Iron and Virulence in Stenotrophomonas Maltophilia: All We Know So Far
- Department of Medical Microbiology and Parasitology, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Selangor Darul Ehsan, Malaysia
Stenotrophomonas maltophilia is a multi-drug-resistant global opportunistic nosocomial pathogen, which possesses a huge number of virulence factors and antibiotics resistance characteristics. Iron has a crucial contribution toward growth and development, cell growth and proliferation, and pathogenicity. The bacterium found to acquire iron for its cellular process through the expression of two iron acquisition systems. Two distinct pathways for iron acquisition are encoded by the S. maltophilia genome-a siderophore-and heme-mediated iron uptake system. The entAFDBEC operon directs the production of the enterobactin siderophore of catecholate in nature, while heme uptake relies on hgbBC and potentially hmuRSTUV operon. Fur and sigma factors are regulators of S. maltophilia under iron-limited condition. Iron potentially act as a signal which plays an important role in biofilm formation, extracellular polymeric substances (EPS), extracellular enzymes production, oxidative stress response, diffusible signal factor (DSF) and siderophore production in S. maltophilia. This review summarizes the current knowledge of iron acquisition in S. maltophilia and the critical role of iron in relation to its pathogenicity.
Introduction
Stenotrophomonas maltophilia is a Gram-negative, Gammaproteobacteria, that is present ubiquitously in the environment; particularly in the soil and plants rhizospheres (Alavi et al., 2014). Therefore, S. maltophilia has many attributes that could be applied in different biotechnological processes such as bioremediation, phytoremediation, degradation of an organic compound, biocontrol activity and many more (Antonioli et al., 2007; Pages et al., 2008; Mukherjee and Roy, 2016). Despite its biotechnological applications, the bacterium was recently reviewed to gain access into the clinical settings, thus recognized as an important multi-drug-resistant global opportunistic nosocomial pathogen (Brooke et al., 2017). Stenotrophomonas maltophilia is responsible for causing various infections ranging from bacteremia, endocarditis, pneumonia, meningitis, ocular infections, urinary tract infection, enteritis, and skin/soft tissue infections (Senol, 2004; Abbott et al., 2011). A debatable question regarding “S. maltophilia is a colonizer or a pathogenic culprit?” still remains due to the failure in distinguishing colonization and acquired infections, as the microorganism poses a limited pathogenic potential in causing illness in healthy hosts (Neela, 2014; Norton and Dachs, 2015).
The invading pathogen must be able to produce various virulence factors in order to establish infections and this largely depends on environmental conditions and level of micronutrients within the hostile environment (Sritharan, 2006). In such circumstances, S. maltophilia is known to exhibit its pathogenicity through: (1) pili/flagella/fimbrial/adhesins which contributes to adherence, auto-aggregation, colonization of biotic and abiotic surfaces; (2) outer membrane lipopolysaccharide (LPS) plays a role in biofilm formation and resistance to antibiotic as well as complement-mediated cell killing; (3) diffusible signal factor (DSF) plays a huge role in quorum sensing, which in turn mediate motility, extracellular enzymes production, LPS synthesis, microcolony formation, and tolerance toward antibiotics and heavy metal ions; and (4) extracellular enzymes production such as proteases, lipases, esterase, DNase, RNase, and fibrinolysin (Looney, 2005; Abbott et al., 2011; Brooke, 2012).
In general, most of the bacteria can acquire all of the nutrients such as nitrogen, amino acids, nucleotides, phosphates and other inorganic ions for its survival, except for iron as it is not freely available from the host tissue (Ratledge and Dover, 2000). In order to counteract the difficulty to fulfill the iron requirement, the bacteria have evolved numerous mechanisms; particularly by demonstrating efficient iron acquisition systems under iron-limited conditions (Andrews et al., 2003; Thomas and Wigneshweraraj, 2014; Kalidasan et al., 2018b). This phenomenon is not an exception for S. maltophilia, as iron was found to plays a crucial role in the regulation of its virulence activities (García et al., 2015). At this juncture, we highlight the iron acquisition strategies in S. maltophilia focusing on the siderophore- and heme-mediated systems; together describing the regulator involved in iron homeostasis and metabolism. The expression of virulence factors in relation to iron availability in S. maltophilia, is discussed extensively in this review.
Iron Acquisition Systems in S. maltophilia:
Little is known about iron uptake systems in S. maltophilia (Huang and Lee Wong, 2007). However, the iron acquisition strategies in other Gram-negative bacteria have been extensively studied previously (Braun and Hantke, 2013; Runyen-Janecky, 2013). In general, the iron uptake systems in Gram-negative bacteria can be mediated by: (1) transferrin (Tf) or lactoferrin (Lf); (2) heme (Hm) and hemoglobin (Hb); (3) siderophores; and (4) ferrous iron (Fe2+) (Marx, 2002). The bacteria depends on high-affinity surface receptor proteins that potentially bind with ferric iron (Fe3+) loaded to siderophores or heme, and followed by subsequent delivery into the periplasmic space by the TonB–ExbB-ExbD complex (Faraldo-Gómez and Sansom, 2003). The periplasmic-binding proteins and ATP transporters available at the cytoplasmic membrane are used to ensure further transport into the cell. On the other hand, Hm can be obtained from Hb and hemoglobin-haptoglobin (Hb-Hpt) complex by outer membrane proteins (OMPs). Apart from that, some Gram-negative bacteria can utilize Fe3+ bound to transferrin and lactoferrin at the outer membrane, and transported into the cell. Under anaerobic conditions, soluble Fe2+ can diffuse across outer membrane porins, and is subsequently imported by FeoABC system. A model for iron uptake in S. maltophilia can reasonably be proposed based on previous studies (Adamek et al., 2014; Nas and Cianciotto, 2017; Kalidasan et al., 2018a) as shown in Figure 1.
