Structure-based virtual screening identified novel FOXM1 inhibitors as the lead compounds for ovarian cancer
- Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Ovarian cancer (OC) is a gynecological tumor with possibly the worst prognosis, its 5-year survival rate being only 47.4%. The first line of therapy prescribed is chemotherapy consisting of platinum and paclitaxel. The primary reason for treatment failure is drug resistance. FOXM1 protein has been found to be closely associated with drug resistance, and inhibition of FOXM1 expression sensitizes cisplatin-resistant ovarian cancer cells. Combining existing first-line chemotherapy drugs with FOXM1 prolongs the overall survival of patients, therefore, FOXM1 is considered a potential therapeutic target in ovarian cancer. Previous research conducted by our team revealed a highly credible conformation of FOXM1 which enables binding by small molecules. Based on this conformation, the current study conducted virtual screening to determine a new structural skeleton for FOXM1 inhibitors which would enhance their medicinal properties. DZY-4 showed the highest affinity towards FOXM1, and its inhibitory effect on proliferation and migration of ovarian cancer at the cellular level was better than or equal to that of cisplatin, while its efficacy was equivalent to that of cisplatin in a nude mouse model. In this study, the anti-tumor effect of DZY-4 is reported for the first time. DZY-4 shows potential as a drug that can be used for ovarian cancer treatment, as well as a drug lead for future research.
Introduction
Ovarian cancer is a serious health issue that endangers the health of women worldwide. The rate of incidence of ovarian cancer is ranked third among those of gynecological cancers, second to those of cervical cancer and uterine cancer (Momenimovahed, et al., 2019). Globally, 308,069 new cases of ovarian cancer were diagnosed in 2020, resulting in 193,811 cancer-specific deaths (Yang et al., 2020). Ovarian cancer has the highest mortality rate and the worst prognoses among gynecological cancers, with the 5-year survival rate being only 47.4% (Stewart et al., 2019). Due to the absence of formal screening methods and obscure early symptoms, a majority (approximately 80%) of patients are diagnosed at an advanced stage (III/IV) and do not qualify for radical surgery (Gralewska et al., 2020). Although first-line treatment, consisting of surgery followed by combination chemotherapy, usually carboplatin and paclitaxel, is effective (Gadducci et al., 2019), approximately 75% of patients with advanced ovarian cancer experience recurrence within 3 years (Dasari and Tchounwou, 2014). In recent years, despite new advances in drug therapy, such as the combining of bevacizumab, a vascular endothelial growth factor receptor (VEGFR) inhibitor, with carboplatin and paclitaxel chemotherapy, progression free survival (PFS) was prolonged only by 3.5 months, with no significant differences in overall survival (OS) being observed (Lheureux et al., 2019). Recently, poly (ADP-ribose) polymerase (PARP) inhibitors, such as niraparib, became the first targeted agents to be approved by the United States Food and Drug Administration Agency (FDA). PARPs imparted substantial clinical benefits to recurrent ovarian cancer patients following platinum-based chemotherapy, the median PFS in the niraparib group being significantly longer than that in the placebo group (21.9 months vs. 10.4 months, respectively) (Lord and Ashworth, 2017). However, its efficacy is somewhat limited due to the presence of diverse resistance mechanisms, including the loss of PARP1 protein itself, which maintains replication fork stability and mutations in BRCA thereby sufficiently restoring the homologous recombination repair (HRR) function (Jiang et al., 2020). Currently, drug therapy remains the mainstay of ovarian cancer treatment, and the need to identify new targets and more effective drugs against FOXM1 in order to improve treatment has been recognized.
Forkhead box M1 (FOXM1), a member of the Forkhead box (FOX) transcription factor family (Su et al., 2021), is well-known for its imperative role in various physiological functions, including cell cycle regulation, DNA damage repair, and apoptosis. It is ubiquitously expressed in proliferating and regenerating mammalian cells. FOXM1 contributes to oncogenesis (Quan et al., 2013), overexpression of FOXM1 increased invasion (Halasi and Gartel, 2013), angiogenesis (Karadedou et al., 2012), epithelial-mesenchymal transition (Meng et al., 2015), and self-renewal as well as chemotherapy resistance (Gomes et al., 2013). The Cancer Genome Atlas (TCGA) revealed that the FOXM1 transcriptional pathway was eccentrically activated in over 85% of high-grade serous carcinoma (HGSC) cases (Pratheeshkumar et al., 2018) and that high FOXM1 expression was associated with the stage and prognosis of ovarian cancer (Gentles et al., 2015). Other studies have shown that FOXM1 enhances the invasion and metastasis of ovarian cancer cells by upgrading the expression of matrix metalloproteinase (MMP-2) and degrading the extracellular matrix (Gentles et al., 2015). In addition, FOXM1 promotes DNA repair and chemotherapy resistance. Thiostrepton, a small molecule inhibitor (SMI) of FOXM1, reportedly sensitizes ovarian cancer cells to platinum-based chemotherapy (Lin et al., 2013). Furthermore, cisplatin-resistant ovarian cancer tissues and cells display increased FOXM1, which therefore acts as an independent indicator of the rapid progression of platinum-resistant epithelial ovarian cancer (EOC); (Tassi et al., 2017). Investigations of FOXM1 also showed that it plays a role in PARPi and paclitaxel resistance in ovarian cancer (Liu et al., 2021). In conclusion, FOXM1 plays an important role in the development of ovarian cancer as well as in drug resistance in this disease, indicating that inhibition of FOXM1 activity may play a role in ovarian cancer therapy.
