A gene expression profile-based approach to screen the occurrence and predisposed host characteristics of drug-induced liver injury: a case study of Psoralea corylifolia Linn
- 1The Department of Pharmacy, First Affiliated Hospital of Henan University of Traditional Chinese Medicine, Zhengzhou, China
- 2Henan Province Engineering Research Center for Clinical Application, Evaluation and Transformation of Traditional Chinese Medicine, Zhengzhou, China
- 3Henan Provincial Key Laboratory for Clinical Pharmacy of Traditional Chinese Medicine, Zhengzhou, China
- 4Henan Province Engineering Research Center of Safety Evaluation and Risk Management of Traditional Chinese Medicine, Zhengzhou, China
- 5Henan University of Traditional Chinese Medicine, Zhengzhou, China
Drug-induced liver injury (DILI) is one of the most common causes of a drug being withdrawn, and identifying the culprit drugs and the host factors at risk of causing DILI has become a current challenge. Recent studies have found that immune status plays a considerable role in the development of DILI. In this study, DILI-related differentially expressed genes mediated by immunoinflammatory cytokines were obtained from the Gene Expression Omnibus (GEO) database to predict the occurrence of DILI (named the DILI predictive gene set, DILI_PGS), and the predictability of the DILI_PGS was verified using the Connectivity Map (CMap) and LiverTox platforms. The results obtained DILI_PGS from the GEO database could predict 81.25% of liver injury drugs. In addition, the Coexpedia platform was used to predict the DILI_PGS-related characteristics of common host diseases and found that the DILI_PGS mainly involved immune-related diseases and tumor-related diseases. Then, animal models of immune stress (IS) and immunosuppressive (IP) were selected to simulate the immune status of the above diseases. Meanwhile, psoralen, a main component derived from Psoralea corylifolia Linn. with definite hepatotoxicity, was selected as an experimental drug with highly similar molecular fingerprints to three idiosyncratic hepatotoxic drugs (nefazodone, trovafloxacin, and nimesulide) from the same DILI_PGS dataset. The animal experiment results found a single administration of psoralen could significantly induce liver injury in IS mice, while there was no obvious liver function change in IP mice by repeatedly administering the same dose of psoralen, and the potential mechanism of psoralen-induced liver injury in IS mice may be related to regulating the expression of the TNF-related pathway. In conclusion, this study constructed the DILI_PGS with high accuracy to predict the occurrence of DILI and preliminarily identified the characteristics of host factors inducing DILI.
1 Introduction
Drug-induced liver injury (DILI) is a common cause of drug withdrawal, restriction, and termination of marketing (Regev, 2014). It is a rare but potentially severe adverse drug event with serious clinical results, including acute liver failure and the need for liver transplantation (European Association for the Study of the Liver, 2019), and acute DILI accounts for more than 50% of all cases of acute liver failure (Reuben et al., 2016; Donnelly et al., 2017). The elimination of iatrogenic “injury” caused by treatment purposes is a priority of patient care. In this regard, the websites of LiverTox (https://www.ncbi.nlm.nih.go) and DILIrank (https://www.fda.gov/science-research/liver-toxicity-knowledge-base-ltkb/drug-induced-liver-injury-rank-dilirank-dataset) have included more than 1,200 kinds of chemical drugs and dietary supplements that can induce liver injury, providing a certain reference basis for avoiding the occurrence of DILI (Hoofnagle et al., 2013; Chen et al., 2016).
However, DILI is typically classified as either intrinsic or idiosyncratic. In contrast to intrinsic DILI, in which the damage is dose-dependent and occurs transitorily (hours to days) after exposure to the drug with the occurrence mechanism and prevention-control methods clear and predictable, idiosyncratic DILI (IDILI), which occurs without obvious dose dependence and with long latency (weeks to months), is multifactorial and unpredictable (European Association for the Study of the Liver, 2019). Recently, indirect DILI has been described as a third type of DILI caused by the mechanism of action of the drug due to its idiosyncratic properties (Hoofnagle and Björnsson, 2019). At present, it is generally believed that the occurrence of IDILI is closely related to host factors (e.g., genetics and immunity) (Chalasani et al., 2021). Although the occurrence of IDILI is relatively rare, once it occurs, about 10% of patients may experience life-threatening clinical outcomes, such as acute liver failure, with most requiring a liver transplant or ultimately mortality; if not recognized and the drug causing it is not discontinued promptly, some patients will develop chronic DILI (Wei et al., 2007; Reuben et al., 2010; Chalasani et al., 2015). Therefore, identifying the host factors that cause individual susceptibility is the focus and difficulty of the ongoing DILI research.
Some key features of IDILI drugs that induce liver injury are mediated by the adaptive immune system, and almost all forms of idiosyncratic drug-induced hepatitis exhibit features of drug hypersensitivities, such as rash, fever, and eosinophilia (Devarbhavi et al., 2011; Chalasani et al., 2015). Drug-protein adducts formed from drugs or secondary metabolites interacting with host proteins as haptens are generally considered to be presented as neoantigens by the major histocompatibility complex Class II, playing a key role in adaptive immune responses (Hoofnagle and Björnsson, 2019). Studies have shown that the inflammatory microenvironment can enhance the specificity of DILI (Jiang et al., 2017). Thus, the gene spectrum composed of a series of related differentially expressed genes (DEGs) that enhance the specificity of DILI in the inflammatory microenvironment may partially characterize the organism status of susceptible individuals at risk of IDILI.
Connectivity Map (CMap) (https://clue.io/query) is a database platform that contains more than 5,000 marketed drugs or small molecule compounds with potential pharmacological activity that interfere with the expression of perturbed genes in 80 human cell lines, and provides a method to quantify the disease-drug similarity “link score” criteria and online analysis tools based on pattern matching algorithms (Subramanian et al., 2017). Over the past 15 years, drug repurposing and disease redefining conducted based on the database has provided biomedical insights and treatment clues for a variety of diseases, including coronavirus disease 2019 (COVID-19) (Chen et al., 2021; Li et al., 2022). The Coexpedia platform (https://www.coexpedia.org/) is a data mining system that integrates functional hypotheses extracted from transcriptomic data sources in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and provides a new approach for human disease research based on gene expression profile similarity (Yang et al., 2017). Therefore, it may be feasible to first obtain the DILI-related DEG profile induced by inflammatory microenvironments based on the GEO database, repurpose small molecular compounds with similar gene expression profiles for DILI-related DEGs according to the CMap database, verify the prediction ability of the DILI-related DEGs profiles to characterize hepatotoxicity by querying the LiverTox database, and further mine and analyze the characteristics of potential susceptible diseases inducing DILI based on the Coexpedia database.
Psoralea corylifolia Linn. (BGZ), a dried and mature fruit of Psoralea corylifolia L. from a leguminous plant, has been reported to have hepatotoxicity in recent years (Wang et al., 2020a; Ge et al., 2021), but its hepatotoxic components (including bakuchiol and psoralen) are still controversial (Park et al., 2005; Li et al., 2017; Jiang et al., 2022; Ma et al., 2022). Morgan fingerprints are a method of encoding molecular structure and are a highly informative representation for many chemical analog predictions via extended connectivity fingerprinting (Rogers and Hahn, 2010; Chang et al., 2018). Accordingly, the study plans to clarify the feasibility of the DILI-related DEGs profiles to predict the characteristics of DILI and preliminarily identify the hepatotoxic components of BGZ (Figure 1).
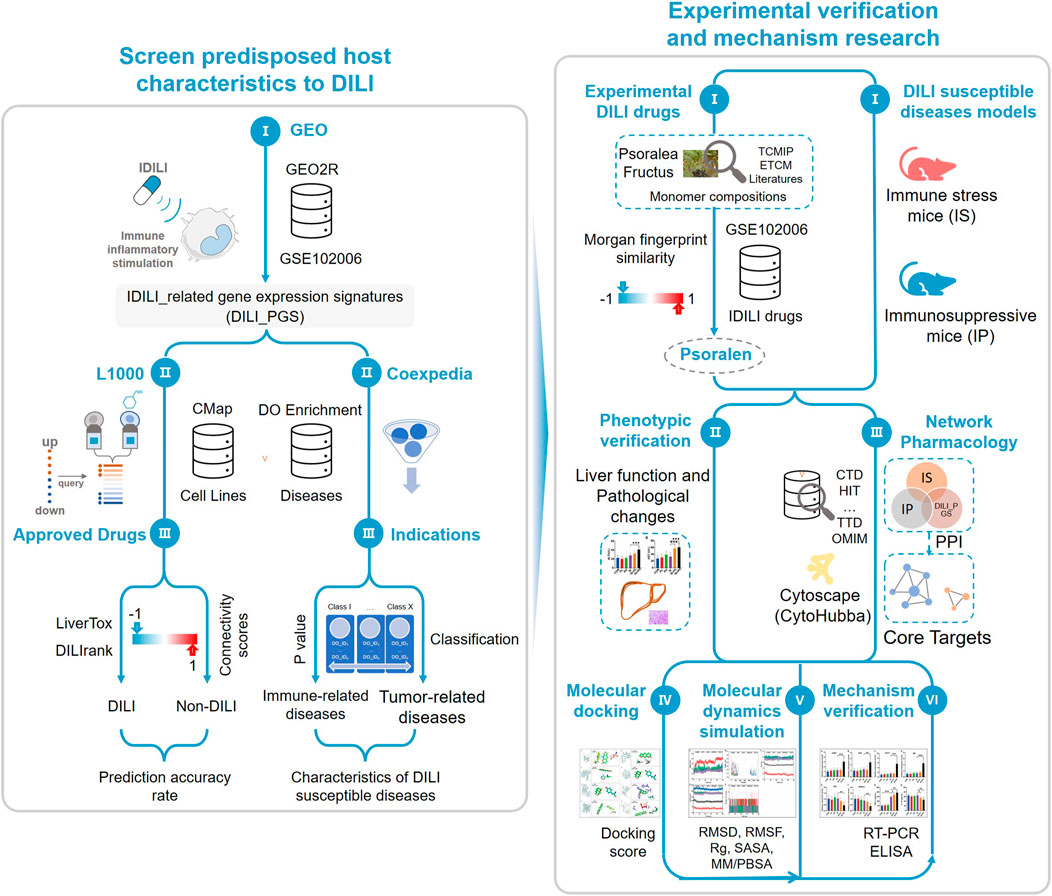
FIGURE 1. Study schematic diagram. By integrating databases of GEO, CMap, LiverTox, DILIrank, and Coexpedia, the study obtained drug-induced liver injury (DILI)-related gene expression signatures with certain accuracy and the host characteristics predisposed to DILI. According to the Morgan molecular fingerprint similarity algorithm, the monomer components with high molecular fingerprint similarity of the idiosyncratic drugs in GSE102006 were selected from the known hepatotoxic traditional Chinese medicine Psoralea corylifolia Linn. (BGZ) as the experimental drugs and the host status of DILI susceptibility was characterized by the immune stress (IS) animal model and the immunosuppressive (IP) animal model, then phenotypic verification was carried out using the above experimental drugs and animal models. Network pharmacology, molecular docking, and molecular dynamics simulation were used to preliminarily screen the core target, and reverse transcription-polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA) were used to verify the above core targets.
2 Materials and methods
2.1 Construct a gene set to predict the occurrence of DILI
2.1.1 Acquisition of the DILI-related DEGs
The gene expression data of DILI were acquired from the GEO database by searching the terms “idiosyncratic drug-induced liver injury”. The collected datasets were further selected if they met the following inclusion criteria: (a) whole-genome transcriptome profiling, and (b) species of origin were “Homo sapiens”. DEGs with an adjusted p value <0.05 and ǀLog2 fold changeǀ≥1 were identified as the DILI predictive gene set (DILI_PGS).
