Genome-Wide Analysis of H3K27me3 in Porcine Embryonic Muscle Development
- 1National Engineering Research Center for Breeding Swine Industry, College of Animal Science, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Agro-Animal Genomics and Molecular Breeding, College of Animal Science, South China Agricultural University, Guangzhou, China
- 3Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction, Huazhong Agricultural University, Ministry of Education, Wuhan, China
The trimethylation of histone H3 lysine 27 (H3K27me3) is one of the most important chromatin modifications, which is generally presented as a repressive mark in various biological processes. However, the dynamic and global-scale distribution of H3K27me3 during porcine embryonic muscle development remains unclear. Here, our study provided a comprehensive genome-wide view of H3K27me3 and analyzed the matching transcriptome in the skeletal muscles on days 33, 65, and 90 post-coitus from Duroc fetuses. Transcriptome analysis identified 4,124 differentially expressed genes (DEGs) and revealed the key transcriptional properties in three stages. We found that the global H3K27me3 levels continually increased during embryonic development, and the H3K27me3 level was negatively correlated with gene expression. The loss of H3K27me3 in the promoter was associated with the transcriptional activation of 856 DEGs in various processes, including skeletal muscle development, calcium signaling, and multiple metabolic pathways. We also identified for the first time that H3K27me3 could enrich in the promoter of genes, such as DES, MYL1, TNNC1, and KLF5, to negatively regulate gene expression in porcine satellite cells (PSCs). The loss of H3K27me3 could promote muscle cell differentiation. Taken together, this study provided the first genome-wide landscape of H3K27me3 in porcine embryonic muscle development. It revealed the complex and broad function of H3K27me3 in the regulation of embryonic muscle development from skeletal muscle morphogenesis to myofiber maturation.
Introduction
Skeletal muscles represent the most abundant tissue of the body, and such muscles are essential for motion and support (Chal and Pourquié, 2017). Compromised muscle function can result in debilitating musculoskeletal disorders from developmental disorders to muscular dystrophies (Chal and Pourquié, 2017; Yue et al., 2021). Embryonic skeletal muscle development (embryonic myogenesis) predominantly determines the growth and mass of skeletal muscle. The total number of muscle fibers is basically fixed during embryonic myogenesis, which determines the postnatal fiber hypertrophy (Rehfeldt et al., 2008; Ali et al., 2021; Yue et al., 2021). In addition, impaired embryonic myogenesis has a long-lasting effect on postnatal muscle growth and animal performance, even after compensatory growth (Ali et al., 2021). Thus, research on embryonic muscle development is critical to reveal the genetic mechanisms affecting muscle development and improve meat production and animal health.
Embryonic myogenesis begins from the proliferation and differentiation of skeletal myogenic progenitors in somites. These cells lose the expression of the transcription factors (Pax3 and Pax7) during the progressive specialization as the expression of myogenic regulatory factors (MRFs), including Myf5, Myod, Myog, and Mrf4, increases. Skeletal muscle formation continues with the differentiation of these specialized progenitors to form muscle fibers (Tu et al., 2016). Pig is an omnivorous, monogastric species with many anatomic and physiological similarities to human, which can serve as an animal model for muscle diseases (Pabst, 2020). Pig embryonic myogenesis is a complex process that consists of two growth waves: the first growth wave occurs during 30–55 days post coitus (dpc) when primary skeletal muscle fibers develop and provide the attached scaffold for the secondary skeletal muscle fibers; the second growth wave occurs from 50 dpc to 90 dpc, when the secondary muscle fibers form and increase in number by many times (Yue et al., 2021). The formation of myofibers ceases and the total number of myofibers is settled by 85-90 dpc (Ali et al., 2021). The continual advances of high-throughput sequencing methods in epigenetics allow lower input material and more accurate examination of the dynamic alterations to histone, DNA, and RNA methylation patterns (Barski et al., 2007; Dominissini et al., 2012; Wang et al., 2013; Abo Alrob and Lopaschuk, 2014). These techniques with increasingly analytical tools have contributed to the investigation of RNA splicing and transcriptional regulation in various processes, and the prediction and therapy of diseases (Heerboth et al., 2014; Gu et al., 2015; Bauer et al., 2016). Myogenesis in pig is regulated by various epigenetic regulations, such as noncoding RNA, RNA methylation, DNA methylation, and histone modifications (Begue et al., 2017; Kudou et al., 2017; Jin et al., 2018; Qin et al., 2019; Wang et al., 2019c; Yonamine et al., 2019). H3K27me3 is one of the most frequent histone modifications that govern chromatin structure and gene expression. EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2), which is responsible for H3K27me3 that leads to gene silencing. Increasing studies have indicated that H3K27me3 plays an important role in various processes of skeletal muscle development. H3K27me3 can regulate the differentiation of satellite cells by altering the gene expression of MRFs, such as Myog (Asp et al., 2011; Faralli et al., 2016; Adhikari and Davie, 2018). The deposition of H3K27me3 in the promoter of Ccnd1 and Ccne1 can inhibit the proliferation of muscle cells (Adhikari et al., 2019). H3K27me3 level increases during muscle regeneration, and the loss of H3K27me3 demethylase UTX or methylase EZH2 activity impairs regeneration and reduces the number of stem cells (Liu et al., 2013; Woodhouse et al., 2013; Faralli et al., 2016). The fiber area and muscle mass of conditional EZH2 knockout mouse decrease as the global H3K27me3 level is significantly reduced (Woodhouse et al., 2013). During embryonic development, abnormal H3K27me3 causes embryonic lethality by knocking out demethylase UTX (Faralli et al., 2016). H3K27me3 can affect the acquisition and repression of defined cell lineage transcriptional programs of embryonic stem cells (Juan et al., 2016). A study on ovine skeletal muscle has presented genome-wide maps of H3K27me3 during the late stage of embryonic development and revealed the important role of H3K27me3 in the early development process and neuron function (Byrne et al., 2014). However, the regulation of H3K27me3 in the skeletal muscle of pigs during embryonic development remains poorly elucidated.
In this study, we performed RNA-seq and chromatin immunoprecipitation (ChIP)-seq to provide the first comprehensive analysis of H3K27me3 and gene expression profile during embryonic muscle development (d33, d65, and d90) in Duroc pig. Our analysis revealed the transcriptome characteristics in the three stages and investigated the dynamic changes of gene expression in muscle development. We highlighted the important role of H3K27me3 in regulating gene expression and identified the essential processes regulated by H3K27me3 during pig embryonic myogenesis. This study can provide the basis for studying and expanding the regulatory mechanisms of muscle fiber development.
Materials and Methods
Sample Collection
Duroc pigs (Sus scrofa) are widely used as terminal boars for its elite growth performance. In this study, Duroc fetuses were obtained from the pig breeding farm of Guangdong Wen’s Foodstuffs Group Co., Ltd. (Yunfu, China). Nine Duroc pig fetuses were divided into three groups, consisting of three fetuses on gestation day 33 (d33), three fetuses on gestation day 65 (d65), and three fetuses on gestation day 90 (d90). The three fetuses on the same gestation day were half-sibs for reducing the influence of individual differences. The longissimus muscle tissues were quickly sectioned in a 2-ml centrifuge tube and stored in liquid nitrogen.
Cell Culture
The isolation of porcine satellite cells (PSCs) was performed as previously described (Wang et al., 2019a). For proliferation, PSCs were cultured in RPMI-1640 medium (11875119, Gibco, United States) containing 20% FBS (10099141, Gibco, United States), 1% non-essential amino acids (Gibco, United States), 0.5% chicken embryo extract (0928501, GEMINI, United States), 1% GlutaMax (35050061, Gibco, United States), 1% antibiotic–antimycotic (15140122, Gibco, United States), and 2.5 ng/ml bFGF (13256029, Gibco, United States) under moist air with 5% CO2 at 37°C. At about 70% confluence, DMEM high-glucose medium (12430054, Gibco, United States) supplemented with 2% horse serum (SH30074.03, HyClone, United States) and 1% antibiotic–antimycotic (15140122, Gibco, United States) was used to induce PSCs to differentiation. GSK343 (SML0766, SIGMA, United States) was dissolved to 2.5 μmol/L with DMSO (D4540, SIGMA, United States) and used for the depletion of H3K27me3 in PSCs. After 48-h incubation with 2.5 μmol/L GSK343 or vehicle 0.1% DMSO in proliferation, PSCs were induced to differentiation and continually treated for 36 h.
Quantitative RT-PCR
Total RNA was extracted by using TRIzol reagent (15596018, Gibco, United States) according to the manufacturer’s instruction. PrimeScript RT reagent kit with gDNA Eraser (RR047A, Takara, Japan) was used to erase genomes and reverse transcription. The reaction mixture and condition for quantitative real-time PCR (RT-qPCR) was described in our published paper (Tan et al., 2020). The qPCR reaction was performed in Quant Studio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, United States). All experiences were carried out with three fully independent biological replicates and three technical repeats. The relative gene expression level was calculated by the Ct (2–ΔΔCt) method. The primers are listed in Supplementary Table S5.
Western Blotting
Porcine satellite cells or muscle tissues were lysed in RIPA buffer containing 1% (v/v) phenylmethylsulfonyl fluoride (PMSF) (ST505, Beyotime, China) to acquired total protein. Approximately 30 μg of protein of each sample was loaded on SDS-PAGE and transferred to PVDF membranes (IPVH85R, Millipore, United States). After blocking with 5% non-fat milk, the proteins in membranes were subjected to immunoblotting analysis with primary and secondary antibodies. Antibodies used in this study were as follows: anti-H3K27me3 (17-622, Millipore, United States), anti-beta-tubulin (GB11017, Servicebio, China), anti-MYOD (sc-760, Santa Cruz Biotechnology, United States), anti-MYH4 (A15293, ABclonal, United States), and HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (SA00001-2, Proteintech, United States). The membranes were developed with ECL (WBULS0500, Millipore, United States) for visualization. ImageJ software was used to determine the bands’ signal intensities. The density value of each band was normalized by corresponding beta-tubulin density value.
Immunofluorescence
Porcine satellite cells were cultured in a six-well plate and differentiated for 1.5 days. The immunofluorescence staining was performed according to our previous studies (Wang et al., 2019b). The antibodies and their dilutions were as follows: MYHC (sc-376157, Santa Cruz Biotechnology, United States) and secondary antibody (A0521, Beyotime, China). Cell nuclei were stained by using 4′,6-diamidino-2-phenylindole (DAPI; C1006, Beyotime, China). The cell differentiation index was calculated by the ratio of the number of nuclei in the myotubes to the total number of nuclei in one field of view.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation experiment was carried out by using ChIP kit (P2078, Beyotime, China) according to the recommended protocol. Briefly, 4 × 106 PSCs were cultured to 90% confluence and fixed for 10 min at room template with 1% formaldehyde. The crosslinking was stopped by adding 125 mM glycine. Cell lysates were sheared by sonication in 1% SDS lysis buffer to generate chromatin fragments, followed in sequence with immunoprecipitation, reversal of cross-links, and DNA purification. Two micrograms of antibody H3K27me3 (17-622, Millipore, United States) or negative control IgG was used to each immunoprecipitated reaction. Fold enrichment was quantified using RT-qPCR. The primers are listed in Supplementary Table S5.