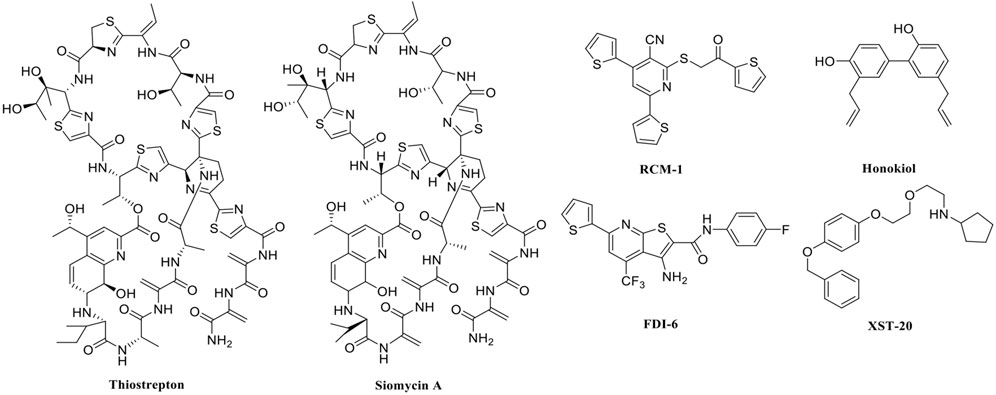
In recent years, FOXM1 inhibitors have been successively reported, and the structures of some are shown (Figure 1). Siomycin A and thiostrepton were reported as inhibitors of the transcriptional activity of FOXM1. However, these two are protease inhibitors, which target multiple signaling pathways in addition to those associated with FOXM1 (Bhat et al., 2009). XST-20 was identified as a leading compound that inhibits the transcriptional activity of FOXM1 and specifically blocks FOXM1-DNA binding. However, it poses certain medicine-related issues, such as poor water solubility and oral absorption difficulties (Zhang et al., 2022). Gormally et al. used a high-throughput screen, to demonstrate that FOXM1 is inhibited by a novel small molecule inhibitor, FDI-6, and they also have demonstrated that the ligand makes direct contact with FOXM1 and inhibits DNA binding in cells. However, its high cytotoxicity limited its use in many biomolecular experiments (Gormally et al., 2014). Shukla et al., also identified a novel small molecule, RCM-1, via high-throughput screening, which can effectively and specifically inhibit nuclear localization of the FOXM1 protein, causing it to be ubiquitinated and degraded by proteasomes. However, the binding mode of RCM-1, FDI-6, and FOXM1 remains unclear (Shukla et al., 2019). The natural product, honokiol reportedly hinders FOXM1 transcriptional activity, disrupting the FOXM1 auto-regulation loop and reducing FOXM1 mRNA and protein expression. However, further clinical application of these inhibitors is hampered by their insolubility in water and resistance to absorption (Halasi et al., 2018). Currently, there is no explicit FOXM1-based drug lead that can be used in subsequent studies. Our group previously used molecular docking to study the binding mode of FDI-6 to FOXM1. We conducted FDI-6 similarity retrieval to establish a derivative library, following which affinities of the compounds in the derivative library were determined via surface plasmon resonance (SPR), at the molecular level, and via IC50, at the cellular level. Based on the relationship between virtual docking energy, affinity, and anti-ovarian cancer activity of compounds, a more reliable binding conformation of FDI-6 and FOXM1 was identified.
In this study, we aimed to explore new structural types of FOXM1 inhibitors utilizing previous binding conformations to identify potential drug candidates for ovarian cancer. Our findings may also be used as a biochemical research tool to identify the molecular mechanism underlying the role of FOXM1 in ovarian cancer, as well as to assess its value as a therapeutic target in this disease.
Results and discussion
Structure-based virtual screening for FOXM1 inhibitors
A search for new FOXM1 inhibitors via structure-based virtual screening requires the identification of the cavity in which FOXM1 binds small molecules. The crystal structure of FOXM1 (PDB ID: 3G73) obtained from the RCSB protein database is used to generate receptor input during the docking process. The position at which FOXM1 binds to small molecules, as indicated by previous studies, is shown (Figure 2A). The surface binding mode is also shown (Figure 2B). The key amino acids involved in the binding between FOXM1 and small molecules are suggested to be ARG236, TYR272 (Figures 2C,D; docking pose). We used this binding site for subsequent screening.
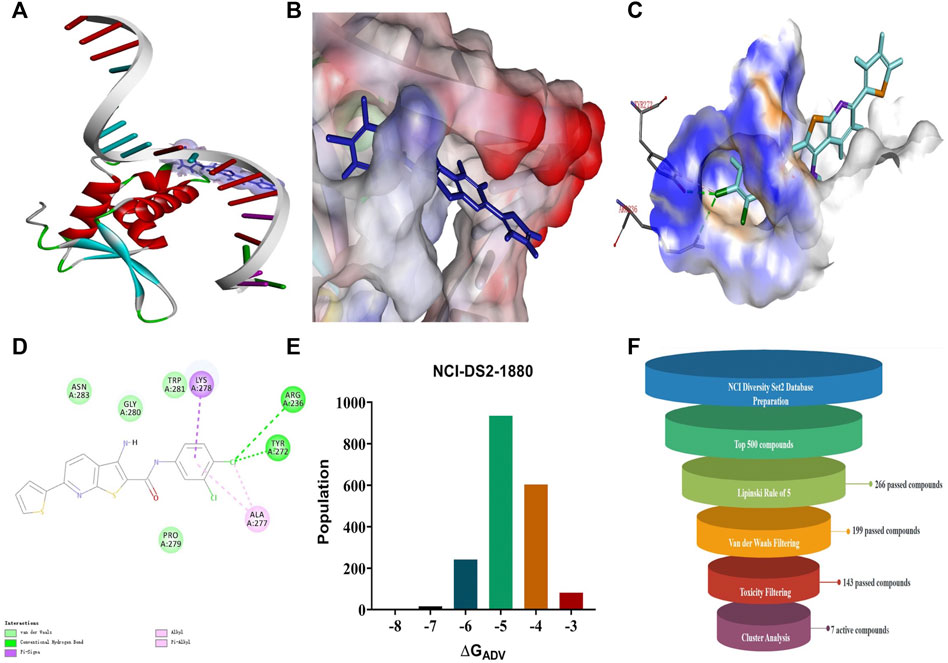
FIGURE 2. Structure-based virtual screening for FOXM1 inhibitors. (A) The position of FOXM1 binding to small molecule. (B) Surface view of a small molecule bound to FOXM1 (C) Close-up view of a small molecule bound to FOXM1.The key residues that contact the inhibitor are labeled. The small molecule is shown in purple. (D) 2D interaction diagram of FOXM1 and the small molecule. (E) Results of the VS. (using ADV) of the NCI Diversity Set 2 against FOXM1. Bars represent numbers of Diversity Set compounds with predicted free energies of binding in indicated 1 kcal/mol bins. (F) Overview of the flowchart for the discovery of FOXM1 inhibitors.