2.1.2 Prediction of DILI-associated drugs by the DILI_PGS
The composition characteristics of the DILI_PGS, including significantly up- and downregulated DEGs, were used as the input data and compared with the gene expression signatures of available CMap drugs by using a pattern-matching algorithm based on the non-parametric rank-ordered Kolmogorov–Smirnov statistic to determine connectivity scores (Subramanian et al., 2017) and obtain the drugs with similar gene expression profiles to the DILI_PGS. The results of signature matching for each pair are shown with connectivity scores (CS) ranging from −1 to +1 (Xing et al., 2022). The closer the CS is to 1, the stronger the positive correlation, and the closer the CS is to −1, the stronger the negative correlation. Among them, drugs with a significant positive CS have the potential to induce DILI, and drugs with a significant negative CS have the potential to reverse DILI.
2.1.3 Feasibility evaluation of the DILI_PGS
The drugs acquired by the CMap platform were further filtered according to |CS)| ≥ 0.60 and inclusion in the Drugbank database (https://go.drugbank.com/), and the percentage of hepatotoxicity drugs was calculated based on the databases containing hepatotoxic drugs (specifications of origin were “Homo sapiens”, excluding cell level), such as LiverTox and DILIrank, to evaluate the feasibility of predicting the host characteristics of patients with DILI based on the DILI_PGS.
2.2 Analysis of disease characteristics inducing DILI
2.2.1 Analysis of the immune cell infiltration for the corresponding DILI_PGS
Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) (https://cibersort.stanford.edu/) was used to comprehensively estimate the infiltration level of each immune cell subtype between the IDILI with inflammatory factor stimulation (IDILI_with) and IDILI without inflammatory factor stimulation (IDILI_without) based on linear support vector regression (Newman et al., 2015). Estimation of STromal and Immune cells in MAlignant Tumours using Expression data (ESTIMATE) (https://bioinformatics.mdanderson.org/estimate/) was used to assess the immune scores, stromal scores, estimate scores, and tumor purity between the IDILI_with group and IDILI_without group (Yoshihara et al., 2013). The analyses of CIBERSORT and ESTIMATE were completed using the Sangerbox tool (http://sangerbox.com/home.html) (Shen et al., 2022).
2.2.2 Gene set enrichment analysis
The study used the Gene Set Enrichment Analysis (GSEA) software (version 3.0) to assess the differentially enriched pathways between the IDILI_with group and IDILI_without group according to inflammatory factor stimulation conditions, and the gene set of ‘c2.cp.kegg.v7.4.symbols.gmt’ was downloaded to evaluate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways from the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/downloads.jsp). The minimum gene set was five, the maximum gene set was 5,000, and 1,000 resampling, p < 0.05, and false discovery rate (FDR) < 0.25 were considered statistically significant (Subramanian et al., 2005). The analysis of GSEA was completed using the Sangerbox tool (Shen et al., 2022).
2.2.3 Analysis of disease characteristics of the corresponding gene set associated with DILI
The DILI_PGS was analyzed using the COEXPEDIA platform for disease ontology (DO) analysis to explore the disease characteristics that potentially induce DILI, which is a database of context-associated co-expression networks inferred from individual series’ of microarray samples for human and mouse samples based on the GEO database (Yang et al., 2017).
2.3 Molecular fingerprint similarity calculation
In the study, the similarity of molecular fingerprints between the related components of BGZ and IDILI-related drugs acquired from the data sets of the GEO database was calculated by Morgan fingerprints. The related components of BGZ were obtained from Traditional Chinese Medicine Systems Pharmacology (TCMSP) (https://tcmsp-e.com/tcmsp.php), Encyclopedia of Traditional Chinese Medicine (ETCM) (http://www.tcmip.cn/ETCM/index.php/Home/Index/), Pubmed (https://pubmed.ncbi.nlm.nih.gov/), and other databases, and the Structure Data File (SDF) structures of BGZ-related monomer compounds and IDILI-related drugs were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Morgan fingerprints were acquired by RDKit (Rogers and Hahn, 2010), and the similarity coefficient ranges from 0 to 1; the larger the value, the higher the similarity. Ultimately, BGZ-related compounds with high similarity were selected for subsequent experimental verification.
2.4 Animal experimental verification
2.4.1 Establishment of animal models and treatment
C57BL/6 female mice, 6–8 weeks of age (16–18 g), were obtained from SPF Biotechnology Co., Ltd. (License No. SCXK20190010, Beijing, China) and housed in the Laboratory Animal Center of the Fifth Medical Center, Chinese PLA General Hospital (Animal Ethics Committee approval No. YFYDW2020017). All mice were raised under specific pathogen-free conditions under a 12 h light/dark cycle, with free access to adequate food and water. All animals were fed adaptively for 1 week before starting the experiments. The mice were randomly divided into the normal group (CON), low-dose psoralen group (PL, 40 mg/kg), high-dose psoralen group (PH, 80 mg/kg), immunological stress group (IS), immunological stress-treated with PL group (IS+PL), immunological stress-treated with PH group (IS+PH), immunosuppressive group (IP), IP-treated with PL group (IP+PL), and IP-treated with PH group (IP+PH). The number of mice in each group was eight.
For the IS model: the mice were administered via tail vein (i.v.) lipopolysaccharide (LPS) (from Escherichia coli O55:B5, Sigma) (2 mg/kg) in sterile saline (Wang et al., 2020b), and, 2 h later, PL, PH or its vehicle (0.5% carboxymethylcellulose sodium, 0.5% CMC-Na) was administered through gastric irrigation, respectively. Mice serum was collected 6 h after PL or PH treatment. For the IP model, the mice were administered intraperitoneal (i.p.) cyclophosphamide (CTX) in sterile saline (80 mg/kg) on days 1, 3, and 5 (Li et al., 2021a), and, from the 8th day, IP mice and normal mice were given PL, PH or its vehicle (0.5% CMC-Na) by gastric irrigation for 7 consecutive days, respectively.
2.4.2 Hematoxylin and eosin staining
The left hepatic lobe was fixed with 4% paraformaldehyde for 48 h, embedded in wax, and sectioned at approximately 5 µm for hematoxylin and eosin (H&E) pathological staining analysis.
2.4.3 Alanine aminotransferase and aspartate transaminase
Serum alanine aminotransferase (ALT) and aspartate transaminase (AST) levels were measured according to the manufacturer’s directions (both were purchased from Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.5 Mechanism exploration and verification
2.5.1 Protein–protein interaction network analysis for the DILI_PGS
The DILI_PGS was imported into the STRING database (https://cn.string-db.org/) for protein–protein interaction (PPI) network analysis, in which the organization was set as human, and an interaction score of more than 0.40 was set as the threshold. The data obtained from the STRING database were imported into Cytoscape (V3.8.0) for network visualization, and the hub targets were screened by eleven topological analysis methods of CytoHubba.
2.5.2 Molecular docking
To analyze the binding affinity between ligands from BGZ and hub target proteins, Maestro software (V11.5) was used to carry out the docking scores. The structure of the ligand was downloaded from the PubChem database, and the protein structures were retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) (https://www.pdbus.org/) and Uniprot databases (https://www.uniprot.org/). For docking analysis, the ligand and proteins were optimized by the LigPrep tool and Protein Preparation Wizard, respectively. In addition, partial atomic charge attribution, protonation states generation at pH 7 ± 2.0, and energy minimization were achieved using the OPLS-2005 force field. To test the docking parameters, the ligand was docked into the catalytic pocket of the core target proteins using Grid-Based Ligand Docking with Energetics (Glide v11.5, Schrödinger) in ‘extra precision’ mode without applying any constraints.
2.5.3 Molecular dynamics simulations
To verify the interactions and stability between the core target proteins and ligands, explicit solvent molecular dynamics (MD) simulations were carried out with GROMACS 2020.3 software. The simulation box size was optimized with the distance between each atom of the protein and the box greater than 1.0 nm. Filling the box with an explicit solvent-simple point chargemodel (SPC216 water molecules) and replacing the water molecules with Na* and Cl counterions to make the simulation system electrically neutral. Following the steepest descent method, energy optimization of 5.0×104 steps was performed to minimize the energy consumption of the entire system and finally to reduce the unreasonable contact or atom overlap in the entire system. After energy minimization, first-phase equilibration was performed with the Canonical ensemble (NVT) ensemble at 300 K for 100 ps to stabilize the temperature of the system. Second-phase equilibration was simulated with the Constant-pressure and Constant-temperature (NPT) ensemble at 1 bar and 100 ps. The primary objective of the simulation was to optimize the interaction between the target proteins and the solvent and ions so that the simulation system was fully pre-equilibrated. All MD simulations were performed for 50 ns. PyMol2.5 was used to visualize the composite PDB format file.
2.5.4 Reverse transcription-polymerase chain reaction (RT-PCR)
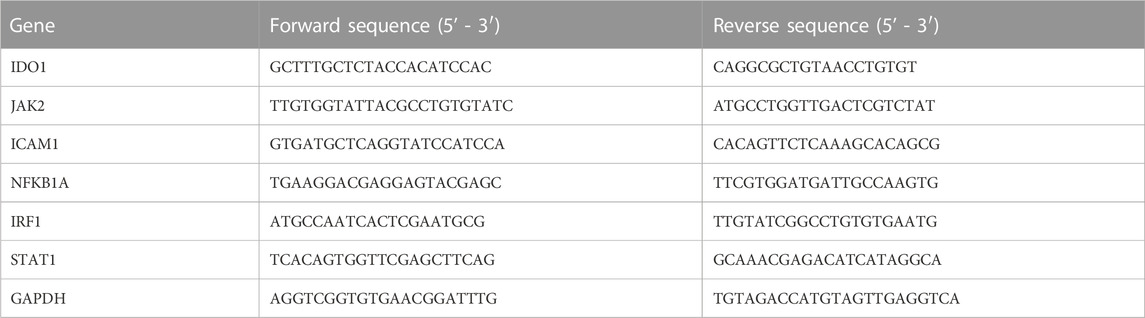
According to the manufacturer’s instructions, the total ribonucleic acid (RNA) in the liver was isolated by RNA-Quick Purification Kit (ES Science, RN001, China) and then synthesized into cDNA by reverse transcriptase (RT) (ES Science, RT001, China). Furthermore, cDNA amplification with appropriate primers to amplification (Table 1) was performed using Super SYBR Green qPCR Master Mix (ES Science, QP002, China) by QuantStudio 6 Flex PCR System (Applied Biosystems, USA). The cycle threshold (Ct) values of indoleamine 2,3-dioxygenase 1 (IDO1), Janus kinase 2 (JAK2), intercellular adhesion molecule 1 (ICAM1), nuclear factor kappa-B inhibitor alpha (NFKBIA), interferon regulatory factor 1 (IRF1), and signal transducer and activator of transcription 1 (STAT1) were obtained and normalized to that of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (all reagents were from TIANYI HUIYUAN, Beijing, China), and the results were analyzed by the 2−ΔΔCT method.
2.5.5 Enzyme-linked immunosorbent assay
Serums of tumor necrosis-α (TNF-α) and interleukin 10 (IL-10) were detected using enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s directions (both were purchased from Mlbio, Shanghai, China).
2.6 Statistical analyses
All data are presented as mean ± SD and analyzed by GraphPad version 8. Multiple comparisons performed using ANOVA were used to analyze the liver function and core target detection by RT-qPCR or ELISA. p < 0.05 was considered statistically significant.
