Extracellular Matrix Composition and Remodeling: Current Perspectives on Secondary Palate Formation, Cleft Lip/Palate, and Palatal Reconstruction
- 1Laboratory of Extracellular Matrix Biology and Cellular Interaction, Department of Anatomy, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
- 2Clinical Research Laboratory in Dentistry, Federal Fluminense University, Niterói, Brazil
- 3Directory of Life Sciences Applied Metrology, National Institute of Metrology, Quality and Technology, Duque de Caxias, Brazil
- 4Center for Craniofacial Research, UTHealth School of Dentistry at Houston, Houston, TX, United States
- 5Pediatric Research Center, UTHealth McGovern Medical School, Houston, TX, United States
- 6Department of Diagnostic and Biomedical Sciences, UTHealth School of Dentistry at Houston, Houston, TX, United States
Craniofacial development comprises a complex process in humans in which failures or disturbances frequently lead to congenital anomalies. Cleft lip with/without palate (CL/P) is a common congenital anomaly that occurs due to variations in craniofacial development genes, and may occur as part of a syndrome, or more commonly in isolated forms (non-syndromic). The etiology of CL/P is multifactorial with genes, environmental factors, and their potential interactions contributing to the condition. Rehabilitation of CL/P patients requires a multidisciplinary team to perform the multiple surgical, dental, and psychological interventions required throughout the patient’s life. Despite progress, lip/palatal reconstruction is still a major treatment challenge. Genetic mutations and polymorphisms in several genes, including extracellular matrix (ECM) genes, soluble factors, and enzymes responsible for ECM remodeling (e.g., metalloproteinases), have been suggested to play a role in the etiology of CL/P; hence, these may be considered likely targets for the development of new preventive and/or therapeutic strategies. In this context, investigations are being conducted on new therapeutic approaches based on tissue bioengineering, associating stem cells with biomaterials, signaling molecules, and innovative technologies. In this review, we discuss the role of genes involved in ECM composition and remodeling during secondary palate formation and pathogenesis and genetic etiology of CL/P. We also discuss potential therapeutic approaches using bioactive molecules and principles of tissue bioengineering for state-of-the-art CL/P repair and palatal reconstruction.
Introduction
The first studies on palate development and cleft lip/palate (CL/P) date back to the beginning of the 20th century (Whitehead, 1902; Fawcett, 1906; Tweedie, 1910). These studies were fundamental to our understanding of the molecular and cellular aspects that drive palate formation, that when disrupted, may explain the occurrence of CL/P. CL/P is the most common craniofacial anomaly occurring in approximately 1 in 700 live births, and representing a substantial burden worldwide (Shaw, 2004; Massenburg et al., 2016). The treatment of this disorder is complex and demands a multiplicity of health professionals to perform numerous interventions throughout the patient’s life (Kantar et al., 2019). Besides the high cost of treatment, CL/P imposes a significant impact on the quality of life of affected children and their families (Macho et al., 2017; Sundell et al., 2017). The primary treatment for CL/P repair is surgical correction, frequently including autologous bone grafts from the iliac crest to repair the palatal bone defect. This increases hospitalization time, pain, and donor site morbidity; hence, new strategies for the use of regenerative therapies and bone graft substitutes are needed to reduce the morbidity associated with the condition and improve treatment outcomes and patient’s quality of life (Sharif et al., 2016). Further, the identification of key factors involved in the etiology of CL/P may provide the foundation for the development of bioactive molecules and precision therapy approaches for CL/P.
Extracellular matrix (ECM) genes, soluble factors, and enzymes responsible for ECM remodeling (e.g., metalloproteinases) are expressed during lip and palate development and suggested to play a role in the etiology of CL/P. However, a comprehensive evaluation of ECM dynamics during palatogenesis is still fragmented. Historically, the ECM was considered to be the scaffold that provided an adequate architecture for tissue structure. Subsequently, knowledge of the soluble factors secreted by the cells into the ECM and its function as storage site for the rapid bioavailability of several molecules demonstrated the role of ECM as a crucial component of the cellular microenvironment (Ricard-Blum and Vallet, 2016, 2019). An intricate balance between proteases that degrade the ECM components and their inhibitors maintain the ECM homeostasis. Abnormal ECM remodeling (excessive or inefficient) is often involved in the development and progression of several pathologies and conditions, including CL/P.
This review focuses on the role of genes involved in ECM composition and remodeling during secondary palate formation and with regard to the genetic etiology of CL/P. It also presents an overview of therapeutic approaches using bioactive molecules and principles of tissue bioengineering for state-of-the-art CL/P repair and palatal reconstruction.
