ULK4 in Neurodevelopmental and Neuropsychiatric Disorders
- 1Department of Pharmacy, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Hunan Provincial Engineering Research Center of Translational Medicine and Innovative Drug, Changsha, China
- 3Department of Psychiatry, National Clinical Research Center for Mental Disorders, The Second Xiangya Hospital of Central South University, Changsha, China
The gene Unc51-like kinase 4 (ULK4) belongs to the Unc-51-like serine/threonine kinase family and is assumed to encode a pseudokinase with unclear function. Recently, emerging evidence has suggested that ULK4 may be etiologically involved in a spectrum of neuropsychiatric disorders including schizophrenia, but the underlying mechanism remains unaddressed. Here, we summarize the key findings of the structure and function of the ULK4 protein to provide comprehensive insights to better understand ULK4-related neurodevelopmental and neuropsychiatric disorders and to aid in the development of a ULK4-based therapeutic strategy.
Introduction
Neuropsychiatric disorders are a wealth of debilitating brain diseases with overlapping etiologies, including genetic variants and environmental stress. The concordance rate is high and the heritability is substantial, although the influence of de novo mutations cannot be ignored especially in autism spectrum disorders (ASDs) (Alonso-Gonzalez et al., 2018). During the past decades, genome-wide association studies (GWASs) have reported numerous genetic alleles with single nucleotide polymorphisms (SNPs) (Uffelmann et al., 2021). In addition, recent progress in whole genome interrogation has also demonstrated massive genetic variants that are not covered by GWAS(Rao et al., 2021). The advances in research methodologies have expanded our understanding of the genetic architecture of psychiatric patients but also revealed further complexity. Hence, it is compelling to identify the predisposing risk alleles and to fully elucidate the associated mechanisms underpinning neuropsychiatric disorders. Unfortunately, thus far, only limited success has been achieved. Intriguingly, recent studies have revealed overwhelming evidence in neurodevelopmental elements in neuropsychiatric disorders (Cristino et al., 2014; Cardoso et al., 2019; Al-Naama et al., 2020). Various genetic alterations that occur during the embryonic stages can lead to pathological brain development and may precipitate the onset of psychosis in adolescence. These developmental insults are believed to disturb the neuronal connectivity and cellular architecture within the brain. The most common neurodevelopmental and neuropsychiatric disorders include depression, schizophrenia, autism spectrum disorders (ASD), bipolar disorder, attention deficit hyperactivity disorder, and X-linked intellectual disability, among others. The prevalence of these disorders is growing rapidly, which has caused a tremendous socioeconomic burden, primarily due to their high incidence in children and adolescents (Androutsos, 2012; Robertson et al., 2015; Hansen et al., 2018; Ghandour et al., 2019; Post and Grunze, 2021). During the past several decades, strenuous research has been performed in these fields. Unfortunately, the etiology and underlying mechanisms remain poorly understood.
In 2014, we first reported that Unc-51-like kinase 4 (ULK4) is crucial for neuritogenesis and neuronal motility and, when defective, may predispose people to neuropsychiatric disorders including schizophrenia (Lang et al., 2014). Since then, accumulating evidence has strongly suggested that ULK4 participates in corticogenesis, cilia maintenance, myelination, and white matter integrity, although the precise downstream signaling pathways and interacting substrates remain elusive. Recently, we have provided evidence that ULK4 deletion can cause decreased intermediate neural progenitors and increased apoptosis, which strongly disrupt normal cortical development (Hu et al., 2021). In addition, ULK4 can form an interactome by physically binding with PP2A and PP1α, the two most abundant phosphatases, and is responsible for over 90% of total Ser/Thr dephosphorylation in eukaryotes. This interactome closely regulates the expression of p-Akt and p-GSK-3α/β, and mice with ULK4-targeted deletion in the excitatory neurons of the forebrain present a spectrum of core features of schizophrenia. These data collectively suggest that ULK4 is a rare susceptibility gene for psychiatric disorders, especially schizophrenia. In this review, we will summarize the current knowledge of the roles of ULK4 in neurodevelopmental and neuropsychiatric disorders.
Main Text
Unc-51-like Serine/Threonine Kinase (ULK) Family
In 1998, a novel mouse ortholog of the Caenorhabditis elegans serine/threonine kinase uncoordinated-51 (UNC-51) was first cloned (Yan et al., 1998), and thereafter, five related genes in total were found and grouped into the UNC-51-like serine/threonine kinase (ULK) family: ULK1, ULK2, ULK3, ULK4, and serine/threonine kinase 36 (STK36). The kinase domains of ULKs are conserved and located at the N-terminus, and the C-terminal region contains protein interaction motifs important for substrate recruitment (Figure 1). In mammals, ULK1 and ULK2 are evolutionarily conserved serine/threonine kinase orthologs of the yeast autophagy-related (ATG) family member ATG1, and play a necessary but somewhat redundant function in proper autophagy initiation (Wang et al., 2018). The high-resolution structure analysis shows that ULK1 and ULK2 share a high degree of conservative domain architecture, including an N-terminal catalytic kinase, extensive middle linker, and C-terminal domain essential for interaction with their binding partners (Lazarus et al., 2015; Chaikuad et al., 2019). During autophagy, the canonical early regulatory complex consists of ULK1/ULK2, ATG13, RB1-inducible coiled-coil protein 1 (RB1CC1, also known as FIP200), and ATG101, which translate upstream nutrient and energy signals (e.g., mTOR and AMPK) into the downstream autophagy pathway (Ganley et al., 2009; Jung et al., 2009; Wong et al., 2013; Lin and Hurley, 2016). Disrupting ULK1 expression in mice leads to defective autophagy-mediated clearance of mitochondria, and mice lacking both ULK1 and ULK2 die shortly after birth due to a defect in glycogen metabolism, which is similar to what occurs with other autophagy-defective mice (Kundu et al., 2008; Cheong et al., 2014). Apart from these processes, ULK1/ULK2 also regulates TrkA receptor trafficking and signaling, which instructs filopodia extension and neurite branching during sensory axon outgrowth (Zhou et al., 2007). Knockdown of ULK2 reduced asymmetric neuropil elaboration and affected habenular development in the brain (Taylor et al., 2011). Recently, Kang et al. revealed an association between ULK2 polymorphisms and schizophrenia in the Korean population (Kang et al., 2022).
