Not a waste: Wastewater surveillance to enhance public health
- 1Epidemiology, Human Genetics, and Environmental Sciences, University of Texas Health Science Center at Houston School of Public Health, Houston, TX, United States
- 2Department of Geography, Oklahoma State University, Stillwater, OK, United States
- 3Department of Pharmaceutical Economics and Policy, School of Pharmacy, Chapman University, Orange, CA, United States
- 4TAILOR Labs, Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX, United States
- 5Texas Epidemic Public Health Institute, Houston, TX, United States
Domestic wastewater, when collected and evaluated appropriately, can provide valuable health-related information for a community. As a relatively unbiased and non-invasive approach, wastewater surveillance may complement current practices towards mitigating risks and protecting population health. Spurred by the COVID-19 pandemic, wastewater programs are now widely implemented to monitor viral infection trends in sewersheds and inform public health decision-making. This review summarizes recent developments in wastewater-based epidemiology for detecting and monitoring communicable infectious diseases, dissemination of antimicrobial resistance, and illicit drug consumption. Wastewater surveillance, a quickly advancing Frontier in environmental science, is becoming a new tool to enhance public health, improve disease prevention, and respond to future epidemics and pandemics.
1 Introduction
The concept of wastewater-based epidemiology (WBE) was first proposed in 2001 to monitor illicit-abused drugs (Daughton, 2001). Recently, the COVID-19 pandemic has helped to renew public health interest in WBE, further facilitating the general recognition that wastewater and population health are interconnected. Wastewater, specifically for municipalities, is a combination of domestic (both gray and black water), industrial and commercial sources. Water is flushed or drained through a network of underground sewer pipes, which carry it to a municipal wastewater treatment facility (WWTF) where it undergoes a multi-stage treatment process (coagulation, flocculation, sedimentation, filtration, and disinfection) before being discharged. This wastewater is a complex matrix, consisting of approximately 99.9% water with the remaining .1% including microbes (e.g., viruses, bacteria, protozoa, fungi, and helminths), organic and inorganic materials, indigestible food matter, and nutrients (Wastewater-What Is It?, 2017) (Figure 1A). After several treatment stages, the majority of solids, microorganisms, and organic materials are removed, resulting in a water product of improved quality that is safe for the environment and human health.
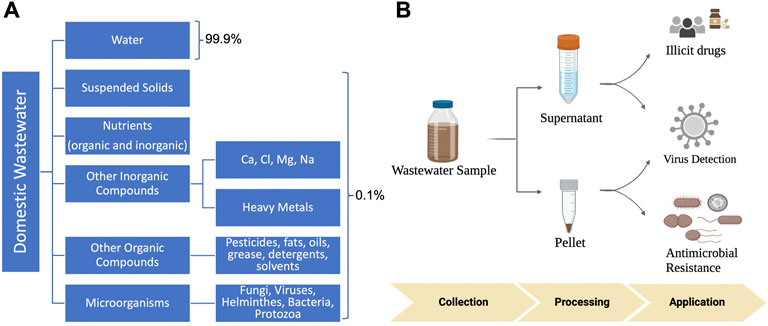
FIGURE 1. (A) Wastewater surveillance for public health. (A) Constituents of wastewater. Wastewater is mostly composed of water (∼99.9%), with .1% of solids, nutrients, organic, and inorganic compounds, and microorganisms. (B) Applications of wastewater in public health. Wastewater sample is collected and processed, with the resulting supernatant and pellet being utilized to detect and quantify human pathogens, illicit drugs, and antimicrobial resistance.
Human pathogen detection in wastewater has been in practice for nearly a century (Paul et al., 1939; Moore, 1951; Kelly et al., 1957). During the poliomyelitis epidemics in 1939, John R. Paul, James D. Trask, and C. S. Culotta demonstrated for the first time the presence of the poliomyelitis virus in wastewater. Two wastewater samples were inoculated intraperitoneally into two monkeys, both of which developed experimental poliomyelitis with an incubation period 7–8 d (Paul et al., 1939). Following that discovery, polioviruses and enteroviruses were monitored and isolated from wastewater samples in multiple countries during 1960s–1990s. Similarly, after successful detection of SAR-CoV-2 in wastewater, surveying wastewater has been utilized as a non-invasive approach to track viral infection trends in a designated sewershed (Ahmed et al., 2020; Medema et al., 2020; Wu et al., 2020). The value of wastewater monitoring data to supplement clinical case data has been investigated globally, with wastewater data typically preceding reported COVID-19 case data by 3 days to 3 weeks (Peccia et al., 2020; Róka et al., 2021; Wurtzer et al., 2021; Galani et al., 2022). Moreover, wastewater surveillance provides an unbiased sampling of infected individuals including asymptomatic and pre-symptomatic cases, and individuals who have the disease but do not seek healthcare. Those cases may be missed or not identified in time by clinical surveillance when testing capacity is limited.
Antimicrobial resistance (AMR), another global crisis, presents a significant threat to human, animal, and environmental health. AMR exposure can result in adverse medical outcomes including increased morbidity and mortality due to the limited number of effective medications available for treatment (World Health Organization, 2020). Bacteria with AMR are resistant to antibiotics and have been found extensively in the environment (soil, water, and air) (Pärnänen et al., 2019). With the increasing emergence of antibiotic resistance, it is estimated that AMR infections will be the most common cause of death by the year 2050 and at a total cost of trillions to world economies (Thompson, 2022).
Besides microbiological agents, the utilization of wastewater for tracking illicit drugs and opioids has also gained attention due to the increased health concerns associated with exposure to the synthetic opioid fentanyl (Gushgari et al., 2019). Illicit drug use is widespread in many countries, including Europe and United States, where opioid overdose related deaths have significantly increased since 2018 (Understanding the Epidemic | Drug Overdose | CDC Injury Center, 2022). Given it is challenging to obtain population-wide data on opioid consumption, which is important for formulating effective strategies to combat this health crisis, wastewater surveillance serves as a complementary tool to track illicit drug consumption within a sewershed (Gushgari et al., 2019; Endo et al., 2020; Kumar et al., 2022) and understand the societal burden at population level.
Here, we briefly review the applications of wastewater in the three public health issues: infectious diseases, antimicrobial resistance, and illicit drugs.
2 Monitoring of viral diseases
Typically, spatial-temporal disease monitoring consists of questionnaires, hospital admissions, mortality and morbidity rates, and clinical and sentinel (by medical professionals) surveillance for specific infectious diseases (Sims and Kasprzyk-Hordern, 2020). However, these tools can have limited success largely due to a lack of responses, dependence on symptomatic cases, and constrained testing capacity, therefore causing underreported data (Diamond et al., 2022). Pathogens which are shed through feces or urine, and can persist in sewage, are optimal targets for wastewater monitoring. Compared to bacterial pathogens, viral pathogens may be a more suitable target given their inability to replicate outside of living cells (Figure 1B). Three viruses of significant public health concern are reviewed including SARS-CoV-2, Monkeypox and enteroviruses.