Craniofacial Development and Palatogenesis
Embryonic development is a precise temporal and spatial multistep process that is coordinated by specific molecular patterns, cell–cell and reciprocal cell–ECM interactions from the totipotent stem cell up to a fully developed organism. The vertebrate craniofacial complex arises from three embryonic tissue layers (endoderm, mesoderm, and ectoderm) and multipotent migrating neural crest cells (NCCs), also known as the “fourth layer.” NCCs are a population of epithelial cells within the neural tube, which migrate and then undergo epithelial–mesenchymal transition (EMT) prior to neural tube closure, delaminating from the neuroepithelium, and migrating toward the growing swellings. NCCs contribute to neural structures found in the whole vertebrate body and ectomesenchyme of the head and neck and originate the mesenchymal/stromal stem cells (MSCs)/progenitors that will differentiate into the dermis, skeletal, and connective tissues of the face and the neck, being the primary source of mesenchymal tissue in this region. They are also responsible for the bones and cartilage of the head and neck, while the trunk and appendicular members come from the mesoderm. Craniofacial development is one of the most complex processes in an organism and one-third of birth defects arise from errors in this process, causing significant infant mortality (Weston and Thiery, 2015; Francis-west and Crespo-Enriquez, 2016; Liu and Cheung, 2016; Dupin et al., 2018; Pla and Monsoro-Burq, 2018; Rothstein et al., 2018).
Lip and palate formation occurs in a series of coordinated steps, which take place between the fourth and 12th gestational week (GW) in humans and between the 11th and 15.5th embryonic day (ED) in mice. Facial development begins by frontonasal (central), maxillary, and mandibular (laterals) prominences growing around the primitive oral cavity, called the stomodeum, to give rise to the face. NCCs from distinct sites of the developing brain, such as the midbrain and forebrain cells (frontonasal), midbrain and hindbrain cells (maxillary), where the mix of the midbrain and hindbrain cells and mesenchyme from the first pharyngeal arch (mandibular) enrich these prominences (Jankowski and Márquez, 2016).
The frontonasal prominence is the most fundamental structure for external nose and the primary palate formation. Between the fifth–seventh GW and 10.0th–11.0th ED, it expands from two ectoderm nasal or olfactory placodes which each enlarge to divide into the nasomedial and nasolateral processes. The nasomedial processes grow downward and merge to originate the globular or intermaxillary process, which will form the philtrum of the upper lip and primary palate. Anatomically, the primary palate is anterior to the incisive foramen and contains the maxillary incisors (Jankowski and Márquez, 2016).
The maxillary prominences drive the formation of the upper part of the face, lip, upper jaw (maxillae), and the secondary palate. The mandibular prominences originate the lower part of the face, lip, and lower jaw (mandible). Briefly, the secondary palate develops from two mesenchymal projections (palatal shelves) derived from maxillary prominences extending inferiorly and bilaterally to the tongue (Figure 1A). With the progressive development of the mandible, these projections assume a horizontal position above the tongue (Figure 1B; Jankowski and Márquez, 2016). Subsequently, the adhesion of the epithelium from both palatal shelves forms a single line, called medial edge epithelia (MEE), which must disappear to allow for palatal tissue confluence and fusion (Figure 1C). Programmed cell death, EMT, and cell migration from the oral to the nasal epithelia, or a combination of these events have been suggested as plausible mechanisms for MEE disintegration, albeit this remains a controversial issue (Figure 1D; Ray and Niswander, 2012; Hammond et al., 2017).
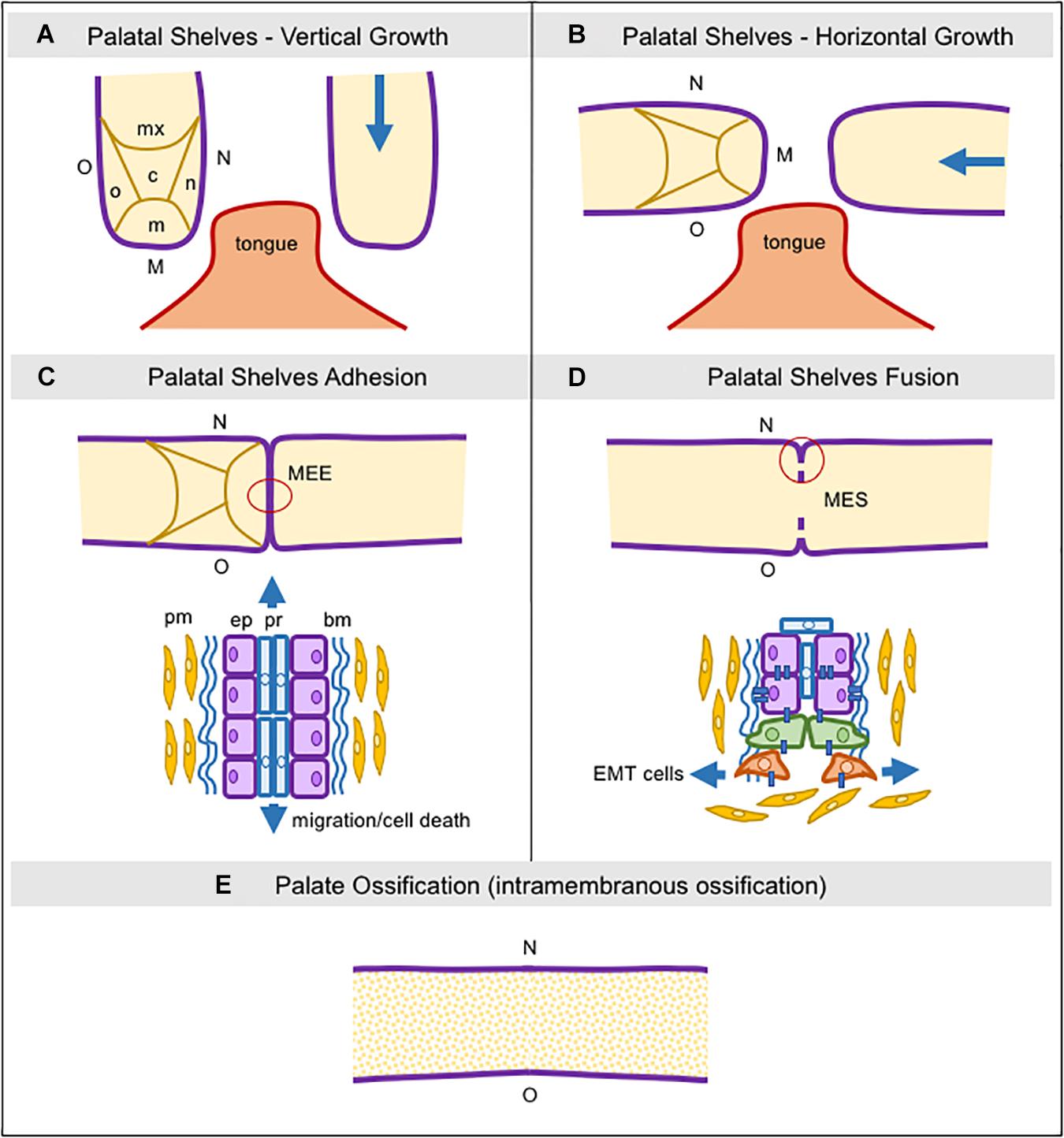
Figure 1. Schematic representation of consecutive steps during secondary palatogenesis. (A) Initially, palatal shelves grow down, surrounding the tongue, and five regions in the palatal mesenchyme can be seen: nasal, medial, oral, central, and maxillary, as well as three regions in the palatal epithelium: nasal, medial, and oral. (B) Palatal shelve orientation switches from vertical to horizontal, toward each other and above the tongue. (C) The medial palatal epithelia from both shelves establish adhesion and it is now named the Media Edge Epithelia (MEE). In this area, we can see two different types of epithelial cells localized in two layers, the peridermal and the non-stratified cuboid epithelium. Peridermal cells start to migrate toward both nasal and oral epithelial sites. (D) The MEE starts the fragmentation since the epithelial layer begins the epithelial-to-mesenchymal transition and these cells then migrate into the palatal mesenchyme. (E) The MES completely disappears and the palatal mesenchymal cells start to differentiate into osteoblasts via intramembranous ossification. bm: basement membrane; c: central region; EMT: epithelial-to-mesenchymal transition; ep: epithelial cells; M: medial site; m: medial region; MEE: medial edge epithelia; MES: medial epithelial seam; mx: maxillary region; n: nasal region; N: nasal site; o: oral region; O: oral site; pm: palatal mesenchyme; pr: peridermal cells.
Once palatal fusion is complete, the anterior two-thirds mineralize by intramembranous ossification (hard palate) (Figure 1E), and the posterior third will give rise to a fibromuscular tissue (soft palate) under the signaling by numerous factors, particularly bone morphogenetic proteins (BMPs), fibroblast growth factors (FGFs), hedgehog (HH), vascular endothelial growth factor (VEGF), and Wnt/β-catenin signaling, which drive the palatal mesenchyme to undergo osteoblast differentiation for mineralization (Wu et al., 2008; Baek et al., 2011; Nelson et al., 2011; Pan et al., 2013; Smith et al., 2013; Hill et al., 2014, 2015; Nassif et al., 2014; Iyyanar and Nazarali, 2017; Zhang et al., 2017; Xu J. et al., 2018; Thompson et al., 2019).
Anatomically, the fusion of primary and secondary palates with the nasal septum originates the palate, a physical barrier that separates the fully developed nasal and oral cavities. Physiologically, it has a function in breathing, speech, and swallowing. The local regulation of palate development depends on a network of several factors, such as transcription factors, signaling molecules, soluble factors, ECM proteins, ECM remodeling enzymes, ECM cross-linkers, and cell adhesion molecules. Disturbance of this tightly controlled network may inhibit the fusion of the palatal shelves and, hence, result in a cleft palate (Funato, 2015).
