Exercise training ameliorates myocardial phenotypes in heart failure with preserved ejection fraction by changing N6-methyladenosine modification in mice model
- 1Department of Biochemistry and Immunology, Capital Institute of Pediatrics-Peking University Teaching Hospital, Beijing, China
- 2Department of Biochemistry and Immunology, Capital Institute of Pediatrics, Beijing, China
- 3Beijing Municipal Key Laboratory of Child Development and Nutriomics, Beijing, China
- 4Graduate School of Peking Union Medical College, Beijing, China
- 5Beijing TongRen Hospital, Capital Medical University, Beijing, China
Heart failure with preserved ejection fraction (HFpEF) shows complicated and not clearly defined etiology and pathogenesis. Although no pharmacotherapeutics have improved the survival rate in HFpEF, exercise training has become an efficient intervention to improve functional outcomes. Here, we investigated N6-methyladenosine (m6A) RNA methylation modification in a “two-hit” mouse model with HFpEF and HFpEF with exercise (HFpEF + EXT). The manner of m6A in HFpEF and HFpEF + EXT hearts was explored via m6A-specific methylated RNA immunoprecipitation followed by high-throughput and RNA sequencing methods. A total amount of 3992 novel m6A peaks were spotted in HFpEF + EXT, and 426 differently methylated sites, including 371 hypermethylated and 55 hypomethylated m6A sites, were singled out for further analysis (fold change >2, p < 0.05). According to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, unique m6A-modified transcripts in HFpEF + EXT were associated with apoptosis-related pathway and myocardial energy metabolism. HFpEF + EXT had higher total m6A levels and downregulated fat mass and obesity-related (FTO) protein levels. Overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy. Totally, m6A is a significant alternation of epitranscriptomic processes, which is also a potentially meaningful therapeutic target.
Introduction
Heart failure (HF) with preserved ejection fraction (HFpEF) is a complicated clinical syndrome associated with poor quality of life, extensive utilization of medical resources and premature death (Dunlay et al., 2017; Mishra and Kass, 2021; Omote et al., 2022). HFpEF affects 50% of patients with HF worldwide and has been increasing in prevalence largely connected with the aging of the population (Mishra and Kass, 2021; Omote et al., 2022). HFpEF is an aging disease characterized by cardiac hypertrophy, myocardial interstitial fibers, and cardiac diastolic dysfunction (Prandi et al., 2022). Considering the complex pathogenesis of HFpEF, it is the most significant unmet medical need in cardiovascular disease (Mishra and Kass, 2021). Previous studies have suggested that HFpEF is a hemodynamic disorder characterized by hypertension, myocardial hypertrophy, myocardial fibrosis and diastolic dysfunction. However, current studies believe that HFpEF is a syndrome with abnormal function of multiple organs, including diseases caused by the comprehensive effects on the heart, lung, kidney, immunity, inflammation, metabolism and others (Figtree et al., 2021; Mishra and Kass, 2021; Omote et al., 2022; Prandi et al., 2022). Cardiac molecular and cellular mechanisms with HFpEF include cardiac hypertrophy, myocardial fibrosis, inflammation, cardiomyocyte sarcomere disfunction and mitochondrial and metabolic defects (Schiattarella et al., 2021a). However, the etiology and pathogenesis of HFpEF are unclear. At present, the effective treatment of HFpEF is very limited and cannot prevent the development of the disease. Urgent problems and effective solutions are needed.
Recent studies have shown that exercise training (EXT) is an important means of nonpharmacological intervention and prevention of cardiovascular diseases. EXT can improve exercise ability and quality of life, and is related to reducing the risk of hospitalization and cardiovascular death (Leggio et al., 2020; Jaconiano and Moreira-Gonçalves, 2022). However, its positive influence basically lacks a physiological explanation in HFpEF. In patients with heart failure with reduced ejection fraction (HFrEF), physical exercise is already a class I level A recommendation (Ho et al., 2019). Concerning HFpEF, systematic reviews and meta-analysis offer a high level of evidence that EXT is a safe and effective strategy improving upon peak VO2, 6 min walk, cardiorespiratory fitness, diastolic function, quality of life, and general health (Ismail et al., 2013; Dieberg et al., 2015; Fukuta et al., 2019). Another research supported that EXT significantly improves the exercise capacity and left ventricular diastolic function, indicating an improvement in ventricular stiffness and filling pressures (Edelmann et al., 2011; Fukuta et al., 2019). Nevertheless, this evidence is not consensual. Therefore, we use a new “two-hit” mouse model of HFpEF (Schiattarella et al., 2019; Schiattarella et al., 2021b), which simulates concomitant metabolic and hypertensive stress in mice to explore the effect mechanism of EXT on HFpEF.
Epigenetic regulation includes DNA methylation, histone modifications, and noncoding RNAs. Their important role in HFpEF has been widely researched, and the effect of RNA alternation on the control of gene expression in HFpEF will be clarified (Handy et al., 2011; Feil and Fraga, 2012; Poller et al., 2018; Zhang et al., 2021a; Hamdani et al., 2021). N6-methyladenosine (m6A) methylation is the most common internal mRNA alternation, which affects the metabolism of RNA throughout its life cycle (Wang et al., 2014a; Wang et al., 2014b; Alarcón et al., 2015). The latest evidence shows that m6A modification not only participates in normal biological processes, but also serves vital functions in the occurrence and progression of different heart diseases (Dorn et al., 2019; Mathiyalagan et al., 2019; Mo et al., 2019; Berulava et al., 2020; Gao et al., 2020; Krüger et al., 2020; Lin et al., 2020). But the reversal of myocardial dysfunction in HFpEF by EXT through m6A modification has not been studied.
The modification of m6A is related to different types of cardiovascular diseases, including cardiac fibrosis, cardiac hypertrophy, myocardial infarction, atherosclerosis, abdominal aortic aneurysm, heart failure and HFpEF (Dorn et al., 2019; Mathiyalagan et al., 2019; Berulava et al., 2020; Gao et al., 2020; Lin et al., 2020). Nevertheless, the transcriptome-wide distribution of m6A in HFpEF + EXT heart samples remains largely unknown. This research tried to clarify the m6A methylation profiles of heart tissue samples from HFpEF + EXT and control (HFpEF) mice and give evidence of highly diverse m6A-modified patterns in both groups. It is displayed that abnormal m6A RNA alternations in HFpEF + EXT samples are possible for modulating myocardial apoptosis and myocardial energy metabolism, close to the cardiac aging phenotype. Our outcomes offer evidence that exercise-induced m6A alternation is closely related to the pathogenesis of HFpEF in the new model and will promote further research on the potential drug targets of m6A alternation in the treatment of HFpEF.
Materials and methods
Animals
The contents of this study had been reviewed and approved by the ethics committee of Capital Institute of Pediatrics, whose approved license number was DWLL2019003. This study’s routines were all in accordance with corresponding ethical standards. C57BL/6J male mice were adopted for wild-type studies. All mice used in the trials were 8 weeks old (Schiattarella et al., 2019) and kept in cages at 24°C with a 12 h alternating light/dark cycle. The HFpEF model was constructed using mice with unrestricted access to HFD (High Fat Diet) food (D12492, Research Diet) and water for 16 weeks. Nω-nitro-l-arginine methyl ester (l-NAME) (0.5 g/L, Sigma-Aldrich) was added in the water, and the pH of the mixture was adjusted to 7.4. Mice in the control group were fed unlimited normal food and water. At the eighth week, mice began to do moderate-intensity running on a treadmill at a 10° gradient for 8 weeks (5 times a week, running for 5 days and rest for 2 days, with 5 min of running and 2 min of rest for 10 cycles, a total of 50 min of running and speed with 10 m/min). After the mice were euthanized, we collected hearts with 1.5 ml RNase-free centrifuge tubes, and stored them immediately at 4°C for 12 h. Samples were finally placed at −80°C for long-term storage to prevent RNA degradation.
Echocardiographic assessment
At 28 weeks of age, cardiac function by echocardiography in all mice in a scrambled order was assessed with a VEVO 3100 instrument, by dedicated personnel. Anesthesia mice were induced by continuous inhalation using 2% isoflurane +2 L/min air flow rate. The probe was placed in the center of the heart to observe the short axis of the heart. Observe the long axis of the heart during the slitting and adjust the heart rate to maintain 415–460 beats/min. Coefficients gathered included: left ventricular end-diastolic diameter, left ventricular end-systolic diameter, heart rate, left ventricular shortening, LVEF, peak doppler flow velocity in early mitral valve diastolic, peak doppler flow velocity across late mitral valve diastolic. All measurements were averages of at least three beats.
Western blotting and antibodies
Our western blotting assays were performed strictly following the standard protocols of our laboratory. Firstly, mouse heart tissues were ground using the tissue crusher with the tissue lysate, and lysates were prepared with RIPA lysis buffer from Beyotime Biotechnology, where protease inhibitor were added. Blots were screened using specific antibodies: GAPDH (5174, 1:10000; CST), Mettl3 (ab195352, 1:1000; Abcam), FTO (ab92821, 1:1000; Abcam), ALKBH5 (ab69325, 1:1000; Abcam), Mettl14 (HPA038002, 1:1000; Sigma), and m6A (ab208577, 1:500; Abcam).