2.1 SARS-CoV-2
The COVID-19 pandemic starkly revealed the impacts a global infectious disease can have on all aspects of society, let alone the cascading effects on public safety, health, and accessibility to healthcare (Daughton, 2020). Tracking the transmission, dispersion, and evolution of COVID-19 at population-wide scales is challenging, especially given the urgent need to evaluate and estimate the distribution of the virus in a timely manner. Detecting SARS-CoV-2 in wastewater is not only a non-invasive tool, but also provides an earlier snapshot of active infections than those in clinics due to virus shedding in stool which may precede the onset of symptoms (Bibby et al., 2021). Various studies have shown that increased SARS-CoV-2 concentrations in wastewater were found to precede spikes in clinical cases (Peccia et al., 2020; Saguti et al., 2021; Wu et al., 2022c; Karthikeyan et al., 2022). SARS-CoV-2 was detected in wastewater from a nursing home in Spain before an outbreak was declared, with the lag time between wastewater data and cases ranging from 5 to 19 days (Bonanno Ferraro et al., 2021). A wastewater surveillance program implemented at the University of California-San Diego used an automatic wastewater notification system to alert residents in buildings of positive wastewater samples (Karthikeyan et al., 2021). The system successfully led to early diagnosis of nearly 85% of all COVID-19 cases on the University campus, and increased testing rates by 1.9 to 13×. However, wastewater data may not serve as a leading indicator when clinical testing capacity is adequate. A study in Massachusetts determined that wastewater data had a lead time before reported cases during the first wave of the pandemic (March-August 2020), but not during the second wave (after August 2020), as testing capacity increased (Xiao et al., 2022).
The emergence of SARS-CoV-2 variants, especially variants of concern (VOC), necessitates the need for variant-targeted wastewater surveillance, such as through next-generation sequencing (NGS) or VOC-specific polymerase chain reaction (PCR)-based assays (Table 1) (Bar-Or et al., 2021; Izquierdo-Lara et al., 2021; Karthikeyan et al., 2021). Genomic sequencing of wastewater samples has detected viral lineages circulating within a population that had not yet been detected by clinical specimen sequencing (Crits-Christoph et al., 2021). A recent study sequenced wastewater samples in New York City and found an increasing frequency of novel cryptic SARS-CoV-2 lineages containing mutations belonging to the Omicron variant as well as mutations that had been rarely observed in clinical samples (Smyth et al., 2022). The SARS-CoV-2 variants, Alpha and Delta, were detected in the University wastewater samples up to 2 weeks before detection through clinical genomic surveillance (Karthikeyan et al., 2022). It is worthwhile to mention that bioinformatic pipelines customized for wastewater sequencing data processing are also important due to the ‘composite’ nature of wastewater samples. Researchers recently developed an end-to-end analysis pipeline to reconstruct the infection dynamics of different VOCs from wastewater sequencing data, and validated on multiple datasets (Schumann et al., 2022). Additionally, allele-specific PCR assays are also widely used to quantify the distribution of VOCs in the sewersheds (Heijnen et al., 2021; Lee et al., 2021; Lee et al., 2022). Thus, wastewater surveillance integrated with genomic sequencing and mutation-specific technologies are critical for tracking specific viral strains and emerging variants in the population.
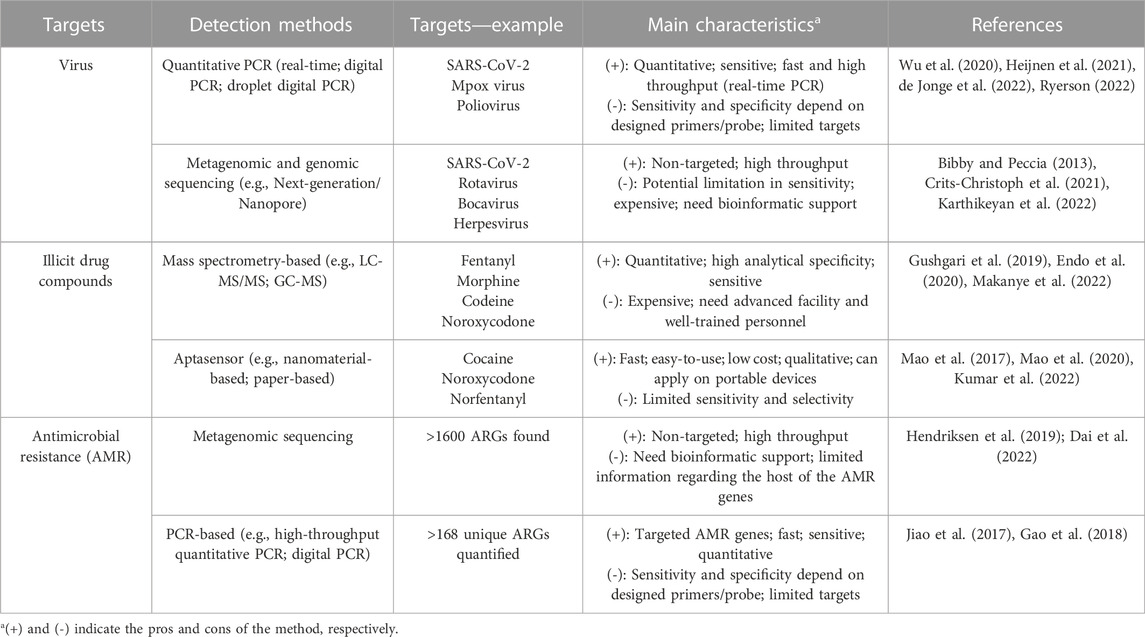
TABLE 1. Main methods for the detection of viral diseases, illicit drugs, and antimicrobial resistance in wastewater.
2.2 Monkeypox virus
Monkeypox virus, recently renamed as “Mpox” by WHO, is an enveloped, double-stranded DNA virus. Mpox has been found in infected individuals’ feces, urine, skin, semen, nasal secretions, and saliva (Peiró-Mestres et al., 2022). Current efforts to use PCR-based tools to detect Mpox DNA in wastewater has been successful (Tiwari et al., 2023). Mpox monitoring in wastewater has the potential to detect trends before the onset of symptoms and laboratory confirmation of cases, even given the limiting factors associated with viral shedding length times, environmental persistence, and sensitivity of analytical methods (Chen and Bibby, 2022). Eight out of nine publicly operated WWTFs (with a total of 287 samples collected) in California had positive detection for Mpox DNA using the CDC’s G2R_G (targeting the OPG002 gene for all Mpox) and G2R_WA (targeting the OPG002 gene for the West Africa clade) assays (Wolfe et al., 2022). A study in Amsterdam qualitatively detected Mpox in 31% of samples from five districts, 56% samples from two WWTFs, and 26% of samples from the Schiphol airport using both assays (de Jonge et al., 2022). Primer and probe mismatches have also been identified in the CDC Mpox generic real time PCR assay and seven other Mpox and orthopoxvirus diagnostic assays (Wu et al., 2022b), suggesting the need for careful evaluation of those assays before testing. Further investigation is warranted to optimize analytical methods in Mpox detection, improve clinical/biological understanding of viral shedding dynamics, evaluate the overall effectiveness of Mpox vaccines, and quantify viral recovery efficiency in wastewater, which are crucial for using WBE data for epidemiological inference.