ECM Structural Molecules and Soluble Factors
Collectively, the secretome is the set of membrane proteins that are tethered on the cell surface and interact with the ECM, secreting molecules into ECM in soluble forms or inside of extracellular vesicles (EVs). Part of the secretome contains the matrisome, which encompasses all ECM-proteins and ECM-associated proteins. The core matrisome is composed mainly of structural proteins encoded by around 300 genes, whereas matrisome-associated proteins are those that modulate ECM functions and are encoded by about 700 genes, corresponding to 4% of the human or mouse genomes. The increasing knowledge regarding specific ECM tissue signatures contributes to the understanding of the role of the ECM in development, homeostasis, tissue repair, and disease (Hynes and Naba, 2012; Figure 2).
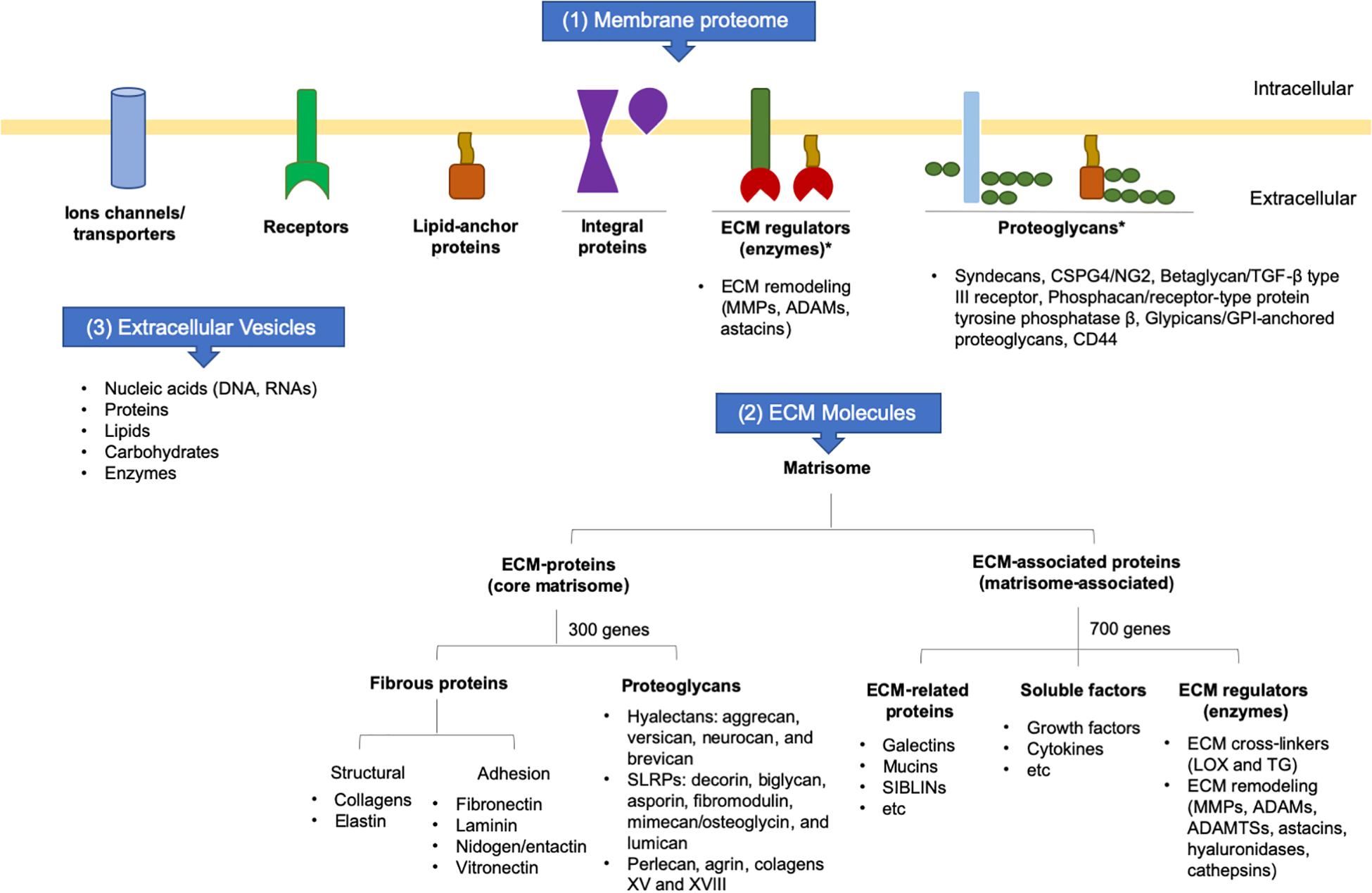
Figure 2. Schematic representation of cell surface molecules and secretome. ADAM: adisintegrin and metalloproteinase; ADAMTS: adisintegrin and metalloproteinase with thrombospondin motifs; ECM: extracellular matrix; GPI: glycosylphosphatidylinositol; LOX: lysyl oxidase; MMP: matrix metalloproteinase; SIBLIN: small integrin-binding ligand n-linked glycoprotein; SLRP: small leucine-rich proteoglycans; TG: transglutaminase; TGF-: transforming growth factor beta.