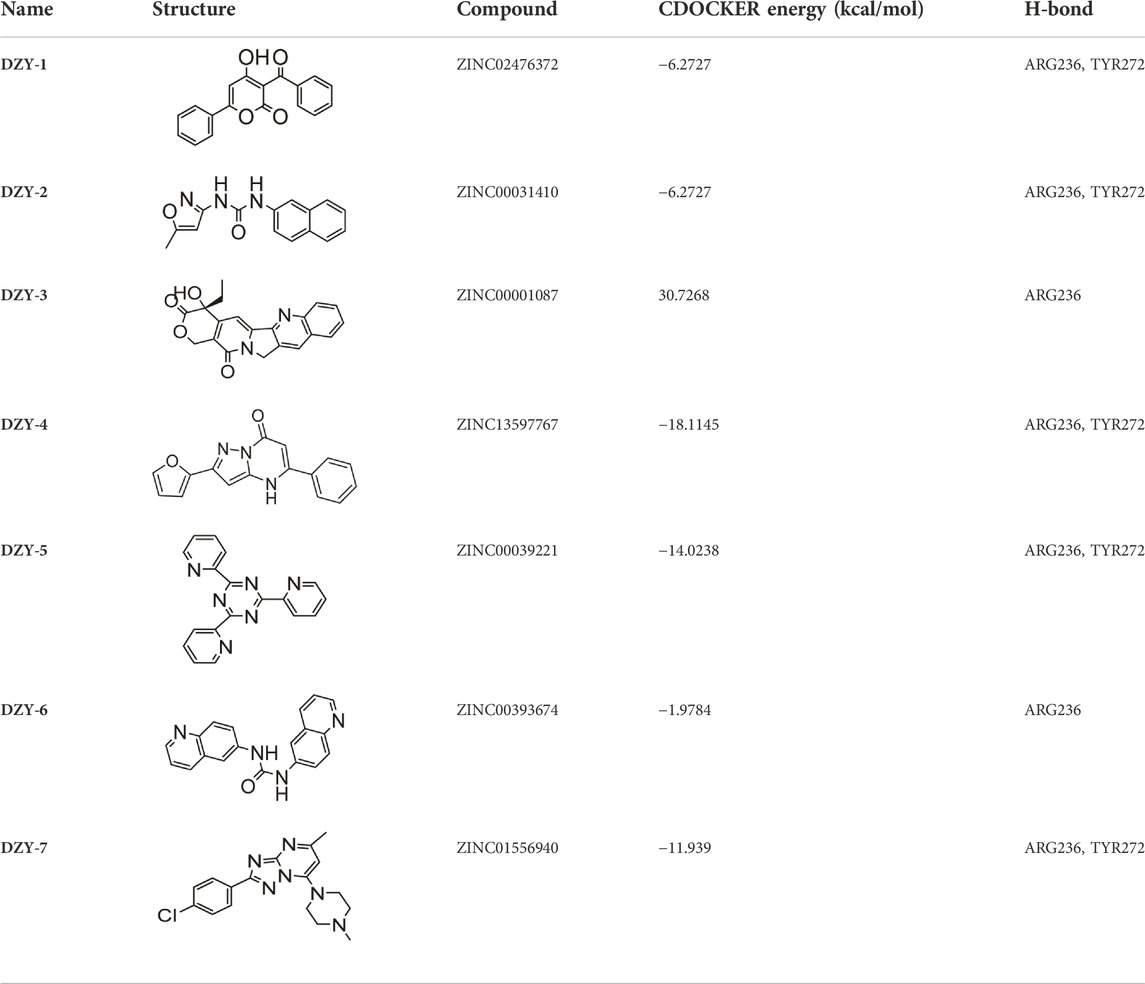
First, we used a structure-based virtual docking approach to screen the National Cancer Institute (NCI) Diversity Set 2, which represent the diversity of 265242 compounds available at NCI. A total of 1880 compounds were docked at FOXM1 according to AutoDock Vina (ADV). The virtual screening (VS) results were sorted based on their predicted binding free energies (GADV). The results indicated that ∆GADV ranged from −2.8 to −7. 8 kcal/mol, where 13.7% of Set 2 had energies lower than −6.0 kcal/mol (GADV of FDI-6); (Figure 2E). To narrow down the selection and eliminate unsuitable molecules in the database, we used the calculated binding energies data to select 500 compounds. These compounds were filtered according to Lipinski’s “Rule of five” and the Veber rule. The remaining compounds were further refined using the “toxicity prediction” tool, implemented by the Discovery Studio (DS), to filter out potentially carcinogenic, mutagenic, or teratogenic compounds. This three-step filtering procedure, generated a library containing a 25 specs drug-like set (Figure 2F). Finally, we comprehensively evaluated binding energy data, interactions with key residues at binding sites, and structural diversity to select 7 compounds for subsequent in vitro biological evaluation, the DS virtual screening scores of which are reported (Table 1).
Structure-based virtual screening aimed at discovering and validating FOXM1 inhibitors
We purchased the seven candidate compounds obtained via virtual screening and evaluated their affinity for FOXM1 at the molecular level via surface plasmon resonance (SPR). Anti-tumor activity was also evaluated at the cellular level using the ovarian cancer cell lines, SKOV3 and A2780. The results showed that DZY-3 and DZY-4 showed better affinity and antitumor activity at a concentration of 100 µM (Figures 3A,B). DZY-3 is a camptothecin, which has been used as an antitumor agent, and its new mechanism of action may be found here. However, the off-target effects of DZY-3 prevent it from acting as a FOXM1-specific inhibitor. The antitumor activity of DZY-4 and its structural analogs remain unreported. DZY-4 activity was further evaluated at multiple concentrations, following which the IC50 of DZY-4 were found to be 28.73 µM for SKOV3 and 37.00 µM for A2780 (Figure 3C). The proliferation inhibitory activity of DZY-4 was better than FDI-6. DZY-4 combined with cisplatin could improve the therapeutic effect of cisplatin (Figure 3D). In SKOV3-DDP cells resistant to cisplatin, DZY-4 remained active while cisplatin activity decreased significantly (Figure 3E). Based on these results, we propose for the first time that the DZY-4 class of compounds may exert antitumor effects by inhibiting FOXM1.