2.3 Enteric viruses
Enteric viruses are also frequently detected in wastewater. Enteric pathogenic viruses are shed in high concentrations (105–1012 viral particles per gram of fecal matter) (Gerba, 2000) and tend to have low infectious doses (e.g., rotavirus and norovirus can be infectious at doses of 10–100 virus particles) (Yezli and Otter, 2011; Santiago-Rodriguez, 2022). Wastewater surveillance, as part of the “Global Polio Eradication Initiative,” was implemented in multiple countries to monitor poliovirus in the population before 2000 (Hovi et al., 2012). However, poliovirus has re-emerged as an infectious disease of concern in the United States after an unvaccinated adult in Rockland County, New York suffering from paralytic poliomyelitis tested positive for the vaccine-derived poliovirus type 2 (VDPV2) in July 2022 (Ryerson, 2022). Wastewater samples from 48 sewersheds in Rockland and surrounding counties were then tested for poliovirus from 9 March to 11 October 2022, with 8.3% of the 1,076 samples (from 10 sewersheds) having detectable concentrations of the virus (Ryerson, 2022). Routine wastewater surveillance (April to July 2022) in Jerusalem, Israel detected and identified an increase of vaccine-originated poliovirus type 2 even in the absence of oral polio vaccine (Zuckerman et al., 2022).
Besides poliovirus, other enteroviruses have also been detected in wastewater. A targeted population study in Clermon-Ferrand, France examined wastewater, using a two-step procedure of polyethylene glycol precipitation and tangential flow ultrafiltration, to detect an array of viruses including norovirus, adenovirus, rotavirus, enterovirus (EV), hepatitis A and E viruses (Bisseux et al., 2018). All 54 samples analyzed were positive for at least one virus; and of the samples EV-positive, it was determined that EV-D68 quietly circulated throughout the community in September 2015 (Bisseux et al., 2018). A study in Naples, Italy identified detectable concentrations of human enteroviruses in treated effluent samples at three WWTFs (Pennino et al., 2018), posing a concern for environmental contamination.
2.4 Public health implications
Virus detection in wastewater or water sources is critical given the potential transmission risks and severe health outcomes associated with the exposure of waterborne pathogens, especially for the elderly, children, and immunocompromised individuals (Teunis et al., 2010; Bogler et al., 2020). Quantitative viral tracking data in wastewater further helps identify or confirm “ongoing,” either silent or recognized, outbreaks and reflect infection patterns in the sewershed. Such data can be further used in dynamic models for epidemiological inference and prediction (McMahan et al., 2021; Phan et al., 2023). On the other hand, deep sequencing of wastewater samples could further provide genetic information, such as single nucleotide polymorphisms to understand viral evolution at the population level. Quantitative wastewater data provide a complementary perspective to understand the transmission of infectious diseases and inform public health decision-making for epidemic and pandemic responses. Thus, we believe that successful public health monitoring of viral pathogens requires a multi-prong approach of both wastewater and clinical surveillance, not only to detect emerging infectious diseases, but to enhance surveillance of seasonal diseases. And integration of wastewater and clinical surveillance will be also cost-effective for mass surveillance (Wu et al., 2022a), disease management, and interventions.
3 Illicit drugs and opioids
Wastewater surveillance has also been used to detect human health-related biomarkers, specifically those that are associated with illicit drugs and opioids (Figure 1B) and evaluate community exposures and health risks. A range of illicit compounds have been detected in wastewater, including cocaine, heroin, MDMA, methamphetamine, cannabis, morphine, oxycodone, fentanyl, and methadone (Gushgari et al., 2019; Endo et al., 2020). The utilization of wastewater for tracking illicit compounds has also gained attention recently due to the increased health concerns associated with exposure to synthetic opioid fentanyl (Gushgari et al., 2019) and the continued increase in drug-related overdose deaths in the United States, especially during the beginning of the COVID-19 pandemic (Friedman and Akre, 2021).
3.1 Detection and analysis of drug metabolites in wastewater
Drugs and their metabolites in wastewater tend to be in low concentration due to dilution, therefore requiring highly sensitive and specific detection methods. High-performance liquid chromatography tandem mass spectrometry (LC-MS/MS) has become the gold standard tool to detect and quantify illicit drug compounds in wastewater (Table 1). LC-MS/MS is regarded as the definitive approach to determine specific drugs because of its high sensitivity, selectivity, and reproductivity (Harper et al., 2017; Liu et al., 2018). However, it also requires sample extraction and preparation, well-trained personnel, and advanced facilities (Harper et al., 2017), making this a challenging method to adopt for high-throughput WBE programs. Other detection techniques include gas chromatography–mass spectrometry (GC-MS), surface-enhanced Raman spectroscopy (SERS), lateral flow immunoassay, and carbon nanotube electrode, which efficiently detected illicit drugs in complex matrices (Angelini et al., 2019; De Rycke et al., 2020; Dragan et al., 2021; Azimi and Docoslis, 2022; Makanye et al., 2022). Recently, advances in sensor technology, specifically with aptamer sensors (aptasensors, Table 1) have shown promise as a new analytical tool that is selective, stable, automatic, and easy to use with low cost (Mao et al., 2020; Kumar et al., 2022). Aptasensor is generally composed of an artificial single stranded nucleic acid for specific binding of the targeted drug compound and a reporter system to signal the binding. The aptasensor-based assay can be further integrated with portable devices for point-of-care detection such as for cocaine and norfentanyl (Mao et al., 2017; Kumar et al., 2022). However, the performance of aptasensors may be impaired by the low concentration and non-specific binding of drug compounds in wastewater, which requires further tests on the assay’s specificity and robustness.
Detection and quantification of drugs in wastewater sheds light on the illicit drug usage and spatiotemporal consumption patterns at the population level. A multi-city study in Croatia analyzed wastewater samples and identified that most illicit drugs consumed included cannabis, heroin and cocaine (Krizman et al., 2016). Drug consumption patterns identified in wastewater data indicated regional and seasonal variations between coastal and continental cities, with associations with summer tourist visits. Such regional and temporal differences of illicit drugs and pharmaceuticals in wastewater were also widely observed in other countries (Zhang et al., 2019; González-Mariño et al., 2020; Guzel, 2022). Oxycodone and tramadol, as opposed to illicit drugs such as heroin and fentanyl, were the most commonly measured opioids within an American Indian Reservation (Driver et al., 2022), suggesting that prescribed opioids may be a greater health concern as opposed to illicit drugs in some areas. Furthermore, the flexibility in sampling offered by wastewater analysis permits specific research scopes for illicit drug use to be pursued, such as detecting trends during weekends, holidays, and social events (Gerrity et al., 2011; Ort et al., 2014; Krizman et al., 2016).