Histological analysis
Mouse hearts were fixed with freshly made 4% paraformaldehyde overnight, dehydrated by a dehydrator, embedded in paraffin, sliced by a paraffin slicer, 7 μm for each section, and stained with hematoxylin and eosin. Furthermore, we previously sectioned frozen heart tissue samples from mice at 7 μm slices, followed by incubation with wheat germ agglutinin (1:100) for 1 h in the dark, and finally washed three times with PBS. Cell nuclei were stained with DAPI (1:1000, 10 min) and washed thrice with PBS. Firstly, we dropped a water-soluble antifade mounting medium on the samples, then we covered the samples with glass cover slips, and finally we investigated the samples with a confocal microscope (Leica Sp8). Using the Image-Pro Plus 6.0 software to calculate the cross-sectional areas of cardiomyocytes. To detect fibrosis in the murine cardiac, we stained heart sections with the usual Masson’s trichrome procedure.
Overexpression of fat mass and obesity-related
Empty adeno-associated virus (AAV-EV) and adeno-associated virus expressing FTO (AAV-FTO) were manufactured by Hanbio Biotechnology Ltd. (Shanghai, China). The viruses were under the control of a heart-specific cTNT promoter with an EGFP tag. The virus titer we used to do the experiment was approximately 1 × 1012 V g/ml. At 8 weeks, we injected 120 μL of AAV-EV and AAV-FTO viruses via tail vein for each mouse.
m6A dot blot assay
We extracted total RNA from experimental mouse hearts with Trizol (Invitrogen). Total RNA was diluted to 1000 ng/μL and denaturated for 3 min at 95°C to break the RNA secondary structure into single strands. Immediately placed on ice to prevent the re-formation of RNA secondary structures. 2 μL RNA was directly dropped onto the Hybond-N+ membrane (GE Healthcare) and incubated at 80°C using an 80°C hybridizer for 1 h. Uncrosslinked RNA was washed with PBS with 0.01% Tween-20 for 5 min. We incubated membranes with anti-m6A antibody (1: 500 in PBS with 0.01% Tween-20) at 4°C for 12 h after blocking with 5% skim milk (in PBS with 0.01% Tween-20) for 1 h. We used horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody to treat membranes for 1 h at room temperature, and rinsed 4 times for 10 min each with PBS followed by chemiluminescence development. To ensure that repeated dots contained the same amount of total RNA, methylene blue staining was utilized.
MeRIP-seq
After three mice in HFpEF and HFpEF + EXT groups were euthanized, the total RNA samples were extracted from heart tissue specimens and quantitatively analyzed applying the NanoDrop ND-1000 (Thermo Fisher Scientific, MA, United States). Next, 20 μg total RNA was collected for interruption and purification to obtain the product after fragmentation treatment, and the Zymo RNA Clean and Concentrator-5 kit was used for the purification and recovery of the fragmented RNA. The resulting product was added with anti-m6A antibody (Sigma-Aldrich: ABE572), protein A-magnetic beads (Invitrogen: 10002D), protein G-magnetic beads (Invitrogen: 10004D); mixed; and incubated overnight. The mixture was subjected to magnetic separation. The supernatant was collected and added with 5× precipitation buffer and RNA enzyme inhibitor, and the mixture was reacted at 4°C for 1–3 h, washed 2–3 times with low-salt precipitation buffer, and washed 2–3 times with high-salt buffer. The RNA was extracted from chloroform lysate to obtain the purified product. Ribosomal RNA removed the obtained products, and the SMART principle synthesized the first-strand cDNA. We amplified enriched library fragments by PCR and purified the library fragments by magnetic beads with DNA to obtain an RNA methylation m6A detection library. Finally, we employed the bioptic qsep100 analyzer to quality check the libraries. The NovaSeq high-throughput sequencing platform and PE150 sequencing mode were adopted for sequencing.
Adapters and filters were trimed with the Cutadapt (v2.5) for sequences, and the Hisat2 aligner (v2.1.0) was adopted to align remaining reads to the human Ensemble genome GRCh38 (mouse Ensemble genome GRCm38). The exomePeak R package was adopted to identify differential m6A peaks. We used the clusterProfiler R package (v3.6.0) for GO and KEGG analyses. And we used the Guitar R package (v1.16.0) to visualize m6A-RNA-related genomic features. We subjected m6A peaks with p value <0.05 for the de novo motif analysis with homer (v4.10.4).
RNA-seq
Total RNA samples were taken from heart tissue specimens after three mice in the HFpEF and HFpEF + EXT groups were euthanized. Trimming adapters and filtering for sequences was done with Cutadapt (v2.5), and the Hisat2 aligner (v2.1.0) was adopted to align the remaining reads to the human Ensemble genome GRCh38 (mouse Ensemble genome GRCm38). The feature Counts were used to determine the reads that mapped the genome (v1.6.3). Differential gene expression analysis was made with the DESeq2 R-package.
Single-base elongation and ligation-based qPCR amplification method validation
The Single-base elongation and ligation-based qPCR amplification method (SELECT) assay was used to monitor site-specific m6A levels, as previously described (Xiao et al., 2018). Briefly, total RNA (2 μg) was combined with 1 μL of dNTP, 2 μL of 10 × CutSmart buffer, and 2 μL of every 400 nmol/L up and down primer (Table.1). RNA free water to the final volume 17 μL. The combination of RNA and primers was annealed at the following temperatures: 90°C for 1 min, 80°C for 1 min, 70°C for 1 min, 60°C for 1 min, 50°C for 1 min, and 40°C for 6 min. Then add 3 μL solution with 0.01 U DNA polymerase, 0.5 U SplintR ligase and 10 nmol ATP. After incubation at 40°C for 20 min and 80°C for 20 min, template abundance was quantified by taking 2 μL of reaction solution for real-time qPCR analysis.
Real-time PCR (RT-PCR)
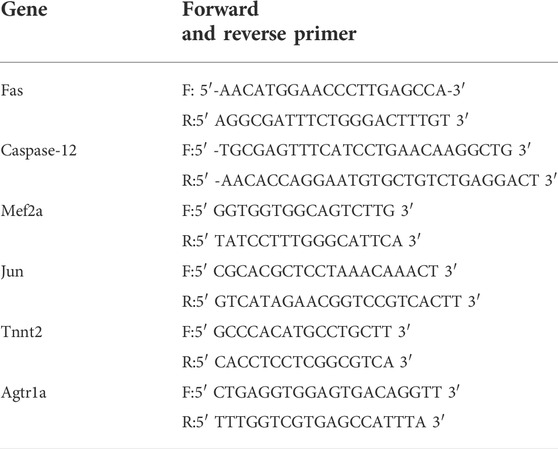
We picked 10 genes with differentially methylated sites based on MeRIP-seq, and tested these genes with Real-time PCR using SYBR-green and standard amplification protocols. The ΔΔCt technique was used to calculate expression levels. Table 2 shows the primer sequences.
Exercise exhaustion test
After 3 days of treadmill adaptation, experimental mice were tested for fatigue. Mice ran upward (20°) on a treadmill at a warm-up rate of 5 m/min for 4 min, which was grown to 14 m/min for 2 min. Subsequently, the speed was grown by 2 m/min every 2 min until the mice were exhausted. Exhaustion was defined as the inability of the mouse to resume running within 10 s of direct contact with the electrical stimulus net. While measuring the running time, the running distance was calculated.
Transferase-mediated dUTP nick-end labeling
TUNEL experiments were made with the In Situ Cell Death Detection Kit (Cat. No.11684795910, Roche). The tissue section was fixed with fixation solution (4% paraformaldehyde in PBS, pH 7.4, freshly prepared) for 20 min at +15°C to +25°C and washed for 30 min with PBS. The 2-minute incubation of slides was made in permeabilization solution (0.1% Triton X-100, 0.1% sodium citrate, freshly prepared) on ice. Next, slides were added with 50 μL TUNEL reaction mixture and incubated in a humidified atmosphere for 60 min at +37°C in the dark. Finally, samples were directly analyzed under a confocal microscope.
Transmission electron microscopy
Mouse hearts were fixed in 2.5% cacodylate buffer with glutaraldehyde, embedded with 2% agarose, fixed in 1% osmium tetroxide buffer, stained in 2% uranyl acetate, dehydrated with ethanol, embedded in resin, sliced by an ultrafine slicing machine, and stained with 2% uranyl acetate and lead citrate. Images for experimental use were obtained on the FEI Tecnai G2 Spirit electron microscope equipped with LaB6.
Statistical analysis
All data for all experiments are from three or more independent experimental manipulations and are shown as mean ± SEM. The GraphPad Prism 6.0 program was used to conduct the statistical analysis. Differences with two groups were used two tailed unpaired Student’s t-test analysis and three or more groups were analyzed with a one- or two-way analysis of variance. Differences with a significance level of p < 0.05 were of statistical significance.
