P14AS upregulates gene expression in the CDKN2A/2B locus through competitive binding to PcG protein CBX7
- Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Division of Etiology, Peking University Cancer Hospital and Institute, Beijing, China
Background: It is well known that P16INK4A, P14ARF, P15INK4B mRNAs, and ANRIL lncRNA are transcribed from the CDKN2A/2B locus. LncRNA P14AS is a lncRNA transcribed from antisense strand of P14ARF promoter to intron-1. Our previous study showed that P14AS could upregulate the expression level of ANRIL and P16INK4A and promote the proliferation of cancer cells. Because polycomb group protein CBX7 could repress P16INK4A expression and bind ANRIL, we wonder whether the P14AS-upregulated ANRIL and P16INK4A expression is mediated with CBX7.
Results: In this study, we found that the upregulation of P16INK4A, P14ARF, P15INK4B and ANRIL expression was induced by P14AS overexpression only in HEK293T and HCT116 cells with active endogenous CBX7 expression, but not in MGC803 and HepG2 cells with weak CBX7 expression. Further studies showed that the stable shRNA-knockdown of CBX7 expression abolished the P14AS-induced upregulation of these P14AS target genes in HEK293T and HCT116 cells whereas enforced CBX7 overexpression enabled P14AS to upregulate expression of these target genes in MGC803 and HepG2 cells. Moreover, a significant association between the expression levels of P14AS and its target genes were observed only in human colon cancer tissue samples with high level of CBX7 expression (n = 38, p < 0.05), but not in samples (n = 37) with low level of CBX7 expression, nor in paired surgical margin tissues. In addition, the results of RNA immunoprecipitation (RIP)- and chromatin immunoprecipitation (ChIP)-PCR analyses revealed that lncRNA P14AS could competitively bind to CBX7 protein which prevented the bindings of CBX7 to both lncRNA ANRIL and the promoters of P16INK4A, P14ARF and P15INK4B genes. The amounts of repressive histone modification H3K9m3 was also significantly decreased at the promoters of these genes by P14AS in CBX7 actively expressing cells.
Conclusions: CBX7 expression is essential for P14AS to upregulate the expression of P16INK4A, P14ARF, P15INK4B and ANRIL genes in the CDKN2A/2Blocus. P14AS may upregulate these genes’ expression through competitively blocking CBX7-binding to ANRIL lncRNA and target gene promoters.
Background
The CDKN2A/2B locus on human chromosome 9p21, known as p15INK4B-P14ARF-P16INK4A gene cluster, encodes three tumor suppressor proteins P15, P16, P14. P16 and P15 play as inhibitors of CDK4/6 kinases that control the CDK4/6-RB-E2F pathway. P14 protects P53 and CDKN1A/P21 proteins from degradation through inhibiting the activity of E3-ubiquitylase MDM2 (Serrano et al., 1993; Gil and Peters, 2006; Sherr, 2012). They control G1-S transition of the cell cycle and prevent the development of cancers. Two long noncoding RNA (lncRNA) ANRIL and P14AS are also transcribed from antisense strand of this gene cluster. While these tumor suppressors are frequently inactivated or downregulated by genetic and epigenetic mechanisms (Zhao et al., 2016; The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium, 2020; Popov and Gil, 2010), oncogenic ANRIL and P14AS are often overexpressed in many human cancers (Lou et al., 2020; Ma et al., 2020). Our and other’s works demonstrated that ANRIL and P14AS could regulate these tumor suppressor expression (Yu et al., 2008; Aguilo et al., 2011; Kotake et al., 2011; Ma et al., 2020). However, the regulatory mechanisms for these lncRNAs are far from clear.
Polycomb group (PcG) proteins are well known transcription repressors for the CDKN2A/2B locus through inducing repressive histone modifications, including H3K27m3 and H3K9m3, subsequently causing condensation of the local chromatin structures (Li et al., 2010; Yap et al., 2010; Laugesen et al., 2019). A recent study demonstrated that lncRNA was required for proper localization of core proteins (such as EZH2 and SUZ12) in polycomb repressive complex 2 (PRC2) to chromatin targets in human pluripotent stem cells (Long et al., 2020). ANRIL was reported to downregulate P15INK4B and P16INK4A expression by interacting with PRC1 and PRC2 proteins (Yu et al., 2008; Kotake et al., 2011). ANRIL could bind to SUZ12 and CBX7 and recruit them to the P15INK4B-P14ARF-P16INK4A locus to promote the formation of H3K27m3 and repress these genes’ transcription (Yap et al., 2010; Laugesen et al., 2019). In our previous work, we found that CBX7, a PRC1 member, was able to inhibit the expression of P16INK4A by inducing H3K9m3 (Li et al., 2010). Recently, we have reported that the novel lncRNA P14AS could upregulate the expression of the P16INK4A gene and lncRNA ANRIL through AUF1-binding and promote the proliferation of colon cancer cells in a P16INK4A expression-independent pattern (Ma et al., 2020). Whether the P14AS regulates these gene expression through CBX7-mediated pathway is unknown.
In the present work, we studied the impact of CBX7 on effect of P14AS on transcription of genes in the CDKN2A/2B locus systemically in details.
Materials and methods
Cell culture and tissue samples
The human cell line HEK293T was kindly provided by Professor Yasuhito Yuasa at Tokyo Medical and Dental University. HCT116 cell line was kindly provided by Professor Yuanjia Chen at Peking Union Medical College Hospital. MGC803 was kindly provided by Professor Yang Ke and HepG2 cell line was kindly provided by Professor Qingyun Zhang at Peking University Cancer Hospital and Institute. The colon cancer (CC) tissues and paired normal tissues from the surgical margin (SM, >5 cm from cancer lesion) from CC patients were collected and stored at −70°C at Peking University Cancer Hospital from 2004 to 2011 (Zheng et al., 2015; Ma et al., 2020). Research protocols were approved by the Institutional Review Board of the Peking University Cancer Hospital and Institute, China. Clinical and histopathological data for each patient were obtained according to approved institutional guidelines.