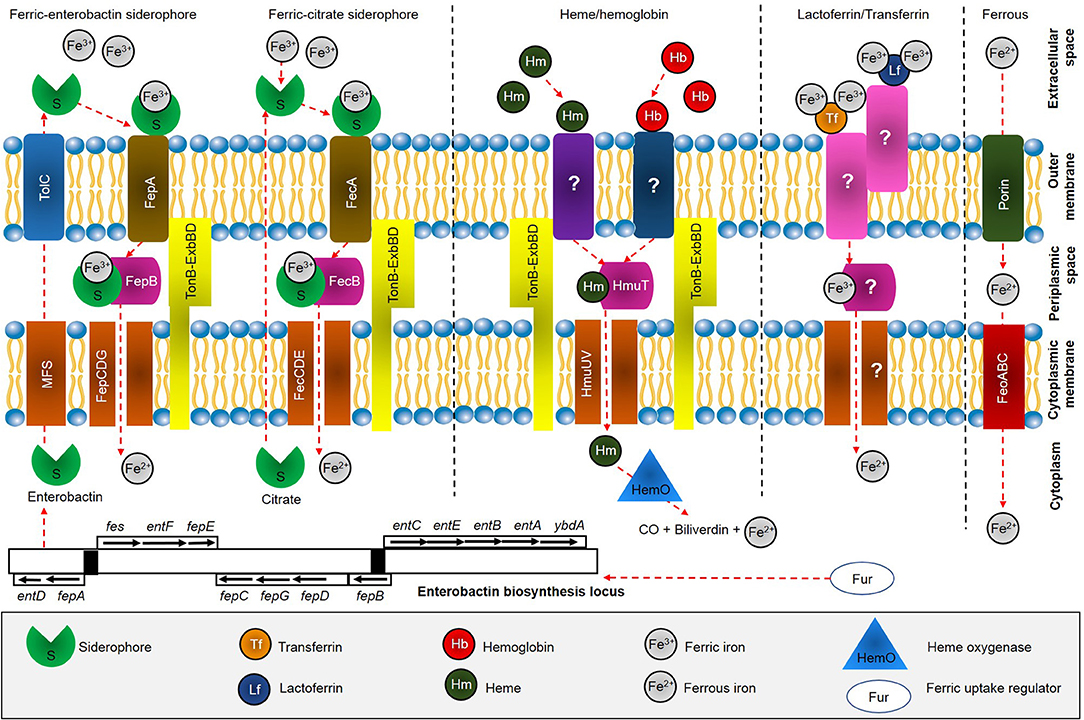
Figure 1. Overview of iron acquisition systems in S. maltophilia. After biosynthesis, siderophore enterobactin is effluxes from cytoplasm through major facilitator superfamily (MFS) protein and further into the extracellular space by outer membrane factor TolC. Enterobactin scavenges free Fe3+ available at the extracellular space and is subsequently recognized and taken up through FepA, which is energized by the TonB-ExbBD machinery. FepB delivers ferric enterobactin from the periplasm by FepCDG transporter into the cytoplasm. On the other hand, ferric citrate is recognized by FecA and further delivered into periplasmic by FecB and transported across the cytoplasm by FecCDE transporter. Heme acquisition is predicted to be taken through receptor at the outer membrane, followed by HmuTUV system. Uptake of iron bound to transferrin and lactoferrin have not been fully identified (marked ?), while Feo system involved in uptake of ferrous iron through action of FeoABC.
Although S. maltophilia was previously reported to uptake iron through pseudobactin (Jurkevitch et al., 1992), a siderophore produced by Pseudomonas strain B10 (Teintze et al., 1981), it was not clear whether the bacterium is capable of producing its own siderophores (Kumar and Audipudi, 2015). Furthermore, the gene(s) responsible for iron acquisition through siderophores is still a question (Adamek et al., 2014). In the study, S. maltophilia isolates K279a and SKK35 (clinical strains), R551-3 (environmental strain), SKA14 (seawater strain), and RA8 (wastewater strain) were found to harbor genes entACF encoding for enterobactin synthetase, that catalyzes the biosynthesis of enterobactin siderophore. However, the siderophore production that can only function in combination with other genes should be interpreted in the context of presence of those other genes; i.e., incomplete gene sets (entBDE) for biosynthesis of enterobactin in S. maltophilia. A recent study revealed the presence of eight loci in S. maltophilia K279a, which are predicted to encode a system for siderophore production, as shown in Figure 1 (Nas and Cianciotto, 2017). The first locus had six open-reading-frames (ORFs) needed to make enterobactin including, EntA, EntF, EntD, EntB, EntE, and EntC, with addition of major superfamily (MFS) membrane transport protein. The second locus encodes TolC which mediates siderophore export across the outer membrane, while the third locus encodes enterobactin receptor FepA. The periplasmic-spanning complex TonB, ExbB, and ExbD proteins were encoded by locus four, five and six, respectively. The seventh locus encodes proteins with similarity to FepC, FepD, and FepG, while the last locus encodes YgiH and ViuB which assist the release of iron from other siderophores. The study concluded S. maltophilia produces an EntC-dependent catecholate siderophore that is distinct from enterobactin, as the siderophore appeared to have a modification at position-3 and/or position-4 in the catecholate structure. The claim was achieved through numerous investigations, such as inability of K279a supernatants to restore growth of Salmonella typhimurium enterobactin-indicator strain (TA2700) on a low-iron medium; ability of K279a siderophore extraction into ethyl acetate but not butanol and dichloromethane; inability of K279a siderophore to migrate as far as enterobactin in thin-layer chromatography (TLC) indicating it is more polar than enterobactin; and a mixture of enterobactin and its monomer did not stimulate the growth of K279a or its entC mutant and fepA mutant derivatives.