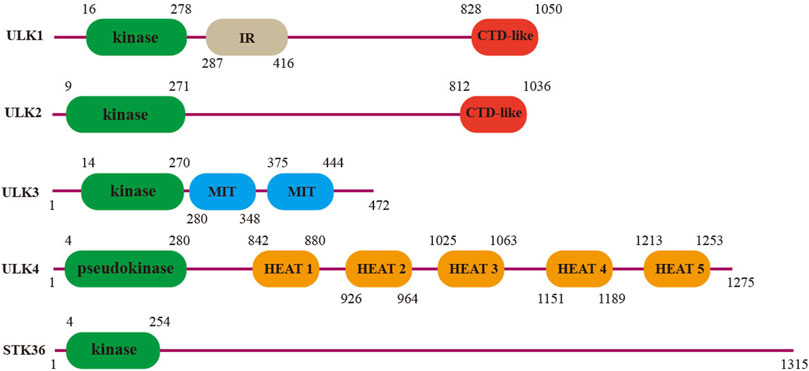
FIGURE 1. Domain architecture of the human ULK family. Protein interaction domains are annotated as interaction domain (IR), C-terminal domain (CTD) (ULK1 and ULK2), microtubule interacting, and trafficking molecule (MIT) (ULK3), and HEAT domains (ULK4).
The other three homologs, ULK3, ULK4, and STK36, contain kinase domains homologous to ULK1/2 but do not have a conserved C-terminal sequence, and they participate in many physiological processes to maintain tissue homeostasis. ULK3 has been reported to be involved in the autophagy induction during senescence (Young et al., 2009). It also has a dual function in the Sonic hedgehog signal transduction pathway, which controls a variety of developmental processes and is implicated in tissue homeostasis and neurogenesis in adults (Fuccillo et al., 2006; Maloverjan et al., 2010). STK36 is essential for the central pair apparatus and cilia orientation of motile cilia in mice. The cilia of STK36−/− mice are stiff and exhibit significantly reduced stroke amplitude or even immotile movement, which eventually promotes the occurrence of hydrocephalus (Merchant et al., 2005; Nozawa et al., 2013). According to the database in the Swiss Institute of Bioinformatics (SIB), ULK4 is widely expressed in different systems, especially in the secretion system, immune system, and nervous system, but its precise function remains largely unclear. Since we first reported that ULK4 may be a rare susceptibility gene for schizophrenia in 2014, research on this gene has been springing up in the neuropsychiatric field.
ULK4 Protein Structure
ULK4 is a large protein (142 kDa) encoded by the gene Unc51-Like Kinase 4, which is located on human chromosome 3p22.1 (Went et al., 2019). Unlike the homolog family member ULK1-3, the ULK4 protein contains a pseudokinase domain at the N-terminus and is thus predicted to be catalytically inactive. There are five HEAT repeats at the C-terminus of ULK4 (842–880, 926–964, 1,025–1,063, 1,151–1,189, and 1,213–1,253) (Figure 1), which are commonly found in large proteins, such as mTOR, and are presumably involved in protein scaffolding or interaction (Andrade et al., 2001; Perry and Kleckner, 2003). The crystallized high-resolution structure of ULK4, including its small-molecule inhibitor and ULK4-ATP-rS, has been recently interpreted by two independent research groups (Khamrui et al., 2020; Preuss et al., 2020). Notably, ULK4 can bind to ATP in an unusual Mg2+-independent manner, and the affinity is higher than that of any known pseudokinase (Khamrui et al., 2020). Because some pseudokinases are capable of binding to ATP and allosterically regulating the catalytic functions of kinases using compensatory motifs, even though ULK4 has no apparent phosphotransferase activity (Zeqiraj and van Aalten, 2010), it is assumed that like many others, ULK4 may work as the sensor of ATP and undergo conformational changes upon the binding which subsequently promotes its roles as a scaffold for substrate recruitment. Indeed, Preuss et al. predicted many ULK4 interacting partners including active kinases and phosphatases, which require further functional validation (Preuss et al., 2020).
Similar to the working mechanism of STRAD/LKB1, the pseudokinase domain of ULK4 specifically interacts with STK36. This strongly indicates that ULK4 can regulate active kinases directly, despite it being deemed catalytically inactive (Zeqiraj et al., 2009). The unique C-terminal HEAT repeats may enable ULK4 to bind to proper substrates or interacting proteins using a similar recruitment mechanism as ULK1/2. This hypothesis was further substantiated by Preuss and his colleagues, who have revealed that these repeated regions interacted uniquely with calmodulin-regulated spectrin-associated protein 1 (CAMSAP1), oral-facial-digital syndrome 1 (OFD1), and poly(A)-specific ribonuclease subunit 2 (PAN2) (Preuss et al., 2020). However, thus far, there has not been any report that there is an interaction partner of the ULK4 HEAT repeats at the C-terminal of STK36. Domain mapping of ULK4 provides a structural framework for its roles in diseases.
ULK4 and Unc-51
The unc-51 gene was initially described in the nematode C. elegans by Brenner in 1974 and showed extensive expression during embryonic brain development when neurons were actively extending their axons, particularly in the head region of late embryos (Brenner, 1974). Surprisingly, worms with the unc-51 mutation were mostly paralyzed, egg-laying defective, and dumpy (McIntire et al., 1992; Ogura et al., 1994). These data strongly suggested that the unc-51 protein is essential for axon maintenance and elongation. In the brains of Drosophila individuals, unc-51-mediated membrane vesicle transport is pivotal in the targeted localization of guidance molecules and organelles that regulate the elongation and compartmentalization of developing neurons as well as motor-cargo assembly (Mochizuki et al., 2011). Similarly, the unc-51 protein was reported to localize in the vesicular structures of growth cones of cerebellar granule cells and spinal sensory neurons in mice, which controls axon formation in granule cells through the endocytic membrane trafficking pathway (Tomoda et al., 1999; Tomoda et al., 2004). As a homologous serine/threonine kinase of unc-51 in humans, ULK4 was initially reported to be associated with blood pressure and hypertension (Levy et al., 2009; Ehret and Caulfield, 2013; Konigorski et al., 2014). Meanwhile, it may be involved in cell cycle control, as its polymorphisms (rs1052501 and rs2272007) were associated with multiple myelomas (Broderick et al., 2011; Greenberg et al., 2013). Inspired by the physiological functions of unc-51, we reanalyzed the common and rare variants of ULK4 in the databases of the International Schizophrenia Consortium (ISC) and among the bipolar Icelandic cases genotyped by deCODE Genetics, and we discovered that it may serve as a rare susceptibility gene for human mental disorders, especially schizophrenia (Lang et al., 2014). Our subsequent functional study further revealed that ULK4 is involved in the remodeling of cytoskeletal components, such as acetylation of α-tubulin, and in this way regulates neurite branching and elongation as well as cell motility.