RNA-Seq and Data Analysis
RNA-seq was performed using three biological replicates. Total RNA was isolated using the TRIzol reagent (15596018, Gibco, United States) according to the manufacturer’s protocol. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (E7775, New England BioLabs, United States). Clean reads were obtained by removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data by using fastp (Chen et al., 2018). BWA mem (Li, 2013) was used to build the index and map the clean reads to reference genome of pig (Ensembl Sscrofa 11.1.94) with default parameters. The gene read counts were calculated by featureCounts (Liao et al., 2014) and normalized to the transcripts per kilobase million (TPM). Differential gene expression analysis between two groups was performed using the R package DESeq2 (Love et al., 2014). Gene ontology (GO) analyses, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, and Gene Set Enrichment Analyses (GSEA) were implemented by R package clusterProfiler (Yu et al., 2012). All gene sets for GSEA were downloaded from MSigDB (Subramanian et al., 2005). R package ggplot2 was contributed to the graphical representation (Ginestet, 2011). Gene with TPM ≥ 0.5 at least in one sample was defined as expressed genes. All genes were classed into four groups according to their expression in each stage: high level (TPM ≥ upper quartile), medium level (lower quartile < TPM < upper quartile), low level (TPM ≤ lower quartile), and silent (never expressed).
Chromatin Immunoprecipitation-Sequencing and Data Analysis
Chromatin immunoprecipitation sample for sequencing was obtained by using the SimpleChIP Enzymatic Chromatin IP kit (9005, Cell Signaling Technologies, United States). Briefly, approximately 25 mg of skeletal muscle tissues was minced with a scalpel, harvested in 1 ml of PBS, cross-linked by 45 μl of 37% formaldehyde for 20 min at room temperature, and quenched with Glycine Solution for 5 min at room temperature. The suspended tissues were centrifuged 500 × g for 5 min at 4°C, washed two times in cold PBS, homogenized using a Dounce homogenizer, and incubated for 20 min at 37°C with micrococcal nuclease. Nuclei were destroyed by sonication, and the debris was removed by centrifugation. The clarified nuclear extracts were incubated overnight with 4 μg of H3K27me3 antibody (17-622, Millipore, United States) and immunoprecipitated with protein G magnetic beads. ChIP DNA library was constructed by Novogene Corporation (Beijing, China). Subsequently, 150-bp pair-end sequencing of sample was performed on Illumina platform. Library quality was determined with the Agilent Bioanalyzer 2100 system. Clean reads were obtained by removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data by using fastp (Chen et al., 2018). Clean reads were mapped to reference genome of pig (Ensembl Sscrofa 11.1) with default parameters by BWA mem (Li, 2013). Duplicated fragments, fragments with a mapping quality of less than 20, were removed by samtools (Danecek et al., 2021). The H3K27me3 peaks were identified by epic2 (Stovner and Sætrom, 2019) with a q-value threshold of 0.05 in broad model and >1-fold enrichment over the background. The R package ChIPseeker (Yu et al., 2015) was used to annotate genomic region of the peak and find the nearest genes around the peaks. The enrichment profiles of H3K27me3 and principal component analysis (PCA) of ChIP-seq data were performed using DeepTools (Ramírez et al., 2014). Read coverage for select genes was visualized in the Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al., 2013). The ChIP-seq peaks that were within −5 kb to 5 kb of transcription start sites (TSSs) were defined as promoter peaks. Read densities (RPKM) were defined as the normalized reads mapping in the promoter per kilobase million. The H3K27me3 level of gene is represented as normalized RPKM using IP/input ratio averaged in the two biological replicates.
Statistical Analysis
Statistical analysis for the results of qPCR and Western blotting was performed by independent sample Student’s t-test in SPSS 20.0 software (IBM, Armonk, NY, United States). A p-value of less than 0.05 was considered as significant difference. Experiment results were presented as the mean ± standard error of mean (SEM).
Results
Gene Expression Dynamics During Porcine Embryonic Muscle Development
The transcriptomes of embryonic porcine longissimus muscle tissues were examined on d33, d65, and d90 to represent three critical time points of porcine embryonic development (Figure 1A and Supplementary Figure S1). Approximately 560 million reads with a unique mapped rate of above 90% were obtained (Supplementary Table S1). The close clustering of biological replicates in the PCA plots indicated high reproducible results (Figure 1B). Upon transcriptome analysis, we detected a total of 4,124 differentially expressed genes (DEGs) (Supplementary Table S2). We explored genes that were upregulated or downregulated among the successive stages and identified those expressed at the highest level in d33 (2,060, namely, “the d33 genes”), d65 (473, namely, “the d65 genes”), or d90 (1,591, namely, “the d90 genes”; Figure 1C). We further hierarchically clustered the expression across three stages and found that all DEGs were primarily clustered into two groups: (1) particularly high expression in d33 and (2) commonly high expression in d65 and d90 (Figure 1D). To preliminarily explore the biological processes with dynamic changes among the successive stages, we generated an alluvial diagram to reflect the degree of differences in expression and found DEGs with dramatical changes in expression (Figure 1E). The expression of DEGs slightly changed from d65 to d90, whereas 23 genes drastically changed between 33 days and 65 days (Supplementary Table S3). Some of these genes, such as TMOD4 and CAVIN4, which are involved in the positive regulation of muscle development, were activated, whereas the expression of neural development-related genes, including WNT6 and CNTN2, decreased. Collectively, these results showed the dynamic changes of gene expression during embryonic muscle development.
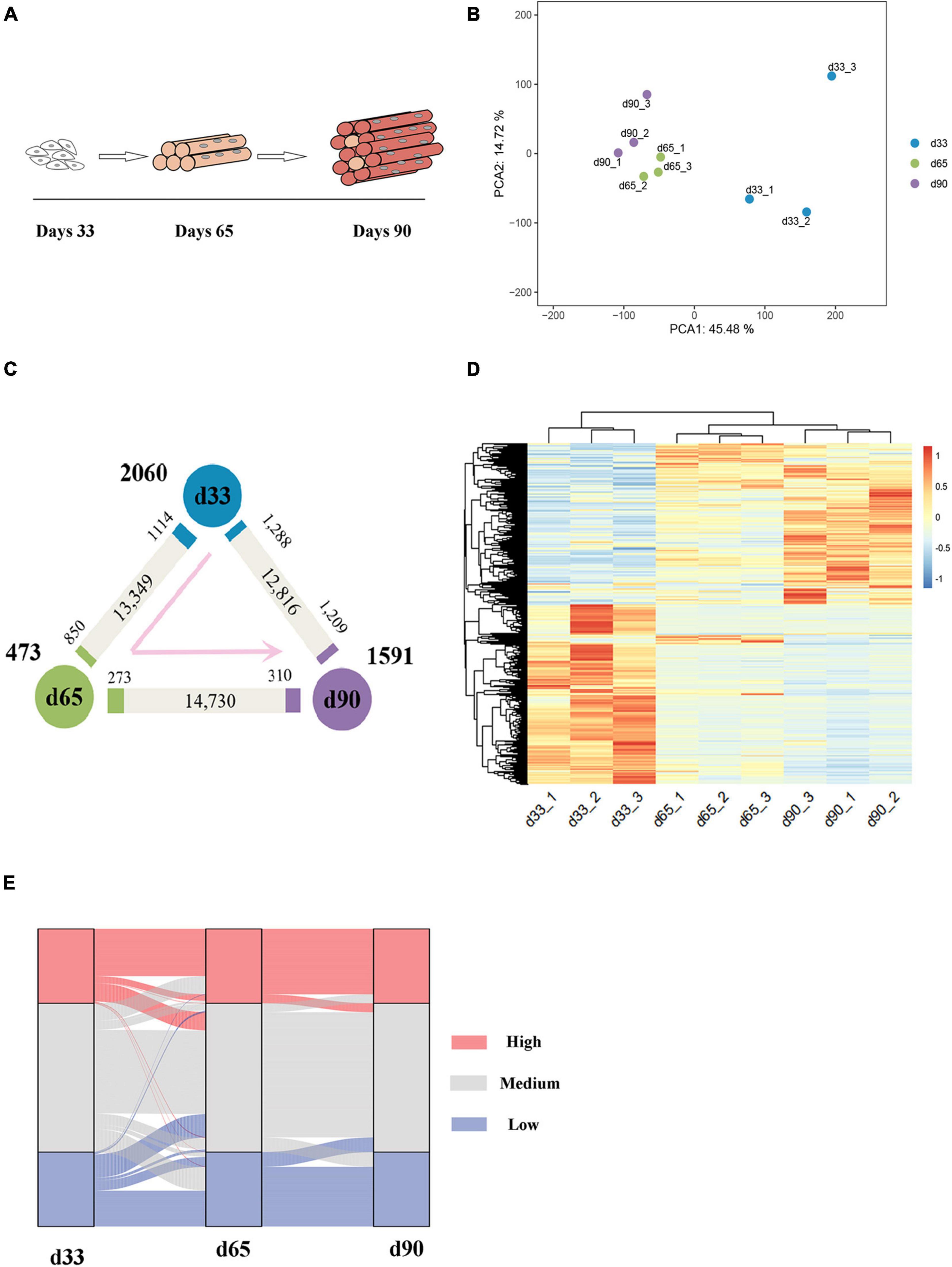
Figure 1. Gene expression dynamics during porcine embryonic muscle development. (A) Scheme for the three key stages in porcine embryonic muscle development. (B) The results of PCA on gene expression data. (C) Gene expression changes. The numbers of gene upregulated (p.adj ≤ 0.05, ≥2-fold change, marked by the corresponding colors) and unchanged (gray) at each stage compared with its neighboring stage are indicated. The numbers of genes expressed at the highest level in each stage are indicated in bold next to each stage. (D) The hierarchical clustering heatmap of DEGs. (E) The alluvial diagram of DEGs. High (TPM ≥ upper quartile); medium (lower quartile < TPM < upper quartile), low (TPM ≤ lower quartile) levels.