3 Results
3.1 DILI-related gene sets
According to the GEO database, the study retrieved a gene expression dataset (numbered GSE102006) (Jiang et al., 2017) of HepG2 injury induced by three idiosyncratic drugs (nefazodone, trovafloxacin, and nimesulide) in the inflammatory microenvironment. With p < 0.05 and ǀLog2 fold changeǀ≥1 as the threshold, a total of 406 upregulated DEGs and 52 downregulated DEGs for nefazodone (Figure 2A), 483 upregulated DEGs and 531 downregulated DEGs for trovafloxacin (Figure 2B), and 331 upregulated DEGs and 86 downregulated DEGs for nimesulide (Figure 2C) were found under the stimulation of inflammatory cytokines. Among the DEGs, a total of 215 DEGs including 203 upregulated DEGs and 12 downregulated DEGs were the co-regulated DEGs among the three idiosyncratic drugs (Figure 2D) with a similar change trend (Figure 2E).
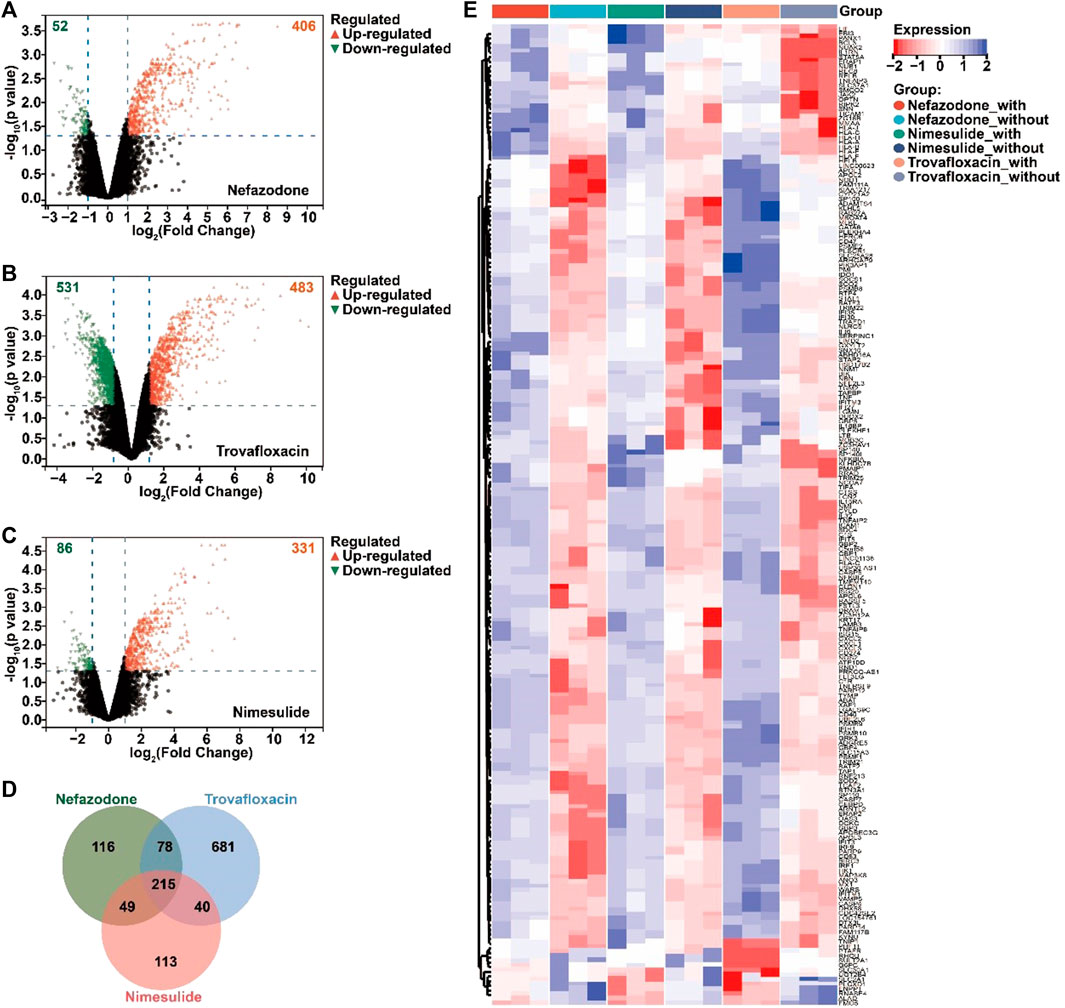
FIGURE 2. Acquired differentially expressed genes (DEGs) of DILI induced by immune inflammation. (A–C) Volcano diagram of the DEGs regulated by nefazodone (A), trovafloxacin (B), and nimesulide (C) under the condition of inflammatory stress in the gene expression dataset of GSE102006 (p < 0.05 and ǀLog2 fold changeǀ≥1); (B) the crosstalk DEGs regulated by nefazodone, trovafloxacin, and nimesulide; (C) heatmap of the crosstalk DEGs. Red represents upregulated DEGs and blue represents downregulated DEGs. (D) Venn analysis of the DEGs regulated by nefazodone, trovafloxacin, and nimesulide. (E) Heat map display of intersecting DEGs regulated by nefazodone, trovafloxacin, and nimesulide.
3.2 DILI risk inquiry of approved drugs
Under the condition of |CS| ≥ 0.60 and being included by the DrugBank database, 40 drugs (including 32 positively related drugs and 8 negatively correlated drugs) were screened with high similarity to the DILI-related gene expression signatures (Table 2). Surprisingly, among the 32 drugs with a positive correlation with DILI-related genes, 26 drugs (including 20 drugs included in the LiverTox database and 6 drugs reported in individual cases (Amitrano et al., 1992; Rank and Olson, 1989; Hannequin et al., 1988; Van Mouwerik et al., 1987; el Saghir and Hawkins, 1984; de Faria et al., 2015)) were reported to have hepatotoxicity, accounting for 81.25%, while among the 8 drugs with a negative correlation with DILI-related genes, only 3 drugs included in the LiverTox database were reported to have hepatotoxicity, accounting for 37.50%. It was suggested that the above 215 DEGs could partially predict the occurrence of DILI, and this gene set was named the DILI predictive gene set (DILI_PGS).
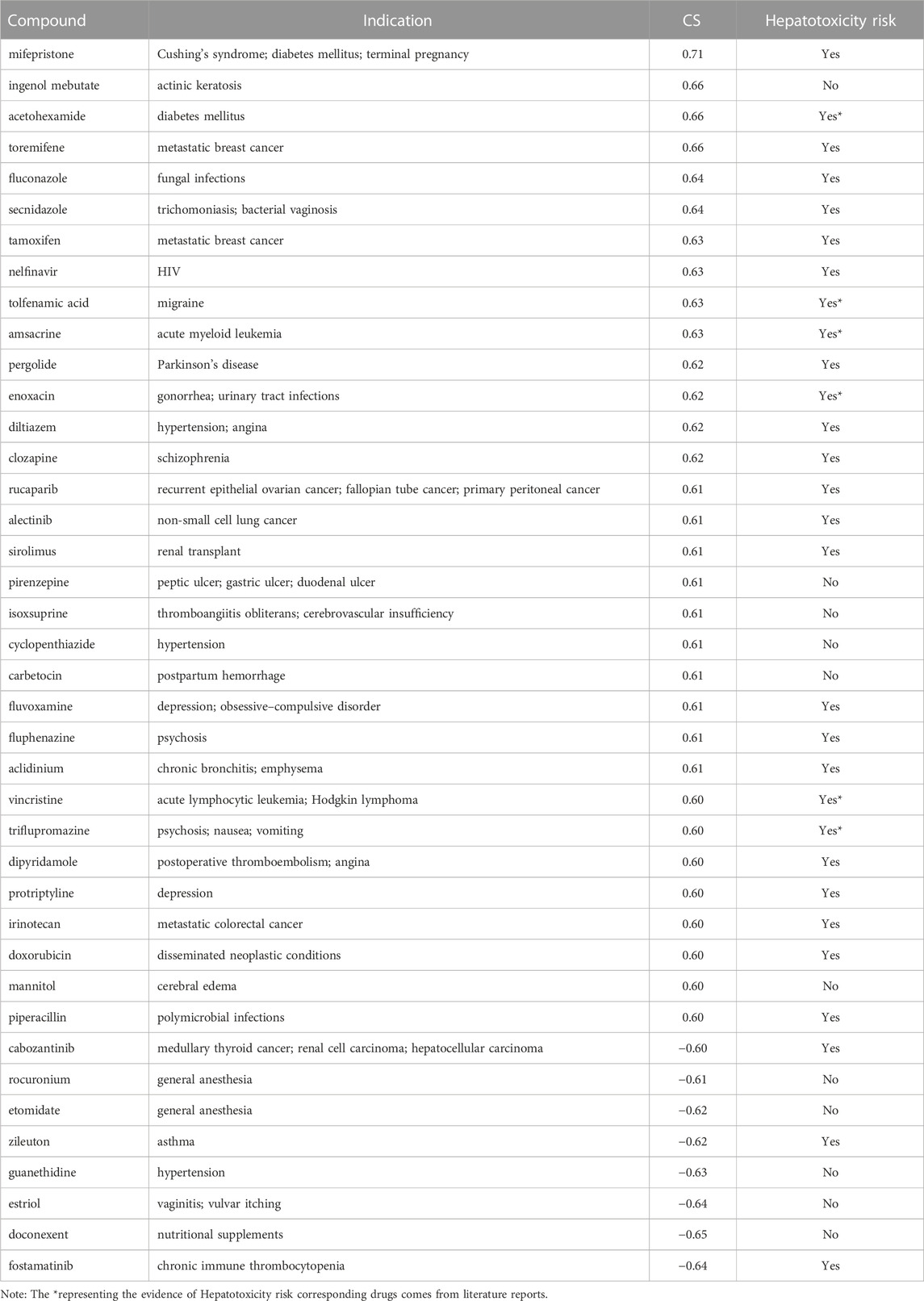
TABLE 2. 40 approved drugs with high similarity to the DILI_PGS expression signatures with |CS| ≥ 0.60.
3.3 Composition of infiltrated immune cells signature for the DILI_PGS
To determine the relevance between the DILI_PGS risk signature and the DILI microenvironment, the CIBERSORT algorithm was used to estimate the difference of 22 types of infiltrating immune cells between the IDILI without inflammatory infiltration (IDILI_without) group and the IDILI with inflammatory infiltration (IDILI_with) group. As shown in Figure 3A, the fraction of immune cells varied significantly among the samples and groups. Meanwhile, compared with the IDILI_without group, the IDILI_with group generally contained a higher fraction of activated dendritic cells (p < 0.001) and a lower fraction of CD4 memory-activated T cells, monocytes, and M2 macrophages (all p < 0.05) (Figure 3B). In addition, the ESTIMATE algorithm was applied to calculate the ESTIMATE score, immune score, stromal score, and tumor purity, representing the HepG2 cells environment, and the result showed immune score, stromal score, and tumor purity were significantly increased in the IDILI_with group (all p < 0.001) (Figure 3C).

FIGURE 3. Estimation of the immune cell infiltration characterization for the occurrence of DILI induced by immune inflammation. (A) Stacked histogram for the relative proportion of 22 tumor-infiltrating immune cells; (B) the box plot of the relative proportion of 22 tumor-infiltrating immune cells estimated by CIBERSORT; (C) the box plot of the ESTIMATE score, immune score, stromal score, and tumor purity estimated by ESTIMATE algorithm. *p < 0.05; **p < 0.01; ***p < 0.001; NA, not significant.