ULK4 and Neurogenesis
Both in vivo and in vitro studies have suggested that ULK4 may play a key role in neurogenesis and corticogenesis during developmental stages. In Xenopus embryos, ULK4 mRNA is mostly expressed in the ventricular (VZ) and subventricular zones (SVZ) zones and distributed throughout the brain after the closure of the neural tube. Constant expression of ULK4 has also been found in neural stem cells in adult Xenopus (Domínguez et al., 2015). Similarly, Ulk4 transcripts are widely found in the VZ, SVZ, and cortical plate in the E15.5 cortex in mice, and ULK4 protein is widely expressed in all cortical layers after postnatal Day 7. Knockdown of ULK4 at E15.5 significantly inhibited cell proliferation and corticogenesis in mice (Lang et al., 2016). Meanwhile, the size of the neural stem cell pool in the forebrain that is important for adult neurogenesis was remarkably reduced in ULK4 null knockout mice at birth (Liu et al., 2016a). Although normal cortical lamination was preserved, the knockout mice showed a thinner cortex due to defective cell proliferation. As abnormal neurogenesis is often associated with neurodevelopmental or neuropsychiatric diseases (Kang et al., 2016; Guarnieri et al., 2018), it is therefore believed that ULK4 may contribute to the development of these diseases. Liu et al. further identified that ULK4 expression was dependent on the cell cycle, with a peak expression in the G2/M phases, and it decreased during both embryonic and adult neurogenesis in ULK4 mutant mice, probably because of a dysregulated Wnt signaling pathway (Liu et al., 2017).
ULK4 and Neurite Arborization
It has been well documented that Unc-51 regulates the dendritic development in the brains of individuals of the genus Drosophila through kinesin-mediated membrane transport (Mochizuki et al., 2011). In C. elegans, Unc-51 mutation often leads to abnormal axonal elongation and structures (Ogura et al., 1994). Consistently, appropriate neurite arborization is important in establishing synaptic connectivity and neuronal plasticity, which is critical for preventing the onset of schizophrenia (Mochizuki et al., 2011; Mizutani et al., 2019). Therefore, it is assumed that the ULK family plays an important role in the establishment of the appropriate neural network and, when defective, may promote the development of neurological diseases. In line with this hypothesis, our data suggest that the proper expression of ULK4 is critical for neurite branching and brain development. Knockdown of ULK4 in SH-SY5Y cells led to less expression of acetylated α-tubulin, which may underlie the reduced dendrite length and/or branching and compromised neuronal migration (Lang et al., 2014). Defective neuritogenesis may involve multiple signaling pathways including protein kinase C (PKC), mitogen-activated protein kinase (MAPK), extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinases (JNK) (Lang et al., 2014). Similarly, our in utero electroporation study in utero also demonstrated that knockdown of ULK4 caused perturbed neurite arborization in the pyramidal neurons of the cortex (Lang et al., 2016).
ULK4 and the Integrity of White Matter
Children’s performance in cognition, intelligence, processing speed, and problem solving is closely associated with the thickness of the white matter, such as the corpus callosum and defective myelination is a hallmark related to neurodevelopmental and neuropsychiatric disorders (Liu et al., 2018b). We previously showed that ULK4 null knockout mice displayed impaired genesis of the corpus callosum (Lang et al., 2014). Liu et al. further reported a 50% decrease in myelination in ULK4−/− mice together with a general reduction in myelin components (Liu et al., 2018b). Myelin is produced by oligodendrocytes and controls impulse conduction speed along the axon, which is important to cognitive performance. Children with a less myelinated white matter in their brains often display developmental delay problems. Meanwhile, ULK4 mutant mice also present thin axons and extensive neuroinflammation, which also promote the occurrence of hypomyelination. In addition, ULK4 deficiency significantly attenuated the enrichment of oligodendrocyte transcription factors, the newly formed oligodendrocytes, and myelinating oligodendrocytes (Liu et al., 2018b). These data collectively indicate that ULK4 may be a crucial factor for the integrity of white matter and myelin.
ULK4 and Ciliopathy
The cilium is an antenna-like structure that protrudes from the surface of almost all mammalian cells. It participates in multiple signaling transduction pathways and when defective, can result in a series of inherited disorders called “ciliopathies”. The most common features of ciliopathy include cystic liver and/or kidney, blindness, neural tube defects, brain anomalies, mental disability, skeletal abnormalities, obesity, and infertility, among others (Oud et al., 2017). Genomic and bioinformatics research has revealed that some primary cilia genes are linked to psychiatric disorders, such as the genes CC2D2A and Disc1, which are involved in ciliogenesis (Shen et al., 2008; Marley and von Zastrow, 2010; Veleri et al., 2014), and their defects can lead to psychiatric disorders, including Joubert syndrome (Bachmann-Gagescu et al., 2012), mental retardation (Noor et al., 2008; Shi et al., 2012), Meckel syndrome (Tallila et al., 2008), and Bardet Biedl syndrome (BBS) (Haq et al., 2019). In addition, several signaling pathways and crucial factors highly associated with schizophrenia, such as Wnt signaling, the fibroblast growth factor signaling system, neuronal migration, and the dopamine hypothesis, are dependent on the complete functionality of the cilium, although the specific mechanism is not yet well understood (Marley and von Zastrow, 2010; Muraki and Tanigaki, 2015; Narla et al., 2017; Hoseth et al., 2018). In the mouse brain, ULK4 is strongly expressed in the choroid plexus and ependymal cells lining the ventricles (Lang et al., 2014). Both ULK4 null knockout and hypomorphic mice present disturbed motile cilia development and disorganized ciliary beating which impair CSF flow and eventually lead to congenital hydrocephalus (Vogel et al., 2012; Liu et al., 2016b). These data strongly indicate the potential connection between ULK4 haploinsufficiency and ciliopathy. Acetylated α-tubulin is an important cytoskeletal component of cilia that is instrumental for cilium assembly. Our study, however, revealed that knockdown of ULK4 in human neuroblastoma cells (SH-SY5Y) and the mouse brain led to reduced expression of acetylated α-tubulin (Lang et al., 2014; Lang et al., 2016). In addition, whole-genome RNA sequencing also revealed massive disruption of genes closely related to ciliogenesis including Foxj1, Pcm1, Tubb4a, Dnah9, Rsph4a, Gsn, Kif5a, Lgals3, Lgals3bp, and Dnal1 in ULK4 mice carrying hypomorphic alleles. Interestingly, it has been reported that Foxj1 may target downstream substrates including Spag6, Rsph9, Rsph4a, Dnah9, Dnal1, Ttll6, and Tekt2 which consequently impairs ciliary development and results in hydrocephalus (Liu et al., 2016b). A recent study also reported that patients with a microdeletion of the ULK4 gene and a microduplication of the BRWD3 gene manifested core features of ciliopathy such as psychomotor delay, epilepsy, autistic features, hearing loss, obesity, minor facial dysmorphisms, peculiar ear malformations, and skeletal abnormalities (such as dorsal kyphosis and/or valgus knees and flat feet) (Tassano et al., 2018). Thus, it is highly likely that ULK4 contributes to ciliopathies. The results demonstrate that ULK4 is crucial for ciliogenesis and ciliopathies.