Fibrous proteins and proteoglycans are the two principal components of the core matrisome. Fibrous proteins are responsible for the matrisome’s supportive function (collagen and elastin) and adhesive functions (fibronectin, laminin, nidogen, and vitronectin). These macromolecules interact with each other and can binding numerous growth factors (Raghunathan et al., 2019).
Proteoglycans are proteins conjugated to GAG chains and are crucial for conferring resistance to compression forces. Most GAGs are highly negatively-charged molecules that attract positively-charged sodium ions and, consequently, water, contributing to the viscosity and hydration within tissues. Among GAGs, chondroitin, dermatan, heparan, and keratan sulfate are the principal GAGs associated with proteoglycans in the connective tissues. High levels of hyaluronic acid or hyaluronan (HA), a non-sulfated GAG, are also found in the ECM (Garantziotis and Savani, 2019). Depending on the type of GAG associated with the proteoglycan, this will determine its location in the cytoplasm (the only member is serglycin), on the cell surface or within the ECM. Most of the heparan sulfate proteoglycans are anchored on the cell membrane via their transmembrane domains or glycosylphosphatidylinositol (GPI) anchors. Thus, proteoglycans can interact with many other molecules, including ECM remodeling enzymes and growth factors, thereby playing an important role in regulating ECM dynamics (Iozzo and Schaefer, 2015).
The provisional matrix is a transitory ECM found during the early stages of development, tissue repair, and disease which is later replaced by a tissue-specific ECM. It is formed by fibrin, fibrinogen, fibronectin, HA, and versican, a large chondroitin sulfate proteoglycan. HA provides a “glue” core to bind all other components and place it in the pericellular space, due to its interaction with a specific membrane receptor, CD44. Several growth factors can stimulate the expression of these macromolecules. The provisional matrix has a viscoelastic property that allows it to create spaces within the ECM, providing means for cell migration. For example, the migratory routes of NCCs during early embryonic development highly express high levels of versican (Barker and Engler, 2017; Chester and Brown, 2017; Wight, 2017) and tenascin (Riou et al., 1992). It is well known that collagens type I, III, IV, V, VI, fibronectin, HSPG, laminin, and tenascin are expressed during palatogenesis in vivo (Ferguson, 1988; Dixon et al., 1993a) and EGF or TGF-α can stimulate their expression on mouse embryonic palatal mesenchymal cells in vitro (Dixon et al., 1993b). The intrinsic “internal shelf force” for palatal elevation has been attributed to HA since it is the most abundant GAG in palatal ECM before shelf elevation (Ferguson, 1988). It is produced on the cell membrane surface by specific enzymes (HA synthases—Has 1-3) and these are differentially expressed in palatal mesenchyme and epithelium during palatogenesis (Galloway et al., 2013). In TGFβ-3 null mice, expression of all Has forms is decreased, leading to reduced amounts of HA and impaired shelf elevation (Galloway et al., 2013). Recently, Has2 has been described to be a crucial HA synthase in NCC-derived mesenchyme during craniofacial development and palatogenesis (Lan et al., 2019). Also, FGFs induce HA synthesis by mouse embryonic palatal mesenchymal cells in vitro (Sharpe et al., 1993). Fibronectin is found during embryonic development in areas characterized by cell migration (Schwarzbauer and DeSimone, 2011). It appears that fibronectin arrangement is vital for cell migration and palatal shelf elevation. In this case, Rac1 and cell density modulates fibronectin deposition in mid-palate (Tang et al., 2015). Moreover, Rac1 is downregulated by retinoic acid, leading to the cleft palate as a consequence of the disarrangement of fibronectin and cell migration as well (Tang et al., 2016).
Cellular communication is a well-known mechanism in which cells can communicate with each other and modify cell behavior through soluble factors. Intercellular communications occur via direct cellular interactions in which cell surface proteins act as mediators able, or not, to bind to the ECM (juxtacrine signaling). Alternatively, cells release local mediators into the ECM to create self-control signals (autocrine signaling) and send information to neighboring cells (paracrine signaling) or reach target cells in long distances via hormones (endocrine signaling) (Ansorge and Pompe, 2018). The local mediators are peptides or growth factors which control many cellular activities. During development, a combination of cell–cell interactions occurs, as well as the secretion of mediators named morphogens, which induce specific cell differentiation in a distinct spatial order and morphogen gradient-dependent manner (Inomata, 2017). The main morphogens are retinoic acid, HH, TGF-β, BMPs, and Wnt/β-catenin.
