB signaling pathway in a negative feedback loop (Yousefzadeh et al., 2015). Several studies showed that IKBKE was a major mediator of NF-κB signaling (Verhelst et al., 2013; Kim et al., 2014). In this study, we identified a potential mmu-miR-155-5p binding site in the 3′ UTR of the IKBKE. Moreover, we observed an inhibition of IKBKE expression in mmu-miR-155-5p mimics-transfected peritoneal macrophages. Interestingly, we also found that NF-κB were upregulated in mmu-miR-155-5p mimics-transfected macrophages, which is consistent with the previous finding (Zhu et al., 2016). It is well-known that NF-κB transcription factor is a major regulator of cytokine and chemokine gene transcription. We observed that upregulation of mmu-miR-155-5p could up-regulate the mRNA expression of IL-12a and IL-12b and downregulate the mRNA expression of IL-6. These above results suggest that mmu-miR-155-5p may modulate the expression of inflammatory cytokines possibly through the IKBKE/NF-κB signaling.
After stimulation with IFN-γ and LPS, macrophages are activated and can produce a large amount of NO (Subbanagounder et al., 2002). Excessive NO has cytotoxic effects, and it can kill intracellular bacteria, parasites and tumor cells (Oswald et al., 1994; Routes et al., 2005; Walch et al., 2015). We found that transfected mmu-miR-155-5p mimics led to an increase in NO secretion and iNOS mRNA expression. There is no a predictive binding site in the 3′ UTR of the iNOS gene, suggesting that mmu-miR-155-5p may indirectly modulate NO production. In macrophages, iNOS expression was induced via the NF-κB signaling (Zhang et al., 2012). In future studies, it is of interest to investigate the mechanism of mmu-miR-155-5p in NO production by macrophages.
Conclusions
In conclusion, our study provides rich and informative data on the miRNA expression profile in peritoneal macrophages of female BALB/c mice responding to E. multilocularis infection. Unveiling the functions of differentially expressed miRNAs in peritoneal macrophages will provide a solid foundation for an in-depth understanding of the interactions between E. multilocularis and its hosts.
Data Availability Statement
The datasets generated in this study have been deposited in the Bioproject database (accession: PRJNA609451).
Ethics Statement
Animal experiments in the study were performed at Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences and handled in accordance with good animal practice according to the Animal Ethics Procedures.
Author Contributions
XG performed the laboratory studies. XG and YZ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by grants from the National Natural Science Foundation of China (31702224, 31472185, and U1703104) and National Key Basic Research Program (973 program) of China (2015CB150300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Miss Jing yang, Yating Li, and Tianyan Lan for help in parasite infection and peritoneal macrophages isolation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00132/full#supplementary-material
Figure S1. GO molecular function annotations of the target genes of differentially expressed miRNAs. According to P value, top15 GO terms of biological process, molecular function, and cellular component were shown.
Figure S2. KEGG pathway analyses of the predicted target genes of differentially expressed miRNAs.
Table S1. Summary of the differentially expressed miRNAs in peritoneal macrophages in mice 30-day post infection.
Table S2. Summary of the differentially expressed miRNAs in peritoneal macrophages in mice 90-day post infection.
Abbreviations
NC, negative control; NO, nitric oxide; SOCS1, suppressor of cytokine signaling1; Ripk1, receptor (TNFRSF)-interacting serine-threonine kinase 1; IKBKE, inhibitor of kappaB kinase epsilon.