3.4 GSEA and DO analysis of the DILI_PGS
To explore the possible pathways and gene sets associated with immune functions, all expression data were divided into the IDILI_with group and IDILI_without group and then subjected to GSEA analysis. Compared with the IDILI_without group, of the 25 pathways screened according to the conditions of p < 0.05 and FDR <0.25 (Figure 4A), 19 pathways were highly related to immune cells and immune-related functions, including the RIG-I-like receptor signaling pathway, leishmania infection, autoimmune thyroid disease, graft-versus-host disease, the toll-like receptor signaling pathway, B cell receptor signaling pathway, cytokine-cytokine receptor interaction, natural killer cell-mediated cytotoxicity, and the chemokine signaling pathway. The details of the pathways are shown in the green font section of Figure 4A.
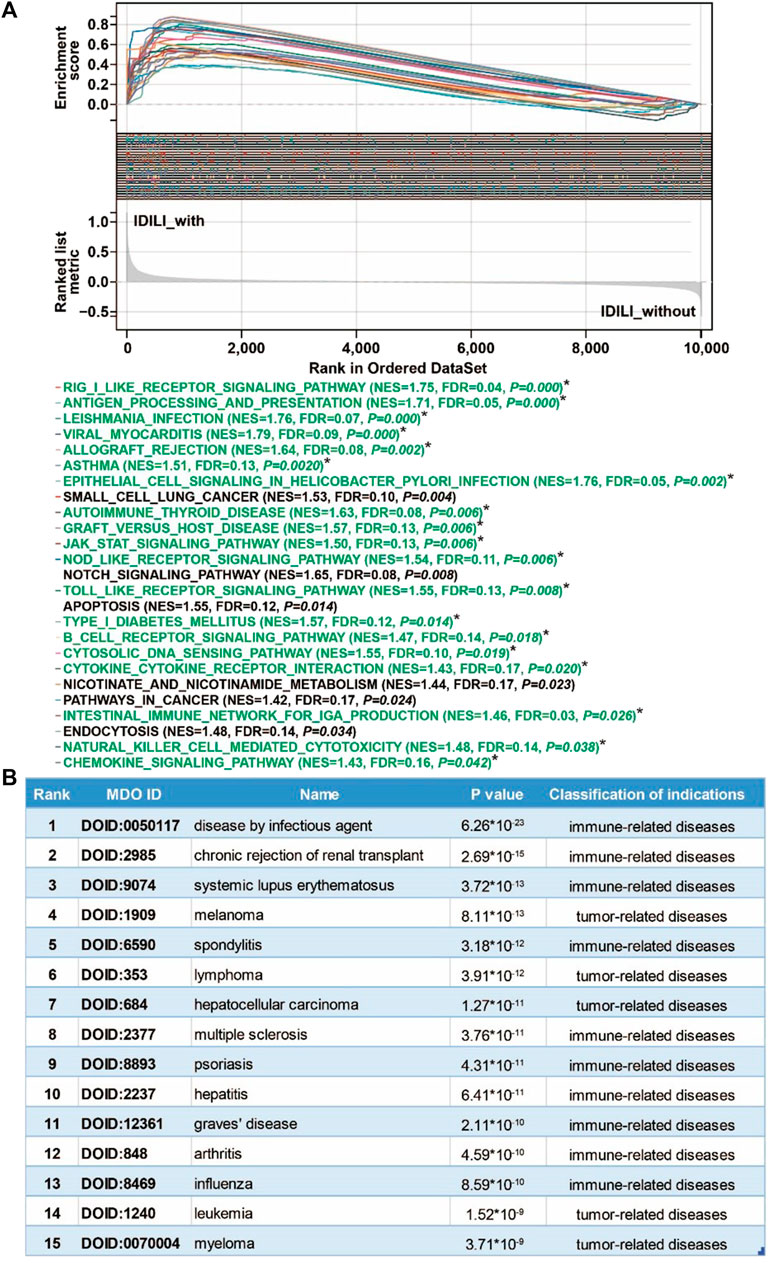
FIGURE 4. Pathways enrichment function analysis and susceptible disease characteristics analysis of the DILI predictive gene set (DILI_PGS). (A) GSEA analysis of the DILI_PGS (pathways marked with * in green font are immune-related); (B) the top 15 disease characteristics acquired using DO analysis of the DILI_PGS.
Based on the DO analysis via the Coexpedia platform, the DILI_PGS are mainly enriched in 300 significant disease entries (p < 0.05), and the top 15 disease entries were screened for display according to the p value. As shown in Figure 4B, the potential indications for the DILI_PGS include 10 immune-related diseases (disease by infectious agent, chronic rejection of renal transplant, systemic lupus erythematosus, spondylitis, multiple sclerosis, psoriasis, hepatitis, Graves’ disease, arthritis, and influenza) and five tumor-related diseases (melanoma, lymphoma, hepatocellular carcinoma, leukemia, and myeloma). Notably, integrating analysis of the above top 15 clinical indications curated using DO analysis and the top 25 pathways analyzed using GSEA, the results revealed that the potential indications of the DILI_PGS are primarily related to immune-related diseases. Therefore, animal models of IS (potential positive correlation) and IP (potential negative correlation) were selected as representative indications to confirm the feasibility of the DILI_PGS.
3.5 Screening of potential DILI risk components in BGZ
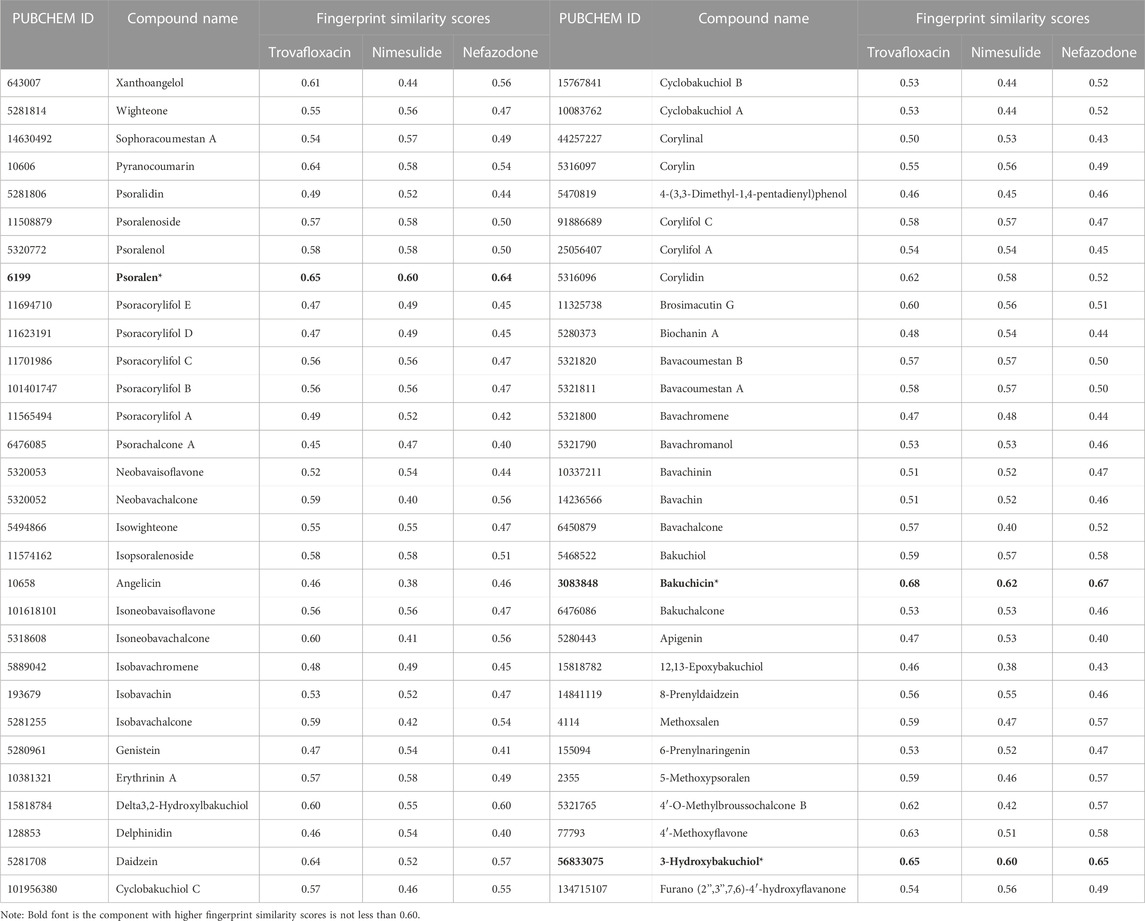
In the study, Morgan fingerprints were used to analyze the structural similarity between the components of BGZ-related monomer compounds and the three idiosyncratic drugs (nefazodone, trovafloxacin, and nimesulide) in the GSE102006 dataset for the potential hepatotoxic components in BGZ. A total of 60 components of BGZ-related monomers were obtained from the databases of TCMSP, ETCM, and the literature search, and among them, psoralen, bakuchin, and 3-Hydroxybakuchiol have high fingerprint similarity scores (all scores≥0.60) compared with the three idiosyncratic drugs (nefazodone, trovafloxacin, and nimesulide) using Morgan molecular fingerprint similarity (Table 3). It is noteworthy that psoralen has been recorded as a hepatotoxicity drug on the LiverTox platform. Therefore, psoralen was used to verify the feasibility of the DILI_PGS in predicting disease characteristics.
3.6 Experimental verification
3.6.1 Different responses of psoralen to liver function and hepatic histological change in IP and IS animal models
In the IS model, as shown in Figure 5A, compared with the CON group, the expression of ALT in PL, PH, and IS groups did not change obviously, while compared with the IS group, it was significantly increased in the IS+PH group (p < 0.05) with no obvious change in the IS+PL group. As for AST expression (Figure 5B), the PL, PH, and IS groups had no obvious changes compared with the CON group, while it was significantly increased in both the IS+PL and IS+PH groups compared with the IS group (both p < 0.05), indicating psoralen may induce liver injury when the body is under the status of immune stress; the H&E staining for pathological changes of the liver also confirmed this phenomenon (Figure 5C).

FIGURE 5. Different effects of psoralen on the liver function of IS and IP model mice. The serum levels of ALT (A) and AST (B) and the H&E staining for pathological changes of the liver (C) in the IS model mice. The serum levels of ALT (D) and AST (E) and the H&E staining for pathological changes of the liver (F) in the IS model mice. *p < 0.05; **p < 0.01; ***p < 0.001.
In the IP model, as shown in Figure 5D, compared with the CON group, the expression of ALT in the PL and IP groups had no obvious changes, while that in the PH group increased significantly (p < 0.05). Although compared with the IP group, the level of ALT in the IP+PH group increased significantly (p < 0.05), no significant change was seen between the IP+PH group and the PH group. As for AST expression (Figure 5E), compared with the CON group, the expression of AST in the PL and PH groups were both significantly increased (both p < 0.05) with no obvious change in the IP group. Although compared with the IP group, the expression of AST in the IP+PL and IP+PH groups were significantly increased (both p < 0.05), no significant changes were seen in the IP+PL and IP+PH groups while comparing with the PL and PH groups, respectively, indicating psoralen has certain liver accumulation toxicity, which is consistent with the literature report (Yang et al., 2020), and the pathological changes stained by H&E also confirmed this phenomenon (Figure 5F). Combined with the different responses of psoralen to the liver function of IS and IP model mice, this suggests that the immune stress status may be the potential host factor of psoralen-induced liver injury, but not the immune suppression status.