The Progress of Current Research on ULK4 in Mental Disorders
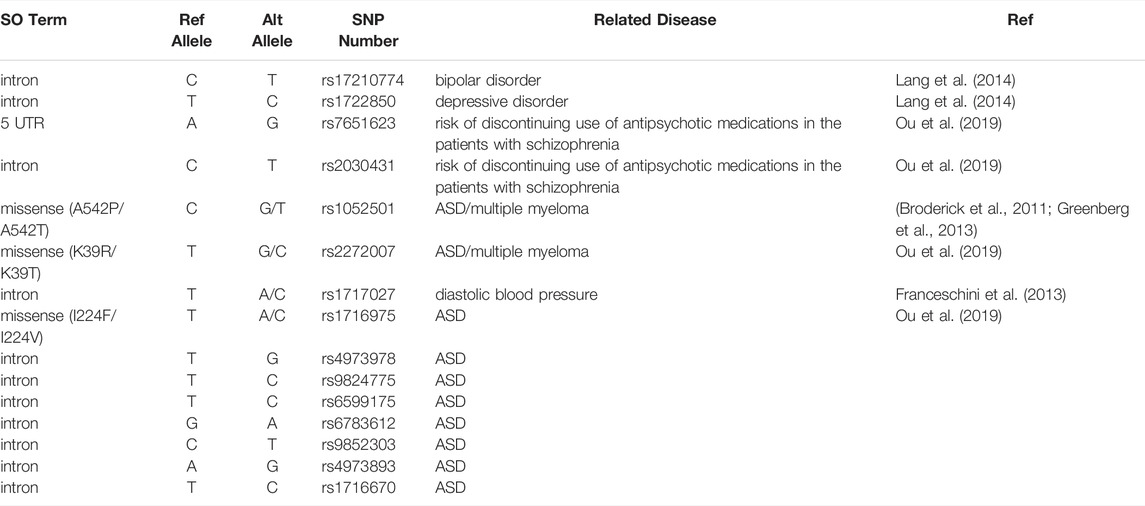
Although previous GWAS studies have suggested that ULK4 is a risk locus for multiple myeloma and interindividual diastolic blood pressure variation, emerging evidence also supports the idea that ULK4 genetic variants may cosegregate people with multiple neuropsychiatric disorders (Levy et al., 2009; Broderick et al., 2011). In our previous research using the cohort data from the International Schizophrenia Consortium, we identified four schizophrenia patients with ULK4 intragenic fragment deletions spanning from exon 21 to exon 34 among 3,391 schizophrenia patients (Lang et al., 2014). Another study implicated that SNPs rs7651623 and rs2030431 of ULK4 are associated with the risk of discontinuing the use of antipsychotics in patients with schizophrenia (Ou et al., 2019). In the Decode database, ULK4 deletion was also enriched in patients with schizophrenia (2/708), bipolar disorder (2/1,136), and autism (1/507) (Lang et al., 2014). In addition, association signals were observed at SNPs rs1052501, rs1716975, and rs2272007, which are located in exons 2, 7, and 17 of ULK4, respectively, for allelic transmission disequilibrium from parents to their children with ASD (Ou et al., 2019). Similarly, SNP rs17210774 of ULK4 is significantly associated with bipolar disorder in Caucasians and another SNP rs1722850, which is close to but downstream of ULK4, is related to major depressive disorders (Lang et al., 2014) (Table 1). A recent study of the brain-body genetic resource exchange (BBGRE) cohort also reported an incidence in a population of 1.2‰, showing ULK4 copy number variation and exhibiting pleiotropic neurodevelopmental problems including learning difficulties and language delay (Liu et al., 2016a). In addition, a recent clinical study revealed 2 cases with ULK4 intragenic microdeletion (together with partial microduplication of BRWD3) that showed autistic features (Tassano et al., 2018).Consistently, in the follow-up functional analysis, we have revealed that knockdown of ULK4 altered the activity of Wnt, PKC, MAPK, ERK1/2, and JNK signaling pathways commonly found in human mental disorders, especially schizophrenia (Figure 2). In addition, both ULK4 knockout and hypomorphic mice presented congenital hydrocephalus featuring dilated ventricles and CSF accumulation. Interestingly, a proportion of schizophrenia patients also display increased global or regional CSF(Vogel et al., 2012; Lang et al., 2014). Moreover, Liu et al. revealed that ULK4 heterozygous mice displayed anxiety-like behavior with reduced GABAergic neurons in the basolateral amygdala and hippocampus (Liu et al., 2018a), and ULK4−/− mice showed a significant hypomyelination phenotype (Liu et al., 2018b). All these studies strongly suggest that ULK4 may be a rare risk factor for neuropsychiatric disorders including schizophrenia but more evidence is warranted in the future.