Results
Exercise reverses heart failure with preserved ejection fraction phenotypes by suppressing myocardial fibrosis and improving cardiac dysfunction with the changes in global m6A levels
In our study, HFpEF + EXT mice evidently ameliorated myocardial fibrosis and myocardial hypertrophy compared with the control (Figures 1A,B) and had evidently decreased heart weight to tibia length proportions (HW/TL, n = 6, p = 0.0005; Figure 1B). Furthermore, EXT clearly ameliorated myocardial interstitial fibrosis (n = 6, p < 0.0001; Figures 1A,C) and markedly reduced the cardiomyocyte cross-sectional region (n = 6, p < 0.0001; Figures 1A,F). In order to prove the improvement of cardiac function in mice, we performed serial echocardiography in HFpEF + EXT mice. Longitudinal echocardiographic evaluation revealed the persistent preservation of LVEF in all groups (n = 6, ns; Figures 1D,E), coupled with significant alterations in degrees of diastolic dysfunction and left ventricular filling pressure in mice exposed to the EXT (n = 6, p < 0.0001; Figures 1G,H). Consistent with the increase of filling pressures reported in the literature, mice exposed to EXT specifically exhibited a significant decrease in lung weight to tibia length ratios (LW/TL, n = 6, p < 0.0001; Figure 1I), indicating improvement in pulmonary function. Furthermore, the running distance was further in HFpEF + EXT mice compared with HFpEF mice (n = 6, p < 0.0001; Figure 1J). Overall, these data demonstrated that EXT reversed HFpEF phenotypes by suppressing myocardial fibrosis and improving cardiac dysfunction.

FIGURE 1. Exercise training reverses HFpEF phenotypes by suppressing myocardial fibrosis and improving cardiac dysfunction with the changes in global m6A levels. (A) Gross morphology of hearts stained with HE, cardiac fibrosis stained with Masson, and cardiomyocyte area stained with wheat germ agglutinin (WGA). Scale bar = 1 mm with HE, 100 μm with Masson, and 50 μm with WGA. (B) Proportion of heart weight to tibia length (n = 6 mice per group). (C) Quantitative analysis of cardiac fibrosis (n = 6 mice per group). (D) Representative M-mode echocardiography images. (E) Left ventricular ejection fraction (LVEF) (n = 6 mice per group). (F) Quantitative analysis of cardiomyocyte area (n = 6 mice per group). (G,H) Representative Doppler echocardiography images and E/E′ ratio (n = 6 mice per group). (I) Ratio of lung weight to tibia length (n = 6 mice per group). (J) Running distance during exercise exhaustion test (n = 6 mice per group). (K,L) Representative dot blot showing global m6A modification levels in hearts and methylene blue (MB) staining and quantification of global m6A modification levels (n = 3 mice per group). Data are mean ± SEM, with all individual data points plotted. Two-way ANOVA and Tukey’s multiple comparison test were used. Numbers above square brackets show significant p values.
The global m6A level is an important feature of m6A modification. Therefore, the global m6A levels of mouse hearts in different groups were detected. Dot blot examination of cardiac samples from separate groups revealed that HFpEF + EXT mice had higher total m6A levels than HFpEF mice. (Figures 1K,L, p = 0.0019). Next, we performed transcriptome-wide MeRIP-seq and mRNA-Seq to analyze the effect of EXT on the m6A methylation of HFpEF.
Exercise training demonstrates differential m6A modification patterns in heart failure with preserved ejection fraction mouse hearts
HFpEF + EXT mouse hearts had specific m6A alternation patterns which were different from those of HFpEF mouse heart samples. We found 26401 m6A peaks, corresponding to 13832 gene transcripts in the HFpEF + EXT group by the model-based analysis through exomePeak v2.13.2 (Figures 2A,B). In the HFpEF group, 27085 m6A peaks, representing 14122 gene transcripts (Figures 2A,B), were confirmed. Compared with the HFpEF group, HFpEF + EXT hearts had 3992 novel peaks, and 4676 peaks were absent. This result indicated that the global m6A alternation types in the HFpEF + EXT group were different from those in the HFpEF group (Figure 2B). We identified a map of m6A methylation using the MEME-ChIP software and confirmed the top consensus motif in m6A peaks as GGACU (Figure 2C). The GGACU was parallel to the previously confirmed RRACH motif (where R = G or A; A = m6A, and H = U, A, or C) (Dominissini et al., 2012; Meyer et al., 2012).
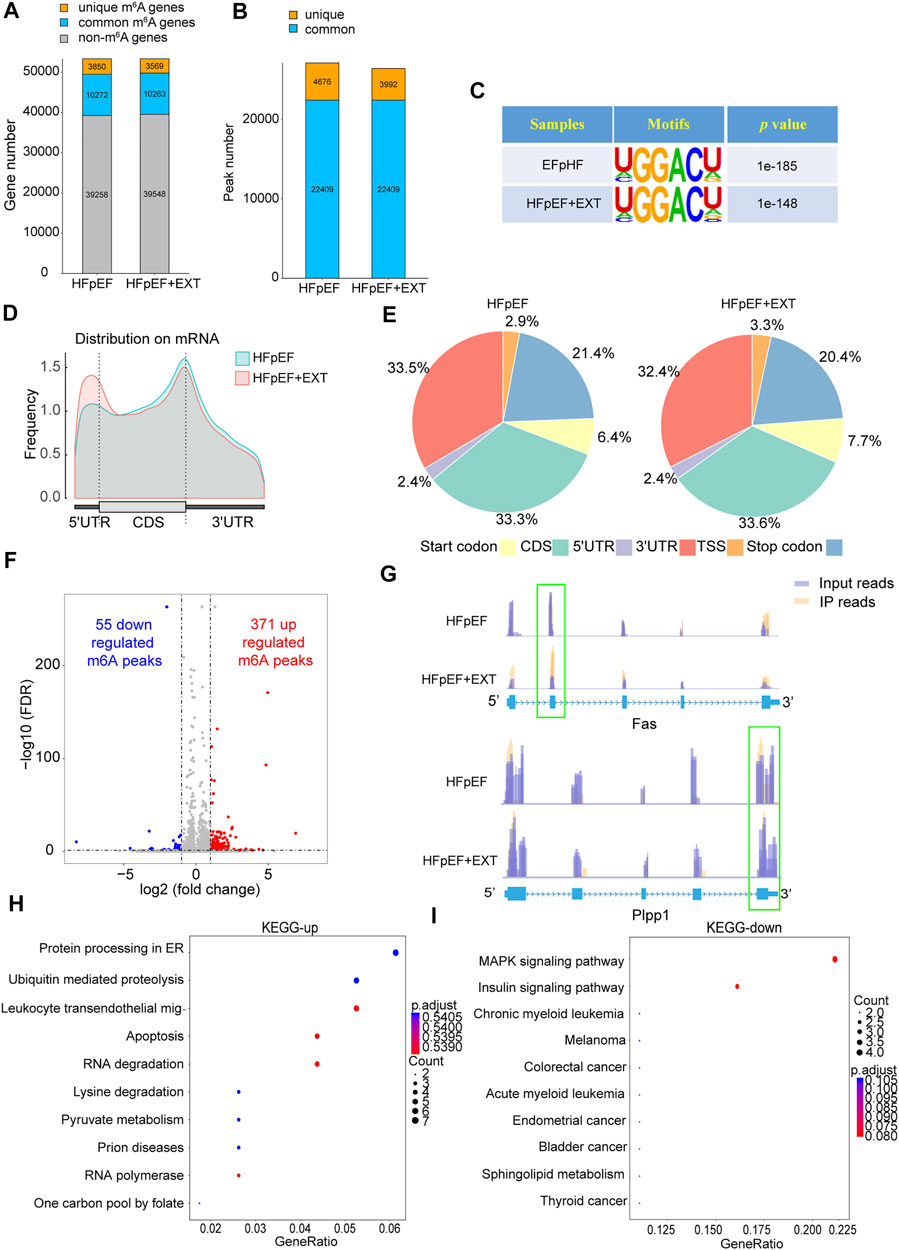
FIGURE 2. Exercise training demonstrates differential m6A modification patterns in HFpEF mouse hearts. (A,B) Summary of genes with identified m6A modification present at m6A-seq. (C) Sequence logo standing for the consensus motif identified by Discriminative Regular Expression Motif Elicitation in the two groups.(D) Distribution of N6-methyladenosine peaks across the length of mRNAs in two groups.(E) Venn diagram showing distribution of m6A peaks in the indicated regions in HFpEF and HFpEF + EXT groups.(F) Identification of 371 hypermethylated and 55 hypomethylated m6A peaks.(G) N6-methyladenosine abundance in Fas and Plpp1 mRNA transcripts in HFpEF and HFpEF + EXT samples (H,I) The top 10 enriched pathways terms from MeRIR-seq for upregulated and downregulated m6A peaks. Leukocyte transendothelial mig-: Leukocyte transendothelial migration.