Functional Enrichment Analysis of Differentially Expressed Genes
We performed GO function category analysis to explore the function of DEGs (Supplementary Table S4). We revealed that the genes for neuron development, embryonic skeletal morphogenesis, and myogenic progenitor development were primarily enriched in the d33 genes, but they were depleted in the d65 genes (Figures 2A,B). By contrast, the genes, including MYOD1, MYOG, MYH7, and TNNC1, for skeletal muscle organ development, myofibril, and myoblast differentiation were prevalent among the d65 genes, which would contribute to the formation of skeletal muscle fiber (Figures 2A,B). The d65 and d90 genes were distinguished by their unique expression pattern in skeletal muscle development, metabolism, immune response, and cell–cell interaction properties. On the one hand, the d90 genes, such as PLIN5 and AK1, were specifically enriched in the multiple metabolic processes (Figures 2A,B), including ATP biosynthesis, fatty acid oxidation, and tricarboxylic acid cycle, indicating that the promoting energy metabolism activity is required for organ maturation in the late stage of embryonic development. On the other hand, the d90 genes were characterized by those genes playing important roles in the skeletal muscle development. The genes for muscle cell differentiation and primary myofibril formation were more prevalent among the d65 genes, whereas the d90 genes were enriched in the development process from primary myofibrils to second myofibrils, including myofibril assembly, myotube development, and skeletal muscle contraction (Figures 2A,B). Several important genes were selected on the basis of our analysis results for validation. The relative expression levels of genes from RT-qPCR were consistent with those obtained from transcriptome sequencing (Figure 2C). These analyses provided a clear delineation of key transcriptional properties of the three stages during embryonic muscle development.
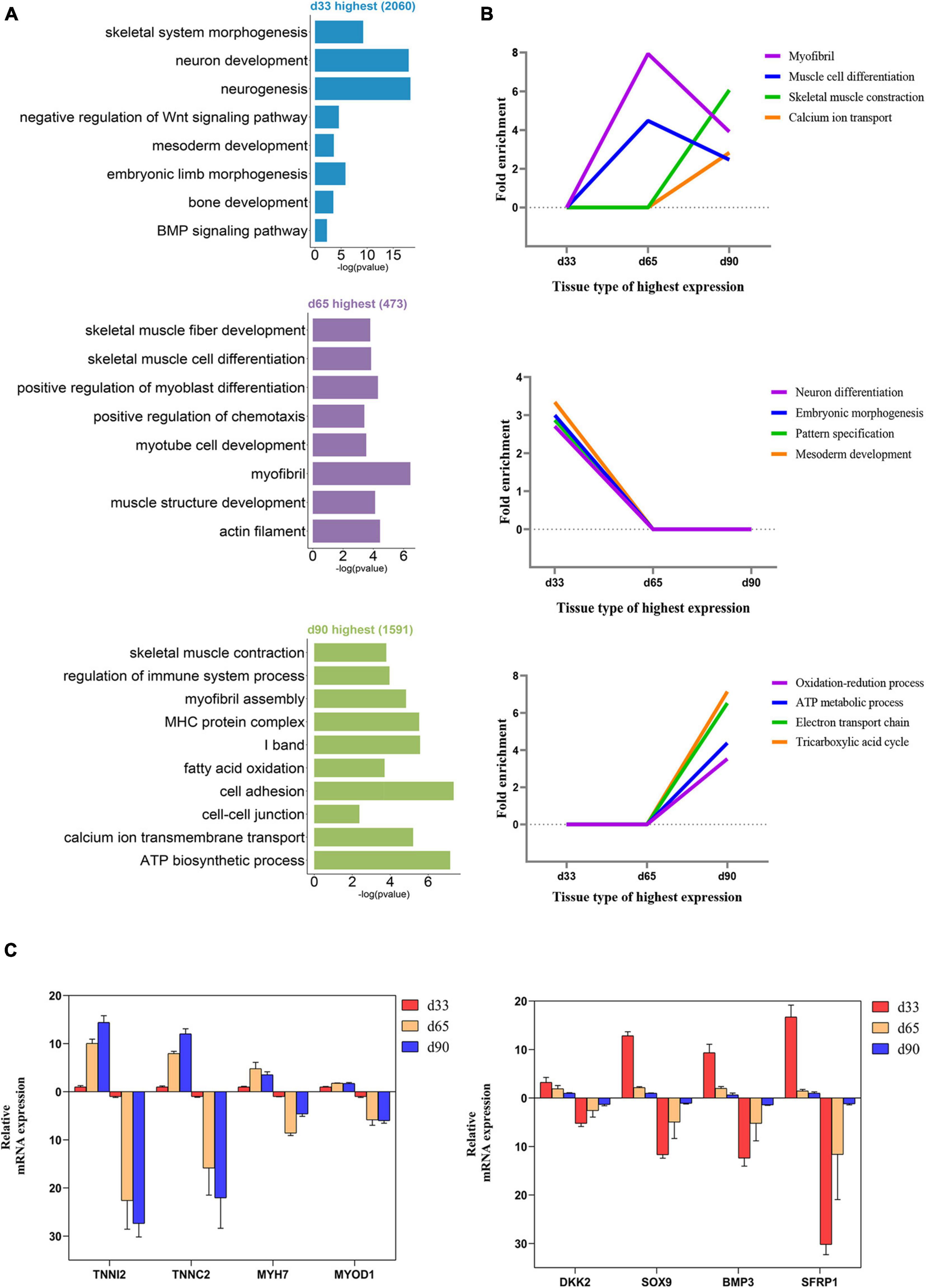
Figure 2. Functional enrichment analysis of differentially expressed genes. (A) The GO enrichment analysis of DEGs expressed at the highest level in the three stages. (B) Fold enrichment of genes for the presented GO terms. Fold enrichment is the proportion of term genes found in the DEGs list compared to the proportion of total term genes found in the background. (C) Validation of the expression of DEGs using RT-qPCR (the above of the x-axis represents the results of RT-qPCR; the below of the x-axis represents the results of RNA-seq).
Characteristic of H3K27me3 During Embryonic Development
Studies have shown that H3K27me3 plays an important role in C2C12 myogenic differentiation (Asp et al., 2011), but few studies have focused on the function of H3K27me3 in porcine embryonic muscle development. Here, we first revealed that the gradual increase in the H3K27me3 level on d33, d65, and d90 indicated that H3K27me3 might regulate the development of porcine embryonic muscle (Figure 3A). The expression levels of genes encoding the core members of PRC2 (EZH2/EED/SUZ12), PRC1(RING1/CBX/BMI1), and demethylases (KDM6B/UTX) were highly dynamic, but they showed similar change pattern during prenatal myogenesis (Supplementary Figure S2). The highest expression in d33 and the sharp decrease from d33 to d65 of H3K27me3 demethylases might contribute to the low H3K27me3 level in d33 and increased H3K27me3 level during porcine embryonic development. We further performed ChIP-seq to gain epigenomic insights into the regulatory mechanisms of H3K27me3 underlying embryonic myogenesis. We obtained >25 million reads for each sample with the above 90% mapping rate, suggesting that the sequencing results of H3K27me3 were reliable (Supplementary Table S1). The PCA results revealed the clear separation of the replicates in different stages, and two biological replicates in each stage were clustered (Figure 3B). We calculated the average signals of H3K27me3 relative to the gene promoter region. Consistent with Western blotting, meta-plot showed that the H3K27me3 level in the region from TSS to TES increased during embryonic development (Supplementary Figure S3). Enrichment heatmap showed the close enrichment level around TSS during d65 to d90 (Figure 3C). Circos plot showed that H3K27me3 was particularly more enriched on the sex chromosome (Supplementary Figure S4).
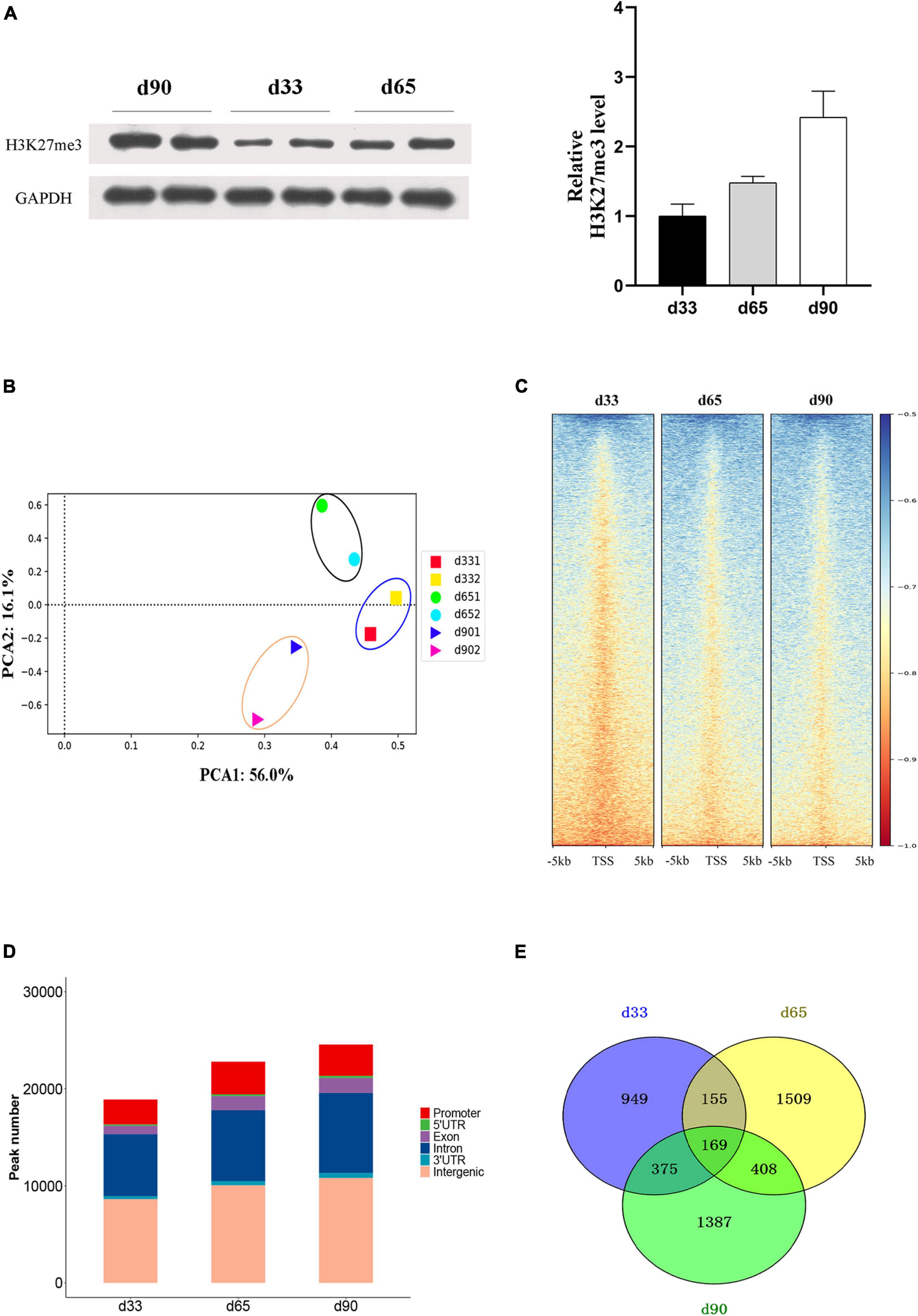
Figure 3. The characteristic of H3K27me3 during embryonic development. (A) Western blotting results showing the dynamics change of H3K27me3 in embryonic muscle development. The protein levels of H3K27me3 were quantified using ImageJ software. (B) Principal component analysis (PCA) of ChIP-seq data of H3K27me3 on d33, d65, and d90. The first and second principal components were depicted as x-axis and y-axis. (C) The enrichment heatmap of H3K27me3 around the 5 kb upstream or downstream of the TSS. (D) The genomic distribution of H3K27me3 region in three stages. (E) Numbers of shared and stage-specific genes associated with H3K27me3 peak in the promoter on d33, d65, and d90.