3.6.2 PPI network analysis and the potential core targets of the DILI_PGS regulated by psoralen in the IS model mice
The combined results of the PPI network analysis and the core target screening by CytoHubba for the DILI_ PGS (Figure 6A) found IRF1, MX dynamin-like GTPase 1 (MX1), interferon regulatory factor 9 (IRF9), interferon-stimulated gene 15 (ISG15), ICAM1, STAT1, and tumor necrosis factor (TNF-α) were the seven core targets with a frequency of no less than 7 calculated by 11 different topological analysis methods (Figure 6B). NFKBIA, ICAM1, IDO1, and JAK2 were the crosstalk targets among the targets of psoralen, IS, and the DILI_PG (Figure 6C).
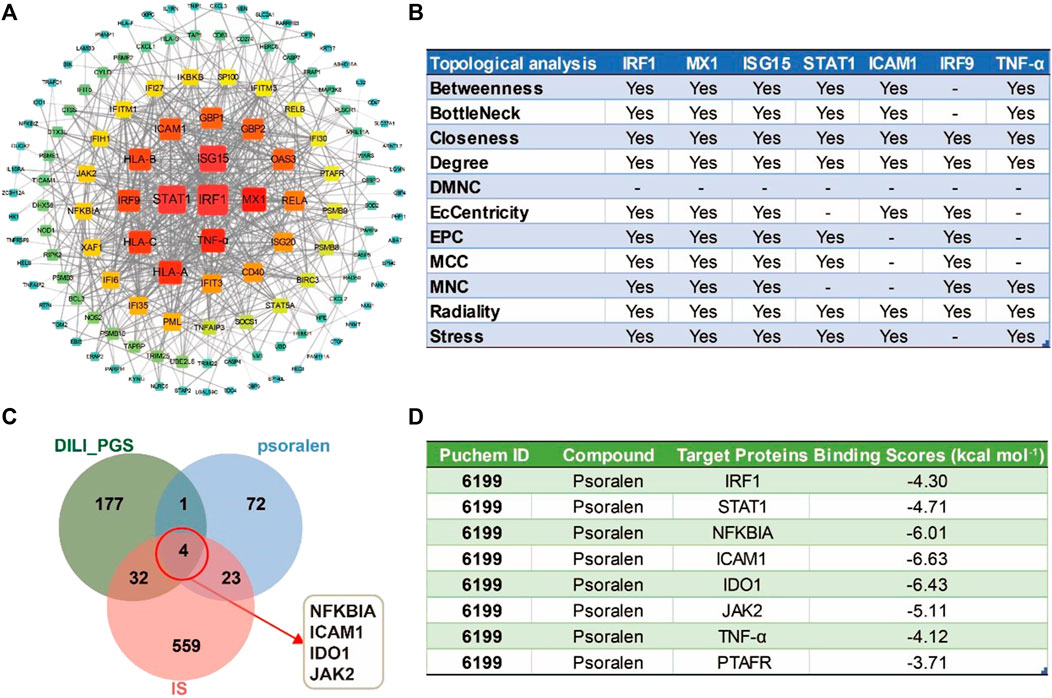
FIGURE 6. The potential core targets regulated by psoralen in IS model mice. (A) The PPI network for the DILI_PGS. (B) The seven core targets were obtained by 11 different topological analysis methods with CytoHubba. (C) Venn diagram of the crosstalk targets among the targets of psoralen, IS, and the DILI_PGS. (D) Docking scores of psoralen with the core targets.
3.6.3 Molecular docking analysis of binding energy between psoralen and potential core targets
The intersection targets (NFKBIA, ICAM1, IDO1, JAK2, and PTAFR) between psoralen and the DILI_PGS, and the core targets (IRF1, TNF-α, and STAT1) acquired by CytoHubb, mainly acted on the TNF signal pathway (So and Ishii, 2019) and play a key role in the immune response of the body. Thus, the above targets were selected as docking proteins, and psoralen was selected as the ligand for molecular docking. The results showed that psoralen was bound to the above proteins through visible hydrogen bonds and strong electrostatic interactions, and had low binding energy of −4.300 (IRF1, PDB: 1IF1), −4.711 (STAT1, PDB: 3WWT), −6.011 (NFKBIA, PDB: 6Y1J), −6.631 (ICAM1, PDB: 3BQN), −6.431 (IDO1, PDB: 2D0T), −5.107 (JAK2, PDB: 3LPB), −4.115 (TNF-α, PDB: 2AZ5), and −3.708 (PTAFR, PDB: 5ZKP) kcal mol-1 (Figure 6D). Furthermore, as shown in Figures 7A–H, psoralen could easily enter and bind to the active pocket of the above proteins, indicating highly stable binding for the complexes. Thus, the above proteins may be the potential proteins regulated by psoralen, especially IDO1, ICAM1, NFKBIA, and JAK2.

FIGURE 7. Molecular docking diagrams of core targets with psoralen. Psoralen binds to the protein (A) IRF1 (1IF1), (B) STAT1 (3WWT), (C) NFKBIA (6Y1J), (D) ICAM1 (3BQN), (E) IDO1 (2D0T), (F) JAK2 (3LPB), (G) TNF-α (2AZ5), and (H) PTAFR (5ZKP).
3.6.4 Molecular dynamics simulation of the interaction between psoralen and potential core targets
3.6.4.1 Conformational stability
The root mean square deviation (RMSD) and root mean square fluctuation (RMSF) plots against simulation time, shown in Figure 8, elucidated smaller fluctuations for all complexes (psoralen/ICAM1, psoralen/IRF1, psoralen/JAK2, psoralen/NFKBIA, and psoralen/STAT1) after 40 ns of the trajectory, indicating attainment of a stable conformation, and the average RMSD values of the above complexes were 0.15 nm, 0.40 nm, 0.23 nm, 0.20 nm, and 0.18 nm, respectively, while the average RMSF values of the above complexes were 0.07 nm, 0.11 nm, 0.09 nm, 0.11 nm, and 0.27 nm, respectively. The lower RMSD and RMSF values of protein–ligand docking demonstrated the structural stabilization and flexibility of the above complexes (Figures 8A, B).
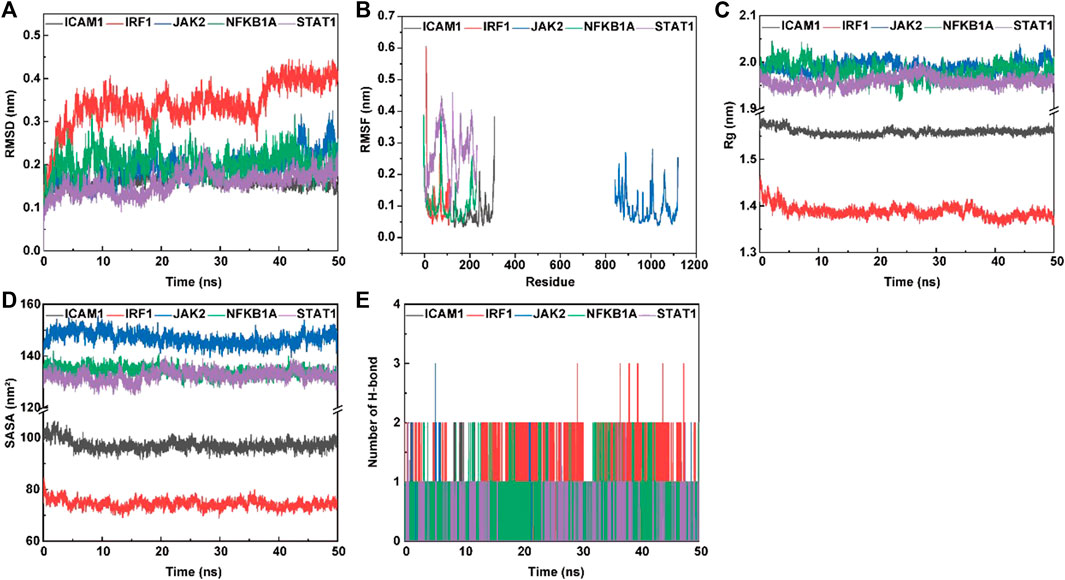
FIGURE 8. The molecular dynamics (MD) simulation of the complexes of ligand (psoralen) with proteins (ICAM1, IRF1, JAK2, NFKBIA, and STAT1). Root mean square deviation (RMSD) value (A), root mean square fluctuation (RMSF) value (B), radius of gyration (Rg) values (C), solvent accessible surface area (SASA) (D), and hydrogen bond numbers (E) of the complex during MD simulation.
3.6.4.2 Compactness analysis
The radius of gyration (Rg) is employed to measure the structural variations (compactness manner and flexibility) of protein–ligand complexes during MD simulations, and the lower value of Rg describes a more rigid structure (namely, a stronger interaction between protein and ligand) in the MD simulation process (Jiang et al., 2019). The plots of Rg versus time (Figure 8C) were quite different for ICAM1 (black), IRF1 (red) JAK2 (blue), NFKB1A (green), and STAT1 (purple) with the mean values at 1.56 nm, 1.38 nm, 1.99 nm, 1.98 nm, and 1.96 nm, respectively.
3.6.4.3 Solvent accessible surface area
Protein solvent accessible surface area (SASA) has been considered a critical element in the study of protein folding and stability, and a lower SASA value indicates greater compactness (Ali et al., 2014). The SASA values are quite different in range for ICAM1 (97.17 nm2, green), IRF1 (73.94 nm2, green), JAK2 (146.75 nm2, green), NFKB1A (133.68 nm2, green), and STAT1 (132.89 nm2, purple), respectively. This observation indicates a similar solvent interaction and also validates the compactness analysis results (Figure 8D).
3.6.4.4 Analysis of hydrogen bonding
Hydrogen bond formation between the ligand and protein shows the binding stability of the complex. The simulation results showed a large number of intermolecular hydrogen bonds formed between protein residues and the ligand (psoralen), indicating a strong interaction (Figure 8E). Figure 8E shows the number of hydrogen bonds formed during the simulations. The average number of hydrogen bonds for the complexes (psoralen/ICAM1, psoralen/IRF1, psoralen/JAK2, psoralen/NFKBIA, and psoralen/STAT1) were 0.32, 1.28, 0.16, 0.77, and 0.06, respectively. The docking studies showed that psoralen formed one hydrogen bond with protease residues Tyr166 in ICAM1 and protease residues Arg70 in STAT1, respectively.
3.6.4.5 Binding free energy and residue interaction energy
Molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) is widely used for free energy calculation based on the MD simulation of complexes (Wang et al., 2019). The binding free energy of all complexes was calculated using the MM/PBSA method from 50 ns extracted at equal intervals of the last 10 ns MD trajectories. The contributions of different interactions were either positive or negative to the binding free energy, as summarized in Table 4 for all complexes. The binding energy of the complexes (psoralen/ICAM1, psoralen/IRF1, psoralen/JAK2, psoralen/NFKBIA, and psoralen/STAT1) were −76.215, −15.363, −9.808, −3.224, and −18.293 kJ/mol, respectively. The binding energy results showed that the psoralen/ICAM1 exhibited more stability than other complexes. Decomposition into separate energy terms revealed that polar solvation energy and comparative enthalpy variation (∆TS) decrease the binding strength of inhibitors to the protease significantly, thereby reducing the total binding energy in both complexes due to the positive energy contributions. Among the various interactions, van der Waals energy showed the most favorable contributions towards the negative binding free energy of both complexes.