We found that m6A peak in the HEpEF + EXT group was mainly increased in the 5′ untranslated region (5′ UTR) and decreased in the 3′ untranslated region (3′ UTR) compared with the HFpEF group. (Figure 2D). According to their precise positions in RNA transcripts, we categorized the total and unique m6A peaks in HFpEF + EXT and HFpEF whole-transcriptome data into 5′ UTR, transcription start site region, coding sequence (CDS), stop codon, and 3′ UTR. Our results showed that the enrichment of m6A peak was mainly concentrated in the region near CDS, 3′ UTR, and stop codon vicinity regions (Figure 2E), which was similar to previous m6A-seq results (Dominissini et al., 2012). HFpEF + EXT-specific m6A peak distributional patterns showed a different style from HFpEF-specific peaks, and which was with a relative increase in CDS, TSS and Start codon regions (33.3% vs. 33.6%, 3.3% vs 2.9%; 7.7% vs. 6.4%; Figure 2E) and a relative decrease in Stop codon and 3′ UTR, (20.4% vs. 21.4%, 32.4% vs. 33.5%; Figure 2E).
Kyoto encyclopedia of genes and genomes pathways enriched for differential m6A methylation transcripts in heart failure with preserved ejection fraction + Exercise training group are mainly concentrated in apoptosis, RNA degradation, and MAPK signaling pathways
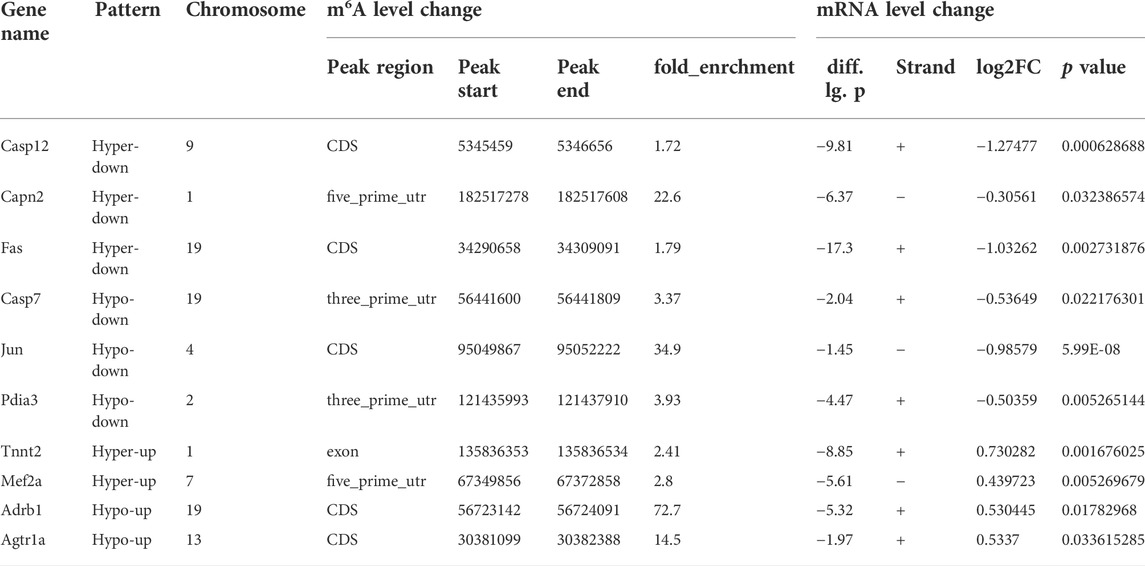
Differential methylation transcripts were identified and analyzed using GO and KEGG pathway analyses. By comparing the abundance of m6A peaks between HFpEF + EXT and HFpEF samples, we found that there were 22409 m6A peaks in both samples. We detected a total of 426 differentially methylated sites and took them as the research target for further research. Among these differentially methylated sites, 371 hypermethylated and 55 hypomethylated m6A sites were observed in the HFpEF + EXT group compared with HFpEF (fold change [FC] > 2, p < 0.05; Figure 2F). When we analyzed the differentially methylated places in both groups using the Integrative Genomics Viewer software, we found that there was significantly altered intensity in the HFpEF + EXT group. Figure 2G showed the representative m6A-methylated mRNA peaks in the Fas/APO-1 receptors (Fas) and phospholipid phosphatase-1 (Plpp1) genes, which were the sites with reduced and grown m6A levels, respectively.
In order to explore the potential biological significance of exercise-related m6A methylation changes in HFpEF, we performed the GO analysis of differentially methylated RNAs. Compared with those in HFpEF mice, our results revealed that hypermethylated and hypomethylated RNAs in HFpEF + EXT mice were especially related to energy metabolism terms, e.g., regulation of carbohydrate catalog, regulation of glycolytic process and carbohydrate catabolic process, suggesting that differentially methylated RNAs were mainly concentrated in energy metabolism (Supplementary Figure S2). Furthermore, the KEGG pathway analysis of differentially methylated RNAs in HFpEF + EXT mice were predominantly concentrated in myocardial cell death and myocardial energy metabolism, e.g., apoptosis, pyruvate metabolism, MAPK signaling pathway, insulin signaling pathway and sphingolipid metabolism pathway, which are mainly and strongly connected with myocardial apoptosis (Figures 2H,I). In conclusion, our findings suggested that these differentially methylated RNAs might take part in the myocardial energy metabolism and apoptosis pathway.
Overall, the transcripts of HFpEF + EXT-specific m6A peaks were mainly concentrated on myocardial cell death and myocardial energy metabolism, which were the main pathological features of HFpEF.
Conjoint analysis of m6A-seq and RNA-Seq data shows that specific m6A-modified transcripts were extremely correlated with the myocardial remodeling pathological characteristics of heart failure with preserved ejection fraction
The reads per kilobase per million mapped reads (FRKM distribution) indicated no difference in gene expression between samples (Figure 3A). RNA-Seq results indicated that 690 mRNAs, including 423 downregulated and 267 upregulated mRNAs, in HFpEF + EXT samples were significantly dysregulated compared with those in HFpEF samples (FC > 2, p < 0.05; Figure 3B). The tendency of differing gene expression between the groups was contemporaneous among individual samples within every group, according to further hierarchical clustering analysis of RNA-Seq data. (n = 3 per group, Figure 3C). Importantly, GO and KEGG pathway analysis displayed that differentially expressed genes were closely connected with apoptosis pathway and myocardial energy metabolism pathway (Figures 3D,E; Supplementary Figure S3), and this finding was coincident with the involvement in myocardial remodeling pathology (Dunlay et al., 2017; Figtree et al., 2021; Mishra and Kass, 2021; Omote et al., 2022; Prandi et al., 2022).

FIGURE 3. Combined analysis of RNA-seq and MeRIP-seq data comparing HFpEF with HFpEF + EXT heart samples. (A) FPKM distribution of two groups. (B) Volcano plots showing different expression mRNAs between HFpEF and HFpEF + RE samples (fold change >2 and p < 0.05). (C) Clustering analysis of differentially expressed mRNAs. (D,E) The top 10 enriched KEGG pathways of differentially mRNA for upregulated and downregulated mRNA. (F) The distribution of transcripts with significantly changed in m6A-modification level and corresponding mRNA expression (p < 0.05) by four-quadrant graph. Red dots represent significant differences in RNA expression and m6A modification, and gray dots do not meet the conditions.
In order to further study the target genes modified by m6A, target genes were idenfieid by the conjoint analysis of m6A-seq and RNA-Seq data. Through bioinformatics analysis, we confirmed 308 hypermethylated m6A peaks in mRNA transcripts, of which 103 and 205 were significantly upregulated and downregulated respectively (Figure 3F). Furthermore, 223 hypomethylated m6A peaks were confirmed in mRNA transcripts, of which 116 and 107 peaks were downregulated and upregulated, respectively (Figure 3F). Furthermore, the integrated analysis of MeRIP-seq and RNA-Seq data and KEGG pathway analysis showed that specific m6A-modified transcripts were closely associated with myocardial energy metabolism, especially myocardial apoptosis. Hence, we identified genes that were critical to these processes and further validated them, including Fas, Capn2, Casp12, Casp7, Jun, Pdia3, Tnnt2, Mef2a, Adrb1 and Agtr1a (Figure 4A; Table 3).