We performed peak calling to identify the H3K27me3 peak and characterized the distribution of H3K27me3 in different stages. A total of 18,947, 22,815, and 26,888 peaks were found on d33, d65, and d90, respectively. The distribution of genomic regions modified by H3K27me3 was classified into six regions (promoter, exon, 3′UTR, 5′UTR, intron, and intergenic; Figure 3D). Consistent with the reported distribution model, we found that the H3K27me3 peak was primarily located in the intergenic and intronic regions (Han et al., 2020). The peaks in the promoter region (about 13.9% of all peaks) were used for further analysis. We found 949, 1,509, and 1,387 specific genes with promoter modified by H3K27me3 on d33, d65, and d90, respectively (Figure 3E). Finally, a total of 4,952 genes with a significant H3K27me3 peak in the promoter were identified, covering about 32% of 15,313 expressed genes, which suggested the broad regulation of H3K27me3 for embryonic muscle development. In summary, we observed that the global level of H3K27me3 dynamically changed across embryonic development and the H3K27me3 peak was primarily located in the intergenic and intronic regions.
Association Analysis Between H3K27me3 and Gene Expression
H3K27me3 is a well-known histone modification that generally represses gene transcription. We clustered all the expressed genes into four groups based on their expression to explore the correlation between H3K27me3 and gene level during embryonic development. The enrichment profile results showed that the gene expression was negatively correlated with H3K27me3 levels around the promoter in three stages (Figure 4A). These results indicated the critical role of the H3K27me3 in repressing gene expression during embryonic development.
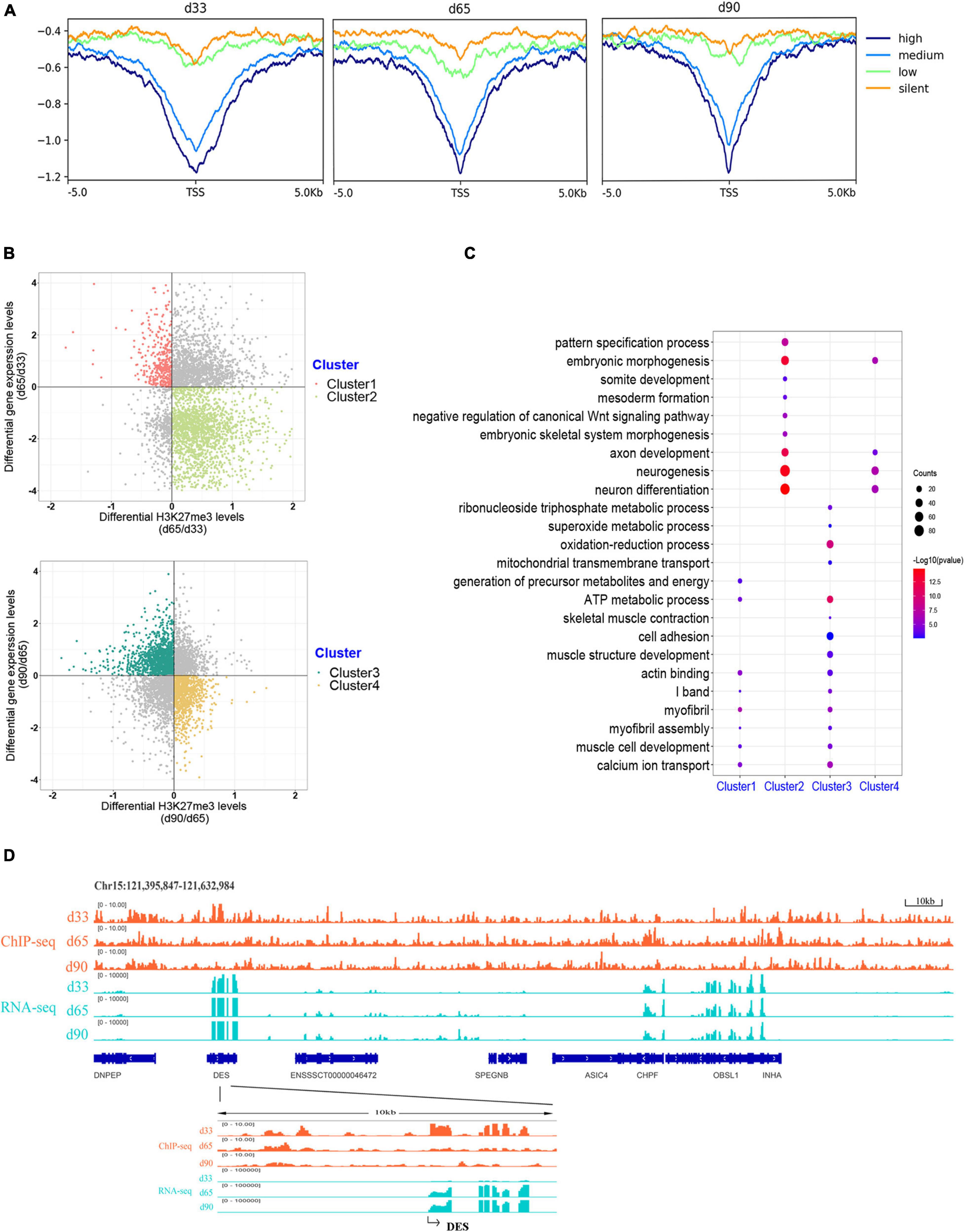
Figure 4. Association analysis between H3K27me3 and gene expression level. (A) The enrichment profile of H3K27me3 in all expressed gene. The y-axis was the average log2 fold change compared with control. High (TPM ≥ upper quartile; medium (lower quartile < TPM < upper quartile); low (TPM ≤ lower quartile); silent (never express). (B) Distribution of different expressed genes comparing the H3K27me3 levels and mRNA level. (C) The GO enrichment analysis of the DEGs exhibiting negative correlation between H3K27me3 and mRNA levels. (D) Genome browser tracks showing H3K27me3 ChIP-seq (orange) and RNA-seq (light blue) data at DES loci during embryonic muscle development. Promoter region (TSS ± 5 kb) of DES was zoomed in.
We focused on the relationship between the gene expression and corresponding H3K27me3 level of 4,124 DEGs to explore whether gene expression was affected by H3K27me3. During d33 to d65, we found 391 upregulated genes with a decreasing H3K27me3 level and 1,907 downregulated genes with an increasing H3K27me3 level (clusters 1 and 2, respectively; Figure 4B). During d65 to d90, we found 1,243 upregulated genes with a decreasing H3K27me3 level and 916 downregulated genes with an increasing H3K27me3 level (clusters 3 and 4, respectively; Figure 4B). A total of 3,373 DEGs exhibited negative correlation between H3K27me3 and mRNA level at least in one period. Furthermore, we conducted GO analysis to reveal the molecular functions of 3,373 genes (Supplementary Table S6). We primarily focused on the dynamic change in H3K27me3 in the genes closely related to muscle development. Based on the GO analysis results of cluster 1 and 3 genes, the upregulated DEGs with loss deposition of H3K27me3 were enriched on the development of myofibril and muscle contraction (e.g., MYH7, DES, MYL1, MYL3, TNNC1, and CAV3). In addition, we found that numerous metabolism-related genes, such as SLN, SLC4A1, and TPI1, were activated with decreasing H3K27me3 level (Figure 4C). For cluster 2 and 4 genes, we found that the repressed state of biological processes, including myogenic progenitor development, embryonic morphogenesis, and neuron development, was associated with the gain deposition of H3K27me3 in the gene-related promoter (Figure 4C). Among these DEGs, DES is an early myogenic marker, and it is important to maintain maturation muscle cytoarchitecture (Soglia et al., 2019). Genome browser tracks of DES clearly revealed that the coverage of H3K27me3 around the promoter significantly decreased from d33 to d65 and remained at low levels in d90, whereas the contrary tendency of changes in the mRNA expression level was observed (Figure 4D). These results demonstrated that H3K27me3 in the promoter of DEGs was dynamically changed during embryonic muscle development, which likely regulated the initiation of myogenesis and the formation and maturation of myofibers by altering the expression of DEGs.
Functional Enrichment Analysis of Genes With Promoter Modified by H3K27me3
Previous studies indicated that H3K27me3 was not strong enough to control gene expression in some cases, although a significant H3K27me3 peak occurred in genes (Sawarkar and Paro, 2010; Grossniklaus and Paro, 2014). Thus, we overlapped all the 4,952 genes modified by H3K27me3 with the 3,373 DEGs exhibiting negative correlation between H3K27me3 and mRNA levels. Consequently, 856 genes (HDEGs) were modified with H3K27me3, and their H3K27me3 level in the promoter was negatively correlated with gene expression (Supplementary Table S7). They were defined as the key H3K27me3-modified gene and used for further analysis in this study. We performed GO and KEGG analysis of HDEGs to explore the biological process regulated by H3K27me3 (Supplementary Table S8). GO analysis indicated that HDEGs were primarily enriched during muscle development, immune response, epithelial and blood development, multiple metabolic processes including lipid biosynthetic process and fatty acid metabolic process, nerve system development, and cell–cell interaction (Figure 5A). KEGG pathway analysis identified several important pathways, including Wnt signaling pathway, MAPK signaling pathway, and calcium signaling pathway (Figure 5B). Wnt signaling pathway is vital in various embryonic muscle development processes, including somitogenesis, dermomyotome specification, and myogenesis (Chal and Pourquié, 2017; Girardi and Le Grand, 2018). We found that the genes in Wnt signaling pathway, such as SFRP1, DKK2, and FZD5, were regulated by H3K27me3, suggesting the important roles of H3K27me3 in regulating muscle development. Our study identified many HDEGs and revealed the essential regulatory processes of H3K27me3 during embryonic muscle development.