The actions of numerous morphogens in palatogenesis have been extensively studied, mainly secreted factors such as HH (Cobourne and Green, 2012; Dworkin et al., 2016; Xavier et al., 2016; Li et al., 2018), FGF (Jiang et al., 2006; Nie et al., 2006; Snyder-Warwick and Perlyn, 2012; Stanier and Pauws, 2012; Prochazkova et al., 2018; Weng et al., 2018), TGF-β (Nawshad et al., 2004; Iwata et al., 2011; Nakajima et al., 2018), BMP (Nie et al., 2006; Parada and Chai, 2012; Graf et al., 2016), and Wnt/β-catenin family proteins (He and Chen, 2012), which are responsible for guiding all steps of palate formation by reciprocal signaling between the embryonic oral epithelium and palatal mesenchyme, as well as transcription factor regulation (Greene and Pisano, 2010; Levi et al., 2011; Bush and Jiang, 2012; Li et al., 2017). Also, other morphogens and growth factors have emerged in palatogenesis, such as connective tissue growth factor (Tarr et al., 2018) and retinoic acid (Okano et al., 2014; Mammadova et al., 2016). Dysregulation of these pathways through genetic variations in individual genes has been suggested as strongly associated with CL/P (Pauws and Stanier, 2007; Krejci et al., 2009; Tang et al., 2013; Okano et al., 2014; Reynolds et al., 2019).
During the last decade, knowledge of new types of RNA with regulatory functions, located in non-coding regions of DNA, has improved our understanding of gene expression regulation (Scherrer, 2018). Many different microRNAs have been identified to temporally and spatially regulate morphogens and transcription factors during palatogenesis (Eberhart et al., 2008; Seelan et al., 2014; Ding et al., 2016; Reiss and Bhakdi, 2017; Schoen et al., 2017). Not surprisingly, microRNAs have been suggested as be new targets for investigating in CL/P studies (Li et al., 2010; Wang et al., 2013, 2017; Ma et al., 2014; Gao et al., 2015; Li D. et al., 2016; Li J. et al., 2016; Schoen et al., 2017, 2018; Chen et al., 2018; Grassia et al., 2018; Pan et al., 2018; Suzuki et al., 2018; Wu N. et al., 2018; Xu M. et al., 2018).
ECM Remodeling
The extracellular microenvironment is dynamically modeled and remodeled by soluble or EV-associated proteases secreted into the ECM or membrane-anchored proteases, which are classified as cross-linkers and remodeling proteases (Sanderson et al., 2019). Of the ECM cross-linkers, lysyl oxidases (LOX) and transglutaminases (TGs) are the major families responsible for establishing cross-links between the central core matrisome proteins. Moreover, an intricate balance between proteases and their inhibitors maintains the ECM homeostasis; abnormal ECM remodeling (excessive or inefficient) is involved in the development and/or progression of several pathologies due to modifications in macromolecule composition (posttranscriptional control and posttranslational modifications), biophysical (architecture), and biomechanical properties (stiffness).
ECM Cross-Linkers
Post-translational modifications (cross-links) in collagen–collagen, collagen–ECM, and ECM–ECM interactions are relevant for the integrity, stiffness, and rigidity of the microenvironment. Once formed, cross-links formed are immature but become more stable due to multivalent cross-links that generate insoluble protein polymers that are resistant to proteolytic degradation, improving the biomechanical properties of the collagen network. Procollagen maturation takes place when both N- and C-termini are classically removed by ADAMTS and BMP-1/TLD (BTPs), respectively, but this cleavage may also be mediated by meprins, forming tropocollagen (Figure 3A). Subsequently, three main pathways promote the final fibrillogenesis: the LOX-mediated, TG-mediated, and sugar-mediated cross-linking pathways (Figure 3B). The latter pathway constitutes a non-enzymatic glycosylation reaction that occurs as the consequence of prolonged exposure to reducing sugars (e.g., ribose and glucose), producing advanced glycation end products (AGEs), which are associated with aging and diabetic complications (Benkovics et al., 2017; Cruz et al., 2018).