Quantitative wastewater data can be further used to estimate the number of illicit drug consumption rate and overdoses. For example, the target compound’s concentration in wastewater (e.g., ng/mL) can be converted to the total load (e.g., ng/day) through multiplying the influent flow, and then utilized to estimate the consumption rates (mg/day/1,000 people) (Gushgari et al., 2019; Endo et al., 2020). The consumption rate can then be used to estimate drug-overdose deaths, which aligned well with the number of reported opioid deaths or prevalence estimates (Zuccato et al., 2008; Gushgari et al., 2019). However, those back-calculation estimates are impacted by multiple factors including loss rate in sewer lines, degradation of drug and corresponding metabolites in wastewater, total flows, population size in the sewershed, and the purity of street products, as discussed elsewhere (European Monitoring Centre for Drugs and Drug Addiction., 2016; Gracia-Lor et al., 2016; Margetts et al., 2020). Thus, further studies in characterizing those factors (e.g., quantification of compounds’ stability and standardization of calculation methods) will improve the inference from wastewater data.
3.2 Public health implications
Due to challenges in gathering data regarding actual opioid and illicit drug usage, wastewater analysis offers a complementing approach to gauge the consumption rates at the population level. These WBE data can further inform new policies and programs to mitigate drug abuse, track the long-term impact of interventions on community health, and perform educational outreach to specific populations (Gushgari et al., 2019; Driver et al., 2022). Accurate detection of which drugs and their concentrations are being used in a specific community may address knowledge gaps and limitations associated with survey health assessments. Longitudinal sampling at upstream locations (e.g., manholes) or WWTFs can further indicate the temporal characteristics of illicit drug and opioid use in the sewershed. Geographical sampling could help identify communities or subgroups with higher consumptions and suggest a more targeted public health intervention. Finally, the WBE data can be further combined with clinical data such as overdose-related hospitalizations and deaths to understand the pattern and burden of opioid and illicit drug use in the sewershed.
4 Antimicrobial resistance
The WWTF is a reservoir and source for antibiotic resistant genes (ARGs). Wastewater received by a WWTF harbors an array of antibiotic resistant bacteria (ARB) and ARGs from both human and animal excretions (e.g., hospitals, industries, livestock, etc.), providing the opportunity for ARGs to congregate closely on mobile genetic elements and generate complex resistance regions. Bacteria can acquire plasmids that may confer multidrug resistance during this process (Nguyen et al., 2021). The influent entering a WWTF is expected to reflect, at least partly, the microbiome characteristics representative of the community served by the WWTF (Pärnänen et al., 2019). Although treatment systems are typically employed in municipal WWTFs to mitigate the environmental release of ARGs, high abundance of core ARGs are still observed in wastewater effluents entering the environment (Raza et al., 2022). Thus, monitoring ARGs (Figure 1B) in wastewater provides a second platform for us to understand the burden, transmission, and persistence of antibiotic resistance in society.
4.1 Diversity of antimicrobial resistance genes in wastewater
Due to the limited scope and time delay of phenotyping pathogens from human clinical infections (Hendriksen et al., 2019), efforts to characterize AMR from wastewater has drastically increased. Metagenomic sequencing techniques, utilizing both long and short reads, facilitate ARGs profiling in the sample (Table 1), compared to culture-based methodologies. Short-read next-generation sequencing data has the ability to identify and quantify thousands of ARGs and associated bacterial taxa, virulence genes, and pathogens (Hendriksen et al., 2019). However, these short reads are susceptible to assembly errors and make it challenging to identify the original organism with specific ARGs (Arango-Argoty et al., 2018; Garner et al., 2021). Long-read sequencing with a typical read length 10–30 kilobases (Ardui et al., 2018) provides a greater insight regarding ARGs genetics such as genome assembly, mapping certainty, and locations (plasmid or chromosome) (Amarasinghe et al., 2020; Dai et al., 2022). Other techniques for evaluating ARGs in wastewater include primarily culture independent methods: single-cell fusion PCR, digital PCR, high-throughput quantitative PCR, 16S rRNA amplicon sequencing and correlation analysis, fluorescence-activated cell sorting and sequencing, and genomic cross-linking (Nguyen et al., 2021). Details on the advantages and disadvantages of each method are discussed elsewhere (Ishii, 2020; Nguyen et al., 2021).
ARG diversity and abundance has been found to vary between different locations and even within WWTF biotreatment compartments. A significant difference in AMR gene abundance was identified between high-income countries (Europe/North and America/Oceania) and low-income countries (Africa/Asia/South America) (Hendriksen et al., 2019). For example, Brazil, Vietnam, and India were identified to have the most divergent distribution of AMR genes, suggesting that these countries could be hot spots for the emergence of new ARGs. AMR gene abundance was found to be associated with several parameters regarding health sanitation, including female child mortality rates, infection and malnutrition, and number of physicians, among others. Another study determined that within the same WWTF, the microbial communities and ARGs were found to be significantly different in both liquid phase effluent and recycled activated sludge (Quintela-Baluja et al., 2019). The recycled activated sludge had a lower ARG richness than the liquid effluent and downstream receiving waters. Spatial differences in ARG abundance and diversity in WWTF effluent have also been reported in China and Hong Kong (Zhang et al., 2016; Yin et al., 2022). The results indicated the spatiotemporal dynamics of ARGs in wastewater and its relevance with socioeconomic, health, and environmental factors.
4.2 Public health implications
WWTFs are considered a critical component in AMR monitoring and mitigation, due to their roles as reservoirs for ARGs and ARG exchange, as well as a barrier by limiting the environmental release of ARGs through treatment processes (Nguyen et al., 2021). The discharge of treated wastewater into the environment also poses an increased risk of exposure for humans via multiple routes such as contaminated food (e.g., irrigated produce) and biosolid reuse in agriculture. The transfer of ARGs into human pathogens is also an emerging public health concern.