Plasmid construction and transfection
The 1043-nt P14AS lentiviral vector was constructed in PCDH-CMV-EF1a-copGFP-T2A-Puro lentiviral vector by Syngen tech Co., Ltd. (Beijing, China) (Ma et al., 2020). The full length of CBX7 cDNA was inserted into expression vector pEGFP-C1. The shRNA sequence (caaag tacag cacgt ggga) targeting CBX7 (accession number XM_066324) was constructed using pGFPU6/Neo vector (GenePharma Company, Shanghai). The scramble shRNA (gttct ccgaa cgtgt cacgt) was used as negative control (Li et al., 2010).
RNA extract and realtime PCR
The total RNA of cell lines and tissues were isolated using Direct-zol RNA MiniPrep Kit (ZYMO Research, USA) and then reverse transcripted into cDNA using TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, China) according to the manufacturer’s instructions. Next, quantitative RT-PCR (qRT-PCR) was performed using the FastStart Universal SYBR Green Master (ROX) (Roche, Germany) on an ABI-7500 Fast system (Applied Biosystems). GAPDH and ALU were used as the endogenous reference genes for the cultured cell lines and tissues, respectively (Zheng et al., 2015). Each sample was quantitatively analyzed in triplicates. The relative expression levels of these genes were determined using the typical ΔΔCt method. Total tissue samples were divided into two groups with low- and high-level of CBX7 expression using the median as the cutoff value. The differences in target gene expression between these two groups were analyzed with Student’s t-test and plotted in GraphPad Prism 9.0. p < 0.05 was considered statistically significant (N.S: not significant. *: p < 0.05. **: p < 0.01). Primer sequences were showed in Supplementary Table S1.
Western blot
Total protein was extracted from cultured cells using RIPA buffer. The primary polyclonal antibody against P14 (ab3648, Abcam), P15 (ab53034, Abcam), P16 (ab81278, Abcam), CBX7 (ab21873, Abcam), GAPDH (60004-1, ProteinTech), or green fluorescent protein (GFP) (ProteinTech, 50430-2-AP; China) was applied at 1:5,000 to 1:1,000 dilutions. The signals were visualized using the Enhanced Chemiluminescence Kit (Millipore) and Alpha Imager system.
RNA immunoprecipitation assay
The RIP assay was carried out using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Sigma) according to the manufacturer’s instructions. Total CBX7-binding RNAs were immunoprecipitated and extracted using anti-CBX7 antibody (ab21873, Abcam). cDNA was synthesized from the RIP-RNAs using random primers, and gene-specific quantitative PCR was then performed using FastStart Universal SYBR Green Master (ROX) (Roche, Germany). 18S rRNA was used as positive and reference control to calculate the enrichment of lncRNA. The qPCR products were analyzed by agarose gel or PAGE gel to detect product size and specificity of amplification.
Chromatin immunoprecipitation assay
ChIP assays were performed and analyzed essentially as described (Li et al., 2010). Rabbit anti-CBX7, rabbit anti-H3K9m3 (07-442; Sigma-Aldrich), and rabbit anit-H3K27m3 (61017; Active Motif) antibodies were used to precipitate endogenous CBX7 and modified histone proteins and the binding chromatin. The enrichment of specific genomic regions was assessed relative to Input DNA. IgG antibody was used as negative control (PP64B, Sigma-Aldrich). Each ChIP experiment was quantitatively analyzed in triplicates. The qPCR products were analyzed by agarose gel or PAGE gel to detect product size and specificity of amplification. Primer sets and annealing temperature used were listed in Supplementary Table S1.
Results
P14AS upregulates the expression of P15INK4B-P14ARF-P16INK4A locus depending on CBX7
We previously reported that P14AS upregulated P16INK4A and ANRIL expression, and that CBX7 downregulated P16INK4A expression through inducing H3K9me3 modification (Li et al., 2010; Ma et al., 2020). To study the effect of CBX7 on P14AS-induced P16INK4A and ANRIL expression, we selected proper cell lines through analyzing the basal expression states of CBX7 and its target genes at first. In Western blot analysis, the amount of endogenous CBX7 protein was much greater in HEK293T and HCT116 cells than in MGC803 and HepG2 cells (Figure 1A). The results of qRT-PCR confirmed the Western blot experiment (Figure 1B). No P16 and P14 proteins were detected in HCT116 cells.