References
Arora, N., Tripathi, S., Singh, A. K., Mondal, P., Mishra, A., and Prasad, A. (2017). Micromanagement of immune system: role of miRNAs in helminthic infections. Front. Microbiol. 8:586. doi: 10.3389/fmicb.2017.00586
Britton, C. (2017). microRNAs-key players in host-parasite interactions. Parasite Immunol. 39:e12418. doi: 10.1111/pim.12418
Cannella, D., Brenier-Pinchart, M. P., Braun, L., van Rooyen, J. M., Bougdour, A., and Bastien, O. (2014). miR-146a and miR-155 delineate a microRNA fingerprint associated with toxoplasma persistence in the host brain. Cell Rep. 6, 928–937. doi: 10.1016/j.celrep.2014.02.002
Craig, P. S. (2006). Epidemiology of human alveolar echinococcosis in China. Parasitol. Int. 55, S221–S225. doi: 10.1016/j.parint.2005.11.034
Elton, T. S., Selemon, H., Elton, S. M., and Parinandi, N. L. (2013). Regulation of the MIR155 host gene in physiological and pathological processes. Gene 532, 1–12. doi: 10.1016/j.gene.2012.12.009
Federer, K., Armua-Fernandez, M. T., Hoby, S., Wenker, C., and Deplazes, P. (2015). In vivo viability of Echinococcus multilocularis eggs in a rodent model after different thermo-treatments. Exp. Parasitol. 154, 14–19. doi: 10.1016/j.exppara.2015.03.016
Guo, X., and Zheng, Y. (2017a). Expression profiling of circulating miRNAs in mouse serum in response to Echinococcus multilocularis infection. Parasitology 144, 1–8. doi: 10.1017/S0031182017000300
Guo, X., and Zheng, Y. (2017b). MicroRNA expression profile in RAW264Â∙7 macrophage cells exposed to Echinococcus multilocularis metacestodes. Parasitology 145, 1–8. doi: 10.1017/S0031182017001652
Han, H., Peng, J., Hong, Y., Zhang, M., Han, Y., Liu, D., et al. (2013). MicroRNA expression profile in different tissues of BALB/c mice in the early phase of Schistosoma japonicum infection. Mol. Biochem. Parasitol. 188, 1–9. doi: 10.1016/j.molbiopara.2013.02.001
Hansen, E. P., Kringel, H., Thamsborg, S. M., Jex, A., and Nejsum, P. (2016). Profiling circulating miRNAs in serum from pigs infected with the porcine whipworm, Trichuris suis. Vet. Parasitol. 223, 30–33. doi: 10.1016/j.vetpar.2016.03.025
He, X., Sai, X., Chen, C., Zhang, Y., Xu, X., Zhang, D., et al. (2013). Host serum miR-223 is a potential new biomarker for Schistosoma japonicum infection and the response to chemotherapy. Parasites Vect. 6, 272–272. doi: 10.1186/1756-3305-6-272
Hong, Y., Fu, Z., Cao, X., and Lin, J. (2017). Changes in microRNA expression in response to Schistosoma japonicum infection. Parasite Immunol. 39:e12416. doi: 10.1111/pim.12416
Hummon, A. B., Lim Difilippantonio, M. J., and Ried, T. (2007). Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 42, 467–470. doi: 10.2144/000112401
Jin, X., Guo, X., Zhu, D., Ayaz, M., and Zheng, Y. (2017). miRNA profiling in the mice in response to Echinococcus multilocularis infection. Acta Tropica 166, 39–44. doi: 10.1016/j.actatropica.2016.10.024
Kamiya, M., and Sato, H. (1990). Complete life cycle of the canid tapeworm, Echinococcus multilocularis, in laboratory rodents. Faseb J. Official Public. Federat. Am. Soc. Exp. Biol. 4:3334. doi: 10.1096/fasebj.4.15.2253847
Kim, D., Guo, J., Challa, S., Coppola, D., and Cheng, J. Q. (2014). Abstract 4416: IKBKE is a key mediator of Ras activation of NF-κB and Ras oncogenic function. Cancer Res. 74, 4416–4416. doi: 10.1158/1538-7445.AM2014-4416
Likun, W., Zhixing, F., Xi, W., Xiaowo, W., and Xuegong, Z. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Lointier, P., Beytout, J., and Dechelotte, P. (1990). Hepatic alveolar echinococcosis: diagnosis and treatment. Rev Prat. 40, 214–219.