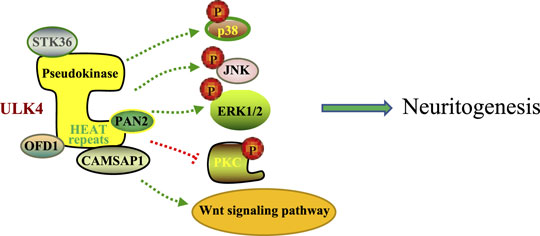
FIGURE 2. A schematic representation of altered activities of multiple signaling pathways including p38 MAPK, JNK, ERK1/2, PKC, and Wnt signaling pathways by ULK4. These alterations contribute to deficient neuritogenesis, a common feature frequently represented by human mental disorders.
Conclusion and Perspectives
Although ULK4 is a member of the Unc-51-like kinase family, unlike its ortholog members ULK1-3 and STK36, it is predicted to be catalytically inactive and to function as a pseudokinase. Initially, ULK4 was found to be associated with blood pressure and hypertension but further research has indicated its important functions during neurodevelopment. Knockdown of ULK4 in vitro also altered the activities of multiple signaling pathways, including Wnt, PKC, p38 MAPK, ERK1/2, and JNK, and mice with ULK4 deletion showed anxiety-like behaviors, perturbed neurogenesis, and decreased myelination. As mentioned above, ULK4 may be a rare risk factor for a range of psychiatric disorders, including schizophrenia, ASD, bipolar disorder, and depression, whose genetic variants were found in relevant patients and are crucial for ciliogenesis and ciliopathies. Further studies are warranted to fully understand the important function of ULK4, especially in neurodevelopment, and the specific underlying mechanisms for psychiatric disorders. With the successful resolution of the protein structure of ULK4 and further elucidation of its function, a series of small molecules targeting ULK4 may be developed to alleviate relevant neurodevelopmental and neuropsychiatric disorders in the future.
Author Contributions
This work was primarily written by SL, NZ, and BL. Figure was produced by SL. All authors read and approved the final manuscript.
Funding
The research was supported by the Research Funds of the National Natural Science Foundation of China (No. 82071507).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Naama, N., Mackeh, R., and Kino, T. (2020). C2H2-Type Zinc Finger Proteins in Brain Development, Neurodevelopmental, and Other Neuropsychiatric Disorders: Systematic Literature-Based Analysis. Front. Neurol. 11. doi:10.3389/fneur.2020.00032
Alonso-Gonzalez, A., Rodriguez-Fontenla, C., and Carracedo, A. (2018). De Novo Mutations (DNMs) in Autism Spectrum Disorder (ASD): Pathway and Network Analysis. Front. Genet. 9, 406. doi:10.3389/fgene.2018.00406
Andrade, M. A., Petosa, C., O’Donoghue, S. I., Müller, C. W., and Bork, P. (2001). Comparison of ARM and HEAT Protein Repeats. J. Mol. Biol. 309 (1), 1–18. doi:10.1006/jmbi.2001.4624
Androutsos, Ch. (2012). Schizophrenia in Children and Adolescents: Relevance and Differentiation from Adult Schizophrenia. Psychiatriki 23 (Suppl. 1), 82–93.
Bachmann-Gagescu, R., Ishak, G. E., Dempsey, J. C., Adkins, J., O'Day, D., Phelps, I. G., et al. (2012). Genotype-phenotype Correlation inCC2D2A-Related Joubert Syndrome Reveals an Association with Ventriculomegaly and Seizures. J. Med. Genet. 49 (2), 126–137. doi:10.1136/jmedgenet-2011-100552
Brenner, S. (1974). The Genetics of Caenorhabditis elegans. Genetics 77 (1), 71–94. doi:10.1093/genetics/77.1.71
Broderick, P., Chubb, D., Johnson, D. C., Weinhold, N., Försti, A., Lloyd, A., et al. (2011). Common Variation at 3p22.1 and 7p15.3 Influences Multiple Myeloma Risk. Nat. Genet. 44 (1), 58–61. doi:10.1038/ng.993
Cardoso, A. R., Lopes-Marques, M., Silva, R. M., Serrano, C., Amorim, A., Prata, M. J., et al. (2019). Essential Genetic Findings in Neurodevelopmental Disorders. Hum. Genomics 13 (1), 31. doi:10.1186/s40246-019-0216-4
Chaikuad, A., Koschade, S. E., Stolz, A., Zivkovic, K., Pohl, C., Shaid, S., et al. (2019). Conservation of Structure, Function and Inhibitor Binding in UNC-51-like Kinase 1 and 2 (ULK1/2). Biochem. J. 476 (5), 875–887. doi:10.1042/bcj20190038
Cheong, H., Wu, J., Gonzales, L. K., Guttentag, S. H., Thompson, C. B., and Lindsten, T. (2014). Analysis of a Lung Defect in Autophagy-Deficient Mouse Strains. Autophagy 10 (1), 45–56. doi:10.4161/auto.26505
Cristino, A. S., Williams, S. M., Hawi, Z., An, J.-Y., Bellgrove, M. A., Schwartz, C. E., et al. (2014). Neurodevelopmental and Neuropsychiatric Disorders Represent an Interconnected Molecular System. Mol. Psychiatry 19 (3), 294–301. doi:10.1038/mp.2013.16
Domínguez, L., Schlosser, G., and Shen, S. (2015). Expression of a Novel Serine/threonine Kinase Gene, Ulk4, in Neural Progenitors during Xenopus laevis Forebrain Development. Neuroscience 290, 61–79. doi:10.1016/j.neuroscience.2014.12.060
Ehret, G. B., and Caulfield, M. J. (2013). Genes for Blood Pressure: an Opportunity to Understand Hypertension. Eur. Heart J. 34 (13), 951–961. doi:10.1093/eurheartj/ehs455