Furthermore, mass spectrometry analysis in S. maltophilia K279a identified SMLT_RS06850 and SMLT_RS19685 encoding for outer membrane receptor FepA and TonB-dependent receptor respectively (García et al., 2015). A BLAST identity revealed SMLT_RS06850 displays similarity of 66% to Xanthomonas citri, while SMLT_RS19685 was found 55% similarity with Pseudomonas putida. In short, genomic investigations suggested S. maltophilia potentially secrete catecholate siderophore and depending on entABCDEF operon for production of distinct enterobactin. On the other hand, plant-associated strains S. maltophilia R551-3 and Stenotrophomonas rhizophila DSM14405 were found to harbor iron uptake locus fcuA and fhuA encoding for ferrichrome receptor proteins, which code for siderophore receptors and the outer membrane adhesin-like gene, respectively (Alavi et al., 2014). It is worthy to noted that, the structure and mechanisms of the outer membrane transporter of enterobactin (fepA), is closely similar to that of FhuA (Marx, 2002).
Siderophores are small molecules and considered to be an important virulence factor, particularly in pathogens that encode multiple siderophores (Holden and Bachman, 2015; Behnsen and Raffatellu, 2016). Any pathogenic strains that are capable of over-producing siderophores are considered to be hypervirulent, whereas strains unable to secrete siderophores have decreased virulence and fitness during infection and colonization. As a far concern, siderophore production in S. maltophilia has been well studied in recent years. Siderophore production among S. maltophilia in the rhizosphere of oilseed rape, showed all isolates investigated were positive for siderophore activity, ranging from 5 to 20 mm orange zone on CAS agar (Berg et al., 1996). In contrast, S. maltophilia strain W81 did not produce prominent fluorescent siderophores (Dunne et al., 1997). The variation in siderophore production, particularly among environmental isolates were also observed in our study (Kalidasan et al., 2018a). We noted the environmental strains did not produce siderophores or produced very minimal amounts compared to clinical isolates investigated. We also observe the percentage of siderophore production investigated through liquid CAS, showed clinical isolate produced a greater amount of siderophore (30.8%) compared to environmental isolate (4%).
Furthermore, an analysis of 50 isolates comprised of clinical and environmental strains was reported to produce minimum siderophore activity, ranging from 5 to 3 mm orange zone on CAS agar (Minkwitz and Berg, 2001). On the hand, analysis of all 32 clinical isolates of S. maltophilia showed siderophore activity ranging from 4.5 to 11 mm orange zone on modified CAS agar and secretion of catechol-type siderophores (Garcia et al., 2012). Similarly, both clinical and environmental isolates produced catechol-type enterobactin (Ryan et al., 2009), also supported by cross-feeding assay in S. maltophilia (Mokracka et al., 2011). Aforementioned, S. maltophilia secretes catecholate siderophore that appears to be novel in structure, rather than enterobactin (or salmochelin) (Nas and Cianciotto, 2017). Although most of the studies reported S. maltphilia is a catecholate-type siderophore producer, a contrary investigation showed S. maltophilia clinical isolates were a hydroxamate-type ornibactin producer, as the study lack estimation of catecholate derivatives (Chhibber et al., 2008). Ornibactin was reported being produced by Burkholderia cepacia complex (BCC) (Sokol et al., 2000; Visser et al., 2004), and such production is possible as S. maltophilia and BCC are a closely related group of non-fermenting gram-negative bacilli (NFGNBs) (Gautam et al., 2009). However, further investigations are required to confirm whether S. maltophilia potentially secretes hydroxamate siderophores under iron limitation.
Even though hemoproteins serve as an iron source for many pathogenic bacteria, heme-acquisition among S. maltophilia has not been fully understood yet. S. maltophilia isolates were found to harbor gene hgbBC encoding hemoglobin binding protein, which suggests potential heme and hemoglobin uptake capability as iron sources (Adamek et al., 2014). However, our previous genotypic and phenotypic investigation identified numerous heme-mediated acquisition system in S. maltophilia including: (1) heme oxygenase, associated with heme uptake (HemO/HO); (2) heme ABC transporter, ATPase component (HmuV); (3) hypothetical protein related to heme utilization (Hyp1); (4) heme ABC transporter, permease protein (HmuU); (5) heme ABC transporter, cell surface heme and hemoprotein receptor (HmuT); (6) hemin uptake protein (Hup); and (7) hemin transport protein (Htp) (Kalidasan et al., 2018a). Furthermore, the growth of clinical (SM77) and environmental (LMG10879) isolates was stimulated with Hb and Tf supplementation, while hemin and Lf having less effect in enhancing the growth of the tested isolates. These findings merit further investigations, to decipher how S. maltophilia could potentially uptake heme and hemin as iron sources, especially when it is associated with bloodstream infection in human host.
Regulator of Iron Acquisition in S. maltophilia
In most Gram-negative bacteria, iron homeostasis, metabolism, and virulence is regulated by the ferric uptake regulator protein (Fur), which potentially represses transcription upon interaction with Fe2+ or causes de-repression in the absence of Fe2+ (Andrews et al., 2003; Troxell and Hassan, 2013). Till date, only study by García et al. (2015) was the first to provide data about the role of iron as a signal, likely through the Fur system in S. maltophilia. The study identified 20 putative Fur boxes using MAST tool. However, it is important to note that, there is no evidence of Fur direct regulation, as the study did not demonstrate the binding of the regulator to the promoters of its putative target genes, either by electrophoretic mobility shift assay (EMSA) or DNase footprinting assay. Moreover, our study has only identified the presence of Fur in clinical and environmental isolates of S. maltophilia through PCR and significant upregulation of Fur under iron-depleted than under iron-replete conditions, suggesting de-repression of Fur (Kalidasan et al., 2018a). In support of these, regulation of iron uptake system in S. maltophilia through Fur was reported in RegPrecise 4.0 database (http://regprecise.lbl.gov/) (Novichkov et al., 2013). The database predicted 17 operons and 39 genes influenced by iron that cater to the pathway for iron homeostasis in S. maltophilia strain K279a as shown in Table 1. It is important to mention that, RegPrecise was constructed and manually curated by utilizing the comparative genomic approach, suggesting further analysis and validation. In spite of, our bioinformatics validation revealed, the regulon showed similarities with P. aeruginosa strain E15_London_28_01_14, which suggests S. maltophilia is closely related to the Pseudomonas species (Calza et al., 2003).