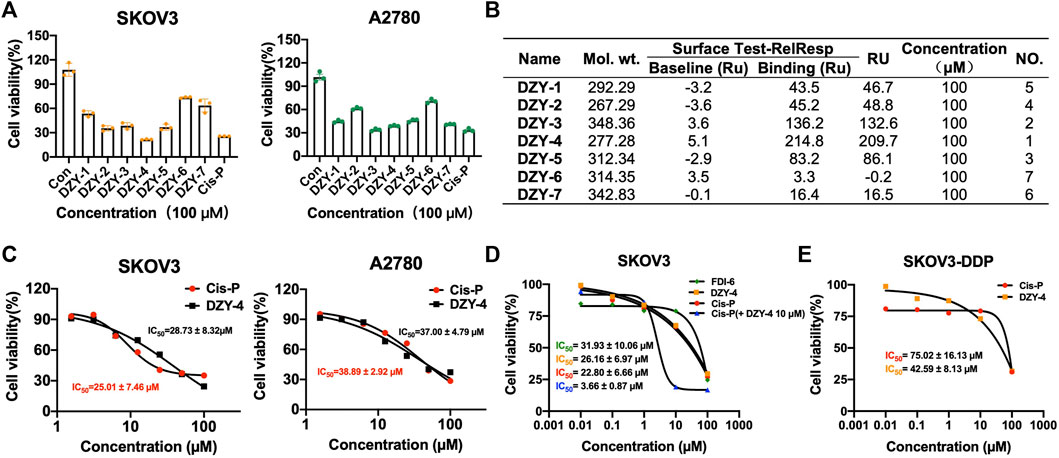
FIGURE 3. Structure-based virtual screening aimed at discovering and validating FOXM1 inhibitors. (A) The effects of FOXM1 inhibitors on the proliferation of SKOV3 and A2780 cells. (B) The kinetic interactions of the FOXM1 protein and seven compounds were determined via surface SPR analyses. Surface test of small molecules on FOXM1 captured by CM5. (C) Effects of DZY-4 on the proliferation of SKOV3 and A2780 cells. (D) IC50 values of FDI-6, DZY-4, Cis-P, and Cis-P combined with DZY-4 (10 M) were detected on SKOV3 cells. Data are representative of 3 wells for more than 3 times. (E) IC50 values of Cis-P and DZY-4 were detected on drug-resistant SKOV3-DDP cells. Data are representative of 3 wells for more than 3 times.
DZY-4 inhibits the migration and invasion of ovarian cancer cells and induces their apoptosis
We evaluated the anti-ovarian cancer effect of DZY-4 at the cellular level, using a wound healing assay, an invasion assay and an apoptosis assay, which assessed the effect of DZY-4 on migration and invasion of SKOV3 cells. Flow cytometry analysis with Annexin V-PI staining was used to evaluate the percentage of apoptotic cells among the SKOV3 cells treated with DZY-4. DZY-4 decreased the migration (Figure 4A) and invasion (Figure 4B) of SKOV3 cells and significantly increased the percentage of apoptotic cells (Figure 4C). The above in vitro results indicated that DZY-4 exerted an anti-ovarian cancer effect which was somewhat superior to that of cisplatin.
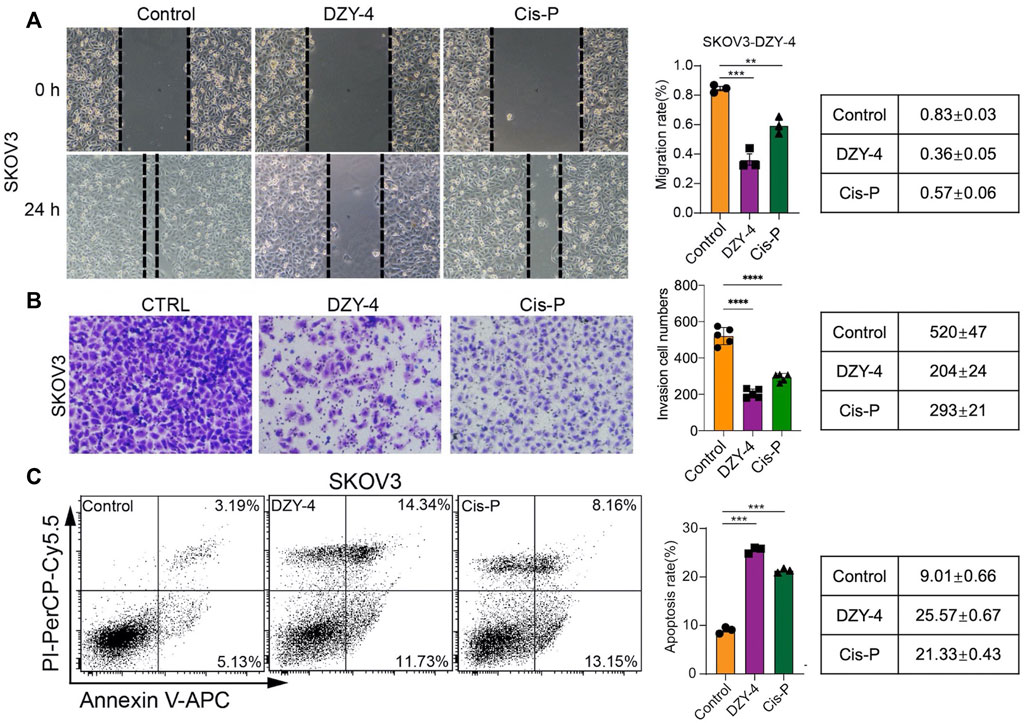
FIGURE 4. DZY-4 inhibits the migration and invasion of OC cells and induces their apoptosis. (A) Representative images of the migration and quantification of SKOV3 cells. SKOV3 cells were treated with vehicle, DZY-4 (10 μM), or Cisplatin (10 μM); Scale bar, 50 μm. (B) Representative images of invasion and quantification of SKOV3 cells. SKOV3 cells were treated with vehicle, DZY-4 (10 μM), or Cisplatin (10 μM); Scale bar, 50 μm. (C) Representative flow cytometry images of Annexin V-PI-based staining and quantification of apoptosis. SKOV3 cells were treated with vehicle, DZY-4 (10 μM), or Cisplatin (10 μM). After treatment for 24 h, apoptotic cells were evaluated via Annexin V-PI staining (left), histograms showing the statistical results of apoptosis rates (right). The data are expressed as mean ± SEM (n = 6 per group). One-way ANOVA was used to compare multiple groups; * = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
DZY-4 suppressed ovarian cancer tumor growth in vivo
Next, we examined the antitumor activity of DZY-4 in nude mice bearing SKOV3 tumor xenografts. Mice were administered with vehicle, DZY-4, or cisplatin via intraperitoneal injection (i.p.) for 21 days after the tumor volume reached approximately 100 mm3. The weight loss induced by DZY-4 in tumor-bearing nude mice was consistent with that in the vehicle group, which may be attributed to tumor burden (Figure 5A) Mice in the cisplatin group lost the most weight, indicating that DZY-4 may be less toxic than cisplatin. Tumor volume was measured during the treatment (Figure 5B). In the in vivo efficacy study, DZY-4 was administered at a significantly higher dose than cisplatin, and the tumor volume on day 21 (Figure 5C) and tumor weight (Figure 5D) were similar in both groups, and both were significantly lower than the blank control group, which had a definite therapeutic effect in ovarian cancer. The final body weight of mice at this dose of DZY-4 was higher than that of cisplatin, and it may be a lower toxicity than cisplatin.