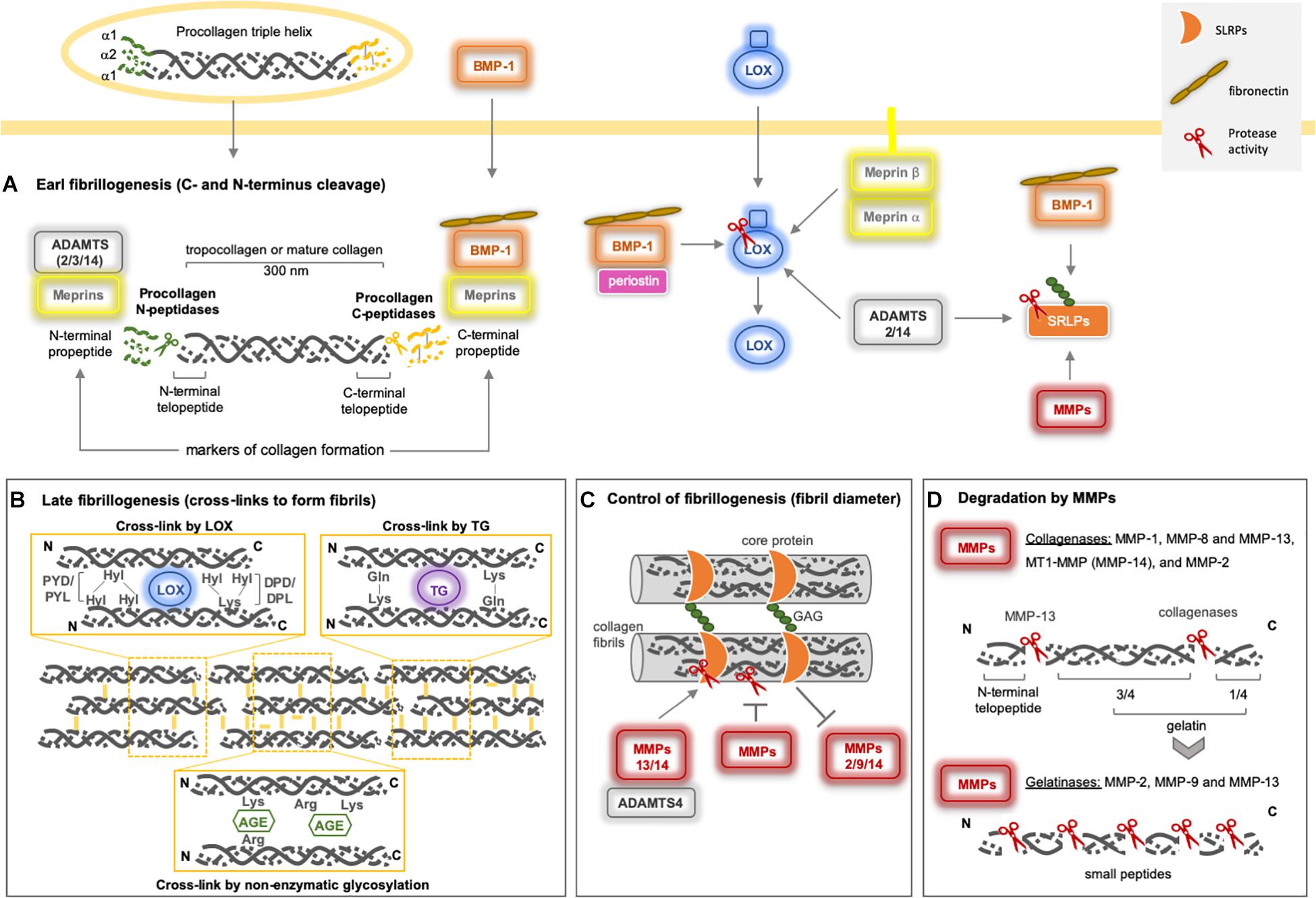
Figure 3. Collagen type I assembly and degradation by metalloproteinases. (A) The early fibrillogenesis process on fibrillar procollagens starts by the excision of both the N- and C- termini by classical Procollagen N-peptidases (sub-group of the ADAMTS family) and Procollagen C-peptidases (BMP-1). They cleave procollagens I, II, III, and V (ADAMTS2/14 and BMP-1), where ADAMTS3 can cleave only procollagen II. Recently, both meprins have been reported to be able to remove both N- and C- terminus as well. (B) After N- and C- termini removal from procollagen, mature collagen is the target for ECM cross-linker enzymes (LOX and TG), which will form collagen fibrils. Under high glucose conditions, a non-enzymatic cross-link takes place, named AGE. (C) The level of collagen fibrillogenesisis controlled by proteoglycans (SRLPs), which can interact with collagen fibrils, resulting in the modification of the diameter of collagen fibers. Fibromodulin, decorin, and lumican prevent or delay the cleavage of collagen by MMP-1 and MMP-13. Lumican acts on MMP-14 enzymatic inhibition (Pietraszek et al., 2014) and gene expression (Niewiarowska et al., 2011; Malinowski et al., 2012). Decorin inhibits MMP-2 and MMP-9 gene expression and activity (Neill et al., 2012). MMP-14 can cleave human decorin (Mimura et al., 2009) and thelumican core (Li et al., 2004), MMP-13 and ADAMTS-4 also cleave decorin (Shu et al., 2019). (D) Among the metalloproteinases, only MMPs can cleave fibrillar collagens. The collagenases generate ¼ and 3/4 fragments (gelatin) that are further cleaved by gelatinases (MMPs-2 and -9) and MMP-13. MMP-13 also removes the N-terminal telopeptide from collagen. Indirectly, metalloproteinases may modulate collagen fibrillogenesis by processing LOX and SRLPs. BMP-1 and ADAMTS2/14 activate proLOX in distinct sites and seem to be essential for LOX-collagen binding (Rosell-García et al., 2019). Periostin increases the proteolytic action of BMP-1 on proLOX (Maruhashi et al., 2010). BMP-1 promotes the maturation of SLRPs (von Marschall and Fisher, 2010), as well as the MMPs, after C-terminal excision. Fibronectin increases BMP-1 activity against biglycan and procollagen I (Huang et al., 2009). AGE: advanced glycation end products; ADAM: adisintegrin and metalloproteinase; ADAMTS: adisintegrin and metalloproteinase with thrombospondin motifs; LOX: lysyl oxidase; MMP: matrix metalloproteinase; SLRP: small leucine-rich proteoglycans; TG: transglutaminase.