While ARG detection and monitoring is critical both nationally and globally for addressing the global burden and transmission of AMR, how to interpret the wastewater data for antibiotic stewardship and infection prevention should be considered. Stewardship can be directed through the ARG data measured in wastewater. If AMR is increasing in certain urban areas, for example, genes associated with extended-spectrum beta-lactamases, physicians might limit or modify their prescriptions accordingly. Additionally, it will alert healthcare systems to be on guard because these infections may be challenging to treat. One example of this importance is the potential detection of an antimicrobial resistance gene of public health importance upon importation into a new geographic area. The early detection of this resistance gene can influence and inform clinical surveillance and may enable public health intervention and mitigation before the wide spread of resistance gene in the sewershed. The rise of novel variants of AMR bacteria are extremely concerning and may be identified in wastewater long before they appear clinically. Analysis of ARG in wastewater used for urban agriculture can be also used to understand the AMR transmission risk among humans and animals (Bougnom et al., 2019), particularly in locations where ARG leaks into densely populated areas. In addition, identifying exposure thresholds, such as through quantitative microbial risk assessment (Pruden et al., 2018), by which ARGs and ARB may adversely impact human health need to be quantified. Public health efforts for managing AMR via wastewater is primarily focused on characterization, but future work incorporating quantitative AMR and ARG data into mathematical modeling could further inform and implement mitigation strategies. While there is plenty work left to do in ARG surveillance in wastewater, it holds great promise in increasing our knowledge of the dissemination and asymptomatic carriage of resistant pathogens.
5 Conclusion
Wastewater surveillance is an advancing Frontier in environmental science to combat the three ongoing epidemics including viral diseases, antibiotic resistance among bacterial pathogens, and opioid misuse and overdoses. This renewed tool can further enhance public health research and practice in multiple facets. First, most wastewater surveillance studies are from developed countries, however, how to develop appropriate WBE programs for areas with limited sewerage connection rates and failing infrastructures remains understudied. Second, analytical methodologies have improved to detect and monitor human-related biomarkers even at low concentrations, but how to use the wastewater data for epidemiological inference still requires considerable efforts to explore and discuss. Third, the connections between wastewater and public health prompts a question about how to treat the wastewater more efficiently and reduce transmission risks in wastewater treatment facilities, which also act as a source of human pathogens and ARGs to the environment. Developing effective experimental protocols, thresholds, and standard analytical methods to examine, monitor, and estimate the societal burdens in the sewershed is a critical step in addressing those public health threats (Nguyen et al., 2021).
WBE campaigns can be costly, time intensive, and require skilled personnel and labor, but the potential benefit of this tool in the “public health toolbox” should not be dismissed. For the three applications discussed above, wastewater data can be integrated with clinical testing results for a better understanding of population health. Such integration of WBE and clinical surveillance can also be more cost-effective for mass surveillance for endemic diseases and future pandemics (Wu et al., 2022a). While rapid advancement has been achieved for retrieving data from wastewater samples, further developments in data validation and translation into real public health actions remain largely unexplored and need wide collaborations with healthcare officials and local public health departments.
In summary, wastewater surveillance holds promise for addressing current public health challenges in infectious diseases and illicit drugs and opioids. Ultimately, within a single sample of wastewater, an array of analytical tests can be conducted to gather health information regarding the entire community, while not being hindered by testing willingness, capacity, or personal consent.
Author contributions
FW conceptualized; AG, JO, and ARG contributed equally to draft the original manuscript; All authors edited the manuscript.
Funding
This work is supported by Faculty Startup funding from the Center for Infectious Diseases at UTHealth, the UT system Rising STARs award, and the Texas Epidemic Public Health Institute (TEPHI).
Acknowledgments
We want to thank Dr. Feng Li at Baylor College of Medicine for reading the manuscript and comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O’Brien, J. W., et al. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728, 138764. doi:10.1016/j.scitotenv.2020.138764
Amarasinghe, S. L., Su, S., Dong, X., Zappia, L., Ritchie, M. E., and Gouil, Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30. doi:10.1186/s13059-020-1935-5
Angelini, D. J., Biggs, T. D., Maughan, M. N., Feasel, M. G., Sisco, E., and Sekowski, J. W. (2019). Evaluation of a lateral flow immunoassay for the detection of the synthetic opioid fentanyl. Forensic Sci. Int. 300, 75–81. doi:10.1016/j.forsciint.2019.04.019
Arango-Argoty, G., Garner, E., Pruden, A., Heath, L. S., Vikesland, P., and Zhang, L. (2018). DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 23. doi:10.1186/s40168-018-0401-z
Ardui, S., Ameur, A., Vermeesch, J. R., and Hestand, M. S. (2018). Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi:10.1093/nar/gky066
Azimi, S., and Docoslis, A. (2022). Recent advances in the use of surface-enhanced Raman scattering for illicit drug detection. Sensors (Basel) 22, 3877. doi:10.3390/s22103877
Bar-Or, I., Weil, M., Indenbaum, V., Bucris, E., Bar-Ilan, D., Elul, M., et al. (2021). Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 789, 148002. doi:10.1016/j.scitotenv.2021.148002
Bibby, K., Bivins, A., Wu, Z., and North, D. (2021). Making waves: Plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 202, 117438. doi:10.1016/j.watres.2021.117438
Bibby, K., and Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 47, 1945–1951. doi:10.1021/es305181x