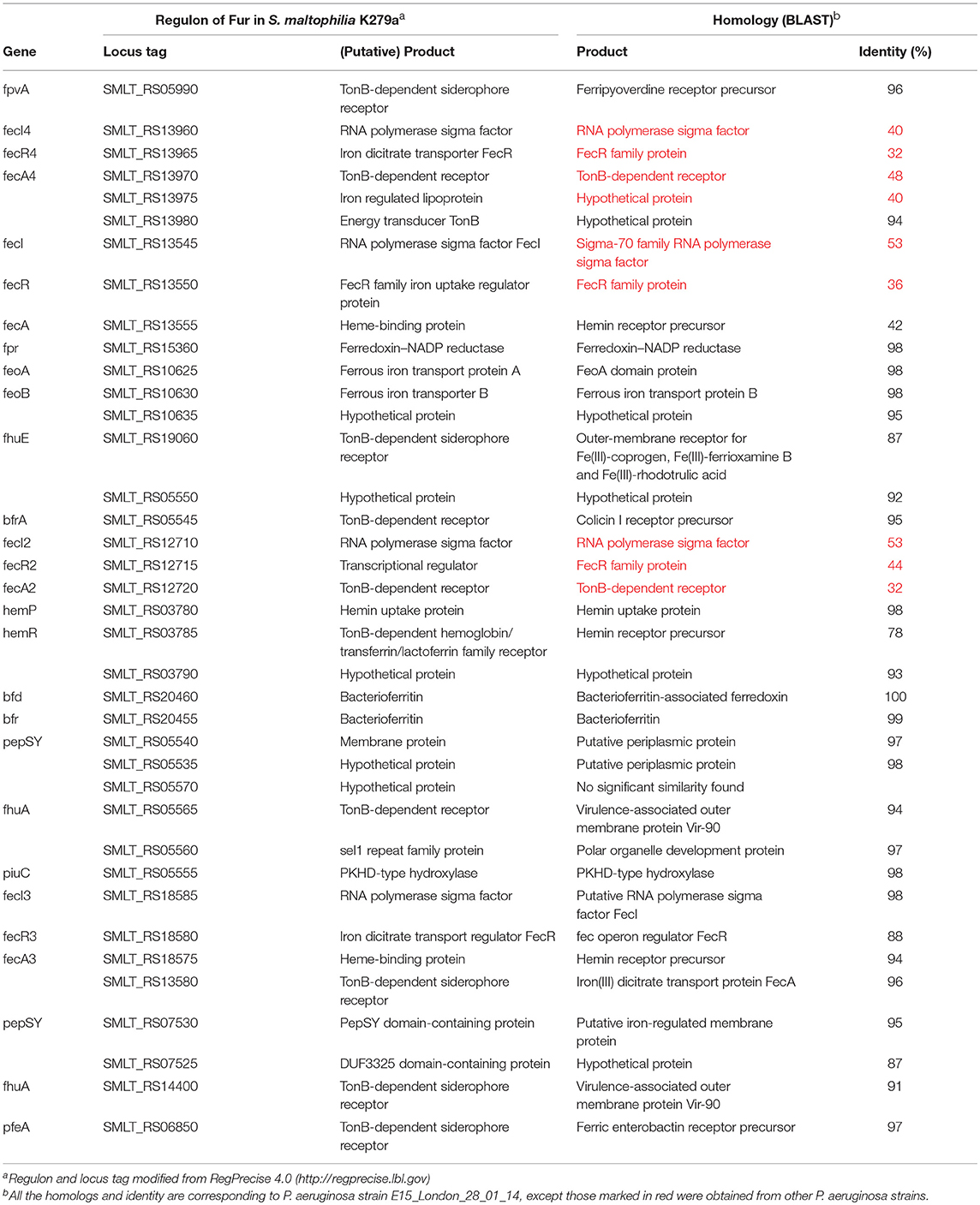
Table 1. Comparison of regulon of Fur in S. maltophilia strain K279a with P. aeruginosa strain E15_London_28_01_14.
Under anaerobic conditions or at low pH, Fe2+ is more abundant and in most bacteria, Feo system is dedicated to transport such iron source into the cell (Lau et al., 2015). The Feo system comprised of mainly of FeoA and FeoB proteins, in which FeoA directs to the inner leaflef of the cytoplasmic membrane, where it could possibly interact with FeoB. In S. maltophilia, the structure of FeoA adopted Src Homology 3 domain (SH3 domain) fold, containing five antiparallel β-strands, additional α-helices at the N-terminal site, RT loop, and C-terminal β-strand (Su et al., 2010). This novel FeoA forms a unique dimer cross-linked by two zinc ions, which was coordinated by His21 in the RT loop of a molecule and Glu52 in the n-Src loop of another molecule. The center of the RT loop was predicted to be favorable for interacting with metal ions. The study also proposed that FeoA may interact with FeoB between the SH3b domain and G-protein domain in order to regulate FeoB-dependent ferrous iron uptake activity as an activating factor. This SH3 domain have been predicted to act as the targeting domains involved in bacterial cell wall recognition and binding as well as involved in metal-binding (Kamitori and Yoshida, 2015).