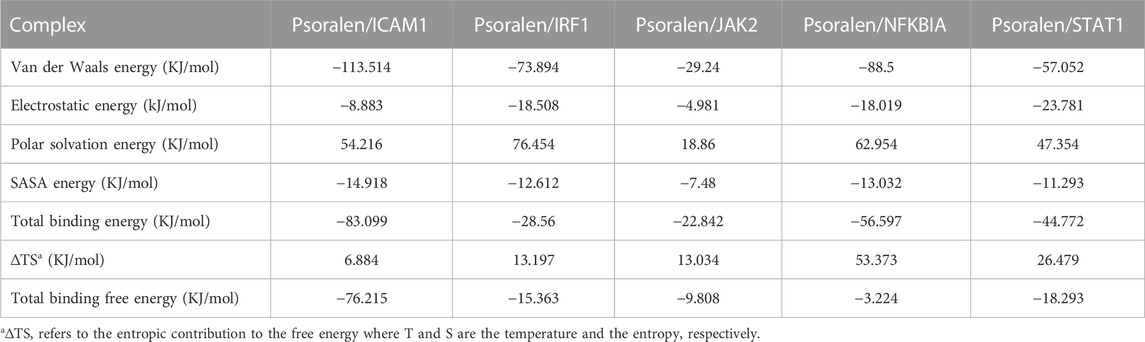
TABLE 4. The binding free energy component of a ligand (psoralen) with proteins (ICAM1, IRF1, JAK2, NFKBIA, and STAT1) based on molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) calculations.
3.6.5 Validation of potential core targets of the DILI_PGS for psoralen in IS model mice
The results of the RT-PCR showed that psoralen significantly increased the mRNA expressions of ICAM1, JAK2, STAT1, and IRF1 (all p < 0.05, Figures 9A–D) and decreased the levels of IDO1 and NFKB1A in the livers of IS model mice (both p < 0.05, Figures 9E, F). In addition, the results of serum ELISA showed that psoralen significantly increased and decreased the protein expression of TNF-α and IL-10 in a dose-dependent manner (all p < 0.05, Figures 9G, H), respectively.
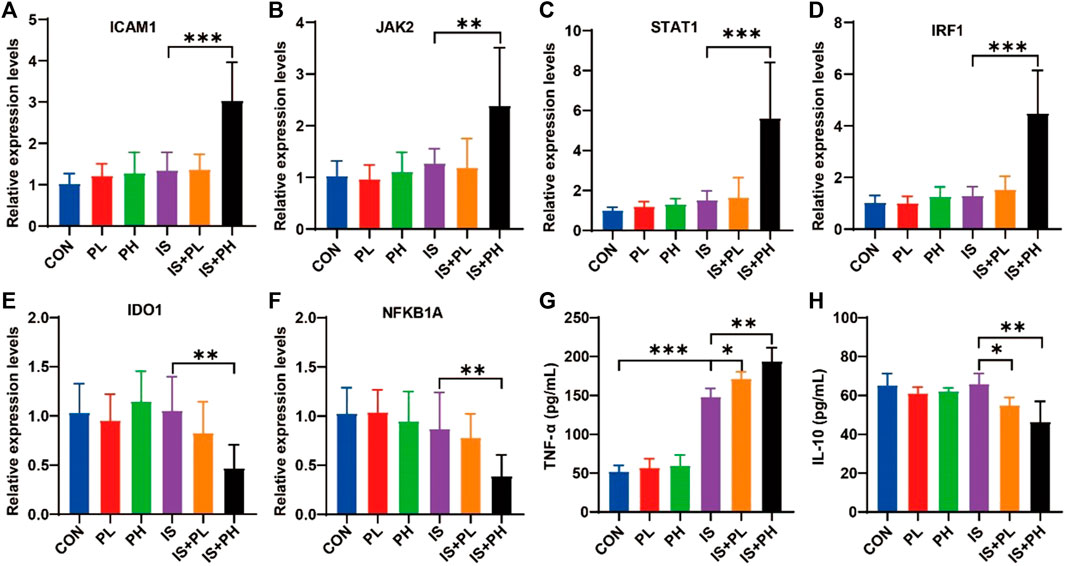
FIGURE 9. Effect of psoralen on the potential core targets expression in IS model mice. (A–F) The mRNA expressions of ICAM1 (A), JAK2 (B), STAT1 (C), IRF1 (D), IDO1 (E), and NFKB1A (F) in the liver of the IS mice were checked by RT-PCR. The protein expressions of TNF-α (G) and IL-10 (H) in the liver of the IS mice were checked by ELISA. *p < 0.05; **p < 0.01; ***p < 0.001.
4 Discussion
In this study, a gene set that can predict the risk of DILI was constructed based on the GEO database, and 32 drugs with high similarity (positive correlation) to the DILI-related DEGs profile were found by the strategy of drug repurposing, with 81.25% of the drugs identified as having hepatotoxicity. However, only three of the eight drugs with a high similarity (negative correlation) to this gene expression profile have been reported to cause liver injury, suggesting that this gene set has a certain ability to predict the risk of DILI.
Based on this gene set, this study also predicted that the basic disease characteristics of patients inducing DILI were mainly immune-related diseases or tumor-related diseases. Among the predicted immune-related diseases, autoimmune diseases including the chronic rejection of kidney transplantation (Wojciechowski and Wiseman, 2021), systemic lupus erythematosus (Fava and Petri, 2019), spondylitis (Ward et al., 2019), multiple sclerosis (Olek, 2021), and arthritis (Burmester and Pope, 2017) are usually treated with immunosuppressants or immunomodulators, such as tacrolimus, cyclosporine, corticosteroids (hydrocortisone, prednisone, and dexamethasone), and cyclophosphamide. However, some of these immunosuppressants (including tacrolimus, cyclophosphamide, cyclosporine, and corticosteroids) were reported to have a risk of hepatotoxicity (Welz et al., 1984; Akay et al., 2006; Ko et al., 2015; Lee et al., 2022), and have been included in the LiverTox platform (https://www.ncbi.nlm.nih.gov). Hepatitis is generally considered to be the critical feature of common underlying diseases inducing DILI (Kleiner et al., 2014; Ettel et al., 2017). The immune system of patients with infectious diseases (including viral hepatitis and influenza) is usually a condition of immune-inflammatory reactions (Knolle and Thimme, 2014; Chen et al., 2018), and previous studies have shown that immune stress is a common feature of DILI (Hoofnagle and Björnsson, 2019). Among the commonly used therapeutic drugs for hyperthyroidism patients, the most obvious side effect of methimazole and propylthiouracil is hepatotoxicity. A Japanese study showed that common drugs such as propylthiouracil and metimazole in patients with Graves’ disease (hyperthyroidism) can induce the occurrence of DILI (Suzuki et al., 2019). For patients with psoriasis and psoriatic arthritis, liver injury also easily occurs when receiving the commonly used treatment drug methotrexate (Gelfand et al., 2021).
For tumor-related diseases predicted by the DILI_PGS, due to the common abnormal immune and metabolic functions in tumor patients, the immune checkpoint inhibitors (PD1 and PDL1) (Peeraphatdit et al., 2020), biosynthesis drugs that interfere with nucleic acid (methotrexate) (Ebbesen et al., 2017), drugs that affect DNA structure and function (cyclophosphamide) (Ming et al., 2019), small-molecule targeted drugs (gefitinib) (Sugiyama et al., 2015) and other commonly used clinical antitumor drugs (Oun et al., 2018) developed based on this have been widely reported to have more side effects. The liver, the most important metabolic organ of the body, has a significantly increased risk of liver injury after taking immune checkpoint inhibitors for hepatocellular carcinoma (Sangro et al., 2020).
For the above immune-related diseases or tumor-related diseases, the body generally exhibits abnormal immune activation or hypoactivation (Khan and Gerber, 2020). To verify the sensitivity of basic disease characteristic information predicted by the DILI_PGS in patients with DILI, animal models of IS and IP were used to simulate the abnormal immune activation or immunosuppressive state, respectively. As for the selection of validation drugs, psoralen, the main monomer component of BGZ which has high molecular fingerprint similarity with the IDILI drugs (nefazodone, trovafloxacin, and nimesulide) from GSE102006 was selected for follow-up study. Psoralen has been documented by the LiverTox database for hepatotoxicity. Consistent with the literature reports (Yang et al., 2020), this study found that psoralen had certain accumulative hepatotoxicity in normal mice, but, surprisingly, in the IS model mice, a single dose of psoralen only could lead to the occurrence of liver damage, while in the IP model mice, multiple doses of psoralen did not significantly increase the severity of liver damage. These results suggest that psoralen is one of the components of BGZ-induced liver injury which occurs under immune stress (Tang et al., 2022). Therefore, the immune stress state may be the potential host factor of psoralen-induced liver injury, rather than an immunosuppressive condition, and the mechanism of psoralen-induced immunogenic liver injury becomes the focus of subsequent studies.
In this study, IDO1 and ICAM1 were screened as potential crosstalk targets of psoralen regulating IS and IP via network pharmacological technology, and the results of molecular docking and molecular dynamics also revealed that psoralen has strong binding energy with IDO1 and ICAM1, respectively. IDO1 is an important immunoregulatory factor and rate-limiting enzyme of the kynurenine pathway of tryptophan metabolism, induced pro-inflammatory cytokines (IFN-γ), and other immune system mediators in response to various immune-related reactions (Merlo et al., 2020). IDO1 is associated with many disorders of immune function, including cancer, HBV infection, allergy, and autoimmune diseases (Ito et al., 2014; Liu et al., 2018; Heidari et al., 2022). Inhibition of IDO1 expression can induce immune-mediated liver injury (Affolter et al., 2019), but it has also been shown that IDO1 deficiency or inhibition can weaken acetaminophen or concanavalin A-induced liver injury (Li et al., 2021b). ICAM1, a cell surface glycoprotein and adhesion receptor, is expressed at a low basic level in immune, endothelial, and epithelial cells, but upregulated in response to inflammatory stimulation, which promotes the recruitment and activation of white blood cells from circulation to inflammatory sites (Bui et al., 2020), playing an important role in the progress of diseases including tumors, hepatitis, colitis, and cardiovascular and cerebrovascular diseases (Chen et al., 2013; Hoke et al., 2015; Zhu et al., 2015; West et al., 2017). As expected, psoralen was found to decrease the expression of IDO1 and increase the expression of ICAM1 in the liver tissue of IS model mice, suggesting that ICAM1 and IDO1 may be potential targets of psoralen-induced immunogenic liver injury.
The core proteins (IRF1, STAT1, and TNF-α) of the DILI_PGS filtered by various topological analyses in the PPI network via CytoHubba are mainly related to the activation of the TNF signaling pathway (So and Ishii, 2019), showing that TNF-α can induce the expression of IRF1 and interferon-beta (IFN-β) by regulating tumor necrosis factor receptor 1 (TNFR1) to participate in the transcriptional activation mediated by NF-κB and STAT1, inducing the increased expression of IRF1 and IFN-β and leading to the prolonged expression of pro-inflammatory chemokines via STAT1 (Yarilina et al., 2008). IRF1 can also act synergistically with STAT1 to induce the expression of IFN-stimulated genes in IFN γ-treated cells (Feng et al., 2021). The results of molecular docking and molecular dynamics in this study showed that psoralen had a good binding ability with IRF1 and STAT1, and the animal experiment further demonstrated that psoralen could significantly increase the expression of IRF1, JAK2, STAT1, and TNF-α and significantly decrease the levels of IL-10 and NFKB1A in IS model mice. The above results suggest that the occurrence of immunogenic liver injury induced by psoralen may be related to the activation of the TNF signaling pathway.