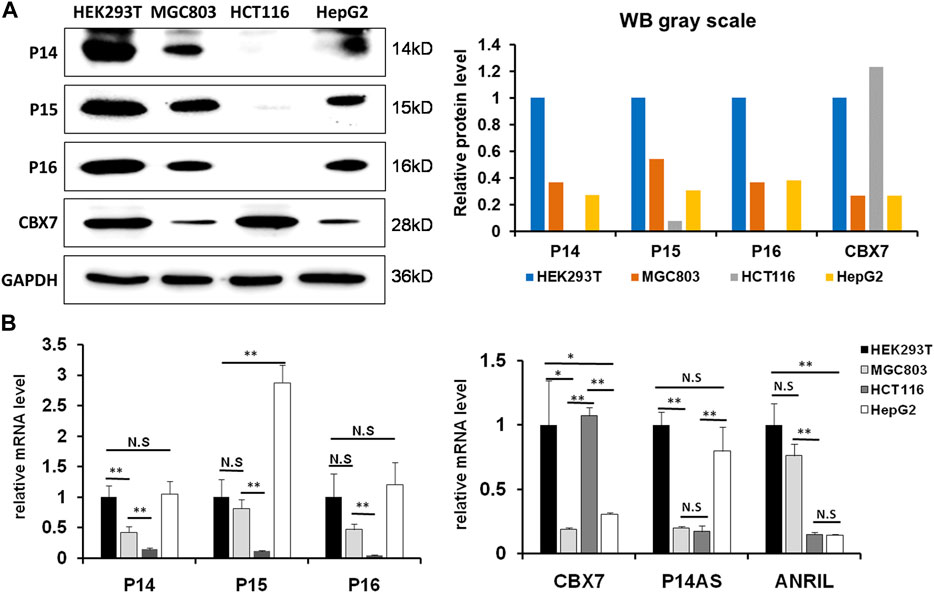
FIGURE 1. The status of basal CBX7, P16INK4A, P14ARF, P15INK4B, P14AS and ANRIL expression in human cell lines. (A) the results of Western blot; (B) qRT-PCR analyses. The relative mRNA levels are presented as mean ± SD. N.S: not significant. *: p < 0.05. **: p < 0.01 in Student’s t-test.
Then, we compared the effect of P14AS on the upregulation of P16INK4A, P14ARF and P15INK4B expression in these cell lines with high and low CBX7 expression. The results showed that transient P14AS overexpression obviously upregulated the expression levels of P16INK4A, P14ARF and P15INK4B only in HEK293T and HCT116 cells with active CBX7 expression, but not in MGC803 and HepG2 cell lines with low CBX7 expression (Figure 2). These results imply that upregulation of P16INK4A, P14ARF and P15INK4B by P14AS might be dependent on active CBX7 expression.
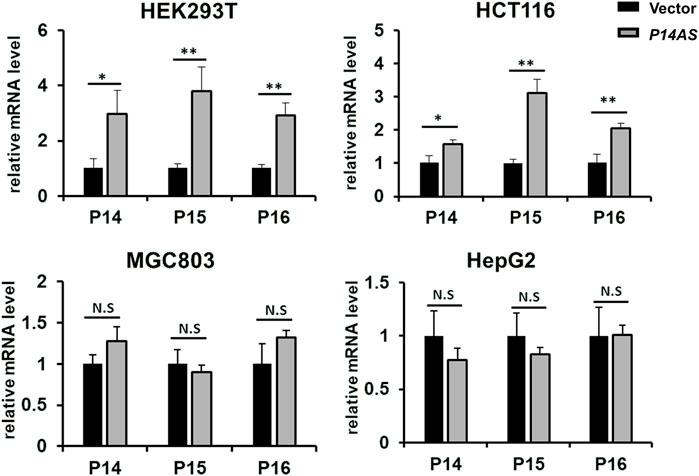
FIGURE 2. Effect of transient P14AS overexpression on the levels of P14ARF, P16INK4A and P15INK4B expression in qTR-PCR analyses.
To clarify the role of CBX7 in the upregulation effect of P14AS on P16INK4A, P14ARF, P15INK4B and ANRIL, the CBX7 expression level was artificially increased in MGC803 and HepG2 cells by transfection of the CBX7 expression vector and decreased in HEK293T and HCT116 cells by transfection of the shR-CBX7 vector, and then the cells were co-transfected with P14AS expression or empty vectors (Supplementary Figure S1). As expected, both the qRT-PCR and Western blot results showed that CBX7 overexpression enabled P14AS to obviously upregulate P16INK4A, P14ARF, P15INK4B and ANRIL expression in the co-transfected MGC803 and HepG2 cells (Figures 3A,B). Overexpressed CBX7 could inhibit the expression of these genes at different level.
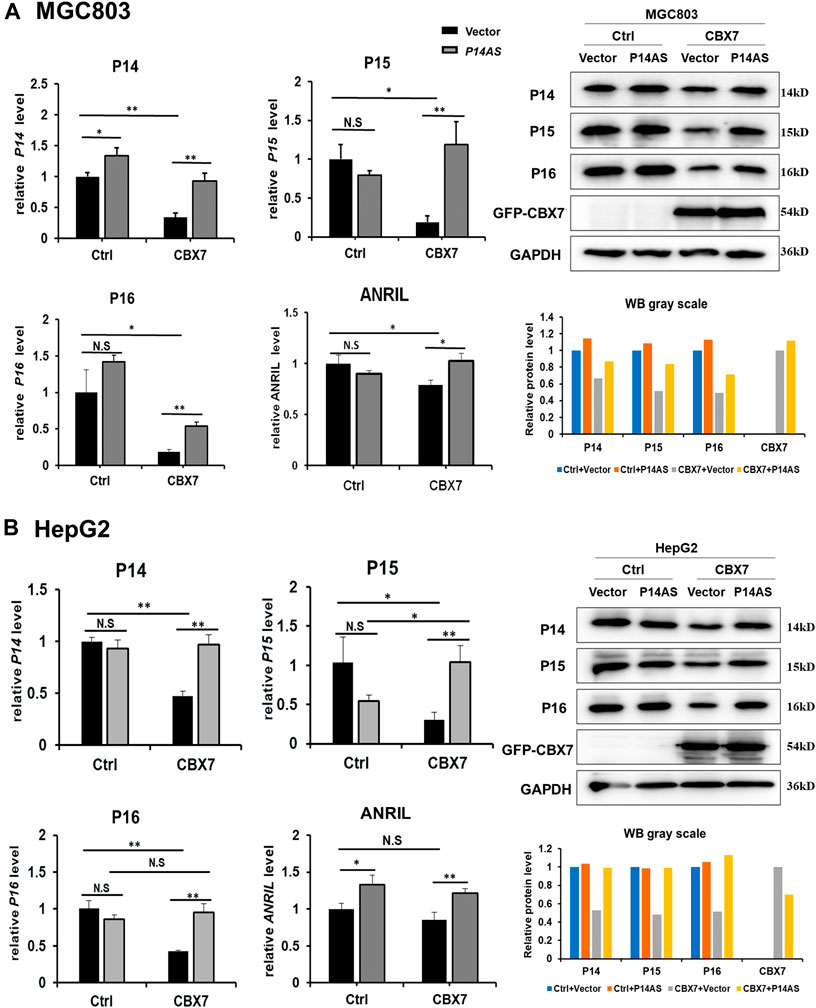
FIGURE 3. Increasing CBX7 expression enables P14AS to upregulate the P14ARF, P15INK4B, P16INK4A expression levels in MGC803 and HepG2 cells with weak basal CBX7 expression in qRT-PCR (left charts for the relative RNA level) and Western blot analyses (right image and chart for the amounts of proteins). (A) MGC803; (B) HepG2 cells.