Bisseux, M., Colombet, J., Mirand, A., Roque-Afonso, A.-M., Abravanel, F., Izopet, J., et al. (2018). Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: A one-year experiment in central France, 2014 to 2015. Euro Surveill. 23, 17–00237. doi:10.2807/1560-7917.es.2018.23.7.17-00237
Bogler, A., Packman, A., Furman, A., Gross, A., Kushmaro, A., Ronen, A., et al. (2020). Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain 3, 981–990. doi:10.1038/s41893-020-00605-2
Bonanno Ferraro, G., Veneri, C., Mancini, P., Iaconelli, M., Suffredini, E., Bonadonna, L., et al. (2021). A state-of-the-art scoping review on SARS-CoV-2 in sewage focusing on the potential of wastewater surveillance for the monitoring of the COVID-19 pandemic. Food Environ. Virol. 14, 315–354. doi:10.1007/s12560-021-09498-6
Bougnom, B. P., Zongo, C., McNally, A., Ricci, V., Etoa, F. X., Thiele-Bruhn, S., et al. (2019). Wastewater used for urban agriculture in West Africa as a reservoir for antibacterial resistance dissemination. Environ. Res. 168, 14–24. doi:10.1016/j.envres.2018.09.022
Chen, W., and Bibby, K. (2022). Model-based theoretical evaluation of the feasibility of using wastewater-based epidemiology to monitor monkeypox. Environ. Sci. Technol. Lett. 9, 772–778. doi:10.1021/acs.estlett.2c00496
Crits-Christoph, A., Kantor, R. S., Olm, M. R., Whitney, O. N., Al-Shayeb, B., Lou, Y. C., et al. (2021). Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio 12, e02703–e02720. doi:10.1128/mBio.02703-20
Dai, D., Brown, C., Bürgmann, H., Larsson, D. G. J., Nambi, I., Zhang, T., et al. (2022). Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements from sewage to activated sludge. Microbiome 10, 20. doi:10.1186/s40168-021-01216-5
Daughton, C. G. (2001). “Illicit drugs in municipal sewage,” in Pharmaceuticals and care products in the environment ACS symposium series (Washington, D.C., USA: American Chemical Society), 348–364. doi:10.1021/bk-2001-0791.ch020
Daughton, C. G. (2020). Wastewater surveillance for population-wide Covid-19: The present and future. Sci. Total Environ. 736, 139631. doi:10.1016/j.scitotenv.2020.139631
de Jonge, E. F., Peterse, C. M., Koelewijn, J. M., van der Drift, A.-M. R., van der Beek, R. F. H. J., Nagelkerke, E., et al. (2022). The detection of monkeypox virus DNA in wastewater samples in The Netherlands. Sci. Total Environ. 852, 158265. doi:10.1016/j.scitotenv.2022.158265
De Rycke, E., Stove, C., Dubruel, P., De Saeger, S., and Beloglazova, N. (2020). Recent developments in electrochemical detection of illicit drugs in diverse matrices. Biosens. Bioelectron. 169, 112579. doi:10.1016/j.bios.2020.112579
Diamond, M. B., Keshaviah, A., Bento, A. I., Conroy-Ben, O., Driver, E. M., Ensor, K. B., et al. (2022). Wastewater surveillance of pathogens can inform public health responses. Nat. Med. 28, 1992–1995. doi:10.1038/s41591-022-01940-x
Dragan, A.-M., Truta, F. M., Tertis, M., Florea, A., Schram, J., Cernat, A., et al. (2021). Electrochemical fingerprints of illicit drugs on graphene and multi-walled carbon nanotubes. Front. Chem., 9, 641147. doi:10.3389/fchem.2021.641147
Driver, E. M., Bowes, D. A., Halden, R. U., and Conroy-Ben, O. (2022). Implementing wastewater monitoring on American Indian reservations to assess community health indicators. Sci. Total Environ. 823, 153882. doi:10.1016/j.scitotenv.2022.153882
Endo, N., Ghaeli, N., Duvallet, C., Foppe, K., Erickson, T. B., Matus, M., et al. (2020). Rapid assessment of opioid exposure and treatment in cities through robotic collection and chemical analysis of wastewater. J. Med. Toxicol. 16, 195–203. doi:10.1007/s13181-019-00756-5
European Monitoring Centre for Drugs and Drug Addiction. (2016). Assessing illicit drugs in wastewater:advances in wastewater based drug epidemiology. LU: Publications Office, Lucknow, India, Available at: https://data.europa.eu/doi/10.2810/017397 [Accessed December 14, 2022].
Friedman, J., and Akre, S. (2021). COVID-19 and the drug overdose crisis: Uncovering the deadliest months in the United States, january‒july 2020. Am. J. Public Health 111, 1284–1291. doi:10.2105/AJPH.2021.306256
Galani, A., Aalizadeh, R., Kostakis, M., Markou, A., Alygizakis, N., Lytras, T., et al. (2022). SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 804, 150151. doi:10.1016/j.scitotenv.2021.150151
Gao, M., Qiu, T., Sun, Y., and Wang, X. (2018). The abundance and diversity of antibiotic resistance genes in the atmospheric environment of composting plants. Environ. Int. 116, 229–238. doi:10.1016/j.envint.2018.04.028
Garner, E., Davis, B. C., Milligan, E., Blair, M. F., Keenum, I., Maile-Moskowitz, A., et al. (2021). Next generation sequencing approaches to evaluate water and wastewater quality. Water Res. 194, 116907. doi:10.1016/j.watres.2021.116907
Gerba, C. P. (2000). Assessment of enteric pathogen shedding by bathers during recreational activity and its impact on water quality. Quant. Microbiol. 2, 55–68. doi:10.1023/A:1010000230103
Gerrity, D., Trenholm, R. A., and Snyder, S. A. (2011). Temporal variability of pharmaceuticals and illicit drugs in wastewater and the effects of a major sporting event. Water Res. 45, 5399–5411. doi:10.1016/j.watres.2011.07.020
González-Mariño, I., Baz-Lomba, J. A., Alygizakis, N. A., Andrés-Costa, M. J., Bade, R., Bannwarth, A., et al. (2020). Spatio-temporal assessment of illicit drug use at large scale: Evidence from 7 years of international wastewater monitoring. Addiction 115, 109–120. doi:10.1111/add.14767
Gracia-Lor, E., Zuccato, E., and Castiglioni, S. (2016). Refining correction factors for back-calculation of illicit drug use. Sci. Total Environ. 573, 1648–1659. doi:10.1016/j.scitotenv.2016.09.179
Gushgari, A. J., Venkatesan, A. K., Chen, J., Steele, J. C., and Halden, R. U. (2019). Long-term tracking of opioid consumption in two United States cities using wastewater-based epidemiology approach. Water Res. 161, 171–180. doi:10.1016/j.watres.2019.06.003
Guzel, E. Y. (2022). Monitoring of changes in illicit drugs, alcohol, and nicotine consumption during Ramadan via wastewater analysis. Environ. Sci. Pollut. Res. 29, 89245–89254. doi:10.1007/s11356-022-22016-w
Harper, L., Powell, J., and Pijl, E. M. (2017). An overview of forensic drug testing methods and their suitability for harm reduction point-of-care services. Harm Reduct. J. 14, 52. doi:10.1186/s12954-017-0179-5
Heijnen, L., Elsinga, G., de Graaf, M., Molenkamp, R., Koopmans, M. P. G., and Medema, G. (2021). Droplet Digital RT-PCR to detect SARS-CoV-2 variants of concern in wastewater. Sci. Total Environ. 799. doi:10.1101/2021.03.25.21254324