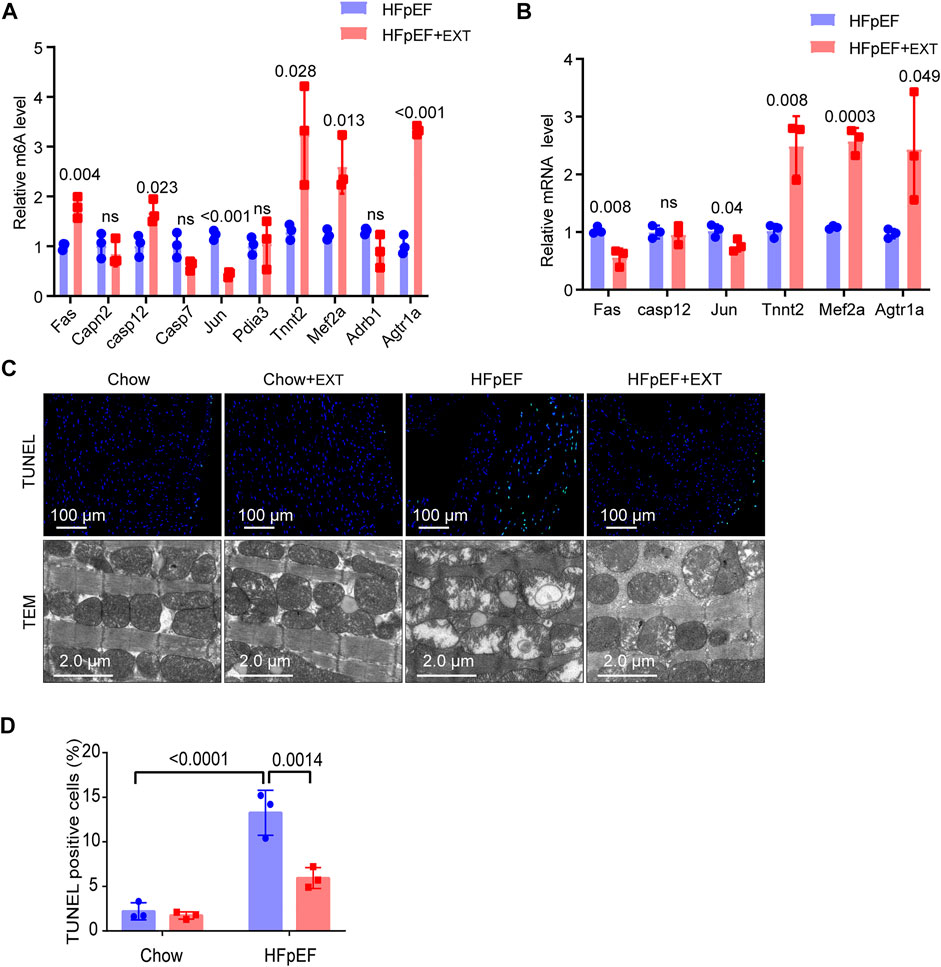
FIGURE 4. Identification of the downstream target genes and mechanisms of exercise improvement of HFpEF. (A) Single-base elongation and ligation-based qPCR amplification method (SELECT) validation of m6A level changes of ten related genes. (B) The relative mRNA levels of six genes determined by real-time PCR in HFpEF and HFpEF + EXT samples. n = 3 mice per group. Data are presented as mean ± SEM. Multiple unpaired t tests were used. (C,D) Representative images of TUNEL and TEM and quantitative analysis of TUNEL-positive cells. n = 3 mice per group. Data are presented as mean ± SEM. Two-way ANOVA and Tukey’s multiple comparison test were used. Numbers above square brackets display great p values.
We used SELECT-PCR to validate the m6A levels of those different critical genes Fas, Capn2, Casp12, Casp7, Jun, Pdia3, Tnnt2, Mef2a, Adrb1 and Agtr1a, which were associated with myocardial apoptosis and myocardial energy metabolism. We found that the m6A methylation levels of Fas, Casp12, Tnnt2, Mef2a and Agtr1a were significantly increased and the m6A methylation of Jun was significantly decreased (Figure 4A). Furthermore, the mRNA levels of Fas, Casp12, Jun, Tnnt2, Mef2a and Agtr1a were measured in HFpEF + EXT and HFpEF hearts (Figure 4B), and results showed that Jun, Tnnt2, Mef2a and Agtr1a had similar mRNA expression tendencies in keeping with their m6A methylation levels, but Fas had opposite mRNA expression tendencies in keeping with their m6A methylation levels (Figure 4A).
Exercise training ameliorates myocardial apoptosis in heart failure with preserved ejection fraction by decreasing apoptosis pathway and increasing myocardial energy metabolism
Apoptosis is one of the most important characteristics of pathological cardiac remodeling, and exercise training is an effective method to reduce cardiomyocyte apoptosis. To investigate the effect of exercise on cardiomyocyte apoptosis in the HFpEF heart, we used terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) to stain cardiac pathological sections. The representative fluorescence photos of TUNEL positive nuclei in the HFpEF heart after exercise are shown in Figure 4C. In the myocardium of HFpEF mice, the proportion of TUNEL positive cardiomyocytes was greatly higher than that of control mice, while a smaller number of TUNEL positive cells could be detected in the myocardium of control or HFpEF mice after exercise (Figures 4C,D).
In order to verify whether exercise can exert its cardioprotective function by maintaining the integrity and function of mitochondria, we measured and detected the mitochondrial structure of the heart of control group and HFpEF mice with or without exercise using an electron microscope. The ultrastructure of heart sections of control group and HFpEF mice with or without exercise was analyzed by an electron microscope to determine the integrity of mitochondria (Figure 4C). In the normal mouse myocardium, mitochondria were well aligned between the longitudinally oriented myocardial myofibrils, and no difference was observed after exercise. However, the abundance of mitochondria in HFpEF appeared to be significantly altered, and the arrangement of mitochondria in HFpEF mice was disorganized, and the mitochondria were swollen and aggregated, and cristae were slightly loosened (Figure 4C). After 2 months of exercise, there was evidence that damage to cardiac mitochondria and disrupted cristae were restored in HFpEF + EXT hearts (Figure 4C).
Overexpression of fat mass and obesity-related cancels out the benefits of exercise in heart failure with preserved ejection fraction + Exercise training mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy
On basis of the analysis outcomes of unique genes in HFpEF + EXT and HFpEF hearts by GO/KEGG, we suspected that the FTO is closely related to the regulation of energy metabolism and might exert a significant effect on the pathogenesis of HFpEF. Therefore, we explored the mechanism of FTO in HFpEF through experiments. The protein level of FTO in HFpEF + EXT mice was downregulated in comparison with that in HFpEF mice. The FTO protein level detected by Western blotting was greatly lower than that in the HFpEF + EXT group in comparison with the HFpEF group (p = 0.001; n = 6 per group; Figures 5A,B). We also looked at the expression of other key methyltransferases and demethylases during m6A modification, including Mettl3, Mettl14, and ALKBH5. Mettl3 was increased, and no significant difference was observed in Mettl4 and ALKBH5 (Figures 5A,C–E).
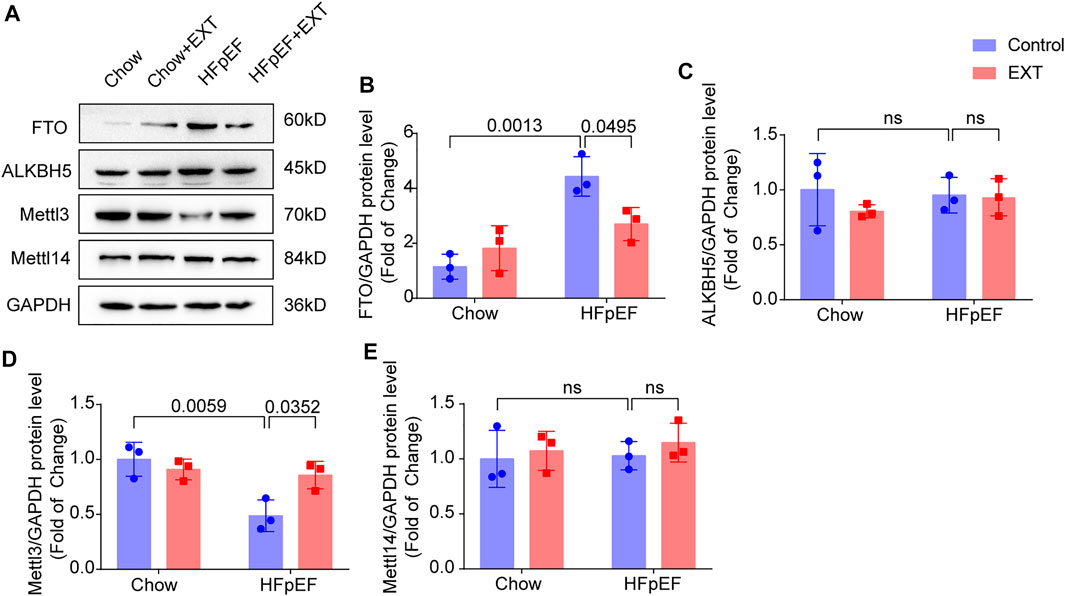
FIGURE 5. FTO is Downregulated in HFpEF + EXT compared to HFpEF. (A–E) Representative Western blots plots of FTO, Mettl3, ALKBH5 and Mettl14, and quantitative analysis of FTO/GAPDH, Mettl3/GAPDH protein, ALKBH5/GAPDH and Mettl14/GAPDH expressions. n = 3 per group. Data are presented as mean ± SEM. Two-way ANOVA and Tukey’s multiple comparison test were used. Numbers above square brackets show significant p values.
Overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy. At the age of 16 weeks, we injected the AAV vector encoding FTO into the HFpEF + EXT mice through the tail vein, and detected the expression of FTO after 8 weeks. After injection of AAV-FTO for 8 weeks, expression of myocardial FTO increased by approximately 7.5-fold in HFpEF + EXT mouse hearts (Supplementary Figures S1A,B). After 8 weeks of adenovirus injection, we found overexpression of FTO increased interstitial fibrosis in HFpEF + EXT mouse hearts (Figures 6A,B, p = 0.0089). Furthermore, the overexpression of FTO efficiently augmented cardiomyocyte hypertrophy, as demonstrated by the increase in cross-sectional area of cardiomyocyte in HFpEF + EXT mouse hearts (Figures 6A,C, p < 0.0001). Moreover, the overexpression of FTO accelerated apoptosis in myocytes and increased the proportion of TUNEL-positive cells in the myocardium (Figures 6A,D, p = 0.0306). The overexpression of FTO significantly destroyed mitochondrial integrity and function by disorganized mitochondrial arrays and aggregates of swollen mitochondria with slight lysis of the cristae (Figure 5A). Furthermore, the doppler echocardiography showed that FTO overexpression exacerbated diastolic dysfunction in HFpEF + EXT mice by increasing the E/E′ ratio (Figure 6E). The overexpression of FTO reduced LW/TL and running distance (n = 6, p = 0.0444 and p < 0.0001; Figures 6F,G). Overexpression of FTO reduced the mRNA levels of Tnnt2, Mef2a and Agtr1a, and increased the mRNA levels of Fas and Jun (Figure 6H). Overall, these data indicated that the overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy.
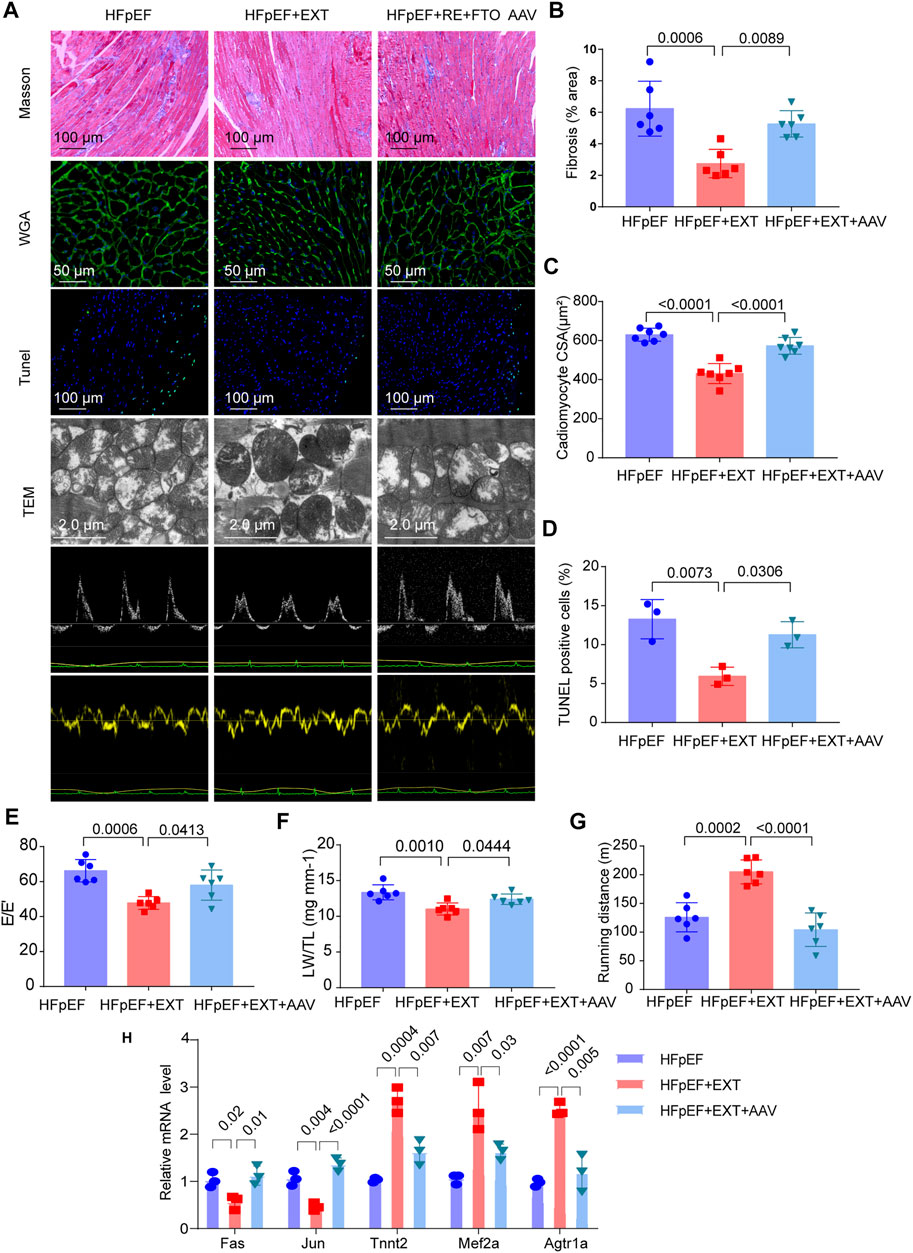
FIGURE 6. Overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice. (A) Gross morphology of hearts stained with Masson, WGA, TUNEL, TEM, and Doppler echocardiography from HFpEF, HFpEF + EXT and HFpEF + EXT + AAV9 mice. (B) Quantitative analysis of interstitial fibrosis (n = 6 mice per group). (C) Quantitative analysis of cardiomyocyte cross-sectional area (n = 6 mice per group). (D) Quantitative analysis of TUNEL-positive cells (n = 3 per group). (E) Quantitative analysis of E/E′ ratio (n = 6 mice per group). (F) Ratio of lung weight to tibia length (n = 6 mice per group). (G) Running distance during exercise exhaustion test (n = 6 mice per group). (H) The relative mRNA levels of six genes determined by real-time PCR in HFpEF, HFpEF + EXT and HFpEF + EXT + AAV9 mice (n = 3 mice per group). Data are presented as mean ± SEM. One-way ANOVA and Tukey’s multiple comparison test were used. Numbers above square brackets show significant p values.
Discussion
HFpEF is an aging disease characterized by cardiac hypertrophy, myocardial interstitial fibers, and cardiac diastolic dysfunction (Prandi et al., 2022). Due to inadequate understanding of the pathophysiology and animal models of HFpEF, it is difficult to develop methods to treat this more prevalent and lethal disease (Schiattarella et al., 2019; Deng et al., 2021). Unlike HFrEF, patients with HFpEF constitute a highly heterogenous population. HFpEF patients usually do not trace back to a primary lesions and injuries of cardiomyocytes but rather to a complex of series of systemic abnormalities, e.g., metabolic dysfunction, hypertension, hyperlipidemia, renal dysfunction and pulmonary dysfunction (Dunlay et al., 2017; Deng et al., 2021; Mishra and Kass, 2021). There are many reasons for the lack of effective therapies of HFpEF, but it is largely because of incompletely understanding its complex pathophysiology and the lack of appropriate animal models to study its pathogenesis. EXT is an important nonpharmacological treatment, which can reduce the risk of hospitalization and cardiovascular mortality risk (Jaconiano and Moreira-Gonçalves, 2022). Although the significance of epigenetic regulation in the control of gene expression to diagnose and treat HFpEF has been widely examined (Hamdani et al., 2021), the effect of RNA alternation on the control of gene expression to affect HFpEF has only recently been investigated. Previous studies demonstrated that the mRNA of m6A writers Mettl3 and Mettl4; m6A eraser FTO; and reader YTHDF2 are elevated in patients with HFpEF (Zhang et al., 2021a). In HFpEF mice, the mRNA of FTO and YTHDC1 is upregulated but Mettl3 is downregulated (Zhang et al., 2021a). In elderly HFpEF mice, EXT could improve exercise capacity, diastolic function, and systolic reserves and reduce pulmonary congestion (Roh et al., 2020). However, the distribution of m6A within transcriptome-wide in HFpEF + EXT samples is still mostly unknown. Our research showed that m6A RNA methylation is changed in HFpEF + EXT mice, indicating specific m6A alternation patterns on the transcriptome-wide and gene-specific scales that were distinct from those of in HFpEF mice. GO and KEGG analyses demonstrated that m6A RNA methylation differential genes between HFpEF + EXT and HFpEF mice are specifically associated with myocardial energy metabolism. HFpEF + EXT mice are observed with downregulated FTO compared with HFpEF mice, and the overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy. FTO is a key m6A demethylase that may regulate cardiomyocyte function through catalyzing the demethylation of m6A on particular mRNAs, such as Fas, Jun, Tnnt2, Mef2a and Agtr1a.