In contrast, the upregulating effects of P14AS overexpression were almost disappeared when endogenous CBX7 expression was stably knocked down by shR-CBX7 treatment in HEK293T and HCT116 cells co-transfected with P14AS (Figures 4A,B).
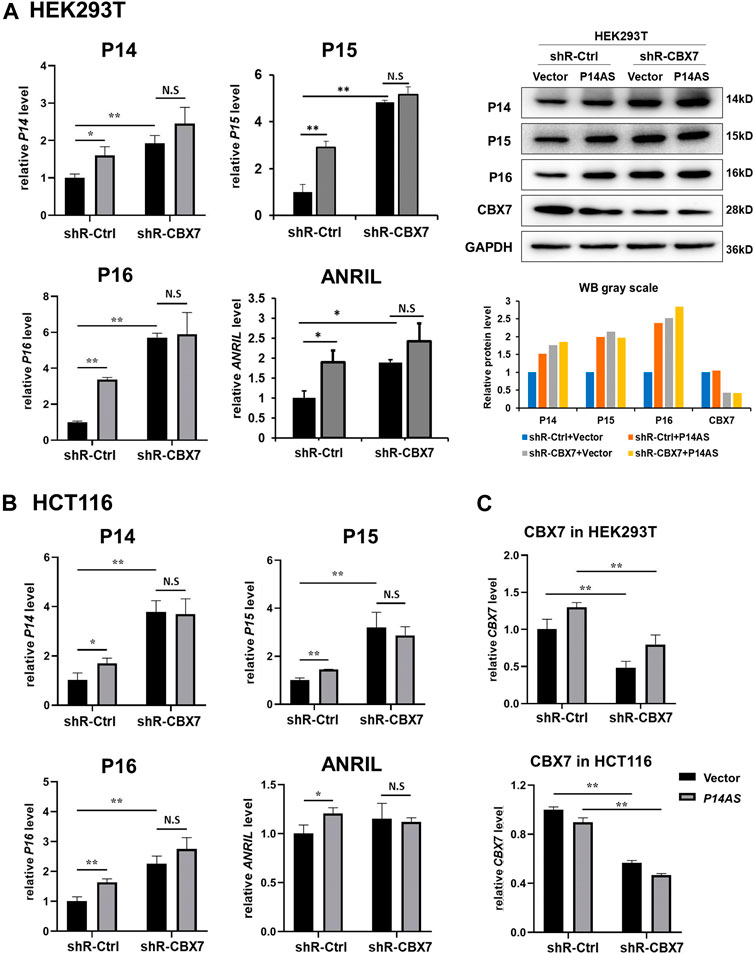
FIGURE 4. Downregulation of CBX7 expression disables P14AS to upregulate the P14ARF, P15INK4B, P16INK4A expression levels in HEK293T and HCT116 cells with active basal CBX7 expression in qRT-PCR and Western blot analyses. (A) HEK293T; (B) HCT116 cells; (C) the efficiency of CBX7 knockdown in HEK293T and HCT116.
All the above results support the hypothesis that P14AS upregulates the expression of these target genes in an active CBX7 expression-dependent pattern.
Coexpression of P14AS with P14ARF and P16INK4A genes in human colon cancer tissues with high CBX7 expression level
To study whether the above CBX7 expression-dependent effect of P14AS on the upregulation of P14ARF, P15INK4B, P16INK4A expression occurs in human tissues, we determined the levels of P14ARF, P15INK4B, P16INK4A, P14AS and CBX7 in colon cancer and paired normal surgical margin tissues from patients (n = 75) by qRT-PCR. Then, these samples were divided into the CBX7 expression-high and -low groups, according to the median CBX7 expression level. As expected, the expression levels of P16INK4A and P14ARF were significantly higher in the P14AS expression-positive (+) samples than in the P14AS expression-negative (−) samples only within the CBX7 high colon cancer group, but not within the CBX7 low colon cancer group (Figure 5A). In addition, no significant difference in these genes’ expression levels was observed between the P14AS expression-positive and expression-negative samples in the paired normal samples (Figure 5B), suggesting a tumor-specific effect of CBX7 on P14AS functions. These results confirm the hypothesis that CBX7 expression is essential for P14AS to regulate P16INK4A, P14ARF and P15INK4B expression in cancer tissues.

FIGURE 5. Coexpression of P14AS with P14ARF, P15INK4B and P16INK4a genes in human colon cancer tissues with high CBX7 expression. (A) Colon cancer tissues; (B) Surgical margin tissues. N.S: not significant. *: p < 0.05. **: p < 0.01.