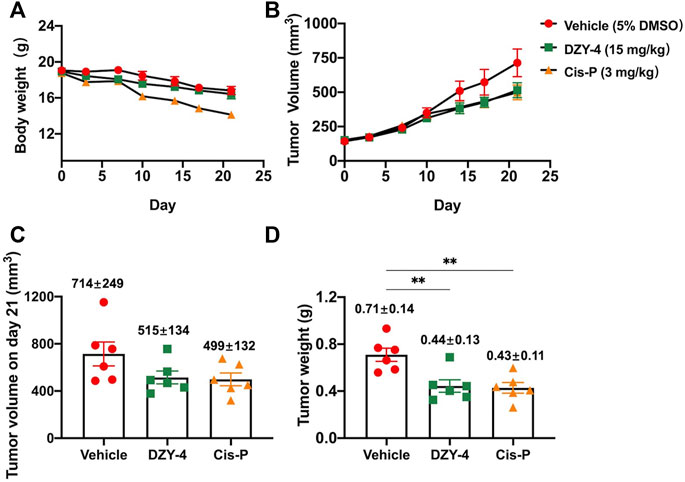
FIGURE 5. DZY-4 suppressed OC tumor growth in vivo. Nude mice were injected subcutaneously with SKOV3 cells. When the tumor volume reached approximately 100 mm3, the mice were treated with vehicle, DZY-4 (15 mg/kg/day), or Cisplatin (3 mg/kg/week). The data represent quantified body weight (A), tumor volume (B), tumor volume on day 21 (C), and tumor weight (D). The data are expressed as mean ± SEM (n = 6 per group). One-way ANOVA was used to compare multiple groups; ** = p < 0.01.
DZY-4 activity was specific to FOXM1 and its homologous proteins
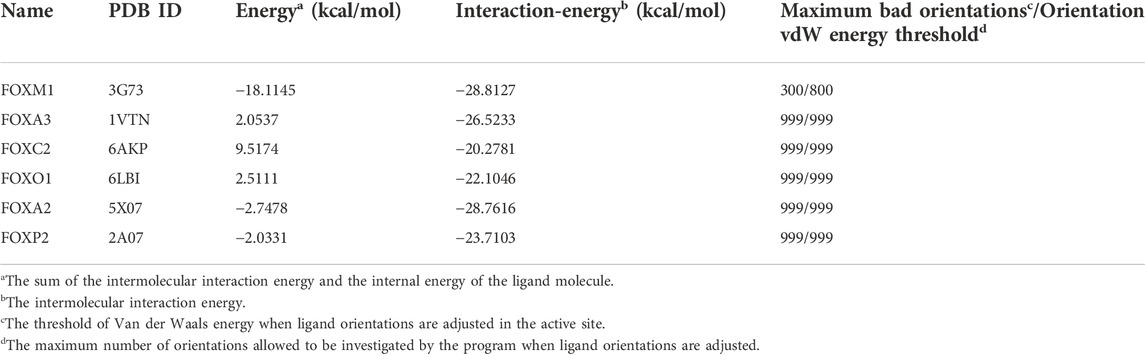
Existing literature shows that the DNA-binding domain of FOXM exhibits 45% identity with those of four of the closest related Forkhead families, namely FOXA, FOXC, FOXO, and FOXP (Laoukili et al., 2007; Li et al., 2017). We selected five proteins with high homology, and based on a literature review, used the conserved residues on H3 helix interacting with DNA as an active site. We also used the “From Receptor Cavities” module based on protein cavities to search for potential binding sites. Then we used LIBDOCK, CDOCKER, and other docking methods to dock DZY-4 to each of these regions, to generate corresponding complexes (Clark et al., 1993; Stroud et al., 2006; Li et al., 2017; Chen et al., 2019; Li et al., 2021). A comparative analysis showed that CDOCKER was the best method for docking DZY-4 and the five homologous proteins. The default parameters of CDOCKER Maximum Bad Orientations and Orientation vdW Energy Threshold were 800 and 300, respectively (Table 2). However, due to poor binding between DZY-4 and the best active site of the FOXM1 homologous protein, it was unable to enter the binding cavity within a general range. Therefore, in order to produce more docking poses, the threshold of the two parameters was adjusted to 999 for CDOCKER docking. Although the threshold for binding between DZY-4 and homologous proteins was adjusted, DZY-4 still showed the lowest interaction energy with FOXM1, indicating that DZY-4 and FOXM1 bind more closely and have a better affinity. In addition, the internal energy of the ligand molecule DZY-4 and FOXM1 was the lowest (10.6982 kcal/mol). The homologous proteins and DZY-4 had twice the internal energy. This indicates that DZY-4 binds more stably to FOXM1. In conclusion, DZY-4 has the best affinity to FOXM1 and binds it stably, whereas it displayed only a slight binding force with its homologous protein, indicating that DZY-4 displays specificity for FOXM1.