In conclusion, based on the strategy of integrating gene expression profile similarities via CMap and Coexpedia, this study constructed a gene set with high accuracy to predict the occurrence of DILI, and this gene set revealed that the immune activation status is one of the main host factor inducing DILI. The study also found and confirmed that psoralen is one of the toxic components of BGZ with immunogenic hepatotoxicity, which extends the cognitive category that psoralen only accumulates hepatotoxicity as previously reported.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by the First Affiliated Hospital of Henan University of Traditional Chinese Medicine. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
M-LZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. W-XL: Formal Analysis, Writing–review and editing. X-YW: Writing–review and editing. HZ: Writing–review and editing. Y-LW: Writing–review and editing. L-QY: Writing–review and editing. X-FC: Writing–review and editing. S-QZ: Writing–review and editing. Y-LC: Writing–review and editing. K-RF: Writing–review and editing. J-FT: Funding acquisition, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Nos. 82173993 and 82004021), the Key Project of Henan Province for Scientific Research of Traditional Chinese Medicine, China (No. 2019ZYBJ08), the Key Scientific Research Projects of Colleges and Universities in Henan Province, China (No. 19A360007), and Science and Technology Innovation Team in Universities of Henan Province (No. 23IRTSTHN026).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1259569/full#supplementary-material
References
Affolter, T., Llewellyn, H. P., Bartlett, D. W., Zong, Q., Xia, S., Torti, V., et al. (2019). Inhibition of immune checkpoints PD-1, CTLA-4, and IDO1 coordinately induces immune-mediated liver injury in mice. PLoS One 14 (5), e0217276. doi:10.1371/journal.pone.0217276
Akay, H., Akay, T., Secilmis, S., Kocak, Z., and Donderici, O. (2006). Hepatotoxicity after low-dose cyclophosphamide therapy. South Med. J. 99 (12), 1399–1400. doi:10.1097/01.smj.0000251467.62842.ad
Ali, S. A., Hassan, M. I., Islam, A., and Ahmad, F. (2014). A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci. 15 (5), 456–476. doi:10.2174/1389203715666140327114232
Amitrano, L., Gigliotti, T., Guardascione, M. A., and Ascione, A. (1992). Enoxacin acute liver injury. J. Hepatol. 15 (1-2), 270. doi:10.1016/0168-8278(92)90050-y
Bui, T. M., Wiesolek, H. L., and Sumagin, R. (2020). ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 108 (3), 787–799. doi:10.1002/jlb.2mr0220-549r
Burmester, G. R., and Pope, J. E. (2017). Novel treatment strategies in rheumatoid arthritis. Lancet 389 (10086), 2338–2348. doi:10.1016/s0140-6736(17)31491-5
Chalasani, N., Bonkovsky, H. L., Fontana, R., Lee, W., Stolz, A., Talwalkar, J., et al. (2015). Features and outcomes of 899 patients with drug-induced liver injury: the DILIN prospective study. Gastroenterology 148 (7), 1340–1352.e7. doi:10.1053/j.gastro.2015.03.006
Chalasani, N. P., Maddur, H., Russo, M. W., Wong, R. J., and Reddy, K. R. (2021). ACG clinical guideline: diagnosis and management of idiosyncratic drug-induced liver injury. Am. J. Gastroenterol. 116 (5), 878–898. doi:10.14309/ajg.0000000000001259
Chang, Y., Park, H., Yang, H. J., Lee, S., Lee, K. Y., Kim, T. S., et al. (2018). Cancer drug response profile scan (CDRscan): A deep learning model that predicts drug effectiveness from cancer genomic signature. Sci. Rep. 8 (1), 8857. doi:10.1038/s41598-018-27214-6
Chen, H., Zhang, D., Wang, S., Wang, X., and Yang, C. (2013). Significance of correlation between interferon-γ and soluble intercellular adhesion molecule-1 and interleukin-17 in hepatitis B virus-related cirrhosis. Clin. Res. Hepatol. Gastroenterol. 37 (6), 608–613. doi:10.1016/j.clinre.2013.05.010
Chen, M., Suzuki, A., Thakkar, S., Yu, K., Hu, C., and Tong, W. (2016). DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today 21 (4), 648–653. doi:10.1016/j.drudis.2016.02.015
Chen, S., Liu, X., Peng, C., Tan, C., Sun, H., Liu, H., et al. (2021). The phytochemical hyperforin triggers thermogenesis in adipose tissue via a Dlat-AMPK signaling axis to curb obesity. Cell Metab. 33 (3), 565–580.e7. doi:10.1016/j.cmet.2021.02.007
Chen, X., Liu, S., Goraya, M. U., Maarouf, M., Huang, S., and Chen, J. L. (2018). Host immune response to influenza A virus infection. Front. Immunol. 9, 320. doi:10.3389/fimmu.2018.00320
de Faria, P. A., Bettanin, F., Cunha, R. L., Paredes-Gamero, E. J., Homem-de-Mello, P., Nantes, I. L., et al. (2015). Cytotoxicity of phenothiazine derivatives associated with mitochondrial dysfunction: a structure-activity investigation. Toxicology 330, 44–54. doi:10.1016/j.tox.2015.02.004
Devarbhavi, H., Karanth, D., Prasanna, K. S., Adarsh, C. K., and Patil, M. (2011). Drug-induced liver injury with hypersensitivity features has a better outcome: a single-center experience of 39 children and adolescents. Hepatology 54 (4), 1344–1350. doi:10.1002/hep.24527
Donnelly, M. C., Davidson, J. S., Martin, K., Baird, A., Hayes, P. C., and Simpson, K. J. (2017). Acute liver failure in scotland: changes in aetiology and outcomes over time (the scottish look-back study). Aliment. Pharmacol. Ther. 45 (6), 833–843. doi:10.1111/apt.13943
Ebbesen, M. S., Nygaard, U., Rosthøj, S., Sørensen, D., Nersting, J., Vettenranta, K., et al. (2017). Hepatotoxicity during maintenance therapy and prognosis in children with acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 39 (3), 161–166. doi:10.1097/mph.0000000000000733
el Saghir, N. S., and Hawkins, K. A. (1984). Hepatotoxicity following vincristine therapy. Cancer 54 (9), 2006–2008. doi:10.1002/1097-0142(19841101)54:9<2006::aid-cncr2820540937>3.0.co;2-f
Ettel, M., Gonzalez, G. A., Gera, S., Eze, O., Sigal, S., Park, J. S., et al. (2017). Frequency and pathological characteristics of drug-induced liver injury in a tertiary medical center. Hum. Pathol. 68, 92–98. doi:10.1016/j.humpath.2017.08.029
European Association for the Study of the Liver (2019). EASL clinical practice guidelines: drug-induced liver injury. J. Hepatol. 70 (6), 1222–1261. doi:10.1016/j.jhep.2019.02.014
Fava, A., and Petri, M. (2019). Systemic lupus erythematosus: diagnosis and clinical management. J. Autoimmun. 96, 1–13. doi:10.1016/j.jaut.2018.11.001
Feng, H., Zhang, Y. B., Gui, J. F., Lemon, S. M., and Yamane, D. (2021). Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 17 (1), e1009220. doi:10.1371/journal.ppat.1009220
Ge, F. L., Niu, M., Han, Z. X., Cao, J. L., Wang, J. B., Bai, Z. F., et al. (2021). Landscape of hepatobiliary adverse drug reactions related to preparations containing psoraleae fructus and its application in pharmacovigilance. Chin. J. Integr. Med. 27, 832–837. doi:10.1007/s11655-021-3442-2
Gelfand, J. M., Wan, J., Zhang, H., Shin, D. B., Ogdie, A., Syed, M. N., et al. (2021). Risk of liver disease in patients with psoriasis, psoriatic arthritis, and rheumatoid arthritis receiving methotrexate: A population-based study. J. Am. Acad. Dermatol 84 (6), 1636–1643. doi:10.1016/j.jaad.2021.02.019
Hannequin, J. R., Doffoel, M., and Schmutz, G. (1988). Hepatitis secondary to current non-steroidal anti-inflammatory agents. Rev. Rhum. Mal. Osteoartic. 55 (12), 983–988.
Heidari, F., Ramezani, A., Erfani, N., and Razmkhah, M. (2022). Indoleamine 2, 3-dioxygenase: A professional immunomodulator and its potential functions in immune related diseases. Int. Rev. Immunol. 41 (3), 346–363. doi:10.1080/08830185.2020.1836176
Hoke, M., Winter, M. P., Wagner, O., Exner, M., Schillinger, M., Arnold, Z., et al. (2015). The impact of selectins on mortality in stable carotid atherosclerosis. Thromb. Haemost. 114 (3), 632–638. doi:10.1160/th14-12-1014
Hoofnagle, J. H., and Björnsson, E. S. (2019). Drug-induced liver injury - types and phenotypes. N. Engl. J. Med. 381 (3), 264–273. doi:10.1056/nejmra1816149
Hoofnagle, J. H., Serrano, J., Knoben, J. E., and Navarro, V. J. (2013). LiverTox: a website on drug-induced liver injury. Hepatology 57 (3), 873–874. doi:10.1002/hep.26175
Ito, H., Ando, T., Ando, K., Ishikawa, T., Saito, K., Moriwaki, H., et al. (2014). Induction of hepatitis B virus surface antigen-specific cytotoxic T lymphocytes can be up-regulated by the inhibition of indoleamine 2, 3-dioxygenase activity. Immunology 142 (4), 614–623. doi:10.1111/imm.12274
Jiang, J., Mathijs, K., Timmermans, L., Claessen, S. M., Hecka, A., Weusten, J., et al. (2017). Omics-based identification of the combined effects of idiosyncratic drugs and inflammatory cytokines on the development of drug-induced liver injury. Toxicol. Appl. Pharmacol. 332, 100–108. doi:10.1016/j.taap.2017.07.014
Jiang, M., Wang, X., Lv, B., Lu, Y., Ma, X., Liu, W., et al. (2022). Psoralen induces hepatotoxicity by covalently binding to glutathione-S-transferases and the hepatic cytochrome P450. Phytomedicine 104, 154165. doi:10.1016/j.phymed.2022.154165
Jiang, Z., You, L., Dou, W., Sun, T., and Xu, P. (2019). Effects of an electric field on the conformational transition of the protein: A molecular dynamics simulation study. Polym. (Basel) 11 (2), 282. doi:10.3390/polym11020282
Khan, S., and Gerber, D. E. (2020). Autoimmunity, checkpoint inhibitor therapy and immune-related adverse events: A review. Semin. Cancer Biol. 64, 93–101. doi:10.1016/j.semcancer.2019.06.012
Kleiner, D. E., Chalasani, N. P., Lee, W. M., Fontana, R. J., Bonkovsky, H. L., Watkins, P. B., et al. (2014). Hepatic histological findings in suspected drug-induced liver injury: systematic evaluation and clinical associations. Hepatology 59 (2), 661–670. doi:10.1002/hep.26709
Knolle, P. A., and Thimme, R. (2014). Hepatic immune regulation and its involvement in viral hepatitis infection. Gastroenterology 146 (5), 1193–1207. doi:10.1053/j.gastro.2013.12.036
Ko, M. S., Choi, Y. H., Jung, S. H., Lee, J. S., Kim, H. S., Lee, C. H., et al. (2015). Tacrolimus therapy causes hepatotoxicity in patients with a history of liver disease. Int. J. Clin. Pharmacol. Ther. 53 (5), 363–371. doi:10.5414/cp202226