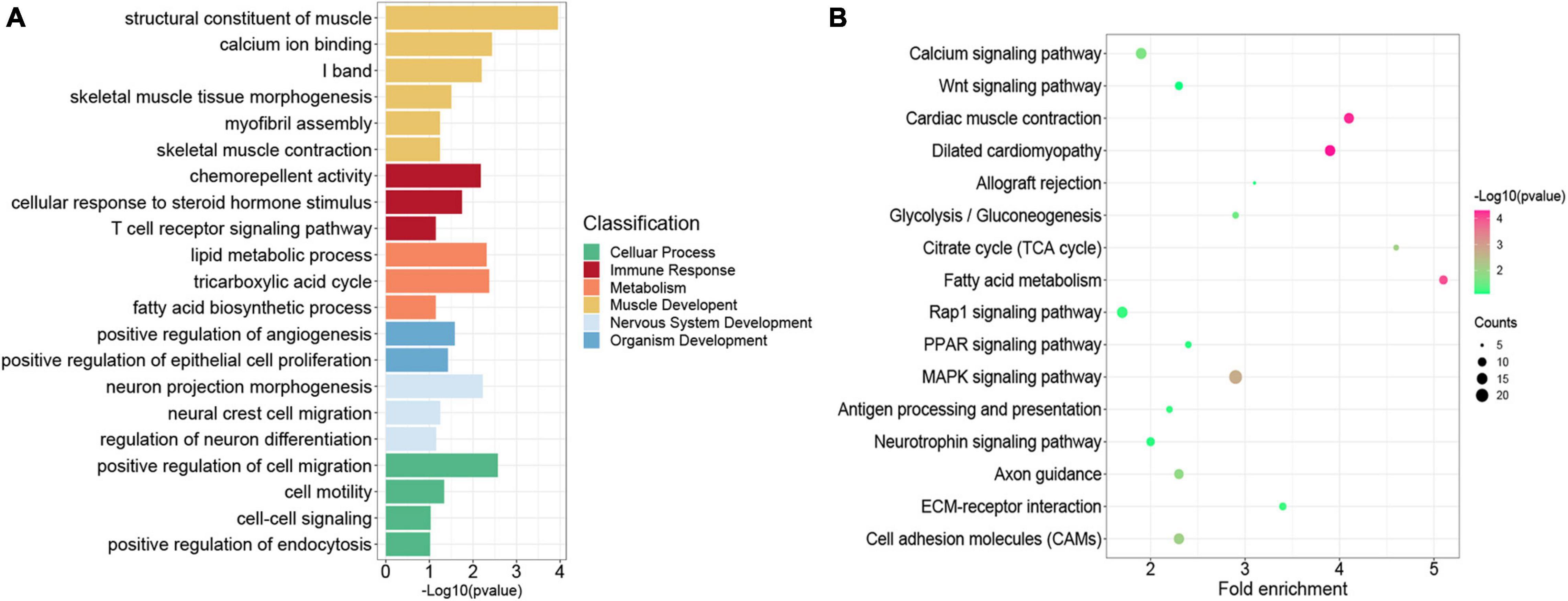
Figure 5. Functional enrichment analysis of genes modified by H3K27me3. (A) The GO enrichment analysis of the HDEGs. The colors represent the different classifications of GO terms. (B) The KEGG pathways analysis of HDEGs.
Regulation of H3K27me3 During Embryonic Muscle Development
Previous studies indicated the important roles of metabolic homeostasis, and proper calcium transport activity in skeletal muscle development (Kraft et al., 2006; Tu et al., 2016). Here, we focused on the regulation of H3K27me3 for the HDEGs related to skeletal muscle organ development, metabolism, and calcium signaling pathways. Gene set enrichment analyses (GSEA) of porcine transcriptome revealed that the expression of the genes for skeletal muscle organ development was primarily higher in d33 than in d90 (Figure 6A). Some of these genes, including MYH7, DES, LMOD2, and TCAP, were modified by H3K27me3 (Figure 6B). These genes were upregulated with decreasing H3K27me3 level, which promoted the development of embryonic muscle after d33 (Figures 6A,B). In addition, we found that calcium signaling pathway was more activated during the late stage of embryonic development, although several genes still exhibited a high mRNA expression level at early stages (Figures 6A,B). With the promotion of the expression of myogenic genes, the transcription inhibition of genes, including RAMP1, HRH1, and ATP2P2, in the calcium signaling pathway disappeared as H3K27me3 in the promoter decreased after d33 (Figure 6B). On the contrary, some genes, such as JPH4 and CEMP, were repressed after d33 (Figure 6B). These results suggested the complex regulatory mechanism of H3K27me3 presented in the regulation of calcium transport in muscle development. In addition, metabolism programming as a novel regulator of skeletal muscle development attracts lots of attention (Ryall, 2013). Thus, we analyzed the regulation of H3K27me3 in the ATP and lipid metabolic process. The GSEA results showed that the expression of the genes related to metabolism was higher in d90 than in d33, suggesting the high energy requirement in the later embryonic stages (Figure 6C). The read coverage of two typical genes, AK1 and SLC25A4, was shown in IGV (Figure 6D). Our study indicated that the regulation of H3K27me3 for embryonic muscle development cooperated with the control for the metabolism and calcium signaling pathway.
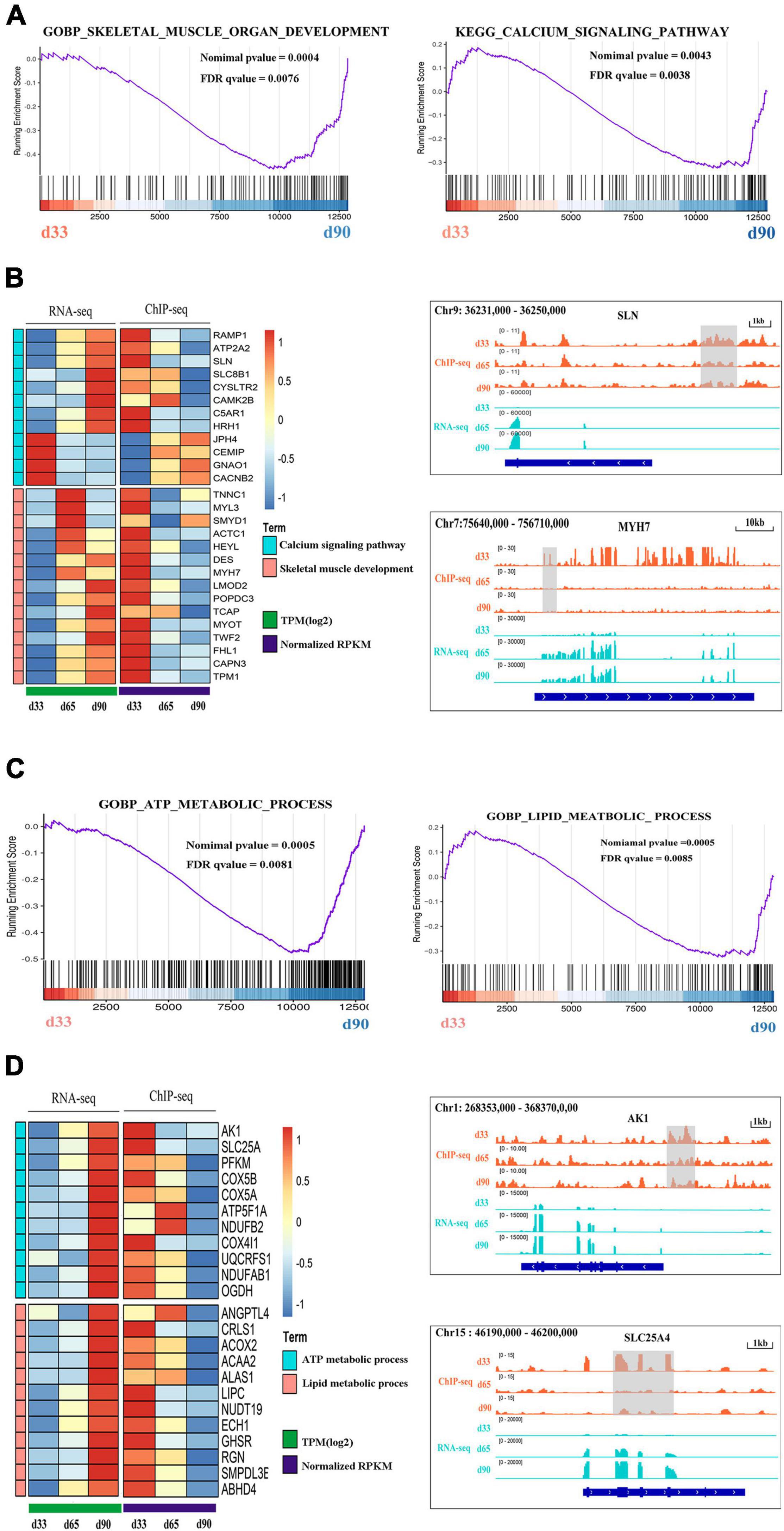
Figure 6. The regulation of H3K27me3 during embryonic muscle development. (A) GSEA plot of genes involved in the skeletal organ muscle development and calcium signaling pathway on d33 and d90. (B) The gene expression heatmap of HDEGs (left) in the skeletal organ muscle development and calcium signaling pathway. Genome browser tracks (right) showing the H3K27me3 ChIP-seq (orange) and RNA-seq (light blue) data at SLN and MYH7 loci during embryonic muscle development. H3K27me3 peak region was colored in gray. (C) GSEA plot of genes involved in the process of ATP metabolic and lipid metabolic on d33 and d90. (D) The gene expression heatmap of HDEGs (left) in the process of ATP metabolic and lipid metabolic. Genome browser tracks (right) showing the H3K27me3 ChIP-seq (orange) and RNA-seq (light blue) data at AK1 and SLC25A4 loci during embryonic muscle development. H3K27me3 peak region was colored in gray.
Exploration of H3K27me3 Function in Muscle Development in vitro
Numerous studies have demonstrated that embryonic myogenesis and regeneration in adult skeletal muscle share many transcription factors and signaling molecules (Bentzinger et al., 2012). Satellite cells are a heterogeneous population of stem and progenitor muscle cells, which are capable of self-renewal and differentiation during muscle regeneration (Wang and Rudnicki, 2011). Thus, we used porcine satellite cells (PSCs) to verify the function of H3K27me3 in vitro. Instead of blocking the expression of EZH2, GSK343 molecule can directly inhibit enzymatic activity of EZH2 to reduce H3K27me3 through the S-adenosyl-L-methionine-competitive pathway (Liu et al., 2015). We used GSK343 to diminish the modification level of H3K27me3 in PSCs during cell differentiation. Western blotting result showed a significant decrease of H3K27me3 after GSK343 treatment in PSCs (Figure 7A). Myogenic marker genes (MYOD1 and MYH4) were upregulated in 1.5d differentiated cells at mRNA and protein levels after the depletion of H3K27me3 (Figures 7A,B). We performed ChIP assay and found that MYOD1 and MYH4 were targeted by H3K27me3 following proliferation to differentiation (Figure 7C). Immunofluorescence staining of terminal differentiation marker (MYHC) revealed an increased number of MYHC+ cells, suggesting that H3K27me3 depletion could promote myogenic differentiation (Figure 7D). Collectively, these results indicated that H3K27me3 could target myogenic genes to reduce gene expression and regulate myogenic differentiation. Then, we selected several HDEGs involved in skeletal muscle development, calcium signaling pathways, and metabolism to detect the effect of H3K27me3 on these genes. WNT10B is modified by H3K27me3 and used as positive control in this study (Zhao et al., 2018). RT-qPCR results showed that genes such as TNNC1, MYL1, and DES for the positive regulation of muscle development; genes such as ABHD4 and CYP1B1 for metabolic process; and genes such as SLN and RAMP1 for calcium transport were upregulated after GSK343 treatment (Figure 7E). ChIP-qPCR revealed that H3K27me3 was significantly enriched in the promoter of these genes, and the enrichment of H3K27me3 decreased during differentiation (Figure 7F). Furthermore, the H3K27me3 level had a strong negative correlation with mRNA expression levels following proliferation to differentiation (Supplementary Figure S5). In summary, these results demonstrated the promoting differentiation of PSCs after the depletion of H3K27me3 and verified the function of H3K27me3 in the regulation of muscle development, multiple metabolism, and calcium signaling pathway.