Conclusion
Ovarian cancer is the third most common gynecologic malignancy in terms of incidence; however, it has the highest mortality rate among gynecological malignancies. Most ovarian cancer patients receive first-line chemotherapy consisting of platinum and paclitaxel regardless of whether they qualify for surgery. The occurrence of drug resistance is the main reason for chemotherapy failures. Studies have shown that FOXM1 expression is increased in cisplatin-resistant cancer tissues, and that FOXM1 promotes the malignant behavior of ovarian cancer and induces cisplatin chemoresistance. FOXM1 expression is increased in cisplatin-resistant ovarian cancer cells, and inhibition of FOXM1 expression sensitizes cisplatin-resistant ovarian cancer cells. In summary, ovarian cancer requires urgent improvements in the area of patient prognoses. FOXM1 shows potential as a new therapeutic target in ovarian cancer, where inhibition of FOXM1 is expected to reverse or delay platinum resistance, thereby prolonging PFS and overall survival (OS) in patients. Therefore, the development of high-quality FOXM1 inhibitors as drug leads should be regarded as an important health need.
The specific small molecule FOXM1 inhibitors that have been studied most extensively at the clinical level are FDI-6 and RCM-1, both of which were identified via high-throughput screening. FDI-6 is highly cytotoxic which limits its use as a drug lead. Since the mode via which a ligand binds to FOXM1 remains unclear, structural modification of a drug lead cannot be guided. Our team determined the SPR affinity and antitumor activity of FDI-6 derivatives and combined these with the energy value of docking conformation, to obtain the conformation of a small molecule binding to FOXM1 with high confidence, by analyzing the correlation between these three factors. Based on the above conformation, we conducted virtual screening of over 260,000 compounds in the NCI library, while considering the properties under Lipinski and Veber rules as well as toxicity prediction results of DS software, in an attempt to obtain FOXM1 inhibitors with new structural backbones and better drug-forming properties. The results showed that six of the seven selected compounds showed an affinity to the FOXM1 protein, corroborating the accuracy of the conformation.
Among the seven compounds selected, DZY-3 and DZY-4 showed the best affinity for FOXM1 and the best inhibitory activity against two ovarian cancer cell lines, SKOV3 and A2780. DZY-3, (camptothecin), which inhibits DNA replication in proliferating tumor cells, has been used as an antitumor drug for many years, and this study was able to propose a new mechanism for its antitumor action, by demonstrating that it inhibits FOXM1. DZY-4 showed the highest affinity to FOXM1 and its IC50 values for both ovarian cancer cells were comparable to those of cisplatin. DZY-4 reduced tumor cell migration and invasion and increased the percentage of apoptotic cells. Although a dose of DZY-4 greater than that of cisplatin was used to treat the nude mouse subcutaneous transplantation tumor model, weight loss was lower than that of cisplatin and the tumor volume and tumor weight were comparable to those of cisplatin. Virtual molecular docking studies showed that DZY-4 was specific for FOXM1, when compared to its homologous proteins. The antitumor activity of DZY-4 and its structural analogs has not been reported in the literature to date; to the best of our knowledge, the current study is the first to report the FOXM1 inhibition-based antitumor effect of such structures of DZY-4.
In summary, DZY-4 shows potential as a therapeutic agent for ovarian cancer and may be used as a drug lead for further studies and as a biochemical research tool to promote studies investigating FOXM1 as a drug target. In the future, we aim to conduct drug discovery tests based on DZY-4 and search for derivative directions to guide conformational studies that may enhance the affinity of the docking conformation of DZY-4 for FOXM1. This study is expected to promote the search for ovarian cancer therapeutic drugs guided by new mechanisms.
Materials and methods
Molecular docking
Ligands of the NCI diversity Set2 database were docked with FOXM1 using AutoDock Vina package. AutoDock Tools 1.5.6 was used to compile the grid and docking parameter files. The active site was defined as 12.963 Å × 31.496 Å × 2.757 Å. The docking grid size was defined as 26, 16, and 20 Å on the X, Y, and Z coordinates respectively. A hydrogen atom with a pH of 7.4 was added to the ligand through the babel module. Next, the conformation was searched using OBConformer Search and Monte Carlo simulations, following which energy was optimized using the steepest descent method with a maximum of 100 steps.
The X-ray structure of FOXM1 and its homologous proteins (PDB ID: 3G73, 1VTN, 6AKP, 6LBI, 5X07, 2A07) were downloaded from the Protein Data Bank (https://www.rcsb.org). The proteins were prepared according to standard operations. The co-crystallized water molecules were deleted, and the “Clean Protein” and “Prepare Protein” tools of Discovery Studio were used to resolve potential problems in protein structures, such as the model missing loop regions, delete alternative conformations, add hydrogen atoms, and generate the protonation state at pH 7.0. The active site was identified according to previous reports or studies, and the docking radius of the sphere was set to 9 Å. The compounds were “Full Minimization.” Discovery Studio 4.0 was used to carry out the CDOCKER method to perform docking-based virtual screening.
Cells and cell culture
SKOV3 and A2780 cells were purchased from Procell Life Science and Technology Co., Ltd. The cells had recently been authenticated by short tandem repeat (STR) profiling and characterized by mycoplasma and cell viability detection. A2780 cells were cultured and maintained in MEM (Gibco, Catlog No. 11090081) supplemented with 10% fetal bovine serum (FBS; Gibco, Catalog No. 10091148) under 5% carbon dioxide. SKOV3 cells were cultured and maintained in DMEM (Gibco, Catalog No. C11885500BT) supplemented with 10% FBS.
Xenografts
All animal studies were approved by the Animal Experimentation Ethics Committee of the Chinese Academy of Medical Sciences, and all procedures were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committees of the Chinese Academy of Medical Sciences. Five-week-old female BALB/c nude (Weitong Lihua) mice were used to determine the drug effects of DZY-4 in vivo. For subcutaneous xenograft models, 1 × 106 SKOV3 cells were collected, resuspended in PBS, and injected into the left flanks of nude mice. When the tumors reached a volume of 100 mm3, the mice were randomly divided into three groups (n = 6). DZY-4 (15 mg/kg, daily) or cisplatin (3 mg/kg, once per week) in DMSO (5%) was administered via i.p. injection for 21 days. A digital caliper was used to gauge tumor size, and tumor volume was calculated using the formula: tumor volume (mm3) = width2 × (length/2).