Franceschini, N., Fox, E., Zhang, Z., Edwards, T. L., Nalls, M. A., Sung, Y. J., et al. (2013). Genome-wide Association Analysis of Blood-Pressure Traits in African-Ancestry Individuals Reveals Common Associated Genes in African and Non-african Populations. Am. J. Hum. Genet. 93 (3), 545–554. doi:10.1016/j.ajhg.2013.07.010
Fuccillo, M., Joyner, A. L., and Fishell, G. (2006). Morphogen to Mitogen: the Multiple Roles of Hedgehog Signalling in Vertebrate Neural Development. Nat. Rev. Neurosci. 7 (10), 772–783. doi:10.1038/nrn1990
Ganley, I. G., Lam, D. H., Wang, J., Ding, X., Chen, S., and Jiang, X. (2009). ULK1·ATG13·FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy. J. Biol. Chem. 284 (18), 12297–12305. doi:10.1074/jbc.M900573200
Ghandour, R. M., Sherman, L. J., Vladutiu, C. J., Ali, M. M., Lynch, S. E., Bitsko, R. H., et al. (2019). Prevalence and Treatment of Depression, Anxiety, and Conduct Problems in US Children. J. Pediatr. 206, 256–267. e253. doi:10.1016/j.jpeds.2018.09.021
Greenberg, A. J., Lee, A. M., Serie, D. J., McDonnell, S. K., Cerhan, J. R., Liebow, M., et al. (2013). Single-nucleotide Polymorphism Rs1052501 Associated with Monoclonal Gammopathy of Undetermined Significance and Multiple Myeloma. Leukemia 27 (2), 515–516. doi:10.1038/leu.2012.232
Guarnieri, F. C., de Chevigny, A., de Chevigny, A., Falace, A., and Cardoso, C. (2018). Disorders of Neurogenesis and Cortical Development. Dialogues Clin. Neurosci. 20 (4), 255–266. doi:10.31887/DCNS.2018.20.4/ccardoso
Hansen, B. H., Oerbeck, B., Skirbekk, B., Petrovski, B. É., and Kristensen, H. (2018). Neurodevelopmental Disorders: Prevalence and Comorbidity in Children Referred to Mental Health Services. Nordic J. PsychiatryJ. Psychiatry 72 (4), 285–291. doi:10.1080/08039488.2018.1444087
Haq, N., Schmidt-Hieber, C., Sialana, F. J., Ciani, L., Heller, J. P., Stewart, M., et al. (2019). Correction: Loss of Bardet-Biedl Syndrome Proteins Causes Synaptic Aberrations in Principal Neurons. Plos Biol. 17 (10), e3000520. doi:10.1371/journal.pbio.3000520
Hoseth, E. Z., Krull, F., Dieset, I., Mørch, R. H., Hope, S., Gardsjord, E. S., et al. (2018). Exploring the Wnt Signaling Pathway in Schizophrenia and Bipolar Disorder. Transl Psychiatry 8 (1), 55. doi:10.1038/s41398-018-0102-1
Hu, L., Chen, Y., Yang, C.-P., Huang, Y., Song, N.-N., Chen, J.-Y., et al. (2021). Ulk4, a Newly Discovered Susceptibility Gene for Schizophrenia, Regulates Corticogenesis in Mice. Front. Cel Developmental Biol. 9. doi:10.3389/fcell.2021.645368
Jung, C. H., Jun, C. B., Ro, S.-H., Kim, Y.-M., Otto, N. M., Cao, J., et al. (2009). ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. MBoC 20 (7), 1992–2003. doi:10.1091/mbc.e08-12-1249
Kang, E., Wen, Z., Song, H., Christian, K. M., and Ming, G.-l. (2016). Adult Neurogenesis and Psychiatric Disorders. Cold Spring Harb. Perspect. Biol. 8 (9), a019026. doi:10.1101/cshperspect.a019026
Kang, W. S., Lee, S. M., Hwang, D., Park, H. J., and Kim, J. W. (2022). Association between Unc-51-like Autophagy Activating Kinase 2 Gene Polymorphisms and Schizophrenia in the Korean Population. Medicine (Baltimore) 101 (5), e28745. doi:10.1097/md.0000000000028745
Khamrui, S., Ung, P. M. U., Secor, C., Schlessinger, A., and Lazarus, M. B. (2020). High-Resolution Structure and Inhibition of the Schizophrenia-Linked Pseudokinase ULK4. J. Am. Chem. Soc. 142 (1), 33–37. doi:10.1021/jacs.9b10458
Konigorski, S., Yilmaz, Y. E., and Bull, S. B. (2014). Bivariate Genetic Association Analysis of Systolic and Diastolic Blood Pressure by Copula Models. BMC Proc. 8 (Suppl. 1), S72. doi:10.1186/1753-6561-8-s1-s72
Kundu, M., Lindsten, T., Yang, C.-Y., Wu, J., Zhao, F., Zhang, J., et al. (2008). Ulk1 Plays a Critical Role in the Autophagic Clearance of Mitochondria and Ribosomes during Reticulocyte Maturation. Blood 112 (4), 1493–1502. doi:10.1182/blood-2008-02-137398
Lang, B., Pu, J., Hunter, I., Liu, M., Martin-Granados, C., Reilly, T. J., et al. (2014). Recurrent Deletions of ULK4 in Schizophrenia: a Novel Gene Crucial for Neuritogenesis and Neuronal Motility. J. Cel Sci. 127 (Pt 3), 630–640. doi:10.1242/jcs.137604
Lang, B., Zhang, L., Jiang, G., Hu, L., Lan, W., Zhao, L., et al. (2016). Control of Cortex Development by ULK4, a Rare Risk Gene for Mental Disorders Including Schizophrenia. Sci. Rep. 6, 31126. doi:10.1038/srep31126
Lazarus, M. B., Novotny, C. J., and Shokat, K. M. (2015). Structure of the Human Autophagy Initiating Kinase ULK1 in Complex with Potent Inhibitors. ACS Chem. Biol. 10 (1), 257–261. doi:10.1021/cb500835z
Levy, D., Ehret, G. B., Rice, K., Verwoert, G. C., Launer, L. J., Dehghan, A., et al. (2009). Genome-wide Association Study of Blood Pressure and Hypertension. Nat. Genet. 41 (6), 677–687. doi:10.1038/ng.384
Lin, M. G., and Hurley, J. H. (2016). Structure and Function of the ULK1 Complex in Autophagy. Curr. Opin. Cel Biol. 39, 61–68. doi:10.1016/j.ceb.2016.02.010