Exercise modifies the m6A profile of HFpEF, and these differential modification genes are closely associated with myocardial energy metabolism. The pattern of m6A modification in HFpEF + EXT mice was different from those of HFpEF mice on both transcriptome-wide and gene-specific scales. 22 409 m6A peaks in HFpEF + EXT mice were determined, indicating that m6A is a widespread post transcriptional RNA alternation in various heart disorders. In addition, we investigated that differentially methylated m6A peaks in HFpEF + EXT mice are primarily present in 5′UTR, which may promote mRNA translation. Importantly, in our research, we found that in HFpEF + EXT mice, the Fas m6A-modified site is primarily present in the 5′UTR. Therefore, we speculated that the differential methylation of RNAs in HFpEF + EXT might affect post-transcriptional translation levels and translation efficiency of RNA. We found that genes with differential m6A methylation and mRNA expression in HFpEF + EXT mice were mainly enriched in processes and pathways related to myocardial energy metabolism and apoptosis pathway, such as MAPK signaling pathway, insulin signaling pathway, sphingolipid metabolism, pyruvate metabolism, apoptosis, and regulation of intrinsic apoptotic signaling pathway processes (Figures 2H,I). HFpEF is a complex disease that includes cardiac hypertrophy, myocardial fibrosis, inflammation, cardiomyocyte sarcomere disfunction and mitochondrial and metabolic defects, which lead to mitochondrial dysfunction, inflammation and myocardial cardiac diastolic dysfunction (Figtree et al., 2021; Jaconiano and Moreira-Gonçalves, 2022). Recent clinical studies found that a year of committed EXT reverses abnormal left ventricular myocardial stiffness in patients with stage B HFpEF (Hieda et al., 2021). However, because of limited access to cardiac tissue, the molecular mechanisms of HFpEF remained largely unknown. Here, our study indicated that exercise induces similar functional benefits in HFpEF mouse, and the Conjoint analysis of MeRIP-seq and RNA-Seq data revealed that exercise alters the metabolic phenotype of cardiomyocytes and improves the apoptosis of myocardial cells for HFpEF. Indeed, our findings suggested that while EXT only ameliorated the HFpEF phenotypes in this “two-hit” model, it improved the overall cardiac performance and exercise capacity. We propose that, from the mice “two-hit” model with HFpEF, EXT confers benefits. The mechanism of epigenetic regulation thus provides a new therapeutic target for HFpEF and a new approach to studying HFpEF.
Exercise improves mitochondrial function in HFpEF and affects the myocardial energy metabolism phenotype (Vega et al., 2017). Post-exercise hearts exhibited a higher capacity for fatty acid oxidation and ATP generation than non-exercise hearts in response to the stimulation of increased cardiac workload by b EXT (Moreira et al., 2020). The transcriptional coactivator PGC-1α is a core molecule in the regulation of cardiomyocyte metabolism, and it was originally recognized as a cold inducible element in mitochondrial biogenesis and necessary for the metabolic adaptations of the heart to exercise (Moreira et al., 2020). In our study, an important target gene for improving the myocardial phenotype of HFpEF during exercise is Fas. Fas ligand (FasL) is a cell surface molecule of the tumor necrosis factor family binding to its receptor Fas to induce apoptosis of Fas bearing cells (Wajant, 2002). In our study, Fas m6A methylation expression levels are upregulated, but the mRNA and protein expression levels are downregulated, indicating that m6A modification of Fas may influence mRNA degradation.
FTO is downregulated in HFpEF + EXT samples, and overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy. In 2007, the FTO gene was identified in a type Ⅱ diabetes genome-wide relationship research (Dina et al., 2007). Furthermore, a population cohort research showed that the effect of FTO genes was closely associated with energy intake (Scuteri et al., 2007). However, it is not clear how FTO affects the mechanisms of obesity and energy metabolism. Our KEGG pathway analysis showed that differentially methylated RNAs are predominantly associated with the MAPK and insulin signaling pathways in cardiomyocytes. In conclusion, the FTO is critical for energy metabolism. FTO expression is downregulated in mammalian heart failure hearts and hypoxic cardiomyocytes, with higher m6A levels in RNA and impaired cardiomyocyte contractile performance, according to recent investigations (Mathiyalagan et al., 2019). Additionally, in mice with heart failure, the m6A demethylase FTO improves cardiac function via controlling glucose absorption and glycolysis (Zhang et al., 2021b). In this research, the overexpression of FTO deteriorates heart function by growing E/E′ and LW/TL and decreasing the running distance. Furthermore, overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy.
The present study has limitations. Firstly, we did not find human heart samples to validate our experimental results. Therefore, we will next seek human heart samples to validate the important role of m6A modification in the pathogenesis of HFpEF. Secondly, although we found the target genes were modified by m6A, the mechanism of how the binding proteins control the target genes has not been explored. Next, how binding proteins affect target gene stability, translational efficiency, or degradation will be explored. Thirdly, although we demonstrated that overexpressing FTO in the heart worsened cardiac function, future research with cardiac specific knockout FTO mice and HFpEF models will investigate the actual mechanism by which FTO mediates HFpEF.
Consequently, m6A RNA methylation was modified in HFpEF + EXT mice, who displayed distinct m6A alternation patterns at the transcriptome and gene levels compared to HFpEF mice. Differentially methylated genes were found to be linked with myocardial fibrosis, myocyte hypertrophy, myocardial apoptosis, and myocardial energy metabolism in GO and KEGG studies. Exercise improved mitochondrial function in HFpEF and myocardial energy metabolism phenotype to reverse the myocardial phenotypes. FTO was downregulated in HFpEF + EXT mice compared with that in HFpEF mice, and the overexpression of FTO cancels out the benefits of exercise in HFpEF + EXT mice by promoting myocyte apoptosis, myocardial fibrosis and myocyte hypertrophy. FTO is a key m6A demethylase that may regulate cardiomyocyte function through catalyzing the demethylation of m6A on particular mRNAs, such as Fas, Jun, Tnnt2, Mef2a and Agtr1a.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Gene Expression Omnibus accession number: GSE208354.
Ethics statement
The animal study was reviewed and approved by Capital Institute of Pediatrics.
Author contributions
KL and JW designed the experiments and wrote the manuscript. KL, WJ, FH, and JH carried out experiments, and SO analyzed the data. JW, HG and ZL supervised this project. KL and WJ contributed equally to this study. All authors have finally been approved for publication.
Funding
This work is supported by grants from CAMS Innovation Fund for Medical Sciences (CIFMS, 2016-I2M-1-008); Public service development and reform pilot project of Beijing Medical Research Institute (BMR2019-11); The Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Hospitals Authority (No. XTZD20180402); the National Key Research and Development Program of China (2018YFC1002503).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.954769/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Flow chart and adeno-associated virus overexpression FTO. . Flow chart of HFpEF modeling and running method in mice. (B,C) Representative Western blots and quantitative analysis of overexpression of FTO. n = 3 Data are presented as mean ± SEM. One-way ANOVA and Tukey’s multiple comparison test were used. Numbers above square brackets show significant p values.
SUPPLEMENTARY FIGURE S2 | GO analysis of Differential m6A modification genes. Biological Process analysis of up-regulated and down-regulated m6A methylation genes. (C,D) Cellular Component analysis of up-regulated and down-regulated m6A methylation genes. (E,F) Molecular Function analysis of up-regulated and down-regulated m6A methylation genes.
SUPPLEMENTARY FIGURE S3 | GO analysis of Differential mRNA genes. Biological Process analysis of upregulated and downregulated mRNA genes. (C,D) Cellular Component analysis of upregulated and downregulated m6A mRNA genes. (E,F) Molecular Function analysis of upregulated and downregulated mRNA genes.