Hendriksen, R. S., Munk, P., Njage, P., van Bunnik, B., McNally, L., Lukjancenko, O., et al. (2019). Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 10, 1124. doi:10.1038/s41467-019-08853-3
Hovi, T., Shulman, L. M., van der Avoort, H., Deshpande, J., Roivainen, M., and De Gourville, E. M. (2012). Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 140, 1–13. doi:10.1017/S095026881000316X
Ishii, S. (2020). Quantification of antibiotic resistance genes for environmental monitoring: Current methods and future directions. Curr. Opin. Environ. Sci. Health 16, 47–53. doi:10.1016/j.coesh.2020.02.004
Izquierdo-Lara, R., Elsinga, G., Heijnen, L., Munnink, B. B. O., Schapendonk, C. M. E., Nieuwenhuijse, D., et al. (2021). Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, The Netherlands and Belgium. Emerg. Infect. Dis. 27, 1405–1415. doi:10.3201/eid2705.204410
Jiao, Y.-N., Chen, H., Gao, R.-X., Zhu, Y.-G., and Rensing, C. (2017). Organic compounds stimulate horizontal transfer of antibiotic resistance genes in mixed wastewater treatment systems. Chemosphere 184, 53–61. doi:10.1016/j.chemosphere.2017.05.149
Karthikeyan, S., Levy, J. I., De Hoff, P., Humphrey, G., Birmingham, A., Jepsen, K., et al. (2022). Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 609, 101–108. doi:10.1038/s41586-022-05049-6
Karthikeyan, S., Nguyen, A., McDonald, D., Zong, Y., Ronquillo, N., Ren, J., et al. (2021). Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems 6, e0079321–21. doi:10.1128/mSystems.00793-21
Kelly, S., Winsser, J., and Winkelstein, W. (1957). Poliomyelitis and other enteric viruses in sewage. Am. J. Public Health Nations Health 47, 72–77. doi:10.2105/AJPH.47.1.72
Krizman, I., Senta, I., Ahel, M., and Terzic, S. (2016). Wastewater-based assessment of regional and temporal consumption patterns of illicit drugs and therapeutic opioids in Croatia. Sci. Total Environ. 566–567, 454–462. doi:10.1016/j.scitotenv.2016.05.075
Kumar, N., Rana, M., Geiwitz, M., Khan, N. I., Catalano, M., Ortiz-Marquez, J. C., et al. (2022). Rapid, multianalyte detection of opioid metabolites in wastewater. ACS Nano 16, 3704–3714. doi:10.1021/acsnano.1c07094
Lee, W. L., Armas, F., Guarneri, F., Gu, X., Formenti, N., Wu, F., et al. (2022). Rapid displacement of SARS-CoV-2 variant Delta by Omicron revealed by allele-specific PCR in wastewater. Water Res. 221, 118809. doi:10.1016/j.watres.2022.118809
Lee, W. L., Imakaev, M., Armas, F., McElroy, K. A., Gu, X., Duvallet, C., et al. (2021). Quantitative SARS-CoV-2 Alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ. Sci. Technol. Lett. 8, 675–682. doi:10.1021/acs.estlett.1c00375
Liu, L., Wheeler, S. E., Venkataramanan, R., Rymer, J. A., Pizon, A. F., Lynch, M. J., et al. (2018). Newly emerging drugs of abuse and their detection methods: An ACLPS critical review. Am. J. Clin. Pathol. 149, 105–116. doi:10.1093/ajcp/aqx138
Makanye, L., Mwenesongole, E., Gautam, L., and Masamba, W. (2022). Interdisciplinary approach to determining the use of drugs of abuse in wastewater using a survey and gas chromatography-mass spectrometry. Toxicol. Anal. Clinique 34, S135. doi:10.1016/j.toxac.2022.06.224
Mao, K., Zhang, H., Pan, Y., Zhang, K., Cao, H., Li, X., et al. (2020). Nanomaterial-based aptamer sensors for analysis of illicit drugs and evaluation of drugs consumption for wastewater-based epidemiology. Trends Anal. Chem. 130, 115975. doi:10.1016/j.trac.2020.115975
Mao, Y., Li, J., Yan, J., Ma, Y., Song, Y., Tian, T., et al. (2017). A portable visual detection method based on a target-responsive DNA hydrogel and color change of gold nanorods. Chem. Commun. 53, 6375–6378. doi:10.1039/C7CC01360D
Margetts, M., Keshaviah, A., Hu, X. C., Troeger, V., Sykes, J., Bishop, N., et al. (2020). Using wastewater-based epidemiology with local indicators of opioid and illicit drug use to overcome data gaps in Montana. 2020.04.18.20064113. doi:10.1101/2020.04.18.20064113
McMahan, C. S., Self, S., Rennert, L., Kalbaugh, C., Kriebel, D., Graves, D., et al. (2021). COVID-19 wastewater epidemiology: A model to estimate infected populations. Lancet Planet. Health 5, e874–e881. doi:10.1016/S2542-5196(21)00230-8
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., and Brouwer, A. (2020). Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 7, 511–516. doi:10.1021/acs.estlett.0c00357
Moore, B. (1951). The detection of enteric carriers in towns by means of sewage examination. J. R. Sanit. Inst. 71, 57–60. doi:10.1177/146642405107100109
Nguyen, A. Q., Vu, H. P., Nguyen, L. N., Wang, Q., Djordjevic, S. P., Donner, E., et al. (2021). Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 783, 146964. doi:10.1016/j.scitotenv.2021.146964
Ort, C., van Nuijs, A. L. N., Berset, J.-D., Bijlsma, L., Castiglioni, S., Covaci, A., et al. (2014). Spatial differences and temporal changes in illicit drug use in Europe quantified by wastewater analysis. Addiction 109, 1338–1352. doi:10.1111/add.12570
Pärnänen, K. M., Narciso-da-Rocha, C., Kneis, D., Berendonk, T. U., Cacace, D., Do, T. T., et al. (2019). Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5, eaau9124. doi:10.1126/sciadv.aau9124
Paul, J. R., Trask, J. D., and Culotta, C. S. (1939). Poliomyelitic virus in sewage. Science 90, 258–259. doi:10.1126/science.90.2333.258
Peccia, J., Zulli, A., Brackney, D. E., Grubaugh, N. D., Kaplan, E. H., Casanovas-Massana, A., et al. (2020). Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 38, 1164–1167. doi:10.1038/s41587-020-0684-z
Peiró-Mestres, A., Fuertes, I., Camprubí-Ferrer, D., Marcos, M. Á., Vilella, A., Navarro, M., et al. (2022). Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Eurosurveillance 27, 2200503. doi:10.2807/1560-7917.ES.2022.27.28.2200503
Pennino, F., Nardone, A., Montuori, P., Aurino, S., Torre, I., Battistone, A., et al. (2018). Large-scale survey of human enteroviruses in wastewater treatment plants of a metropolitan area of southern Italy. Food Environ. virology 10, 187–192. doi:10.1007/s12560-017-9331-3
Phan, T., Brozak, S., Pell, B., Gitter, A., Xiao, A., Mena, K. D., et al. (2023). A simple SEIR-V model to estimate COVID-19 prevalence and predict SARS-CoV-2 transmission using wastewater-based surveillance data. Sci. Total Environ. 857, 159326. doi:10.1016/j.scitotenv.2022.159326
Pruden, A., Alcalde, R. E., Alvarez, P. J. J., Ashbolt, N., Bischel, H., Capiro, N. L., et al. (2018). An environmental science and engineering framework for combating antimicrobial resistance. Environ. Eng. Sci. 35, 1005–1011. doi:10.1089/ees.2017.0520