Lee, C. C., Peng, Y. J., Lu, C. C., Chen, H. C., and Yeh, F. C. (2022). Corticosteroid-induced liver injury in adult-onset still's disease. Med. Kaunas. 58 (2), 191. doi:10.3390/medicina58020191
Li, Y., Song, Y., Deng, G., Tan, Q., Xu, S., Yang, M., et al. (2021b). Indoleamine 2, 3-dioxygenase 1 aggravates acetaminophen-induced acute liver failure by triggering excess nitroxidative stress and iron accumulation. Free Radic. Biol. Med. 172, 578–589. doi:10.1016/j.freeradbiomed.2021.07.008
Li, Y., Yu, P., Fu, W., Cai, L., Yu, Y., Feng, Z., et al. (2021a). Ginseng-Astragalus-oxymatrine injection ameliorates cyclophosphamide-induced immunosuppression in mice and enhances the immune activity of RAW264.7 cells. J. Ethnopharmacol. 279, 114387. doi:10.1016/j.jep.2021.114387
Li, Z. J., Abulizi, A., Zhao, G. L., Wang, T., Zhou, F., Jiang, Z. Z., et al. (2017). Bakuchiol contributes to the hepatotoxicity of Psoralea corylifolia in rats. Phytother. Res. 31 (8), 1265–1272. doi:10.1002/ptr.5851
Li, Z., Peng, M., Chen, P., Liu, C., Hu, A., Zhang, Y., et al. (2022). Imatinib and methazolamide ameliorate COVID-19-induced metabolic complications via elevating ACE2 enzymatic activity and inhibiting viral entry. Cell Metab. 34 (3), 424–440.e7. doi:10.1016/j.cmet.2022.01.008
Liu, M., Wang, X., Wang, L., Ma, X., Gong, Z., Zhang, S., et al. (2018). Targeting the IDO1 pathway in cancer: from bench to bedside. J. Hematol. Oncol. 11 (1), 100. doi:10.1186/s13045-018-0644-y
Ma, X., Li, H., Gong, Y., Liu, F., Tong, X., Zhu, F., et al. (2022). Psoralen inhibits hepatitis B viral replication by down-regulating the host transcriptional machinery of viral promoters. Virol. Sin. 37 (2), 256–265. doi:10.1016/j.virs.2022.01.027
Merlo, L. M. F., DuHadaway, J. B., Montgomery, J. D., Peng, W. D., Murray, P. J., Prendergast, G. C., et al. (2020). Differential roles of IDO1 and IDO2 in T and B cell inflammatory immune responses. Front. Immunol. 11, 1861. doi:10.3389/fimmu.2020.01861
Ming, Z., Yongqiang, Z., Zijin, Z., Yan, X., Di, C., and Xiaoxin, T. (2019). Severe and prolonged cyclophosphamide-induced hepatotoxicity in a breast cancer patient carrying a CYP2B6*7 variant. Pharmacogenomics 20 (16), 1119–1124. doi:10.2217/pgs-2019-0093
Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., et al. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12 (5), 453–457. doi:10.1038/nmeth.3337
Olek, M. J. (2021). Multiple sclerosis. Ann. Intern Med. 174 (6), Itc81–itc96. doi:10.7326/aitc202106150
Oun, R., Moussa, Y. E., and Wheate, N. J. (2018). The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans. 47 (19), 6645–6653. doi:10.1039/c8dt00838h
Park, E. J., Zhao, Y. Z., Kim, Y. C., and Sohn, D. H. (2005). Protective effect of (S)-bakuchiol from Psoralea corylifolia on rat liver injury in vitro and in vivo. Planta Med. 71 (6), 508–513. doi:10.1055/s-2005-864150
Peeraphatdit, T. B., Wang, J., Odenwald, M. A., Hu, S., Hart, J., and Charlton, M. R. (2020). Hepatotoxicity from immune checkpoint inhibitors: A systematic review and management recommendation. Hepatology 72 (1), 315–329. doi:10.1002/hep.31227
Rank, J. M., and Olson, R. C. (1989). Reversible cholestatic hepatitis caused by acetohexamide. Gastroenterology 96 (6), 1607–1608. doi:10.1016/0016-5085(89)90535-0
Regev, A. (2014). Drug-induced liver injury and drug development: industry perspective. Semin. Liver Dis. 34 (2), 227–239. doi:10.1055/s-0034-1375962
Reuben, A., Koch, D. G., and Lee, W. M. (2010). Drug-induced acute liver failure: results of a U.S. multicenter, prospective study. Hepatology 52 (6), 2065–2076. doi:10.1002/hep.23937
Reuben, A., Tillman, H., Fontana, R. J., Davern, T., McGuire, B., Stravitz, R. T., et al. (2016). Outcomes in adults with acute liver failure between 1998 and 2013: an observational cohort study. Ann. Intern Med. 164 (11), 724–732. doi:10.7326/m15-2211
Rogers, D., and Hahn, M. (2010). Extended-connectivity fingerprints. J. Chem. Inf. Model 50 (5), 742–754. doi:10.1021/ci100050t
Sangro, B., Chan, S. L., Meyer, T., Reig, M., El-Khoueiry, A., and Galle, P. R. (2020). Diagnosis and management of toxicities of immune checkpoint inhibitors in hepatocellular carcinoma. J. Hepatol. 72 (2), 320–341. doi:10.1016/j.jhep.2019.10.021
Shen, W., Song, Z., Zhong, X., Huang, M., Shen, D., Gao, P., et al. (2022). Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 1 (3), e36. doi:10.1002/imt2.36
So, T., and Ishii, N. (2019). The TNF-TNFR family of Co-signal molecules. Adv. Exp. Med. Biol. 1189, 53–84. doi:10.1007/978-981-32-9717-3_3
Subramanian, A., Narayan, R., Corsello, S. M., Peck, D. D., Natoli, T. E., Lu, X., et al. (2017). A next generation connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell 171 (6), 1437–1452.e17. doi:10.1016/j.cell.2017.10.049
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Sugiyama, E., Umemura, S., Nomura, S., Kirita, K., Matsumoto, S., Yoh, K., et al. (2015). Impact of single nucleotide polymorphisms on severe hepatotoxicity induced by EGFR tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring EGFR mutations. Lung Cancer 90 (2), 307–313. doi:10.1016/j.lungcan.2015.08.004
Suzuki, N., Noh, J. Y., Hiruma, M., Kawaguchi, A., Morisaki, M., Ohye, H., et al. (2019). Analysis of antithyroid drug-induced severe liver injury in 18,558 newly diagnosed patients with Graves' disease in Japan. Thyroid 29 (10), 1390–1398. doi:10.1089/thy.2019.0045
Tang, J. F., Niu, L., Wang, X. Y., Zhang, H., Zhang, M. L., Wu, Y. L., et al. (2022). Comparison on distribution of 8 components from ethanol extract of Psoraleae Fructus between normal and lipopolysaccharide-induced model rats. Zhongguo Zhong Yao Za Zhi 47 (24), 6763–6779. doi:10.19540/j.cnki.cjcmm.20220823.501
Van Mouwerik, T. J., Caines, P. M., and Ballentine, R. (1987). Amsacrine evaluation. Drug Intell. Clin. Pharm. 21 (4), 330–334. doi:10.1177/106002808702100403
Wang, E., Sun, H., Wang, J., Wang, Z., Liu, H., Zhang, J. Z. H., et al. (2019). End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 119 (16), 9478–9508. doi:10.1021/acs.chemrev.9b00055
Wang, L., Wang, Y., Wee, A., Soon, G., Gouw, A. S. H., Yang, R., et al. (2020a). Clinicopathological features of Bu Gu Zhi-induced liver injury, a long-term follow-up cohort study. Liver Int. 40 (3), 571–580. doi:10.1111/liv.14306
Wang, L., Xu, G., Wang, H., Zhan, X., Gao, Y., Chen, N., et al. (2020b). Icariside Ⅱ, a main compound in Epimedii Folium, induces idiosyncratic hepatotoxicity by enhancing NLRP3 inflammasome activation. Acta Pharm. Sin. B 10 (9), 1619–1633. doi:10.1016/j.apsb.2020.03.006
Ward, M. M., Deodhar, A., Gensler, L. S., Dubreuil, M., Yu, D., Khan, M. A., et al. (2019). 2019 update of the American college of rheumatology/spondylitis association of America/spondyloarthritis research and treatment network recommendations for the treatment of ankylosing spondylitis and nonradiographic axial spondyloarthritis. Arthritis Care Res. Hob. 71 (10), 1285–1299. doi:10.1002/acr.24025
Wei, G., Bergquist, A., Broomé, U., Lindgren, S., Wallerstedt, S., Almer, S., et al. (2007). Acute liver failure in Sweden: etiology and outcome. J. Intern Med. 262 (3), 393–401. doi:10.1111/j.1365-2796.2007.01818.x
Welz, A., Reichart, B., Uberfuhr, P., Kemkes, B., and Klinner, W. (1984). Cyclosporine as the main immunosuppressant in clinical heart transplantation: correlation of hepatotoxicity and nephrotoxicity. Transpl. Proc. 16 (5), 1212–1213.
West, N. R., Hegazy, A. N., Owens, B. M. J., Bullers, S. J., Linggi, B., Buonocore, S., et al. (2017). Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 23 (5), 579–589. doi:10.1038/nm.4307
Wojciechowski, D., and Wiseman, A. (2021). Long-Term immunosuppression management: opportunities and uncertainties. Clin. J. Am. Soc. Nephrol. 16 (8), 1264–1271. doi:10.2215/cjn.15040920
Xing, X., Xia, Q., Gong, B., Shen, Z., and Zhang, Y. (2022). Identification of tissue-specific expressed hub genes and potential drugs in rheumatoid arthritis using bioinformatics analysis. Front. Genet. 13, 855557. doi:10.3389/fgene.2022.855557
Yang, L., Yu, Y. L., Cheng, L. Y., Zhang, P. Y., Zhang, Y., and Zhou, K. (2020). The accumulation of psoralen contributes to its hepatotoxicity revealed by pharmacokinetic and toxicokinetic study after repeated administration. ACS Omega 5 (13), 7307–7315. doi:10.1021/acsomega.9b04153
Yang, S., Kim, C. Y., Hwang, S., Kim, E., Kim, H., Shim, H., et al. (2017). COEXPEDIA: exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH). Nucleic Acids Res. 45 (D1), D389–d396. doi:10.1093/nar/gkw868
Yarilina, A., Park-Min, K. H., Antoniv, T., Hu, X., and Ivashkiv, L. B. (2008). TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 9 (4), 378–387. doi:10.1038/ni1576
Yoshihara, K., Shahmoradgoli, M., Martínez, E., Vegesna, R., Kim, H., Torres-Garcia, W., et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612. doi:10.1038/ncomms3612
Keywords: drug-induced liver injury, differentially expressed genes, predict, host characteristics, psoralen
Citation: Zhang M-L, Li W-X, Wang X-Y, Zhang H, Wu Y-L, Yang L-Q, Chen X-F, Zhang S-Q, Chen Y-L, Feng K-R and Tang J-F (2023) A gene expression profile-based approach to screen the occurrence and predisposed host characteristics of drug-induced liver injury: a case study of Psoralea corylifolia Linn. Front. Chem. 11:1259569. doi: 10.3389/fchem.2023.1259569
Received: 16 July 2023; Accepted: 12 September 2023;
Published: 06 October 2023.
Edited by:
Jun-sheng Tian, Shanxi University, ChinaReviewed by:
Dingkun Zhang, Chengdu University of Traditional Chinese Medicine, ChinaWeijun Kong, Capital Medical University, China
Copyright © 2023 Zhang, Li, Wang, Zhang, Wu, Yang, Chen, Zhang, Chen, Feng and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Fa Tang, a0519@163.com
 Ming-Liang Zhang
Ming-Liang Zhang Wei-Xia Li
Wei-Xia Li Xiao-Yan Wang
Xiao-Yan Wang Hui Zhang
Hui Zhang Ya-Li Wu
Ya-Li Wu Liu-Qing Yang
Liu-Qing Yang Xiao-Fei Chen
Xiao-Fei Chen Shu-Qi Zhang
Shu-Qi Zhang Yu-Long Chen
Yu-Long Chen Ke-Ran Feng
Ke-Ran Feng Jin-Fa Tang
Jin-Fa Tang