Cell Counting Kit-8 assay
For the Cell Counting Kit-8 (CCK-8) assay, A2780 and SKOV3 cells were seeded in 96-well plates at a density of 3,000 cells/well. For verification of FOXM1 inhibitors regarding the proliferation of A2780 and SKOV3 cells, Cisplatin (100 μM) and FOXM1 inhibitor (100 μM) treated cells were cultured for 24 h. For the effects of DZY-4 on the proliferation of A2780 and SKOV3 cells, DZY-4 (0.78, 1.56, 3.125, 6.25, 12.5, 25, 50, 100 μM) or Cisplatin (0.78, 1.56, 3.125, 6.25, 12.5, 25, 50, 100 μM) treated cells were cultured for 24 h. Subsequently, 100 μl of complete medium supplemented with 10 μl of CCK-8 solution (TOPSCIENCE, C0005) was added to each well, and the plates were incubated for 2 h. Finally, absorbance was measured at 450 nm.
Surface plasmon resonance analysis
Surface plasmon resonance (SPR) assays were performed using a Biacore T200 (GE Healthcare) at 25°C. Recombinant FOXM1 (amino acids 222–360) protein [Purity (SDS page) > 90%] was purchased from Genescript USA Inc. (NJ, China) and covalently immobilized on a CM5 sensor chip (GE Healthcare) via amine coupling. Running buffer was 1.0 × PBS + P (GE Healthcare). When the affinity screen of fragments was run, 5% DMSO was added to the buffer. Compounds were injected into the surface of the protein-coupled chip channels at the flow rate of 30 µl/min. The affinities of the fragments were tested once in this study.
Wound healing assays
SKOV3 cells were harvested and seeded into 12-well plates at 2 × 105 cells per well. When cells reached 90% confluence, they were scratched with a 200 μl pipette tip to create a wound and washed with PBS to remove cells that had detached. The cells were then treated with DZY-4 (10 μM) or Cisplatin (10 μM), respectively, for 24 h. Cells were imaged with an inverted microscope (Olympus, Tokyo, Japan). Scratch healing efficiency was analyzed by calculating trauma distance.
Invasion assays
A transwell chamber (Millipore, SCWP04700) with an 8 μm pore size filter membrane was used for the invasion experiments. The Matrigel (40 μl per well, BD Matrigel Matrix, 354230) was coated with 10 μg/ml FN1 (fibronectin 1; MERCK, F0635), polycarbonate filter. Subsequently, the chambers were inserted into 24-well culture plates. Single-cell suspensions were inoculated into the upper chamber (5 × 104 cells per well in 0.4% FBS in DMEM). Twelve hours later, uninvaded cells in the upper part of the filter were removed with a cotton swab. Invading cells were fixed in 4% paraformaldehyde in PBS, stained with 0.1% crystal violet, and counted in six random fields via bright-field microscopy.
Apoptosis
Cell apoptosis was analyzed using a FITC Annexin V Apoptosis Detection Kit I (catalog 55647, BD Pharmingen). SKOV3 cells were treated with DZY-4, Cisplatin, or DMSO. A total of 105 cells were collected after DZY-4 exposure and resuspended in a 1 × binding buffer. Five microliters of FITC Annexin V and 5 μl PI were then added to the cell suspension and incubated for 15 min at room temperature in the dark. The results were determined via flow cytometry within 1 h.
Statistical analysis
Data are expressed as the mean ± SEM. For experiments involving only 2 comparisons, a 2-tailed Student’s t-test was used. For experiments involving multiple comparisons, the significance of differences between treatment means was determined using ANOVA followed by a Bonferroni multiple-comparison test. All analyses were performed using GraphPad Prism 9 (GraphPad Software). p values less than 0.05 were considered statistically significant. For all figures, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Animal Experimentation Ethics Committee of the Chinese Academy of Medical Sciences.
Author contributions
Z-YZ completed the molecular biology experiments. X-YH completed the molecular docking. L-QS and S-YL assisted with the experiments. S-TX designed the study and wrote the manuscript. Z-RL established and directed this subject.
Funding
This work was supported by grants from the CAMS Innovation Fund for Medical Sciences (2022-I2M-2-002), National Natural Science Foundation of China (82073711), and Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (2020-JKCS-017).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bhat, U. G., Halasi, M., and Gartel, A. L. (2009). Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One 4, e5592. doi:10.1371/journal.pone.0005592
Chen, X., Wei, H., Li, J., Liang, X., Dai, S., Jiang, L., et al. (2019). Structural basis for DNA recognition by FOXC2. Nucleic Acids Res. 47, 3752–3764. doi:10.1093/nar/gkz077
Clark, K. L., Halay, E. D., Lai, E., and Burley, S. K. (1993). Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420. doi:10.1038/364412a0
Dasari, S., and Tchounwou, P. B. (2014). Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 740, 364–378. doi:10.1016/j.ejphar.2014.07.025
Gadducci, A., Guarneri, V., Peccatori, F. A., Ronzino, G., Scandurra, G., Zamagni, C., et al. (2019). Current strategies for the targeted treatment of high-grade serous epithelial ovarian cancer and relevance of BRCA mutational status. J. Ovarian Res. 12, 9. doi:10.1186/s13048-019-0484-6
Gentles, A. J., Newman, A. M., Liu, C. L., Bratman, S. V., Feng, W., Kim, D., et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945. doi:10.1038/nm.3909
Gomes, A. R., Zhao, F., and Lam, E. W. (2013). Role and regulation of the forkhead transcription factors FOXO3a and FOXM1 in carcinogenesis and drug resistance. Chin. J. Cancer 32, 365–370. doi:10.5732/cjc.012.10277