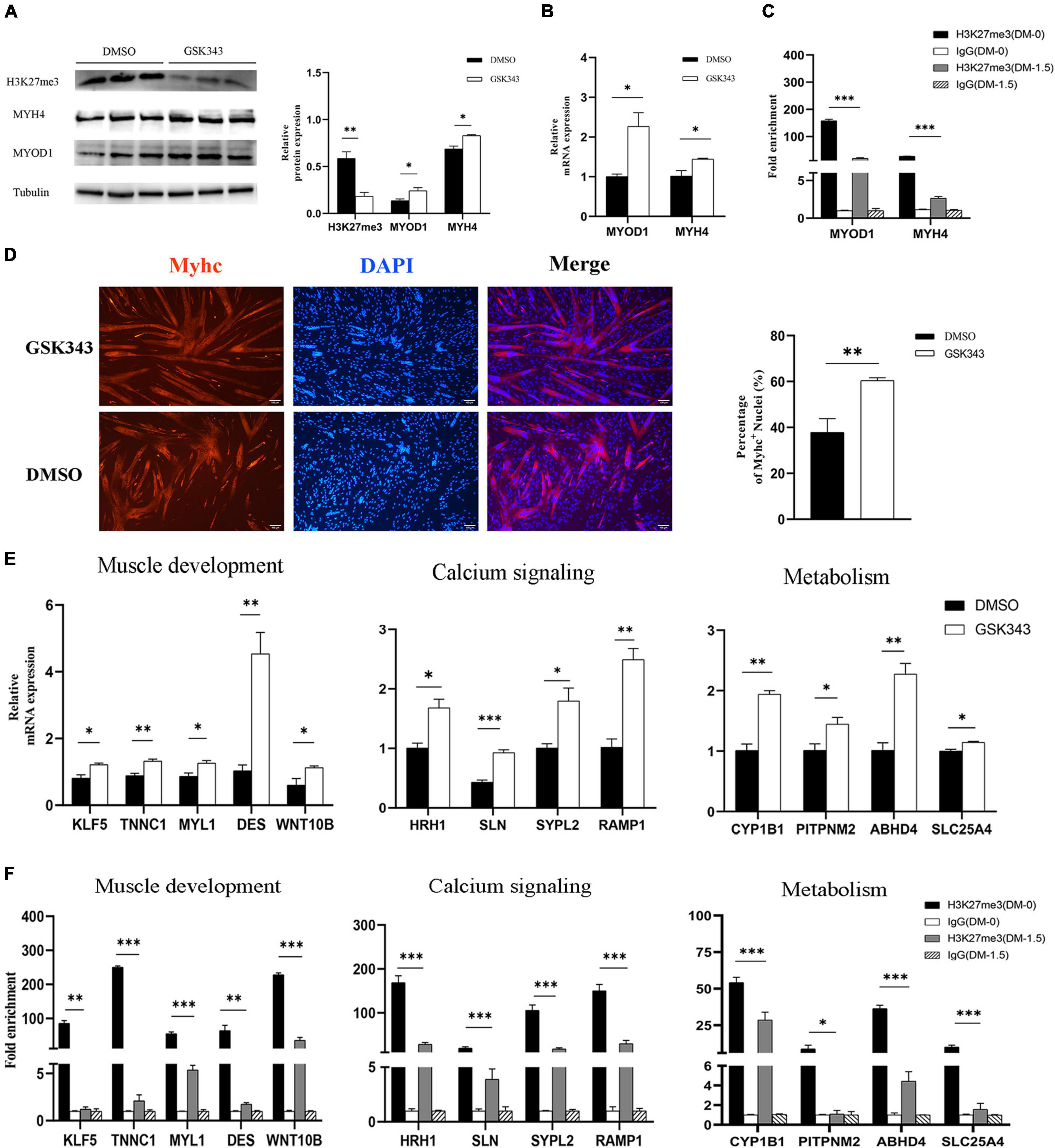
Figure 7. The exploration of H3K27me3 function in muscle development in vitro. (A) Western blotting results showing that the relative protein expression of MYOD1 and MYH4 was increased after GSK343 treatment on day 1.5 post differentiation compared with controls (0.1% DMSO). The protein levels of these genes were quantified using ImageJ software. (B) RT-qPCR results showing that the relative mRNA expression of MYOD1 and MYH4 was increased after GSK343 treatment on day 1.5 post differentiation compared with controls (0.1% DMSO). (C) ChIP-qPCR results showing that the enrichment of H3K27me3 in the MYOD1 and MYH4 were decreased following proliferation to differentiation. (D) Immunofluorescence staining of MYHC showing significant upregulation of PSCs differentiation after GSK343 treatment on day 1.5 post differentiation compared with controls (0.1% DMSO). The number of MYHC+ cells was quantified using ImageJ software. (E) RT-qPCR results showing that the mRNA expression of selected HDEGs was increased after GSK343 treatment on day 1.5 post differentiation compared with controls (0.1% DMSO). (F) ChIP-qPCR results showing that the enrichment of H3K27me3 at the promoter of selected HDEGs were decreased following proliferation to differentiation. All values represent the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns indicates not significant.
Discussion
Skeletal muscle is a highly complex and heterogeneous tissue with a multitude of functions in the organism (Bentzinger et al., 2012). Embryonic muscle development shares a common process consisting of the proliferation of mesodermal stem cells, and progressive specialization into skeletal muscle progenitors, followed by the differentiation of muscle cells and the spatial arrangement of muscle cells to form the primary musculature. Furthermore, muscle cells initially fuse into multinucleated nascent myotubes and then further fuse into myofibers (Tu et al., 2016). A broad spectrum of signaling molecules instructs myogenesis during embryonic development. In this study, we focused on the regulatory mechanism of repressive histone modification, H3K27me3, in the development of porcine embryonic muscle. We comprehensively analyzed the transcriptome and chromatin modification of muscle tissues at specific stages and revealed the essential biological processes regulated by H3K27me3.
Our study initially investigated the dynamic change in gene expression at d33, d65 and d90 of porcine embryonic muscle development. We found that myogenic progenitor development and embryonic skeletal system morphogenesis were still activated on d33. During embryonic development, Wnt signals control the expression of MRFs and participate in myogenic lineage progression (von Maltzahn et al., 2012; Pan et al., 2015). On d33, the negative regulation of Wnt signaling pathway genes, including WNT5A and DACT3, were highly expressed, which can inhibit myogenesis by decreasing the gene expression of MYOD1 (Adhikari and Davie, 2018). The MRFs (e.g., MYOD1 and MYOG) and myofibril-formation-related genes (e.g., TNNC1 and MYH7) were highly expressed on d65. This finding suggested the dynamic activation of the terminal differentiation of muscle cells, myotube fusion, and myofiber formation from d33 to d65. On d90, we found that the highly expressed genes were more enriched in myofibril assembly, muscle contraction, and associated calcium signaling pathway rather than the increasing myofibril formation and muscle differentiation. These results suggested that the period of the formation of primary muscle fibers was from d33 to d65, whereas the period of the development of second muscle fibers and muscle function was from d65 to d90 (e.g., muscle contraction). This finding was similar to that of the previous reports in pig (Picard et al., 2010; Qin et al., 2013). We observed that the changes in gene transcription from d33 to d65 were more dramatic compared with those from d65 to d90, which indicated that the regulatory mechanisms of the primary myofibers might be more complicated than those of second myofibers. We revealed the characteristics of the transcriptome on d33, d65, and d90 and demonstrated that these stages were suitable for the further investigation on the regulation of H3K27me3 for embryonic porcine muscle development.
The epigenetic mark H3K27me3 is one of the repressive histone modifications. Genome-wide mapping has revealed that H3K27me3 occupies in a large set of genes related to cell fate and embryonic development, including developmental transcription factors (such as Hox genes) and cell-surface or extracellular proteins (such as Wnt) (Boyer et al., 2006; Gao et al., 2010). Previous studies indicated the important roles of H3K27me3 in the regulation of muscle cells from proliferation to differentiation (Asp et al., 2011; Adhikari and Davie, 2018; Wang et al., 2019a). In our study, we found that the H3K27me3 level progressively increased during porcine embryonic development from 33d to 90d. The H3K27me3 levels around the TSS of all the expressed genes were negatively correlated with the gene expression levels at all stages. This result suggested that H3K27me3 might widely participate in the regulation of gene expression during embryonic muscle development. We obtained 856 DEGs, which possessed H3K27me3 enrichment peak, and the changes in the modification level were negatively correlated with gene expression. The functional enrichment analysis indicated the diverse function of these genes, implying that the effects of H3K27me3 covered a broad spectrum to modulate different physiological and developmental mechanisms during embryonic myogenesis. Previous studies pointed out that H3K27me3 always colocalized with activating histone modification H3K4me3 during embryonic development (Pan et al., 2007; Liu et al., 2016, 2019). This special modified state, consisting of two interrelated histone marks, is known as the “bivalent domain,” which could rapidly activate or inhibit gene transcription through the “winner-take-all” principle (Cui et al., 2012). In this study, 212 DEGs that were enriched by H3K27me3 but were not correlated with loss of transcript might be caused by the existence of bivalent domain in the gene body. This finding revealed the complex regulatory mechanism of H3K27me3 in governing gene expression during porcine embryonic development. We reported that muscle-fiber-formation-related genes, such as MYH7, MYL3, TNNC1, and KLF5, were modified by H3K27me3. These genes were activated to promote myofibril formation as the H3K27me3 level in the promoter decreased after d33. Kruppel-like factor 5 (KLF5), a zinc-finger transcription factor, is involved in the regulation of muscle differentiation coordinated with myogenic transcription factors such as Myod and Mef2 (Hayashi et al., 2016). Previous studies demonstrated that the Wnt antagonist SFRP1 can block MYOD1 by inhibiting the translocation of β-catenin to the nucleus (Adhikari and Davie, 2018). Our studies indicated that SFRP1 was modified by H3K27me3 and inhibited to express at d65. These results indicated that H3K27me3 could regulate muscle cell differentiation by indirectly regulating myogenic regulatory factors. In PSCs, we revealed that myogenic marker genes, MYH4 and MYOD1, were targeted by H3K27me3, which was consistent with previous studies (Adhikari and Davie, 2018; Wang et al., 2020). The depletion of H3K27me3 could promote muscle cell differentiation by increasing the expression level of myogenic marker genes and the number of MYHC+ cells. These findings were similar with the previous studies (Caretti et al., 2004; Seenundun et al., 2010; Asp et al., 2011). Therefore, these data suggested that porcine myogenesis was required for the regulation of H3K27me3 to properly control gene expression.