P14AS competitively binds to CBX7 and prevents the binding of CBX7 to ANRIL lncRNA and P16INK4A, P14ARF and P15INK4B promoter DNA
To explore the molecular mechanisms of CBX7 affecting the functions of P14AS in the regulation of these target genes, we conducted qPCR analysis based on RIP and ChIP assays using anti-CBX7 antibody. The RIP-qPCR results showed that native CBX7 directly bound not only to lncRNA P14AS, but also to lncRNA ANRIL. In the HEK293T and HCT116 cells with P14AS overexpression, the CBX7-P14AS protein-RNA binding was increased, while the CBX7-ANRIL protein-RNA binding was decreased relative to the vector control cells (Figure 6A). Furthermore, the ChIP-qPCR results showed that native CBX7 directly bound to promoter DNA of these target genes. Notably, the protein-DNA bindings of CBX7-promoters were also decreased in the P14AS overexpression cells (Figure 6B). Together, these results indicated that P14AS could competitively bind to CBX7, which consequently prevented the CBX7-ANRIL and CBX7-promoter bindings and upregulated the expression levels of these target genes.
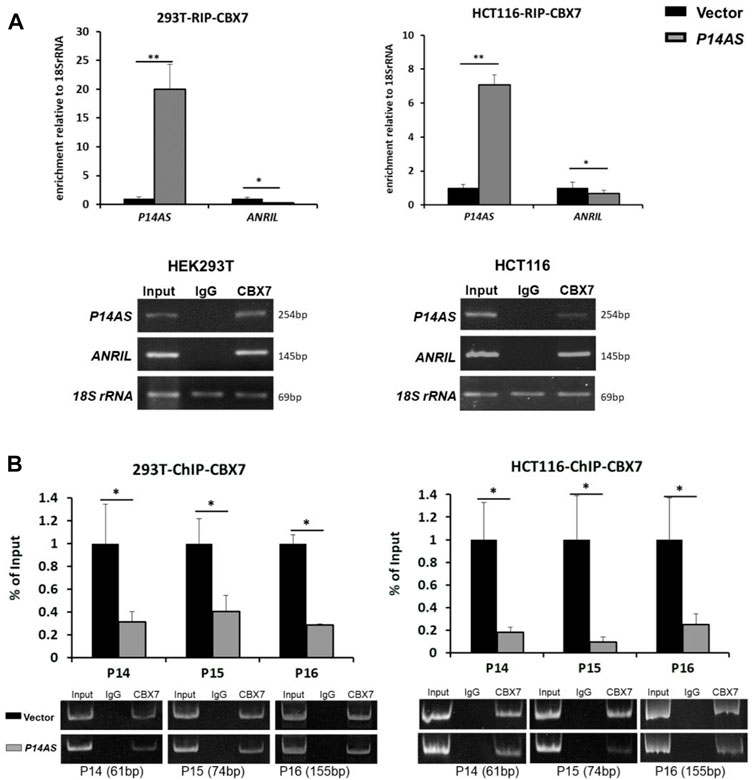
FIGURE 6. P14AS competitively binds to CBX7 and prevents the binding of CBX7 to ANRIL and to P14ARF, P15INK4B, P16INK4A promoters. (A) Levels of P14AS and ANRIL binding to endogenous CBX7, as determined by RIP-qPCR analyses; and (B) binding of gene promoters to endogenous CBX7, as determined by ChIP-qPCR analyses. The experiments were repeated using both HEK293T and HCT116 cell lines.
In addition, we checked the status of histone modifications on these gene promoters in ChIP-qPCR analysis. We found that P14AS overexpression significantly decreased the recruitment of trimethylation of both H3K9 and H3K27 (H3K9m3 and H3K27m3) at these genes’ promoters in HCT116 cells with active CBX7 expression (Figure 7A), but not in MGC803 cells with low CBX7 expression (Figure 7B in the first two columns in Ctrl cells). However, in the MGC803 cells with enforced CBX7 overexpression, P14AS overexpression decreased the recruitment of H3K9m3 at these genes’ promoters (Figure 7B, in the last two columns in CBX7 overexpressed cells), The enforced CBX7 overexpression could also increase the recruitment of H3K9m3 at these genes’ promoters (Figure 7B, in the two black bars in the same panel), which was consistent with the repressive effects of CBX7 on these genes. However, the recruitments of H3K27m3 at these promoters were not changed as obviously as H3K9m3.