References
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Berulava, T., Buchholz, E., Elerdashvili, V., Pena, T., Islam, M. R., Lbik, D., et al. (2020). Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur. J. Heart Fail 22 (1), 54–66. doi:10.1002/ejhf.1672
Deng, Y., Xie, M., Li, Q., Xu, X., Ou, W., Zhang, Y., et al. (2021). Targeting mitochondria-inflammation circuit by β-hydroxybutyrate mitigates HFpEF. Circ. Res. 128 (2), 232–245. doi:10.1161/circresaha.120.317933
Dieberg, G., Ismail, H., Giallauria, F., and Smart, N. A. (2015). Clinical outcomes and cardiovascular responses to exercise training in heart failure patients with preserved ejection fraction: A systematic review and meta-analysis. J. Appl. Physiol. (1985) 119 (6), 726–733. doi:10.1152/japplphysiol.00904.2014
Dina, C., Meyre, D., Gallina, S., Durand, E., Körner, A., Jacobson, P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39 (6), 724–726. doi:10.1038/ng2048
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dorn, L. E., Lasman, L., Chen, J., Xu, X., Hund, T. J., Medvedovic, M., et al. (2019). The N 6 -methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation 139 (4), 533–545. doi:10.1161/circulationaha.118.036146
Dunlay, S. M., Roger, V. L., and Redfield, M. M. (2017). Epidemiology of heart failure with preserved ejection fraction. Nat. Rev. Cardiol. 14 (10), 591–602. doi:10.1038/nrcardio.2017.65
Edelmann, F., Gelbrich, G., Düngen, H. D., Fröhling, S., Wachter, R., Stahrenberg, R., et al. (2011). Exercise training improves exercise capacity and diastolic function in patients with heart failure with preserved ejection fraction: Results of the ex-DHF (exercise training in diastolic heart failure) pilot study. J. Am. Coll. Cardiol. 58 (17), 1780–1791. doi:10.1016/j.jacc.2011.06.054
Feil, R., and Fraga, M. F. (2012). Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 13 (2), 97–109. doi:10.1038/nrg3142
Figtree, G. A., Broadfoot, K., Casadei, B., Califf, R., Crea, F., Drummond, G. R., et al. (2021). A call to action for new global approaches to cardiovascular disease drug solutions. Eur. Heart J. 42 (15), 1464–1475. doi:10.1093/eurheartj/ehab068
Fukuta, H., Goto, T., Wakami, K., Kamiya, T., and Ohte, N. (2019). Effects of exercise training on cardiac function, exercise capacity, and quality of life in heart failure with preserved ejection fraction: A meta-analysis of randomized controlled trials. Heart Fail Rev. 24 (4), 535–547. doi:10.1007/s10741-019-09774-5
Gao, X. Q., Zhang, Y.-H., Liu, F., Ponnusamy, M., Zhao, X.-M., Zhou, L.-Y., et al. (2020). The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nat. Cell Biol. 22 (11), 1319–1331. doi:10.1038/s41556-020-0576-y
Hamdani, N., Costantino, S., Mügge, A., Lebeche, D., Tschöpe, C., Thum, T., et al. (2021). Leveraging clinical epigenetics in heart failure with preserved ejection fraction: A call for individualized therapies. Eur. Heart J. 42 (20), 1940–1958. doi:10.1093/eurheartj/ehab197
Handy, D. E., Castro, R., and Loscalzo, J. (2011). Epigenetic modifications. Circulation 123 (19), 2145–2156. doi:10.1161/circulationaha.110.956839
Hieda, M., Sarma, S., Hearon, C. M., MacNamara, J. P., Dias, K. A., Samels, M., et al. (2021). One-year committed exercise training reverses abnormal left ventricular myocardial stiffness in patients with stage B heart failure with preserved ejection fraction. Circulation 144 (12), 934–946. doi:10.1161/circulationaha.121.054117
Ho, J. E., Zern, E. K., Wooster, L., Bailey, C. S., Cunningham, T., Eisman, A. S., et al. (2019). Differential clinical profiles, exercise responses, and outcomes associated with existing HFpEF definitions. Circulation 140 (5), 353–365. doi:10.1161/circulationaha.118.039136
Ismail, H., McFarlane, J. R., Nojoumian, A. H., Dieberg, G., and Smart, N. A. (2013). Clinical outcomes and cardiovascular responses to different exercise training intensities in patients with heart failure. JACC Heart Fail. 1 (6), 514–522. doi:10.1016/j.jchf.2013.08.006
Jaconiano, E., and Moreira-Gonçalves, D. (2022). Unveiling the role of exercise training in targeting the inflammatory paradigm of heart failure with preserved ejection fraction: A narrative review. Heart Fail Rev. 27 (1), 163–190. doi:10.1007/s10741-021-10138-1
Krüger, N., Biwer, L. A., Good, M. E., Ruddiman, C. A., Wolpe, A. G., DeLalio, L. J., et al. (2020). Loss of endothelial FTO antagonizes obesity-induced metabolic and vascular dysfunction. Circ. Res. 126 (2), 232–242. doi:10.1161/circresaha.119.315531
Leggio, M., Fusco, A., Loreti, C., Limongelli, G., Bendini, M. G., Mazza, A., et al. (2020). Effects of exercise training in heart failure with preserved ejection fraction: An updated systematic literature review. Heart Fail Rev. 25 (5), 703–711. doi:10.1007/s10741-019-09841-x
Lin, J., Zhu, Q., Huang, J., Cai, R., and Kuang, Y. (2020). Hypoxia promotes vascular smooth muscle cell (VSMC) differentiation of adipose-derived stem cell (ADSC) by regulating Mettl3 and paracrine factors. Stem Cells Int. 2020, 2830565. doi:10.1155/2020/2830565
Mathiyalagan, P., Adamiak, M., Mayourian, J., Sassi, Y., Liang, Y., Agarwal, N., et al. (2019). FTO-dependent N 6 -methyladenosine regulates cardiac function during remodeling and repair. Circulation 139 (4), 518–532. doi:10.1161/circulationaha.118.033794
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mishra, S., and Kass, D. A. (2021). Cellular and molecular pathobiology of heart failure with preserved ejection fraction. Nat. Rev. Cardiol. 18 (6), 400–423. doi:10.1038/s41569-020-00480-6
Mo, X. B., Lei, S.-F., Zhang, Y.-H., and Zhang, H. (2019). Examination of the associations between m6A-associated single-nucleotide polymorphisms and blood pressure. Hypertens. Res. 42 (10), 1582–1589. doi:10.1038/s41440-019-0277-8
Moreira, J. B. N., Wohlwend, M., and Wisløff, U. (2020). Exercise and cardiac health: Physiological and molecular insights. Nat. Metab. 2 (9), 829–839. doi:10.1038/s42255-020-0262-1
Omote, K., Verbrugge, F. H., and Borlaug, B. A. (2022). Heart failure with preserved ejection fraction: Mechanisms and treatment strategies. Annu. Rev. Med. 73, 321–337. doi:10.1146/annurev-med-042220-022745
Poller, W., Dimmeler, S., Heymans, S., Zeller, T., Haas, J., Karakas, M., et al. (2018). Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 39 (29), 2704–2716. doi:10.1093/eurheartj/ehx165
Prandi, F. R., Evangelista, I., and Sergi, D. (2022). Mechanisms of cardiac dysfunction in diabetic cardiomyopathy: Molecular abnormalities and phenotypical variants. Dordrecht, Netherlands: Heart Fail Rev. doi:10.1007/s10741-021-10200-y
Roh, J. D., Houstis, N., Yu, A., Chang, B., Yeri, A., Li, H., et al. (2020). Exercise training reverses cardiac aging phenotypes associated with heart failure with preserved ejection fraction in male mice. Aging Cell 19 (6), e13159. doi:10.1111/acel.13159
Schiattarella, G. G., Altamirano, F., Kim, S. Y., Tong, D., Ferdous, A., Piristine, H., et al. (2021). Xbp1s-FoxO1 axis governs lipid accumulation and contractile performance in heart failure with preserved ejection fraction. Nat. Commun. 12 (1), 1684. doi:10.1038/s41467-021-21931-9
Schiattarella, G. G., Altamirano, F., Tong, D., French, K. M., Villalobos, E., Kim, S. Y., et al. (2019). Nitrosative stress drives heart failure with preserved ejection fraction. Nature 568 (7752), 351–356. doi:10.1038/s41586-019-1100-z
Schiattarella, G. G., Sequeira, V., and Ameri, P. (2021). Distinctive patterns of inflammation across the heart failure syndrome. Heart Fail Rev. 26 (6), 1333–1344. doi:10.1007/s10741-020-09949-5
Scuteri, A., Sanna, S., Chen, W.-M., Uda, M., Albai, G., Strait, J., et al. (2007). Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3 (7), e115. doi:10.1371/journal.pgen.0030115
Vega, R. B., Konhilas, J. P., Kelly, D. P., and Leinwand, L. A. (2017). Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metab. 25 (5), 1012–1026. doi:10.1016/j.cmet.2017.04.025
Wajant, H. (2002). The Fas signaling pathway: More than a paradigm. Science 296 (5573), 1635–1636. doi:10.1126/science.1071553
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Xiao, Y., Wang, Y., Tang, Q., Wei, L., Zhang, X., and Jia, G. (2018). An elongation‐ and ligation‐based qPCR amplification method for the radiolabeling‐free detection of locus‐specific N 6 ‐methyladenosine modification. Angew. Chem. Int. Ed. 57 (49), 15995–16000. doi:10.1002/anie.201807942
Zhang, B., Jiang, H., Wu, J., Cai, Y., Dong, Z., Zhao, Y., et al. (2021). m6A demethylase FTO attenuates cardiac dysfunction by regulating glucose uptake and glycolysis in mice with pressure overload-induced heart failure. Sig Transduct. Target Ther. 6 (1), 377. doi:10.1038/s41392-021-00699-w
Keywords: HFPEF, m6A, apoptosis, FTO, myocardial energy metabolism
Citation: Liu K, Ju W, Ouyang S, Liu Z, He F, hao J, Guan H and Wu J (2022) Exercise training ameliorates myocardial phenotypes in heart failure with preserved ejection fraction by changing N6-methyladenosine modification in mice model. Front. Cell Dev. Biol. 10:954769. doi: 10.3389/fcell.2022.954769
Received: 27 May 2022; Accepted: 08 August 2022;
Published: 02 September 2022.
Edited by:
Liang Zhou, Southern Medical University, ChinaReviewed by:
Juan Jin, Anhui Medical University, ChinaFang Fang, Peking Union Medical College Hospital (CAMS), China
Copyright © 2022 Liu, Ju, Ouyang, Liu, He, hao, Guan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxin Wu, cipbiolab@163.com
†These authors share first authorship
 Kai Liu
Kai Liu Wenhao Ju
Wenhao Ju Shengrong Ouyang2,3
Shengrong Ouyang2,3  Feng He
Feng He Hongyan Guan
Hongyan Guan