Many of the myogenic factors that are operated during embryonic development are recruited for the growth of mature or maturing skeletal muscles, but myogenesis and plasticity are distinguished by different intrinsic and extrinsic environments and distinct competencies of cells. As a ubiquitous intracellular signal factor that regulates a series of cellular processes, Ca2+ participates in the regulation of muscle formation, growth, and regeneration (Tu et al., 2016). The identified molecular mechanisms underlying Ca2+ participation in muscle development include the regulation of stored–operated calcium entry (Stiber et al., 2008; Seth et al., 2012) and a number of signaling elements immediately downstream of the Ca2+ signal (Lu et al., 2000a, b). Our analysis showed the important role of H3K27me3 in the regulation of calcium signaling pathways, which are consistent with observation in the alcohol use disorder and chicken skin (Chang et al., 2015; Gavin et al., 2018). We first identified several genes, such as SLN, RAMP1, and HRH1, for calcium signaling pathways were regulated by H3K27me3 during embryonic muscle development. Sarcolipin (SLN), an inhibitor of the sarco/endoplasmic reticulum (SR) calcium pump, can reduce the Ca2+ content of SR and delay muscle differentiation (Seth et al., 2012). Our study demonstrated that H3K27me3 deposition is responsible for the repressed gene expression of SLN from d33 to d65, which promotes muscle cell differentiation. Mitogen-activated protein kinases (MAPK) signaling pathway is one of the downstream signaling of Ca2+ signal, which functions in the muscle progenitor proliferation and myoblast differentiation by, respectively, regulating the gene expression of Ccnd1 and MRFs (Bennett and Tonks, 1997; Zetser et al., 2001). Based on the pathway analysis of H3K27me3, we found that MAPK signaling emerged as an overrepresented pathway, suggesting that the coordinating regulation with Ca2+ signaling in the development of skeletal muscle is also affected by H3K27me3 modification. These findings indicated the complex and broad functions of H3K27me3 in the regulation of spatiotemporal changes in Ca2+ dynamics and their signaling partners in embryonic muscle development.
Similar to all other cell types, muscle cells require energy to carry out the reactions necessary for life. The predominant source of cellular energy used to drive enzymatic reactions is the breakdown of ATP into ADP or AMP and inorganic phosphate (Ryall, 2013). ATP is primarily produced in the mitochondria via oxidative phosphorylation (OXPHOS). The two-carbon units are continuously eliminated from the β-position of fatty acyl-CoA molecule, thereby producing acetyl CoA to sustain OXPHOS and tricarboxylic acid cycle in cells during lipid oxidation (Yan et al., 2019). Our analysis found that multiple metabolic processes were more dramatic at the late stage of embryonic development, which were characterized by the high expression of genes for ATP metabolism and lipid metabolism. These results were associated with the fact that the density of mitochondria, and oxidative phosphorylation activity in muscle tissue dramatically increased following differentiation and fusion into myotube (Kraft et al., 2006). Few studies have focused on the relationships between H3K27me3 and metabolism during embryonic myogenesis. Our study found that H3K27me3 played complex and broad roles in the regulation of H3K27me3 in multiple metabolic processes and pathways. We identified that numerous metabolism-related genes, including ABHD4 and PORCN for lipid metabolic process and AK1 and SLC25A4 for ATP metabolic process, were regulated by H3K27me3. SLC25A4 is a mitochondrial ADP/ATP carrier, which is responsible for the ATP export from the mitochondria to the cytoplasm and provides energy to cells (Clémençon et al., 2013). We observed that the gene expression of SLC25A4 was inhibited by H3K27me3 on d33 and upregulated in contracted muscle fiber to satisfy the high-energy-consuming reactions. In addition, our study showed that H3K27me3 was marked in the genes for glycolysis that is necessary for the differentiation of C2C12 cells (Fulco et al., 2008). Therefore, H3K27me3 could control the intensity of metabolism activity by regulating the proper transcript programming of metabolism-related genes during embryonic muscle development.
Conclusion
In our study, we provided the first genome-wide profiling of H3K27me3 during porcine embryonic muscle development and revealed its important role in the calcium signaling pathway and metabolism coordinated with the regulation of myogenesis. This study could serve as the basis for analyzing not only animal breeding, human muscle disease, and embryonic development but also the regulatory mechanism of H3K27me3.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/, PRJNA556496 and https://www.ncbi.nlm.nih.gov/sra/, PRJNA741351.
Ethics Statement
The animal study was reviewed and approved by Animal Care Committee of South China Agricultural University, Guangzhou, China.
Author Contributions
TG, SW, and BT conceived and designed the research and wrote and revised the manuscript. BT and SW conducted the sequencing data analysis. BT, JZ, and SSW performed the experiments. TG, LH, ZL, JY, and GC collected the samples. EZ administered the project. TG and ZW applied for the funding. All authors have read and approved the final submitted manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (31802036) and the Natural Science Foundation of Guangdong Province, China (2019B1515210014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Guangdong Wen’s pig breeding company in China for providing the experiment samples in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.739321/full#supplementary-material
References
Abo Alrob, O., and Lopaschuk, G. D. (2014). Role of CoA and acetyl-CoA in regulating cardiac fatty acid and glucose oxidation. Biochem. Soc. Trans. 42, 1043–1051. doi: 10.1042/BST20140094
Adhikari, A., and Davie, J. (2018). JARID2 and the PRC2 complex regulate skeletal muscle differentiation through regulation of canonical Wnt signaling. Epigenetics Chromatin 11:46. doi: 10.1186/s13072-018-0217-x
Adhikari, A., Mainali, P., and Davie, J. K. (2019). JARID2 and the PRC2 complex regulate the cell cycle in skeletal muscle. J. Biol. Chem. 294, 19451–19464. doi: 10.1074/jbc.RA119.010060
Ali, A., Murani, E., Hadlich, F., Liu, X., Wimmers, K., and Ponsuksili, S. (2021). Prenatal skeletal muscle transcriptome analysis reveals novel MicroRNA-mRNA networks associated with intrauterine growth restriction in pigs. Cells 10:1007. doi: 10.3390/cells10051007
Asp, P., Blum, R., Vethantham, V., Parisi, F., Micsinai, M., Cheng, J., et al. (2011). Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 108, E149–E158. doi: 10.1073/pnas.1102223108
Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Schones, D. E., Wang, Z., et al. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837.
Bauer, T., Trump, S., Ishaque, N., Thürmann, L., Gu, L., Bauer, M., et al. (2016). Environment-induced epigenetic reprogramming in genomic regulatory elements in smoking mothers and their children. Mol. Syst. Biol. 12:861. doi: 10.15252/msb.20156520
Begue, G., Raue, U., Jemiolo, B., and Trappe, S. (2017). DNA methylation assessment from human slow- and fast-twitch skeletal muscle fibers. J. Appl. Physiol. (1985) 122, 952–967. doi: 10.1152/japplphysiol.00867.2016
Bennett, A. M., and Tonks, N. K. (1997). Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science (New York, N.Y.) 278, 1288–1291.
Bentzinger, C. F., Wang, Y. X., and Rudnicki, M. A. (2012). Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4:a008342. doi: 10.1101/cshperspect.a008342
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353.
Byrne, K., McWilliam, S., Vuocolo, T., Gondro, C., Cockett, N. E., and Tellam, R. L. (2014). Genomic architecture of histone 3 lysine 27 trimethylation during late ovine skeletal muscle development. Anim. Genet. 45, 427–438. doi: 10.1111/age.12145
Caretti, G., Di Padova, M., Micales, B., Lyons, G. E., and Sartorelli, V. (2004). The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18, 2627–2638.
Chal, J., and Pourquié, O. (2017). Making muscle: skeletal myogenesis and. Development (Cambridge, England) 144, 2104–2122. doi: 10.1242/dev.151035
Chang, K.-W., Huang, N. A., Liu, I. H., Wang, Y.-H., Wu, P., Tseng, Y.-T., et al. (2015). Emergence of differentially regulated pathways associated with the development of regional specificity in chicken skin. BMC Genomics 16:22. doi: 10.1186/s12864-014-1202-9
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Clémençon, B., Babot, M., and Trézéguet, V. (2013). The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction. Mol. Aspects Med. 34, 485–493. doi: 10.1016/j.mam.2012.05.006
Cui, P., Liu, W., Zhao, Y., Lin, Q., Zhang, D., Ding, F., et al. (2012). Comparative analyses of H3K4 and H3K27 trimethylations between the mouse cerebrum and testis. Genomics Proteomics Bioinformatics 10, 82–93. doi: 10.1016/j.gpb.2012.05.007
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10:giab008. doi: 10.1093/gigascience/giab008
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Faralli, H., Wang, C., Nakka, K., Benyoucef, A., Sebastian, S., Zhuang, L., et al. (2016). UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 126, 1555–1565. doi: 10.1172/JCI83239
Fulco, M., Cen, Y., Zhao, P., Hoffman, E. P., McBurney, M. W., Sauve, A. A., et al. (2008). Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 14, 661–673. doi: 10.1016/j.devcel.2008.02.004
Gao, Y., Hyttel, P., and Hall, V. J. (2010). Regulation of H3K27me3 and H3K4me3 during early porcine embryonic development. Mol. Reprod. Dev. 77, 540–549. doi: 10.1002/mrd.21180
Gavin, D. P., Hashimoto, J. G., Lazar, N. H., Carbone, L., Crabbe, J. C., and Guizzetti, M. (2018). Stable histone methylation changes at proteoglycan network genes following ethanol exposure. Front. Genet. 9:346. doi: 10.3389/fgene.2018.00346
Girardi, F., and Le Grand, F. (2018). Wnt signaling in skeletal muscle development and regeneration. Prog. Mol. Biol. Transl. Sci. 153, 157–179. doi: 10.1016/bs.pmbts.2017.11.026
Grossniklaus, U., and Paro, R. (2014). Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 6:a019331. doi: 10.1101/cshperspect.a019331
Gu, L., Frommel, S. C., Oakes, C. C., Simon, R., Grupp, K., Gerig, C. Y., et al. (2015). BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet. 47, 22–30. doi: 10.1038/ng.3165
Han, B., Xu, W., Ahmed, N., Yu, A., Wang, Z., and Liu, A. (2020). Changes and associations of genomic transcription and histone methylation with salt stress in castor bean. Plant Cell Physiol. 61, 1120–1133. doi: 10.1093/pcp/pcaa037
Hayashi, S., Manabe, I., Suzuki, Y., Relaix, F., and Oishi, Y. (2016). Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. eLife 5:e17462. doi: 10.7554/eLife.17462
Heerboth, S., Lapinska, K., Snyder, N., Leary, M., Rollinson, S., and Sarkar, S. (2014). Use of epigenetic drugs in disease: an overview. Genet. Epigenetics 6, 9–19. doi: 10.4137/GEG.S12270
Jin, J. J., Lv, W., Xia, P., Xu, Z. Y., Zheng, A. D., Wang, X. J., et al. (2018). Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. U.S.A. 115, E9802–E9811. doi: 10.1073/pnas.1801471115
Juan, A. H., Wang, S., Ko, K. D., Zare, H., Tsai, P.-F., Feng, X., et al. (2016). Roles of H3K27me2 and H3K27me3 examined during fate specification of embryonic stem cells. Cell Rep. 17, 1369–1382. doi: 10.1016/j.celrep.2016.09.087
Kraft, C. S., LeMoine, C. M. R., Lyons, C. N., Michaud, D., Mueller, C. R., and Moyes, C. D. (2006). Control of mitochondrial biogenesis during myogenesis. Am. J. Physiol. Cell Physiol. 290, C1119–C1127.