Mackenstedt, U., Jenkins, D., and Romig, T. (2015). The role of wildlife in the transmission of parasitic zoonoses in peri-urban and urban areas. Int. J. Parasitol. Parasites Wildl. 4, 71–79. doi: 10.1016/j.ijppaw.2015.01.006
Malone, C., Brennecke, J., Czech, B., Aravin, A., and Hannon, G. J. (2012). Preparation of small RNA libraries for high-throughput sequencing. Cold Spring Harbor Protocols 2012, 1067–1077. doi: 10.1101/pdb.prot071431
Manzanoromán, R., and Sileslucas, M. (2012). MicroRNAs in parasitic diseases: potential for diagnosis and targeting. Mol. Biochem. Parasitol. 186, 81–86. doi: 10.1016/j.molbiopara.2012.10.001
O'Connell, R. M., Rao, D. S., Chaudhuri, A. A., and David, B. (2010). Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122. doi: 10.1038/nri2708
Oswald, I. P., Wynn, T. A., Sher, A., and James, S. L. (1994). NO as an effector molecule of parasite killing: modulation of its synthesis by cytokines. Compar. Biochem. Physiol. 108, 11–18. doi: 10.1016/1367-8280(94)90083-3
Reyes, J. L., and Terrazas, L. I. (2010). The divergent roles of alternatively activated macrophages in helminthic infections. Parasite Immunol. 29, 609–619. doi: 10.1111/j.1365-3024.2007.00973.x
Routes, J. M., Morris, K., Ellison, M. C., and Ryan, S. (2005). Macrophages kill human papillomavirus type 16 E6-expressing tumor cells by tumor necrosis factor alpha- and nitric oxide-dependent mechanisms. J. Virol. 79, 116–123. doi: 10.1128/JVI.79.1.116-123.2005
Sonkoly, E., and Pivarcsi, A. (2009). microRNAs in inflammation. Int. Rev. Immunol. 28, 535–561. doi: 10.3109/08830180903208303
Subbanagounder, G., Wong, J. W., Lee, H., Faull, K. F., Miller, E., Witztum, J. L., et al. (2002). Epoxyisoprostane and epoxycyclopentenone phospholipids regulate monocyte chemotactic protein-1 and interleukin-8 synthesis: formation of these oxidized phospholipids in response to interleukin-1β. J. Biol. Chem. 277, 7271–7281. doi: 10.1074/jbc.M107602200
Testa, U., Pelosi, E., Castelli, G., and Labbaye, C. (2017). miR-146 and miR-155: two key modulators of immune response and tumor development. Non-Coding RNA 3:22. doi: 10.3390/ncrna3030022
Verhelst, K., Verstrepen, L., Carpentier, I., and Beyaert, R. (2013). IκB kinase ε (IKKε): a therapeutic target in inflammation and cancer. Biochem. Pharmacol. 85, 873–880. doi: 10.1016/j.bcp.2013.01.007
Walch, M., Dotiwala, F., Mulik, S., Thiery, J., Kirchhausen, T., Clayberger, C., et al. (2015). Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell 161, 1229–1229. doi: 10.1016/j.cell.2015.05.021
Weiss, A. T., Bauer, C., and Köhler, K. (2010). Canine alveolar echinococcosis: morphology and inflammatory response. J. Compar. Pathol. 143, 233–238. doi: 10.1016/j.jcpa.2010.03.004
Yahiro, K., Satoh, M., Nakano, M., Hisatsune, J., Isomoto, H., Sap, J., et al. (2012). Low-density lipoprotein receptor-related protein-1 (LRP1) mediates autophagy and apoptosis caused by Helicobacter pylori VacA. J. Biol. Chem. 287, 31104–31115. doi: 10.1074/jbc.M112.387498
Yousefzadeh, N., Alipour, M. R., and Soufi, F. G. (2015). Deregulation of NF-κB-miR-146a negative feedback loop may be involved in the pathogenesis of diabetic neuropathy. J. Physiol. Biochem. 71, 1–8. doi: 10.1007/s13105-014-0378-4
Zhang, G., He, J. L., Xie, X. Y., and Yu, C. (2012). LPS-induced iNOS expression in N9 microglial cells is suppressed by geniposide via ERK, p38 and nuclear factor-κB signaling pathways. Int. J. Mol. Med. 30:561. doi: 10.3892/ijmm.2012.1030
Zheng, Y., Guo, X., He, W., Shao, Z., Zhang, X., Yang, J., et al. (2016). Effects of Echinococcus multilocularis miR-71 mimics on murine macrophage RAW264.7 cells. Int. Immunopharmacol. 34, 259–262. doi: 10.1016/j.intimp.2016.03.015
Zhou, R., Hu, G., Liu, J., Gong, A.-Y., Drescher, K. M., and Chen, X.-M. (2009). NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 5:e1000681. doi: 10.1371/journal.ppat.1000681
Keywords: Echinococcus multilocularis, peritoneal macrophages, microRNA, mmu-miR-155-5p, IKBKE
Citation: Guo X and Zheng Y (2020) Profiling of miRNAs in Mouse Peritoneal Macrophages Responding to Echinococcus multilocularis Infection. Front. Cell. Infect. Microbiol. 10:132. doi: 10.3389/fcimb.2020.00132
Received: 02 October 2019; Accepted: 12 March 2020;
Published: 03 April 2020.
Edited by:
Emily Derbyshire, Duke University, United StatesReviewed by:
Isabel Mauricio, New University of Lisbon, PortugalCésar López-Camarillo, Universidad Autónoma de la Ciudad de México, Mexico
Copyright © 2020 Guo and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaola Guo, guoxiaola@caas.cn; Yadong Zheng, zhengyadong@caas.cn
 Xiaola Guo
Xiaola Guo Yadong Zheng
Yadong Zheng