Quintela-Baluja, M., Abouelnaga, M., Romalde, J., Su, J.-Q., Yu, Y., Gomez-Lopez, M., et al. (2019). Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters. Water Res. 162, 347–357. doi:10.1016/j.watres.2019.06.075
Raza, S., Shin, H., Hur, H.-G., and Unno, T. (2022). Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Res. 208, 117882. doi:10.1016/j.watres.2021.117882
Róka, E., Khayer, B., Kis, Z., Kovács, L. B., Schuler, E., Magyar, N., et al. (2021). Ahead of the second wave: Early warning for COVID-19 by wastewater surveillance in Hungary. Sci. Total Environ. 786, 147398. doi:10.1016/j.scitotenv.2021.147398
Ryerson, A. B., Lang, D., Alazawi, M. A., Neyra, M., Hill, D. T., St. George, K., et al. (2022). Wastewater testing and detection of poliovirus type 2 genetically linked to virus isolated from a paralytic polio case — New York, March 9–october 11, 2022. MMWR Morb. Mortal. Wkly. Rep. 71, 1418–1424. doi:10.15585/mmwr.mm7144e2
Saguti, F., Magnil, E., Enache, L., Churqui, M. P., Johansson, A., Lumley, D., et al. (2021). Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 189, 116620. doi:10.1016/j.watres.2020.116620
Santiago-Rodriguez, T. M. (2022). The detection of SARS-CoV-2 in the environment: Lessons from wastewater. Water 14, 599. doi:10.3390/w14040599
Schumann, V.-F., de Castro Cuadrat, R. R., Wyler, E., Wurmus, R., Deter, A., Quedenau, C., et al. (2022). SARS-CoV-2 infection dynamics revealed by wastewater sequencing analysis and deconvolution. Sci. Total Environ. 853, 158931. doi:10.1016/j.scitotenv.2022.158931
Sims, N., and Kasprzyk-Hordern, B. (2020). Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 139, 105689. doi:10.1016/j.envint.2020.105689
Smyth, D. S., Trujillo, M., Gregory, D. A., Cheung, K., Gao, A., Graham, M., et al. (2022). Publisher Correction: Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 13, 1836–1839. doi:10.1038/s41467-022-29573-1
Teunis, P. F. M., Xu, M., Fleming, K. K., Yang, J., Moe, C. L., and LeChevallier, M. W. (2010). Enteric virus infection risk from intrusion of sewage into a drinking water distribution network. Environ. Sci. Technol. 44, 8561–8566. doi:10.1021/es101266k
Thompson, T. (2022). The staggering death toll of drug-resistant bacteria. Nature. doi:10.1038/d41586-022-00228-x
Tiwari, A., Adhikari, S., Kaya, D., Islam, M. A., Malla, B., Sherchan, S. P., et al. (2023). Monkeypox outbreak: Wastewater and environmental surveillance perspective. Sci. Total Environ. 856, 159166. doi:10.1016/j.scitotenv.2022.159166
Understanding the Epidemic | Drug Overdose | CDC Injury Center (2022). Understanding drug overdoses and deaths. Available at: https://www.cdc.gov/drugoverdose/epidemic/index.html [Accessed June 21, 2022].
Wastewater - What Is It? (2017). UNL water. Available at: https://water.unl.edu/article/wastewater/wastewater-what-it [Accessed October 17, 2022].
Wolfe, M. K., Duong, D., Hughes, B., Chan-Herur, V., White, B. J., and Boehm, A. B. (2022). Detection of monkeypox viral DNA in a routine wastewater monitoring program. Infect. Dis. Except. HIV/AIDS). doi:10.1101/2022.07.25.22278043
World Health Organization (2020). Antibiotic resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance [Accessed October 20, 2022].
Wu, F., Lee, W. L., Chen, H., Gu, X., Chandra, F., Armas, F., et al. (2022a). Making waves: Wastewater surveillance of SARS-CoV-2 in an endemic future. Water Res. 219, 118535. doi:10.1016/j.watres.2022.118535
Wu, F., Oghuan, J., Gitter, A., Mena, K. D., and Brown, E. L. (2022b). Wide mismatches in the sequences of primers and probes for Monkeypox virus diagnostic assays. medRxiv.
Wu, F., Xiao, A., Zhang, J., Gu, X., Lee, W. L., Kauffman, K., et al. (2020). SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 5, 2020. doi:10.1128/mSystems.00614-20
Wu, F., Xiao, A., Zhang, J., Moniz, K., Endo, N., Armas, F., et al. (2022c). SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 805, 150121. doi:10.1016/j.scitotenv.2021.150121
Wurtzer, S., Waldman, P., Ferrier-Rembert, A., Frenois-Veyrat, G., Mouchel, J. M., Boni, M., et al. (2021). Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res. 198, 117183. doi:10.1016/j.watres.2021.117183
Xiao, A., Wu, F., Bushman, M., Zhang, J., Imakaev, M., Chai, P. R., et al. (2022). Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 212, 118070. doi:10.1016/j.watres.2022.118070
Yezli, S., and Otter, J. A. (2011). Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 3, 1–30. doi:10.1007/s12560-011-9056-7
Yin, X., Yang, Y., Deng, Y., Huang, Y., Li, L., Chan, L. Y. L., et al. (2022). An assessment of resistome and mobilome in wastewater treatment plants through temporal and spatial metagenomic analysis. Water Res. 209, 117885. doi:10.1016/j.watres.2021.117885
Zhang, B., Xia, Y., Wen, X., Wang, X., Yang, Y., Zhou, J., et al. (2016). The composition and spatial patterns of bacterial virulence factors and antibiotic resistance genes in 19 wastewater treatment plants. PLOS ONE 11, e0167422. doi:10.1371/journal.pone.0167422
Zhang, X., Huang, R., Li, P., Ren, Y., Gao, J., Mueller, J. F., et al. (2019). Temporal profile of illicit drug consumption in Guangzhou, China monitored by wastewater-based epidemiology. Environ. Sci. Pollut. Res. 26, 23593–23602. doi:10.1007/s11356-019-05575-3
Zuccato, E., Chiabrando, C., Castiglioni, S., Bagnati, R., and Fanelli, R. (2008). Estimating community drug abuse by wastewater analysis. Environ. Health Perspect. 116, 1027–1032. doi:10.1289/ehp.11022
Keywords: wastewater-based epidemiology, infectious diseases, illicit drugs, antimicrobial resistance, viruses, public health
Citation: Gitter A, Oghuan J, Godbole AR, Chavarria CA, Monserrat C, Hu T, Wang Y, Maresso AW, Hanson BM, Mena KD and Wu F (2023) Not a waste: Wastewater surveillance to enhance public health. Front. Chem. Eng. 4:1112876. doi: 10.3389/fceng.2022.1112876
Received: 30 November 2022; Accepted: 21 December 2022;
Published: 09 January 2023.
Edited by:
Xianzhen Xu, Qingdao University, ChinaCopyright © 2023 Gitter, Oghuan, Godbole, Chavarria, Monserrat, Hu, Wang, Maresso, Hanson, Mena and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuqing Wu, fuqing.wu@uth.tmc.edu
†These authors have contributed equally to this work
 Anna Gitter1†
Anna Gitter1†  Tao Hu
Tao Hu Anthony W. Maresso
Anthony W. Maresso Blake M. Hanson
Blake M. Hanson Kristina D. Mena
Kristina D. Mena Fuqing Wu
Fuqing Wu