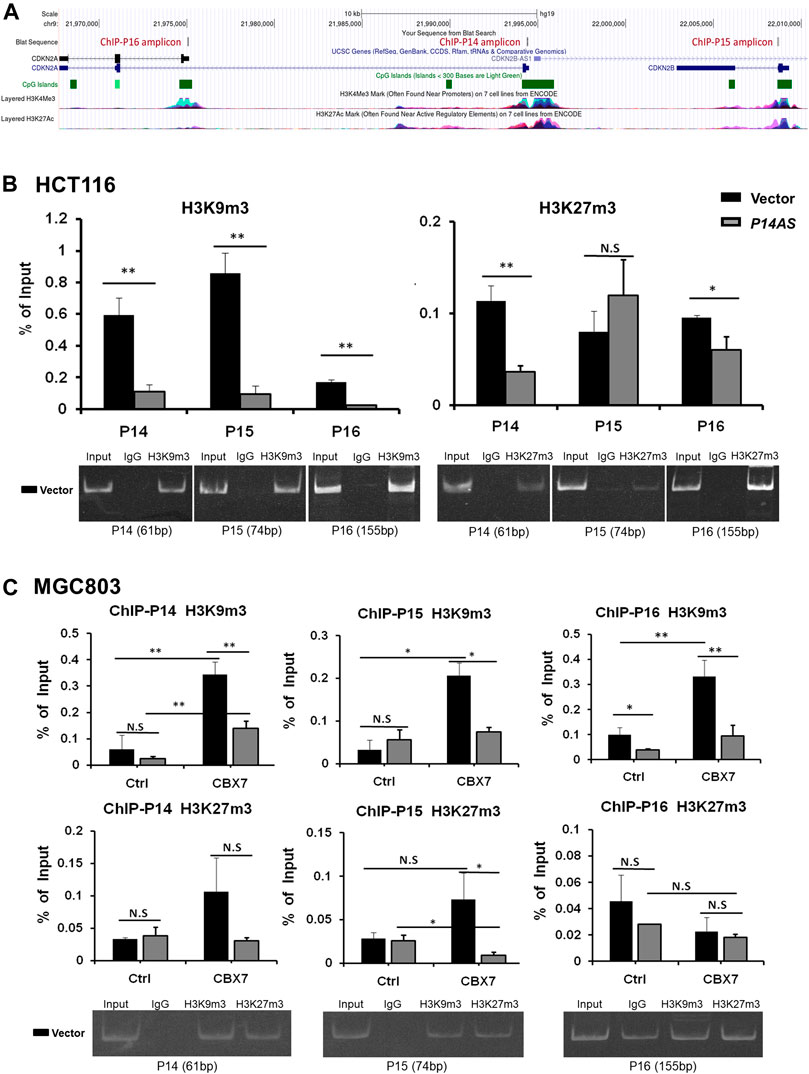
FIGURE 7. Inhibition of repressive histone modifications at promoters of P14ARF, P15INK4B, and P16INK4A by P14AS is CBX7-dependent, as determined by ChIP-qPCR analyses. (A) The ChIP-qPCR fragments and histone modifications within the locus, a screenshot from UCSC Genome Browser; (B) The levels of H3K9m3 and H3K27m3 at the three target genes promoters in HCT116 cells stably transfected with the P14AS or empty vector; (C) The levels of H3K9m3 and H3K27m3 at the three genes’ promoters in MGC803 cells cotransfected with P14AS and CBX7 or empty control vectors. The gel pictures show the size and specificity of qPCR products.
These results illustrated that inhibition of repressive histone modifications by P14AS is also CBX7 expression-dependent.
Discussion
It has been reported that both lcnRNA ANRIL and promoters of P14ARF, P15INK4B, P16INK4A can directly bind to the PcG proteins including CBX7, and form heterochromatin surrounding the CDKN2A/2B locus, which leads to the repression of P15INK4B expression (Yu et al., 2008; Aguilo et al., 2011; Kotake et al., 2011). PcG proteins such as CBX7 have also been reported to repress P16INK4A transcription (Gil et al., 2004; Li et al., 2010; Yap et al., 2010). In our recent study, we reported that lncRNA P14AS transcribed from the antisense strand of P14ARF promoter to intron-1 could directly upregulate the expression of ANRIL-CDKN2A/2B locus (Ma et al., 2020). Here, we further show that P14AS may act as cis-activator to increase these genes’ expression in a CBX7-binding-dependent pattern.
ANRIL contains 19–21 reported exons over a 126 kb region franking the P15INK4B gene (Yap et al., 2010). P14AS is transcribed from the upstream of ANRIL and there is a 79-nt complete overlap between its exon-3 and ANRIL exon-1 (Ma et al., 2020). We wonder that P14AS might be an isoform of ANRIL but use different transcription start site. However, they have distinct effects on the expression regulation of their host genes and are likely to contribute to the concerto of transcription regulation. Recently, the splicing variants of ANRIL were reported to exert opposing effects on endothelial cell activities associated with coronary artery disease (Cho et al., 2020). And truncated isoforms of ANRIL were reported to be overexpressed in bladder cancer and differently correlated with the expression of the P14ARF, P15INK4B, and P16INK4A genes (Hoffmann et al., 2015). Circular ANRIL isoforms could switch from repressors of CDKN2B to activators of CDKN2A and change EZH2 localization during RAF1 oncogene-induced senescence (Muniz et al., 2021). Although we found that total ANRIL expression is regulated by the CBX7-P14AS binding, which ANRIL isoforms’ expression may be affected by the CBX7-P14AS binding is worthy of further study.
AUF1 is a member of RNA binding proteins family and play roles in directing RNA decay and translation at post-transcription stage (White et al., 2017). AUF1 was also identified as an important destabilizer for P16INK4A mRNA and thereby influencing cell senescence (Chang et al., 2010). In our previous study, we reported P14AS could competitively bind with AUF1 and decreased the AUF1-ANRIL/P16INK4A RNA interaction (Ma et al., 2020). Here, we further verified P14AS could also competitively bind to CBX7. The overexpression of P14AS could also inhibit the repressive histone modifications, including H3K9m3 and H3K27m3, within the CDKN2A/2B locus, through inhibiting the CBX7 recruitment, and consequently lead to the upregulation of gene transcription. These results imply that lncRNA may interact with different transcription factors or RNA binding proteins to regulate the gene expression at both transcription and post-transcription level.