A recent investigation using MALDI-TOF fingerprinting found that S. maltophilia strain OK-5 harbored anti-FecI sigma factor (FecR) (Lee et al., 2017). On the other hand, a study identified a homolog of the ferric citrate receptor (FecA) in S. maltophilia strain WR-C (Huang and Lee Wong, 2007). Interestingly, the study found that unlike other Gram-negative bacteria such as Escherichia coli the fecIR regulatory genes are not located upstream of fecA. This suggest that the ferric citrate transport system in S. maltophilia may be regulated differently or the location of the regulators could be somewhere else. Our sequencing results revealed the “iron siderophore sensor protein (FeSS)” is corresponding to “iron dicitrate transport regulator FecR” (SMLT_RS18580) and “sigma factor ECF subfamily” is corresponding to “RNA polymerase sigma factor” (SMLT_RS12950) in strain K279a (Kalidasan et al., 2018a). Overall, iron regulation in S. maltophilia is potentially depended on Fur and sigma factors. However, it is essential to validate using expression profiles of regulatory knockout mutants or any other suitable approaches, to decipher on how these regulators directly control iron acquisition strategies in S. maltophila.
Iron Uptake and Pathogenesis of S. maltophilia
Numerous studies have been reported on virulence properties, specifically investigating biofilm formation in S. maltophilia under normal nutritional status (Crossman and Dow, 2004; Huang et al., 2006; Passerini de Rossi et al., 2007; Pompilio et al., 2008, 2011; Biočanin et al., 2017; Liu et al., 2017; An and Tang, 2018). However, the correlation between iron and expression of virulence profiles among S. maltophilia has not been discussed extensively. Iron limitation was found to stimulate biofilm and extracellular polymeric substances (EPS) formation in S. maltophilia, resulting in less reactive oxygen species (ROS) production. Moreover, the study reported iron negatively regulates DSF production through Fur interaction and proved the expression of two iron-repressed OMPs (IROMPs), FepA, and TonB-dependent siderophore receptor. The killing assay using Galleria mellonella infection model showed spontaneous fur mutant was more virulent compared to wild-type (wt) strain S. maltophilia K279a. This contradicts with another study which revealed that iron repletion neither inhibits nor increases biofilm formation by S. maltophilia strain X26332 (Martinez et al., 2010). Such discrepancy in biofilm formation does not associate either with the phylogenetic connection or with the origin of isolates of S. maltophilia (Steinmann et al., 2018).
A study revealed that production of extracellular protease and chitinase by environmental S. maltophilia strain W81, were not altered even when the iron level was increased (Dunne et al., 1997). This showed the expression of extracellular enzyme among environmental strains are not affected by iron availability, due to the fact that soil contains a high amount of iron that are insoluble and not bioavailable (Berg et al., 1999). A similar trend can be observed in our study, whereby the environmental isolates did not show any significant differential expression for the iron acquisition targets when grown under both iron-depleted and iron-repleted conditions (Kalidasan et al., 2018a). It is important to note that, the amount of siderophore production and the strategies by which plants and microorganisms obtain iron from different sources, is likely to be highly variable under different environmental conditions or seasonally influenced by changes in carbon inputs into the rhizosphere during plant growth (Crowley, 2006). On the other hand, S. maltophilia was found to secrete hemolysin (Hly) (Garcia et al., 2002; Travassos et al., 2004; Thomas et al., 2014) which is important in the lysis of erythrocytes, thereby promoting the release of heme as iron sources for cellular growth (Runyen-Janecky, 2013). The hemolysin activity of Hly positive S. maltophila strains was inhibited with supplementation of ferric chloride (FeCl3) and the hemolytic activities were found similar to those of Aeromonas caviae and Plesiomonas shigelloides (Figueiredo et al., 2006). Furthermore, the study showed hemolysin production to be stimulated by Ca2+ ions but inhibited by EDTA, and in an overall modulated by iron. This finding suggests that synthesis of hemolysin is found to be iron regulated in most Gram-negative bacteria (Kim et al., 2009).
Under low iron level, it was found that regulation of pathogenic f actors (rpf) cluster, rpfF, and rpfB in S. maltophilia strain WR-C are activated to synthesize DSFs, which stimulates iron uptake by FecA (Huang and Lee Wong, 2007). However, the study found that DSF has no effect on biofilm formation and synthesis of LPS, similarly reported in Xanthomonas campestris (Torres et al., 2007). Protease production and hemolytic activity in S. maltophilia were not modulated by DSF, but controlled by cyclic AMP (cAMP) receptor protein (CRP) (Kim et al., 2013). CRP responds to environmental changes, such as iron and glucose levels, and binds to the predicted CRP binding site upstream of rpfF, activating the rpf system. Moreover, rpfF was shown to affect siderophore production in Xanthomonas oryzae pv. oryzae, whereby the rpfF mutant strains were found unable to survive under low iron concentration (Chatterjee and Sonti, 2002). The FeoA family protein was found positively regulated by DSF in S. maltophilia R551-3, which plays important role in Feo system (Alavi et al., 2013). In short, the rpf and/or DSF system are involved in regulating various functional activities in X. campestris pv. campestris, including modulating iron uptake TonB-dependent proteins encoded by tonB, bfeA, fepA, cirA, fyuA, iroN, while exbB, exbD1, exbD2, Xcc3216 are important for accessory proteins production (He et al., 2006).
Conclusion and Future Directions
This review is important for understanding the mechanisms behind iron acquisition in S. maltophilia, it is, to our knowledge, the first of its kind to describe how S. maltophilia efficiently support its lifestyle as multi-drug-resistant global opportunistic nosocomial pathogen under iron availability. S. maltophilia potentially express three iron acquisition pathways which include, siderophore- and heme-mediated and Feo system under iron-limited condition. We regarded S. maltophilia as the “innocent culprit” as its represent potential benefits for biotechnological applications and simultaneously found to be associated with human and plant host. Iron was found to a crucial micronutrient for expression of various virulence profiles in S. maltophilia. Elaboration of these virulence factors may have clinical significance to the human host, especially among patient with immunocompromised conditions, increasing the difficulty in therapeutic approaches. In order to decipher complete iron acquisition systems in S. maltophilia, knockout mutants should be considered to understand the roles of differentially expressed targets during iron limitation. The effect of iron limitation on the proteome of S. maltophilia and mechanisms of Fur regulation are also interesting questions for future investigations.