Liu, M., Fitzgibbon, M., Wang, Y., Reilly, J., Qian, X., O’Brien, T., et al. (2018a). Ulk4 Regulates GABAergic Signaling and Anxiety-Related Behavior. Transl Psychiatry 8 (1), 43. doi:10.1038/s41398-017-0091-5
Liu, M., Guan, Z., Shen, Q., Flinter, F., Domínguez, L., Ahn, J. W., et al. (2016a). Ulk4 Regulates Neural Stem Cell Pool. Stem Cells 34 (9), 2318–2331. doi:10.1002/stem.2423
Liu, M., Guan, Z., Shen, Q., Lalor, P., Fitzgerald, U., O'Brien, T., et al. (2016b). Ulk4 Is Essential for Ciliogenesis and CSF Flow. J. Neurosci. 36 (29), 7589–7600. doi:10.1523/jneurosci.0621-16.2016
Liu, M., Xu, P., Guan, Z., Qian, X., Dockery, P., Fitzgerald, U., et al. (2018b). Ulk4deficiency Leads to Hypomyelination in Mice. Glia 66 (1), 175–190. doi:10.1002/glia.23236
Liu, M., Xu, P., O'Brien, T., and Shen, S. (2017). Multiple Roles of Ulk4 in Neurogenesis and Brain Function. Neurogenesis 4 (1), e1313646. doi:10.1080/23262133.2017.1313646
Maloverjan, A., Piirsoo, M., Kasak, L., Peil, L., Østerlund, T., and Kogerman, P. (2010). Dual Function of UNC-51-like Kinase 3 (Ulk3) in the Sonic Hedgehog Signaling Pathway. J. Biol. Chem. 285 (39), 30079–30090. doi:10.1074/jbc.M110.133991
Marley, A., and von Zastrow, M. (2010). DISC1 Regulates Primary Cilia that Display Specific Dopamine Receptors. PLoS ONE 5 (5), e10902. doi:10.1371/journal.pone.0010902
McIntire, S. L., Garriga, G., White, J., Jacobson, D., and Robert Horvitz, H. (1992). Genes Necessary for Directed Axonal Elongation or Fasciculation in C. elegans. Neuron 8 (2), 307–322. doi:10.1016/0896-6273(92)90297-q
Merchant, M., Evangelista, M., Luoh, S.-M., Frantz, G. D., Chalasani, S., Carano, R. A. D., et al. (2005). Loss of the Serine/threonine Kinase Fused Results in Postnatal Growth Defects and Lethality Due to Progressive Hydrocephalus. Mol. Cel. Biol. 25 (16), 7054–7068. doi:10.1128/mcb.25.16.7054-7068.2005
Mizutani, R., Saiga, R., Takeuchi, A., Uesugi, K., Terada, Y., Suzuki, Y., et al. (2019). Three-dimensional Alteration of Neurites in Schizophrenia. Transl Psychiatry 9 (1), 85. doi:10.1038/s41398-019-0427-4
Mochizuki, H., Toda, H., Ando, M., Kurusu, M., Tomoda, T., and Furukubo-Tokunaga, K. (2011). Unc-51/ATG1 Controls Axonal and Dendritic Development via Kinesin-Mediated Vesicle Transport in the Drosophila Brain. PLoS ONE 6 (5), e19632. doi:10.1371/journal.pone.0019632
Muraki, K., and Tanigaki, K. (2015). Neuronal Migration Abnormalities and its Possible Implications for Schizophrenia. Front. Neurosci. 9, 74. doi:10.3389/fnins.2015.00074
Narla, S. T., Lee, Y.-W., Benson, C. A., Sarder, P., Brennand, K. J., Stachowiak, E. K., et al. (2017). Common Developmental Genome Deprogramming in Schizophrenia - Role of Integrative Nuclear FGFR1 Signaling (INFS). Schizophrenia Res. 185, 17–32. doi:10.1016/j.schres.2016.12.012
Noor, A., Windpassinger, C., Patel, M., Stachowiak, B., Mikhailov, A., Azam, M., et al. (2008). CC2D2A, Encoding a Coiled-Coil and C2 Domain Protein, Causes Autosomal-Recessive Mental Retardation with Retinitis Pigmentosa. Am. J. Hum. Genet. 82 (4), 1011–1018. doi:10.1016/j.ajhg.2008.01.021
Nozawa, Y. I., Yao, E., Lin, C., Yang, J.-H., Wilson, C. W., Gacayan, R., et al. (2013). Fused (Stk36) Is a Ciliary Protein Required for central Pair Assembly and Motile Cilia Orientation in the Mammalian Oviduct. Dev. Dyn. 242 (11), 1307–1319. doi:10.1002/dvdy.24024
Ogura, K., Wicky, C., Magnenat, L., Tobler, H., Mori, I., Müller, F., et al. (1994). Caenorhabditis elegans Unc-51 Gene Required for Axonal Elongation Encodes a Novel Serine/threonine Kinase. Genes Dev. 8 (20), 2389–2400. doi:10.1101/gad.8.20.2389
Ou, J. J., Li, K. K., Guo, H., Xia, K., Hu, Z. M., Zhao, J. P., et al. (2019). ULK4 Genetic Variants Have Pleiotropic Effect on Risk of Autism, Associated with Brain mRNA Expression and Antipsychotic Treatment Response. J. Psychiatry Brain Sci. 4, 190010.:e:
Oud, M., Lamers, I., and Arts, H. (2017). Ciliopathies: Genetics in Pediatric Medicine. J. Pediatr. Genet. 06 (1), 018–029. doi:10.1055/s-0036-1593841
Perry, J., and Kleckner, N. (2003). The ATRs, ATMs, and TORs Are Giant HEAT Repeat Proteins. Cell 112 (2), 151–155. doi:10.1016/s0092-8674(03)00033-3
Post, R. M., and Grunze, H. (2021). The Challenges of Children with Bipolar Disorder. Medicina 57 (6), 601. doi:10.3390/medicina57060601
Preuss, F., Chatterjee, D., Mathea, S., Shrestha, S., St-Germain, J., Saha, M., et al. (2020). Nucleotide Binding, Evolutionary Insights, and Interaction Partners of the Pseudokinase Unc-51-like Kinase 4. Structure 28 (11), 1184–1196. e1186. doi:10.1016/j.str.2020.07.016
Rao, S., Yao, Y., and Bauer, D. E. (2021). Editing GWAS: Experimental Approaches to Dissect and Exploit Disease-Associated Genetic Variation. Genome Med. 13 (1), 41. doi:10.1186/s13073-021-00857-3
Robertson, J., Hatton, C., Emerson, E., and Baines, S. (2015). Prevalence of Epilepsy Among People with Intellectual Disabilities: A Systematic Review. Seizure 29, 46–62. doi:10.1016/j.seizure.2015.03.016