We are still at a very early stage of understanding the functions of most lncRNAs. There are a number of existing models to illustrate the principles of lncRNA regulation. Interactions between loops of lncRNAs and transcription factors have been suggested to be important in maintaining promoters in an accessible status to allow for transcription complex interactions (Kung and Lee, 2013; Laugesen et al., 2019; Long et al., 2020). Enhancers generally contain a low-proportion of CpG sites and active enhancers are often associated with H2A.Z replacement and formation of H3K4m1 and H3K27ac (Calo and Wysocka, 2013; Core et al., 2014). According to the publicly available ENCODE database, the P14AS-transcribed region matches the typical enhancer location, lying between the mammalian-wide interspersed repeat (MIR) element and the CpG island in the P14ARF promoter (Supplementary Figure S2, highlighted in light-blue). Our recent work also showed that there was a loop between the P14AS-transcribed region and P16INK4A promoter region (unpublished data). This suggests that P14AS may be an enhancer lncRNA.
In addition, formation of repressive H3K27m3 and active H3K4m3 have been recognized in embryonic stem cells to be a poised pattern of histone modifications in chromatin of bivalent genes (such as P16INK4A) (Liu et al., 2016; Kumar et al., 2021). H3K27m3, H3K9m3 and H3K4m3 are the main tri-methylation modifications involved in the formation of “bivalent domains”, which is a unique chromatin region consisting of two histone tri-methylation modifications and plays a vital regulatory role in the differentiation of various stem cell systems (Sun et al., 2022). Among the enriched genes in differentiation process of hepatic stem/progenitor cells, the CDKN2A gene was upregulated and exhibiting bivalent domains within 2 kb of transcription start site (Kanayama et al., 2019). The P14AS-transcribed region is also a poised PcG responsive element (PRE) region. This implies that the P14AS may play a role in maintaining the accessibility to the flanking promoter region for transcription factor complexes.
It is generally considered that the P16INK4A, P14ARF, P15INK4B genes at 9p21.3 locus should not be simultaneously silenced in normally differentiated cells. More evidences suggest that these genes provide an important check and balance system that is essential for controlling the G1→S phase transition in cell cycle and that the dysfunctions of these genes play crucial roles in the malignant transformation of human cells (Krimpenfort et al., 2007; Leong et al., 2009). P16 is a weakly expressed nucleic protein in normal cells and long-term high P16INK4A expression will promotes cells to enter senescence. Importantly, inactivity of P16INK4A is one of the most frequent events in many kinds of human tumors (LaPak and Burd, 2014). Therefore, the regulation of P15INK4B-P14ARF-P16INK4A genome locus maybe a precise and complex process and essential to maintain the coordinated balance between tumor suppression and aging. There are numerous advances in understanding these genes’ functions and regulation since their importance was detected almost 40 years ago. However, the underlying mechanisms of their regulation as well as the coordinated expression patterns of these genes are not yet fully understood. Here we provided a new clue that lncRNA P14AS could competitively bind to CBX7 and reduce the repressive histone modifications of this gene cluster region. It might play roles in modifying local genome structure and the precise regulation of the expression of these genes.
Conclusion
LncRNA P14AS can competitively bind to CBX7 and reduce the binding of CBX7 to lncRNA ANRIL and the promoters of P16INK4A, P14ARF and P15INK4B genes, which results in their up-regulation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Review Board of the Peking University Cancer Hospital and Institute, China. The patients/participants provided their written informed consent to participate in this study.
Author contributions
ZL performed qRT-PCR, WB, RIP, ChIP experiments. JQ and WM constructed the vectors and stably transfected cells. JZ and LG collected the colon cancer tissues and prepared RNA and cDNA. DD designed the study and revised the manuscript. BZ analyzed the data, wrote the manuscript draft and obtained financial support. All authors read and approved the final manuscript.
Funding
This study was supported by grants to BZ from the National Natural Science Foundation of China (No. 82073107 and 81773036) and Beijing Municipal Natural Science Foundation (No. 7222022).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.993525/full#supplementary-material
Reference
Aguilo, F., Zhou, M. M., and Walsh, M. J. (2011). Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 71 (16), 5365–5369. doi:10.1158/0008-5472.CAN-10-4379
Calo, E., and Wysocka, J. (2013). Modification of enhancer chromatin: What, how, and why? Mol. Cell. 49, 825–837. doi:10.1016/j.molcel.2013.01.038
Chang, N., Yi, J., Guo, G., Liu, X., Shang, Y., Tong, T., et al. (2010). HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol. Cell. Biol. 30, 3875–3886. doi:10.1128/MCB.00169-10
Cho, H., Li, Y. B., Archacki, S., Wang, F., Yu, G., Chakrabarti, S., et al. (2020). Splice variants of lncRNA RNA ANRIL exert opposing effects on endothelial cell activities associated with coronary artery disease. RNA Biol. 17 (10), 1391–1401. doi:10.1080/15476286.2020.1771519
Core, L. J., Martins, A. L., Danko, C. G., Waters, C. T., Siepel, A., and Lis, J. T. (2014). Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320. doi:10.1038/ng.3142