Author Contributions
VK performs the literature search and wrote the manuscript. NJ proofreads the manuscript. SK, RA, and VN outline the idea and approve the final manuscript.
Funding
This work was supported by Ministry of Higher Education, Malaysia through Fundamental Research Grant Scheme [04-01-14-53FR] and Universiti Putra Malaysia through Geran Putra—Inisiatif Putra Siswazah (IPS) [GP-IPS/2016/9478200].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbott, I. J., Slavin, M. A., Turnidge, J. D., Thursky, K. A., and Worth, L. J. (2011). Stenotrophomonas maltophilia: emerging disease patterns and challenges for treatment. Expert Rev. Anti. Infect. Ther. 9, 471–488. doi: 10.1586/eri.11.24
Adamek, M., Linke, B., and Schwartz, T. (2014). Virulence genes in clinical and environmental Stenotrophomas maltophilia isolates: a genome sequencing and gene expression approach. Microb. Pathog. 67–68, 20–30. doi: 10.1016/j.micpath.2014.02.001
Alavi, P., Müller, H., Cardinale, M., Zachow, C., Sánchez, M. B., Martínez, J. L., et al. (2013). The DSF quorum sensing system controls the positive influence of Stenotrophomonas maltophilia on plants. PLoS ONE 8:e67103. doi: 10.1371/journal.pone.0067103
Alavi, P., Starcher, M. R., Thallinger, G. G., Zachow, C., Müller, H., and Berg, G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics 15:482. doi: 10.1186/1471-2164-15-482
An, S. Q., and Tang, J. L. (2018). The Ax21 protein influences virulence and biofilm formation in Stenotrophomonas maltophilia. Arch. Microbiol. 200, 183–187. doi: 10.1007/s00203-017-1433-7
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Antonioli, P., Lampis, S., Chesini, I., Vallini, G., Rinalducci, S., Zolla, L., et al. (2007). Stenotrophomonas maltophilia SeITE02, a new bacterial strain suitable for bioremediation of selenite-contaminated environmental matrices. Appl. Environ. Microbiol. 73, 6854–6863. doi: 10.1128/AEM.00957-07
Behnsen, J., and Raffatellu, M. (2016). Siderophores: more than stealing iron. MBio 7:e01906–16. doi: 10.1128/mBio.01906-16
Berg, G., Marten, P., and Ballin, G. (1996). Stenotrophomonas maltophilia in the rhizosphere of oilseed rape — occurrence, characterization and interaction with phytopathogenic fungi. Microbiol. Res. 151, 19–27. doi: 10.1016/S0944-5013(96)80051-6
Berg, G., Roskot, N., and Smalla, K. (1999). Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 37, 3594–3600.
Biočanin, M., Madi, H., Vasiljević, Z., Kojić, M., Jovčić, B., and Lozo, J. (2017). Temperature, pH and trimethoprim-sulfamethoxazole are potent inhibitors of biofilm formation by Stenotrophomonas maltophilia clinical isolates. Polish, J. Microbiol. 66, 433–438. doi: 10.5604/01.3001.0010.6996
Braun, V., and Hantke, K. (2013). “The tricky ways bacteria cope with iron limitation,” in Iron Uptake in Bacteria with Emphasis on E. coli and Pseudomonas, eds R. Chakraborty, V. Braun, K. Hantke, and P. Cornelis (New York, NY; London: Springer), 31–66.
Brooke, J. S. (2012). Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25, 2–41. doi: 10.1128/CMR.00019-11
Brooke, J. S., Di Bonaventura, G., Berg, G., and Martinez, J.-L. (2017). Editorial: a multidisciplinary look at Stenotrophomonas maltophilia: an emerging multi-drug-resistant global opportunistic pathogen. Front. Microbiol. 8:1511. doi: 10.3389/fmicb.2017.01511
Calza, L., Manfredi, R., and Chiodo, F. (2003). Stenotrophomonas (Xanthomonas) maltophilia as an emerging opportunistic pathogen in association with HIV infection: a 10-year surveillance study. Infection 31, 155–161.
Chatterjee, S., and Sonti, R. V. (2002). rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant Microbe Interact. 15, 463–471. doi: 10.1094/MPMI.2002.15.5.463
Chhibber, S., Gupta, A., Sharan, R., Gautam, V., and Ray, P. (2008). Putative virulence characteristics of Stenotrophomonas maltophilia: a study on clinical isolates. World J. Microbiol. Biotechnol. 24, 2819–2825. doi: 10.1007/s11274-008-9812-5
Crossman, L., and Dow, J. M. (2004). Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 6, 623–629. doi: 10.1016/j.micinf.2004.01.013
Crowley, D. E. (2006). “Microbial siderophores in the plant rhizosphere,” in Iron Nutrition in Plants and Rhizospheric Microorganisms, eds L. L. Barton and J. Abadia (Dordrecht: Springer), 169–198.
Dunne, C., Crowley, J. J., Moenne-Loccoz, Y., Dowling, D. N., De Bruijn, F. J., and O'Gara, F. (1997). Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 143, 3921–3931.
Faraldo-Gómez, J. D., and Sansom, M. S. (2003). Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116. doi: 10.1038/nrm1015
Figueiredo, P. M. S., Furumura, M. T., Santos, A. M., Sousa, A. C. T., Kota, D. J., Levy, C. E., et al. (2006). Cytotoxic activity of clinical Stenotrophomonas maltophilia. Lett. Appl. Microbiol. 43, 443–449. doi: 10.1111/j.1472-765X.2006.01965.x
García, C. A., Alcaraz, E. S., Franco, M. A., and De Rossi, B. N. (2015). Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front. Microbiol. 6:926. doi: 10.3389/fmicb.2015.00926
Garcia, C. A., Passerini De Rossi, B., Alcaraz, E., Vay, C., and Franco, M. (2012). Siderophores of Stenotrophomonas maltophilia: detection and determination of their chemical nature. Rev. Argent. Microbiol. 44, 150–154.