Shen, S., Lang, B., Nakamoto, C., Zhang, F., Pu, J., Kuan, S.-L., et al. (2008). Schizophrenia-related Neural and Behavioral Phenotypes in Transgenic Mice Expressing Truncated Disc1. J. Neurosci. 28 (43), 10893–10904. doi:10.1523/jneurosci.3299-08.2008
Shi, Z.-Y., Li, Y.-J., Zhang, K.-J., Gao, X.-C., Zheng, Z.-J., Han, N., et al. (2012). Positive Association ofCC2D1AandCC2D2AGene Haplotypes with Mental Retardation in a Han Chinese Population. DNA Cel Biol. 31 (1), 80–87. doi:10.1089/dna.2011.1253
Tallila, J., Jakkula, E., Peltonen, L., Salonen, R., and Kestilä, M. (2008). Identification of CC2D2A as a Meckel Syndrome Gene Adds an Important Piece to the Ciliopathy Puzzle. Am. J. Hum. Genet. 82 (6), 1361–1367. doi:10.1016/j.ajhg.2008.05.004
Tassano, E., Uccella, S., Giacomini, T., Striano, P., Severino, M., Porta, S., et al. (2018). Intragenic Microdeletion of ULK4 and Partial Microduplication of BRWD3 in Siblings with Neuropsychiatric Features and Obesity. Cytogenet. Genome Res. 156, 14–21. doi:10.1159/000491871
Taylor, R. W., Qi, J. Y., Talaga, A. K., Ma, T. P., Pan, L., Bartholomew, C. R., et al. (2011). Asymmetric Inhibition of Ulk2 Causes Left-Right Differences in Habenular Neuropil Formation. J. Neurosci. 31 (27), 9869–9878. doi:10.1523/jneurosci.0435-11.2011
Tomoda, T., Bhatt, R. S., Kuroyanagi, H., Shirasawa, T., and Hatten, M. E. (1999). A Mouse Serine/threonine Kinase Homologous to C. elegans UNC51 Functions in Parallel Fiber Formation of Cerebellar Granule Neurons. Neuron 24 (4), 833–846. doi:10.1016/s0896-6273(00)81031-4
Tomoda, T., Kim, J. H., Zhan, C., and Hatten, M. E. (2004). Role of Unc51.1 and its Binding Partners in CNS Axon Outgrowth. Genes Dev. 18 (5), 541–558. doi:10.1101/gad.1151204
Uffelmann, E., Huang, Q. Q., Munung, N. S., de Vries, J., Okada, Y., Martin, A. R., et al. (2021). Genome-wide Association Studies. Nat. Rev. Methods Primers 1 (1), 59. doi:10.1038/s43586-021-00056-9
Veleri, S., Manjunath, S. H., Fariss, R. N., May-Simera, H., Brooks, M., Foskett, T. A., et al. (2014). Ciliopathy-associated Gene Cc2d2a Promotes Assembly of Subdistal Appendages on the Mother Centriole during Cilia Biogenesis. Nat. Commun. 5, 4207. doi:10.1038/ncomms5207
Vogel, P., Read, R. W., Hansen, G. M., Payne, B. J., Small, D., Sands, A. T., et al. (2012). Congenital Hydrocephalus in Genetically Engineered Mice. Vet. Pathol. 49 (1), 166–181. doi:10.1177/0300985811415708
Wang, B., Iyengar, R., Li-Harms, X., Joo, J. H., Wright, C., Lavado, A., et al. (2018). The Autophagy-Inducing Kinases, ULK1 and ULK2, Regulate Axon Guidance in the Developing Mouse Forebrain via a Noncanonical Pathway. Autophagy 14 (5), 796–811. doi:10.1080/15548627.2017.1386820
Went, M., Kinnersley, B., Sud, A., Johnson, D. C., Weinhold, N., Försti, A., et al. (2019). Transcriptome-wide Association Study of Multiple Myeloma Identifies Candidate Susceptibility Genes. Hum. Genomics 13 (1), 37. doi:10.1186/s40246-019-0231-5
Wong, P.-M., Puente, C., Ganley, I. G., and Jiang, X. (2013). The ULK1 Complex. Autophagy 9 (2), 124–137. doi:10.4161/auto.23323
Yan, J., Kuroyanagi, H., Kuroiwa, A., Matsuda, Y.-i., Tokumitsu, H., Tomoda, T., et al. (1998). Identification of Mouse ULK1, a Novel Protein Kinase Structurally Related toC. elegansUNC-51. Biochem. Biophysical Res. Commun. 246 (1), 222–227. doi:10.1006/bbrc.1998.8546
Young, A. R. J., Narita, M., Ferreira, M., Kirschner, K., Sadaie, M., Darot, J. F. J., et al. (2009). Autophagy Mediates the Mitotic Senescence Transition. Genes Dev. 23 (7), 798–803. doi:10.1101/gad.519709
Zeqiraj, E., Filippi, B. M., Deak, M., Alessi, D. R., and van Aalten, D. M. F. (2009). Structure of the LKB1-STRAD-MO25 Complex Reveals an Allosteric Mechanism of Kinase Activation. Science 326 (5960), 1707–1711. doi:10.1126/science.1178377
Zeqiraj, E., and van Aalten, D. M. (2010). Pseudokinases-remnants of Evolution or Key Allosteric Regulators? Curr. Opin. Struct. Biol. 20 (6), 772–781. doi:10.1016/j.sbi.2010.10.001
Keywords: ULK4, neurodevelopmental disorder, neuropsychiatric disorder, pseudokinase, schizophrenia
Citation: Luo S, Zheng N and Lang B (2022) ULK4 in Neurodevelopmental and Neuropsychiatric Disorders. Front. Cell Dev. Biol. 10:873706. doi: 10.3389/fcell.2022.873706
Received: 11 February 2022; Accepted: 29 March 2022;
Published: 12 April 2022.
Edited by:
Weihua Yue, Peking University Sixth Hospital, ChinaReviewed by:
Konstantinos Zarbalis, University of California, Davis, United StatesXueqin Song, First Affiliated Hospital of Zhengzhou University, China
Copyright © 2022 Luo, Zheng and Lang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nanxi Zheng, 208211058@csu.edu.cn; Bing Lang, bing.lang@csu.edu.cn
 Shilin Luo
Shilin Luo Nanxi Zheng3*
Nanxi Zheng3*  Bing Lang
Bing Lang