Kudou, K., Komatsu, T., Nogami, J., Maehara, K., Harada, A., Saeki, H., et al. (2017). The requirement of Mettl3-promoted mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol. 7:170119. doi: 10.1098/rsob.170119
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] arXiv: 1303.3997,
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, J., An, L., Wang, J., Liu, Z., Dai, Y., Liu, Y., et al. (2019). Dynamic patterns of H3K4me3, H3K27me3, and Nanog during rabbit embryo development. Am. J. Transl. Res. 11, 430–441.
Liu, L., Cheung, T. H., Charville, G. W., Hurgo, B. M. C., Leavitt, T., Shih, J., et al. (2013). Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204. doi: 10.1016/j.celrep.2013.05.043
Liu, T.-P., Lo, H.-L., Wei, L.-S., Hsiao, H. H.-Y., and Yang, P.-M. (2015). S-Adenosyl-L-methionine-competitive inhibitors of the histone methyltransferase EZH2 induce autophagy and enhance drug sensitivity in cancer cells. Anti Cancer Drugs 26, 139–147. doi: 10.1097/CAD.0000000000000166
Liu, X., Wang, C., Liu, W., Li, J., Li, C., Kou, X., et al. (2016). Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562. doi: 10.1038/nature19362
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Lu, J., McKinsey, T. A., Nicol, R. L., and Olson, E. N. (2000a). Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 4070–4075.
Lu, J., McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2000b). Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6, 233–244.
Pabst, R. (2020). The pig as a model for immunology research. Cell Tissue Res. 380, 287–304. doi: 10.1007/s00441-020-03206-9
Pan, G., Tian, S., Nie, J., Yang, C., Ruotti, V., Wei, H., et al. (2007). Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1, 299–312. doi: 10.1016/j.stem.2007.08.003
Pan, Y. C., Wang, X. W., Teng, H. F., Wu, Y. J., Chang, H. C., and Chen, S. L. (2015). Wnt3a signal pathways activate MyoD expression by targeting cis-elements inside and outside its distal enhancer. Biosci. Rep. 35:e00180. doi: 10.1042/BSR20140177
Picard, B., Berri, C., Lefaucheur, L., Molette, C., Sayd, T., and Terlouw, C. (2010). Skeletal muscle proteomics in livestock production. Brief. Funct. Genomics 9, 259–278. doi: 10.1093/bfgp/elq005
Qin, J., Sun, Y., Liu, S., Zhao, R., Zhang, Q., and Pang, W. (2019). MicroRNA-323-3p promotes myogenesis by targeting Smad2. J. Cell. Biochem. 120, 18751–18761. doi: 10.1002/jcb.29187
Qin, L., Chen, Y., Liu, X., Ye, S., Yu, K., Huang, Z., et al. (2013). Integrative analysis of porcine microRNAome during skeletal muscle development. PLoS One 8:e72418. doi: 10.1371/journal.pone.0072418
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A., and Manke, T. (2014). deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191. doi: 10.1093/nar/gku365
Rehfeldt, C., Henning, M., and Fiedler, I. (2008). Consequences of pig domestication for skeletal muscle growth and cellularity. Livest. Sci. 116, 30–41.
Ryall, J. G. (2013). Metabolic reprogramming as a novel regulator of skeletal muscle development and regeneration. FEBS J. 280, 4004–4013. doi: 10.1111/febs.12189
Sawarkar, R., and Paro, R. (2010). Interpretation of developmental signaling at chromatin: the polycomb perspective. Dev. Cell 19, 651–661. doi: 10.1016/j.devcel.2010.10.012
Seenundun, S., Rampalli, S., Liu, Q.-C., Aziz, A., Palii, C., Hong, S., et al. (2010). UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 29, 1401–1411. doi: 10.1038/emboj.2010.37
Seth, M., Li, T., Graham, V., Burch, J., Finch, E., Stiber, J. A., et al. (2012). Dynamic regulation of sarcoplasmic reticulum Ca(2+) stores by stromal interaction molecule 1 and sarcolipin during muscle differentiation. Dev. Dyn. 241, 639–647. doi: 10.1002/dvdy.23760
Soglia, F., Mazzoni, M., Zappaterra, M., Di Nunzio, M., Babini, E., Bordini, M., et al. (2019). Distribution and expression of vimentin and desmin in broiler affected by the growth-related muscular abnormalities. Front. Physiol. 10:1581. doi: 10.3389/fphys.2019.01581
Stiber, J., Hawkins, A., Zhang, Z.-S., Wang, S., Burch, J., Graham, V., et al. (2008). STIM1 signalling controls store-operated calcium entry required for development and contractile function in skeletal muscle. Nat. Cell Biol. 10, 688–697. doi: 10.1038/ncb1731
Stovner, E. B., and Sætrom, P. (2019). epic2 efficiently finds diffuse domains in ChIP-seq data. Bioinformatics (Oxford, England) 35, 4392–4393. doi: 10.1093/bioinformatics/btz232
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Tan, B., Hong, L., Qiao, J., Zhou, J., Xing, P., Yan, G., et al. (2020). Identification and expression pattern of EZH2 in pig developing fetuses. BioMed Res. Int. 2020:5315930. doi: 10.1155/2020/5315930
Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics 14, 178–192. doi: 10.1093/bib/bbs017
Tu, M. K., Levin, J. B., Hamilton, A. M., and Borodinsky, L. N. (2016). Calcium signaling in skeletal muscle development, maintenance and regeneration. Cell Calcium 59, 91–97. doi: 10.1016/j.ceca.2016.02.005
von Maltzahn, J., Chang, N. C., Bentzinger, C. F., and Rudnicki, M. A. (2012). Wnt signaling in myogenesis. Trends Cell Biol. 22, 602–609. doi: 10.1016/j.tcb.2012.07.008
Wang, Q., Gu, L., Adey, A., Radlwimmer, B., Wang, W., Hovestadt, V., et al. (2013). Tagmentation-based whole-genome bisulfite sequencing. Nat. Protoc. 8, 2022–2032. doi: 10.1038/nprot.2013.118
Wang, Y., Li, M., Wang, Y., Liu, J., Zhang, M., Fang, X., et al. (2019c). A Zfp609 circular RNA regulates myoblast differentiation by sponging miR-194-5p. Int. J. Biol. Macromol. 121, 1308–1313. doi: 10.1016/j.ijbiomac.2018.09.039
Wang, S., Sun, Y., Ren, R., Xie, J., Tian, X., Zhao, S., et al. (2019a). H3K27me3 depletion during differentiation promotes myogenic transcription in porcine satellite cells. Genes 10:231. doi: 10.3390/genes10030231
Wang, S., Zuo, H., Jin, J., Lv, W., Xu, Z., Fan, Y., et al. (2019b). Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 10:505. doi: 10.1038/s41419-019-1742-7
Wang, S., Xu, X., Liu, Y., Jin, J., Zhu, F., Bai, W., et al. (2020). RIP-Seq of EZH2 identifies as a regulator of myogenesis in pigs. Front. Cell Dev. Biol. 8:618617. doi: 10.3389/fcell.2020.618617
Wang, Y. X., and Rudnicki, M. A. (2011). Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 13, 127–133. doi: 10.1038/nrm3265
Woodhouse, S., Pugazhendhi, D., Brien, P., and Pell, J. M. (2013). Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J. Cell Sci. 126(Pt 2) 565–579. doi: 10.1242/jcs.114843
Yan, D., Adeshakin, A. O., Xu, M., Afolabi, L. O., Zhang, G., Chen, Y. H., et al. (2019). Lipid metabolic pathways confer the immunosuppressive function of myeloid-derived suppressor cells in tumor. Front. Immunol. 10:1399. doi: 10.3389/fimmu.2019.01399
Yonamine, C. Y., Alves-Wagner, A. B., Esteves, J. V., Okamoto, M. M., Correa-Giannella, M. L., Giannella-Neto, D., et al. (2019). Diabetes induces tri-methylation at lysine 9 of histone 3at Slc2a4 gene in skeletal muscle: a new target to improve glycemic control. Mol. Cell Endocrinol. 481, 26–34. doi: 10.1016/j.mce.2018.11.006
Yu, G., Wang, L.-G., and He, Q.-Y. (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics (Oxford, England) 31, 2382–2383. doi: 10.1093/bioinformatics/btv145
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. doi: 10.1089/omi.2011.0118
Yue, J., Hou, X., Liu, X., Wang, L., Gao, H., Zhao, F., et al. (2021). The landscape of chromatin accessibility in skeletal muscle during embryonic development in pigs. J. Anim. Sci. Biotechnol. 12:56. doi: 10.1186/s40104-021-00577-z
Zetser, A., Frank, D., and Bengal, E. (2001). MAP kinase converts MyoD into an instructive muscle differentiation factor in Xenopus. Dev. Biol. 240, 168–181.
Keywords: H3K27me3, skeletal muscle, embryonic development, pig, ChIP-seq
Citation: Tan B, Wang S, Wang S, Zeng J, Hong L, Li Z, Yang J, Cai G, Zheng E, Wu Z and Gu T (2021) Genome-Wide Analysis of H3K27me3 in Porcine Embryonic Muscle Development. Front. Cell Dev. Biol. 9:739321. doi: 10.3389/fcell.2021.739321
Received: 10 July 2021; Accepted: 11 October 2021;
Published: 05 November 2021.
Edited by:
Kyoko Yokomori, University of California, Irvine, United StatesReviewed by:
Lei Gu, Max Planck Institute for Heart and Lung Research, GermanyKrishna Mohan Parsi, University of Massachusetts Medical School, United States
Copyright © 2021 Tan, Wang, Wang, Zeng, Hong, Li, Yang, Cai, Zheng, Wu and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenfang Wu, wzf@scau.edu.cn; Ting Gu, tinggu@scau.edu.cn
†These authors have contributed equally to this work and share first authorship
 Baohua Tan
Baohua Tan Sheng Wang3†
Sheng Wang3†  Linjun Hong
Linjun Hong Zicong Li
Zicong Li Jie Yang
Jie Yang Ting Gu
Ting Gu