Garcia, D. D. O., Timenetsky, J., Martinez, M. B., Francisco, W., Sinto, S. I., and Yanaguita, R. M. (2002). Proteases (caseinase and elastase), hemolysins, adhesion and susceptibility to antimicrobials of Stenotrophomonas maltophilia isolates obtained from clinical specimens. Braz. J. Microbiol. 33, 157–162. doi: 10.1590/S1517-83822002000200012
Gautam, V., Ray, P., Vandamme, P., Chatterjee, S. S., Das, A., Sharma, K., et al. (2009). Identification of lysine positive non-fermenting gram negative bacilli (Stenotrophomonas maltophilia and Burkholderia cepacia complex). Indian J. Med. Microbiol. 27, 128–133. doi: 10.4103/0255-0857.49425
He, Y. W., Xu, M., Lin, K., Ng, Y. J. A., Wen, C. M., Wang, L. H., et al. (2006). Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol. Microbiol. 59, 610–622. doi: 10.1111/j.1365-2958.2005.04961.x
Holden, V. I., and Bachman, M. A. (2015). Diverging roles of bacterial siderophores during infection. Metallomics 7, 986–995. doi: 10.1039/C4MT00333K
Huang, T., Somers, E. B., and Wong, A. C. L. (2006). Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol. 188, 3116–3120. doi: 10.1128/JB.188.8.3116-3120.2006
Huang, T. P., and Lee Wong, A. C. (2007). A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 73, 5034–5040. doi: 10.1128/AEM.00366-07
Jurkevitch, E., Hadar, Y., and Chen, Y. (1992). Differential siderophore utilization and iron uptake by soil and rhizosphere bacteria. Appl. Environ. Microbiol. 58, 119–124.
Kalidasan, V., Azman, A., Joseph, N., Kumar, S., Awang Hamat, R., and Neela, V. (2018a). Putative iron acquisition systems in Stenotrophomonas maltophilia. Molecules 23:E2048. doi: 10.3390/molecules23082048
Kalidasan, V., Joseph, N., Kumar, S., Hamat, R. A., and Neela, V. K. (2018b). The “Checkmate” for iron between human host and invading bacteria: chess game analogy. Indian J. Microbiol. 58, 257–267. doi: 10.1007/s12088-018-0740-2
Kamitori, S., and Yoshida, H. (2015). “Structure-function relationship of bacterial SH3 domains,” in SH Domains, ed N. Kurochkina (New York, NY: Springer Cham), 71–89.
Kim, C., Chung, Y., and Shin, S. (2009). Iron differentially regulates gene expression and extracellular secretion of Vibrio vulnificus cytolysin-hemolysin. J. Infect. Dis. 200, 582–589. doi: 10.1086/600869
Kim, Y. R., Lee, S. E., Kim, B., Choy, H., and Rhee, J. H. (2013). A dual regulatory role of cyclic adenosine monophosphate receptor protein in various virulence traits of Vibrio vulnificus. Microbiol. Immunol. 57, 273–280. doi: 10.1111/1348-0421.12031
Kumar, N. P., and Audipudi, A. V. (2015). Exploration of a novel plant growth promoting bacteria Stenotrophomonas maltophilia AVP27 isolated from the chilli rhizosphere soil. Int. J. Eng. Res. Gen. Sci. 3, 265–276.
Lau, C. K. Y., Krewulak, K. D., and Vogel, H. J. (2015). Bacterial ferrous iron transport : the Feo system. FEMS Microbiol. Rev. 40, 273–298. doi: 10.1093/femsre/fuv049
Lee, B. U., Choi, M. S., Kim, D. M., and Oh, K. H. (2017). Genome Shuffling of Stenotrophomonas maltophilia OK-5 for Improving the Degradation of Explosive RDX (Hexahydro-1,3,5-trinitro-1,3,5-triazine). Curr. Microbiol. 74, 268–276. doi: 10.1007/s00284-016-1179-5
Liu, W., Tian, X. Q., Wei, J. W., Ding, L. L., Qian, W., Liu, Z., et al. (2017). BsmR degrades c-di-GMP to modulate biofilm formation of nosocomial pathogen Stenotrophomonas maltophilia. Sci. Rep. 7:4665. doi: 10.1038/s41598-017-04763-w
Looney, W. J. (2005). Role of Stenotrophomonas maltophilia in hospital-acquired infection. Br. J. Biomed. Sci. 62, 145–154. doi: 10.1080/09674845.2005.11732702
Martinez, R., Kopp, D., Mangat, R., Snouffer, A., and Brooke, J. (2010). “Effect of ferric chloride on biofilm formation of Stenotrophomonas maltophilia,” in 60th Annual Conference Canadian Society of Microbiologists (Hamilton, ON: Canadian Society of Microbiologists), abstr II24.