Gil, J., Bernard, D., Martínez, D., and Beach, D. (2004). Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell. Biol. 6 (1), 67–72. doi:10.1038/ncb1077
Gil, J., and Peters, G. (2006). Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell. Biol. 7 (9), 667–677. doi:10.1038/nrm1987
Hoffmann, M. J., Dehn, J., Droop, J., Niegisch, G., Niedworok, C., Szarvas, T., et al. (2015). Truncated isoforms of lncRNA ANRIL are overexpressed in bladder cancer, but do not contribute to repression of INK4 tumor suppressors. Noncoding. RNA 1 (3), 266–284. doi:10.3390/ncrna1030266
Kanayama, K., Chiba, T., Oshima, M., Kanzaki, H., Koide, S., Saraya, A., et al. (2019). Genome-wide mapping of bivalent histone modifications in hepatic stem/progenitor cells. Stem Cells Int. 2019, 9789240. doi:10.1155/2019/9789240
Kotake, Y., Nakagawa, T., Kitagawa, K., Suzuki, S., LiuN., , KitagawaM., , et al. (2011). Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30 (16), 1956–1962. doi:10.1038/onc.2010.568
Krimpenfort, P., Ijpenberg, A., Song, J., van der Valk, M., Nawijn, M., Zevenhoven, J., et al. (2007). p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature 448, 943–946. doi:10.1038/nature06084
Kumar, D., Cinghu, S., Oldfield, A. J., Yang, P., and Jothi, R. (2021). Decoding the function of bivalent chromatin in development and cancer. Genome Res. 31 (12), 2170–2184. doi:10.1101/gr.275736.121
Kung, J. T., and Lee, J. T. (2013). RNA in the loop. Dev. Cell. 24, 565–567. doi:10.1016/j.devcel.2013.03.009
LaPak, M., and Burd, C. E. (2014). The molecular balancing act of p16(INK4a) in cancer and aging. Mol. Cancer Res. 12 (2), 167–183. doi:10.1158/1541-7786.MCR-13-0350
Laugesen, A., Hojfeldt, J. W., and Helin, K. (2019). Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell. 74, 8–18. doi:10.1016/j.molcel.2019.03.011
Leong, W., Chau, J., and Li, B. (2009). p53 Deficiency leads to compensatory up-regulation of p16INK4a. Mol. Cancer Res. 7, 354–360. doi:10.1158/1541-7786.MCR-08-0373
Li, Q., Wang, X., Lu, Z., Zhang, B., Guan, Z., Liu, Z., et al. (2010). Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PLoS One 5, e13732. doi:10.1371/journal.pone.0013732
Liu, X., Wang, C., Liu, W., Li, J., Li, C., Kou, X., et al. (2016). Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537 (7621), 558–562. doi:10.1038/nature19362
Long, Y., Hwang, T., Gooding, A. R., Goodrich, K. J., Rinn, J. L., and Cech, T. R. (2020). RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 52, 931–938. doi:10.1038/s41588-020-0662-x
Lou, N., Liu, G., and Pan, Y. (2020). Long noncoding RNA ANRIL as a novel biomarker in human cancer. Future Oncol. 16 (35), 2981–2995. doi:10.2217/fon-2020-0470
Ma, W. R., Qiaog, J. L., Zhou, J., Gu, L., and Deng, D. (2020). Characterization of novel LncRNA P14AS as a protector of ANRIL through AUF1 binding in human cells. Mol. Cancer 19 (1), 42. doi:10.1186/s12943-020-01150-4
Muniz, L., Lazorthes, S., Delmas, M., Ouvrard, J., Aguirrebengoa, M., Trouche, D., et al. (2021). Circular ANRIL isoforms switch from repressors to activators of p15/CDKN2B expression during RAF1 oncogene-induced senescence. RNA Biol. 18 (3), 404–420. doi:10.1080/15476286.2020.1812910
Popov, N., and Gil, J. (2010). Epigenetic regulation of the INK4b-ARF-INK4a locus: In sickness and in health. Epigenetics 5 (8), 685–690. doi:10.4161/epi.5.8.12996
Serrano, M., Hannon, G. J., and Beach, D. (1993). A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366, 704–707. doi:10.1038/366704a0
Sherr, C. J. (2012). Ink4-Arf locus in cancer and aging. Wiley Interdiscip. Rev. Dev. Biol. 1 (5), 731–741. doi:10.1002/wdev.40
Sun, H., Wang, Y., Wang, Y., Ji, F., Wang, A., Yang, M., et al. (2022). Bivalent regulation and related mechanisms of H3K4/27/9me3 in stem cells. Stem Cell. Rev. Rep. 18 (1), 165–178. doi:10.1007/s12015-021-10234-7
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (2020). Pan-cancer analysis of whole genomes. Nature 578, 82–93. doi:10.1038/s41586-020-1969-6
White, E. J., Matsangos, A. E., and Wilson, G. M. (2017). AUF1 regulation of coding and noncoding RNA. WIREs. RNA 8 (2), e1393. doi:10.1002/wrna.1393
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 38 (5), 662–674. doi:10.1016/j.molcel.2010.03.021
Yu, W., Gius, D., Onyango, P., Muldoon-Jacobs, K., Karp, J., Feinberg, A. P., et al. (2008). Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206. doi:10.1038/nature06468
Zhao, R., Choi, B. Y., Lee, M. H., Bode, A. M., and Dong, Z. (2016). Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine 8, 30–39. doi:10.1016/j.ebiom.2016.04.017
Keywords: lncRNA, P14AS, CBX7, p16INK4A, p14ARF, p15INK4B
Citation: Li Z, Qiao J, Ma W, Zhou J, Gu L, Deng D and Zhang B (2022) P14AS upregulates gene expression in the CDKN2A/2B locus through competitive binding to PcG protein CBX7. Front. Cell Dev. Biol. 10:993525. doi: 10.3389/fcell.2022.993525
Received: 13 July 2022; Accepted: 24 August 2022;
Published: 13 September 2022.
Edited by:
Beisi Xu, St. Jude Children’s Research Hospital, United StatesReviewed by:
Zhilian Jia, City of Hope National Medical Center, United StatesSanqi An, Guangxi Medical University, China
Yanbin Zheng, University of Texas Southwestern Medical Center, United States
Copyright © 2022 Li, Qiao, Ma, Zhou, Gu, Deng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dajun Deng, dengdajun@bjmu.edu.cn; Baozhen Zhang, zhangbaozhen@bjmu.edu.cn
 Zhuoqi Li
Zhuoqi Li Juanli Qiao
Juanli Qiao Wanru Ma
Wanru Ma  Dajun Deng
Dajun Deng Baozhen Zhang
Baozhen Zhang