Gormally, M. V., Dexheimer, T. S., Marsico, G., Sanders, D. A., Lowe, C., Matak-Vinković, D., et al. (2014). Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat. Commun. 5, 5165. doi:10.1038/ncomms6165
Gralewska, P., Gajek, A., Marczak, A., and Rogalska, A. (2020). Participation of the ATR/CHK1 pathway in replicative stress targeted therapy of high-grade ovarian cancer. J. Hematol. Oncol. 13, 39. doi:10.1186/s13045-020-00874-6
Halasi, M., and Gartel, A. L. (2013). FOX(M1) news-it is cancer. Mol. Cancer. Ther. 12, 245–254. doi:10.1158/1535-7163.MCT-12-0712
Halasi, M., Hitchinson, B., Shah, B. N., Váraljai, R., Khan, I., Benevolenskaya, E. V., et al. (2018). Honokiol is a FOXM1 antagonist. Cell Death Dis. 9, 84. doi:10.1038/s41419-017-0156-7
Jiang, Y., Wang, C., and Zhou, S. (2020). Targeting tumor microenvironment in ovarian cancer: Premise and promise. Biochimica Biophysica Acta - Rev. Cancer 1873, 188361. doi:10.1016/j.bbcan.2020.188361
Karadedou, C. T., Gomes, A. R., Chen, J., Petkovic, M., Ho, K. K., Zwolinska, A. K., et al. (2012). FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene 31, 1845–1858. doi:10.1038/onc.2011.368
Laoukili, J., Stahl, M., and Medema, R. H. (2007). FoxM1: At the crossroads of ageing and cancer. Biochimica Biophysica Acta - Rev. Cancer 1775, 92–102. doi:10.1016/j.bbcan.2006.08.006
Lheureux, S., Gourley, C., Vergote, I., and Oza, A. M. (2019). Epithelial ovarian cancer. Lancet 393, 1240–1253. doi:10.1016/S0140-6736(18)32552-2
Li, J., Dai, S., Chen, X., Liang, X., Qu, L., Jiang, L., et al. (2021). Mechanism of forkhead transcription factors binding to a novel palindromic DNA site. Nucleic Acids Res. 49, 3573–3583. doi:10.1093/nar/gkab086
Li, J., Dantas Machado, A. C., Guo, M., Sagendorf, J. M., Zhou, Z., Jiang, L., et al. (2017). Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus site. Biochemistry 56, 3745–3753. doi:10.1021/acs.biochem.7b00211
Lin, J., Zheng, Y., Chen, K., Huang, Z., Wu, X., and Zhang, N. (2013). Inhibition of FOXM1 by thiostrepton sensitizes medulloblastoma to the effects of chemotherapy. Oncol. Rep. 30, 1739–1744. doi:10.3892/or.2013.2654
Liu, C., Barger, C. J., and Karpf, A. R. (2021). FOXM1: A multifunctional oncoprotein and emerging therapeutic target in ovarian cancer. Cancers 13, 3065. doi:10.3390/cancers13123065
Lord, C. J., and Ashworth, A. (2017). PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158. doi:10.1126/science.aam7344
Meng, F. D., Wei, J. C., Qu, K., Wang, Z. X., Wu, Q. F., Tai, M. H., et al. (2015). FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J. Gastroenterol. 21, 196–213. doi:10.3748/wjg.v21.i1.196
Momenimovahed, Z., Tiznobaik, A., Taheri, S., and Salehiniya, H. (2019). <p>Ovarian cancer in the world: Epidemiology and risk factors</p>. Int. J. Womens Health. 11, 287–299. doi:10.2147/IJWH.S197604
Pratheeshkumar, P., Divya, S. P., Parvathareddy, S. K., Alhoshani, N. M., Al-Badawi, I. A., Tulbah, A., et al. (2018). FoxM1 and β-catenin predicts aggressiveness in Middle Eastern ovarian cancer and their co-targeting impairs the growth of ovarian cancer cells. Oncotarget 9, 3590–3604. doi:10.18632/oncotarget.23338
Quan, M., Wang, P., Cui, J., Gao, Y., and Xie, K. (2013). The roles of FOXM1 in pancreatic stem cells and carcinogenesis. Mol. Cancer 12, 159. doi:10.1186/1476-4598-12-159
Shukla, S., Milewski, D., Pradhan, A., Rama, N., Rice, K., Le, T., et al. (2019). The FOXM1 inhibitor RCM-1 decreases carcinogenesis and nuclear β-Catenin. Mol. Cancer Ther. 18, 1217–1229. doi:10.1158/1535-7163.mct-18-0709
Stewart, C., Ralyea, C., and Lockwood, S. (2019). Ovarian cancer: An integrated review. Seminars Oncol. Nurs. 35, 151–156. doi:10.1016/j.soncn.2019.02.001
Stroud, J. C., Wu, Y., Bates, D. L., Han, A., Nowick, K., Paabo, S., et al. (2006). Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14, 159–166. doi:10.1016/j.str.2005.10.005
Su, X., Yang, Y., Yang, Q., Pang, B., Sun, S., Wang, Y., et al. (2021). NOX4-derived ROS-induced overexpression of FOXM1 regulates aerobic glycolysis in glioblastoma. BMC Cancer 21, 1181. doi:10.1186/s12885-021-08933-y
Tassi, R. A., Todeschini, P., Siegel, E. R., Calza, S., Cappella, P., Ardighieri, L., et al. (2017). FOXM1 expression is significantly associated with chemotherapy resistance and adverse prognosis in non-serous epithelial ovarian cancer patients. J. Exp. Clin. Cancer Res. 36, 63. doi:10.1186/s13046-017-0536-y
Yang, C., Xia, B. R., Zhang, Z. C., Zhang, Y. J., Lou, G., and Jin, W. L. (2020). Immunotherapy for ovarian cancer: Adjuvant, combination, and neoadjuvant. Front. Immunol. 11, 577869. doi:10.3389/fimmu.2020.577869
Keywords: ovarian cancer, FOXM1, virtual screening, camptothecin, inhibitors
Citation: Zhou Z-Y, Han X-Y, Sun L-Q, Li S-Y, Xue S-T and Li Z-R (2022) Structure-based virtual screening identified novel FOXM1 inhibitors as the lead compounds for ovarian cancer. Front. Chem. 10:1058256. doi: 10.3389/fchem.2022.1058256
Received: 30 September 2022; Accepted: 08 November 2022;
Published: 24 November 2022.
Edited by:
Kailin Yang, Cleveland Clinic, United StatesReviewed by:
Anish K. Vadukoot, Vanderbilt Ingram Cancer Center, United StatesSourav Saha, National Cancer Institute (NIH), United States
Copyright © 2022 Zhou, Han, Sun, Li, Xue and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Si-Tu Xue, xuesitu@imb.pumc.edu.cn; Zhuo-Rong Li, lizhuorong@imb.pumc.edu.cn
†These authors have contributed equally to this work and share first authorship
 Zi-Ying Zhou†
Zi-Ying Zhou†  Si-Tu Xue
Si-Tu Xue Zhuo-Rong Li
Zhuo-Rong Li