Marx, J. J. (2002). Iron and infection: competition between host and microbes for a precious element. Best Pract. Res. Clin. Haematol. 15, 411–426. doi: 10.1053/beha.2002.0001
Minkwitz, A., and Berg, G. (2001). Comparison of antifungal activities and 16S ribosomal DNA sequences of clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 39, 139–145. doi: 10.1128/JCM.39.1.139-145.2001
Mokracka, J., Cichoszewska, E., and Kaznowski, A. (2011). Siderophore production by Gram-negative rods isolated from human polymicrobial infections. Biol. Lett. 48, 147–157. doi: 10.2478/v10120-011-0012-x
Mukherjee, P., and Roy, P. (2016). Genomic potential of Stenotrophomonas maltophilia in bioremediation with an assessment of its multifaceted role in our environment. Front. Microbiol. 7:967. doi: 10.3389/fmicb.2016.00967
Nas, M. Y., and Cianciotto, N. P. (2017). Stenotrophomonas maltophilia produces an EntC-dependent catecholate siderophore that is distinct from enterobactin. Microbiology 163, 1590–1603. doi: 10.1099/mic.0.000545
Neela, V. K. (2014). “Could clinical Stenotrophomonas maltophilia be a potential pathogen in clinical setting?” in 3rd International Conference on Clinical Microbiology and Microbial Genomics (Valencia), 53.
Norton, J., and Dachs, R. (2015). “Stenotrophomonas maltophilia : culprit or colonizer?” in Southern Medical Association (Destin, FL), 30.
Novichkov, P. S., Kazakov, A. E., Ravcheev, D. A., Leyn, S. A., Kovaleva, G. Y., Sutormin, R. A., et al. (2013). RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. doi: 10.1186/1471-2164-14-745
Pages, D., Rose, J., Conrod, S., Cuine, S., Carrier, P., Heulin, T., et al. (2008). Heavy metal tolerance in Stenotrophomonas maltophilia. PLoS ONE 3:e1539. doi: 10.1371/journal.pone.0001539
Passerini de Rossi, B., Calenda, M., Vay, C., and Franco, M. (2007). Biofilm formation by Stenotrophomonas maltophilia isolates from device-associated nosocomial infections. Rev. Argent. Microbiol. 39, 204–212.
Pompilio, A., Piccolomini, R., Picciani, C., D'Antonio, D., Savini, V., and Di Bonaventura, G. (2008). Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 287, 41–47. doi: 10.1111/j.1574-6968.2008.01292.x
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11:159. doi: 10.1186/1471-2180-11-159
Ratledge, C., and Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941. doi: 10.1146/annurev.micro.54.1.881
Runyen-Janecky, L. J. (2013). Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol. 3:55. doi: 10.3389/fcimb.2013.00055
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Senol, E. (2004). Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J. Hosp. Infect. 57, 1–7. doi: 10.1016/j.jhin.2004.01.033
Sokol, P. A., Darling, P., Lewenza, S., Corbett, C. R., and Kooi, C. D. (2000). Identification of a siderophore receptor required for ferric ornibactin uptake in Burkholderia cepacia. Infect. Immun. 68, 6554–6560. doi: 10.1128/IAI.68.12.6554-6560.2000
Steinmann, J., Mamat, U., Abda, E. M., Kirchhoff, L., Streit, W. R., Schaible, U. E., et al. (2018). Analysis of phylogenetic variation of Stenotrophomonas maltophilia reveals human-specific branches. Front. Microbiol. 9:806. doi: 10.3389/fmicb.2018.00806
Su, Y. C., Chin, K. H., Hung, H. C., Shen, G. H., Wang, A. H., and Chou, S. H. (2010). Structure of Stenotrophomonas maltophilia FeoA complexed with zinc: a unique prokaryotic SH3 - Domain protein that possibly acts as a bacterial ferrous iron-transport activating factor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 636–642. doi: 10.1107/S1744309110013941
Teintze, M., Hossain, M. B., Barnes, C. L., Leong, J., and van der Helm, D. (1981). Structure of ferric pseudobactin, a siderophore from a plant growth promoting Pseudomonas. Biochem. 20, 6446–6457. doi: 10.1021/bi00525a025
Thomas, M. S., and Wigneshweraraj, S. (2014). Regulation of virulence gene expression. Virulence 5, 832–834. doi: 10.1080/21505594.2014.995573
Thomas, R., Hamat, R. A., and Neela, V. (2014). Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence 5, 326–330. doi: 10.4161/viru.27724
Torres, P. S., Malamud, F., Rigano, L. A., Russo, D. M., Marano, M. R., Castagnaro, A. P., et al. (2007). Controlled synthesis of the DSF cell-cell signal is required for biofilm formation and virulence in Xanthomonas campestris. Environ. Microbiol. 9, 2101–2109. doi: 10.1111/j.1462-2920.2007.01332.x
Travassos, L. H., Pinheiro, M. N., Coelho, F. S., Sampaio, J. L. M., Merquior, V. L. C., and Marques, E. A. (2004). Phenotypic properties, drug susceptibility and genetic relatedness of Stenotrophomonas maltophilia clinical strains from seven hospitals in Rio de Janeiro, Brazil. J. Appl. Microbiol. 96, 1143–1150. doi: 10.1111/j.1365-2672.2004.02248.x
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Keywords: S. maltophilia, iron-depleted, Fur, siderophore, microbial iron acquisition, virulence factors
Citation: Kalidasan V, Joseph N, Kumar S, Awang Hamat R and Neela VK (2018) Iron and Virulence in Stenotrophomonas Maltophilia: All We Know So Far. Front. Cell. Infect. Microbiol. 8:401. doi: 10.3389/fcimb.2018.00401
Received: 27 July 2018; Accepted: 23 October 2018;
Published: 12 November 2018.
Edited by:
Vincenzo Scarlato, Università degli Studi di Bologna, ItalyReviewed by:
Susu M. Zughaier, Qatar University, QatarGianni Prosseda, Università degli Studi di Roma La Sapienza, Italy
Copyright © 2018 Kalidasan, Joseph, Kumar, Awang Hamat and Neela. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vasantha Kumari Neela, neela2000@hotmail.com; vasantha@upm.edu.my
 V. Kalidasan
V. Kalidasan Narcisse Joseph
Narcisse Joseph  Suresh Kumar
Suresh Kumar Rukman Awang Hamat
Rukman Awang Hamat Vasantha Kumari Neela
Vasantha Kumari Neela