The LOX and LOX-like (1–4) proteins are a family of copper-dependent amine oxidase enzymes that catalyze the formation of unstable aldehydes by the oxidation of the ε-amino groups of lysine/hydroxylysine in collagens and lysine in elastin, forming covalent cross-linkages in collagen–collagen and collagen–elastin complexes, respectively. LOX is secreted as a proenzyme into the ECM, but is also found intracellularly, and is then processed by BMP-1, the same enzyme that cleaves fibrillar procollagens (Figure 3). As such, there is a direct relationship between the collagen process and its cross-linking, suggesting a major role for LOX in ECM orientation (Rodriguez-Pascual and Rosell-Garcia, 2018).
The TGs [formed of nine members, including tissue TG (tTG) or TG2, which is the most abundant in tissues] belong to a multi-functional family of calcium-dependent acyl-transferase enzymes that catalyze transamidation of glutamine and lysine to form covalent bonds both inside and outside of the cell. These cross-links form between collagen–collagen, collagen–ECM, or ECM–ECM and can involve fibronectin, mainly, and also nidogen/entactin, osteonectin, osteopontin, laminin, vitronectin, fibrinogen, and heparan sulfate proteoglycan. Initially, the TG catalyzes the formation of an isopeptide bond between y-carboxamide groups of glutamine residue side chains and the ε-amino groups of lysine residue side chains with subsequent release of ammonia. Subsequently, glutamine and lysine residues are available to bind with peptides or proteins, and the intra- or inter-molecular ε-(γ-glutamyl)lysine cross-links take place. In the presence of water, TGs are also able to hydrolyze glutamine residues to glutamic acid by deamidation (Figure 3B). These cross-links exhibit high resistance to physical, chemical, and proteolytic degradation, mainly by matrix metalloproteinases (MMPs). Physiologically, TGs generate biological polymers that are indispensable for creating barriers and stable structures in several systems, while pathologically, they contribute to generating fibrotic matrices (Eckert et al., 2014; Savoca et al., 2018).
ECM Remodeling Enzymes: Metalloproteinases
Most of the ECM remodeling enzymes belong to the metzincin family (Stöcker et al., 1995), which share numerous similarities, including multiple domains, zinc-dependent endopeptidases, a typical structural profile and tertiary configurations of the catalytic domain (the secondary structure contains three histidines bound to zinc, at the catalytic site, and a methionine, or “Met-turn”). This family comprehends vertebrate matrixins (MMPs), adamalysins (ADAMs—a family of disintegrin and metalloproteinase, mainly sheddases; and ADAMTSs—a family of disintegrins and metalloproteinases with thrombospondin-like motifs, mainly formed of proteoglycanases and procollagen N-propeptidases), astacins (BMP-1/Tolloid-like protease 1 and Meprins, mainly formed of procollagen N- and C-propeptidases), and pappalysins (main bioavailability of IGFs) (Cerdà-Costa and Gomis-Rüth, 2014) and encodes around 200 genes, identified in humans and mice, comprising around 1/3 of proteases, the largest proteolytic enzyme group existent (López-Otín and Bond, 2008). Among them, MMPs are classically recognized to degrade all ECM components, but other metalloproteinases have been recognized to play essential roles in ECM maturation and to generate bioactive molecules. As a result of the extensive study of ECM remodeling enzymes over the last six decades, other biological functions have also been attributed to them, due to their broad spectrum of substrates, identified in both subcellular and extracellular compartments (Bond, 2019). Other enzymes, such as urokinase-type plasminogen activator, cathepsins, and heparanases, are also indispensable in these processes, and growth factor bioavailability within the ECM is protease-dependent. As a result of the extensive study of ECM remodeling enzymes over the last six decades, other biological functions have also been attributed to metalloproteinases, due to their broad spectrum of substrates, identified in both subcellular and extracellular compartments (Bond, 2019). We will focus only on metalloproteinases currently known to have a role in palatogenesis and CL/P.
Among metzincins, ADAMs, ADAMTSs, and MMPs are closely related in structure, regulation, and activation. However, they have different substrates and, therefore, distinct functions under physiological and pathological conditions. Structurally, the N-terminal ectodomain of most secreted MMPs is composed of pre-, pro-, and catalytic domains (metalloproteinase domain) and contains a furin region in all ADAM, ADAMTS, and membrane-anchorage MMPs. Complementary domains confer proteolytic specificity and localization. As such, MMPs are the most studied metalloproteinases and act in many cellular functions (e.g., proliferation, migration, differentiation, among others) due to their cleavage of substrates in the ECM, on the cell surface and intracellularly (cytoplasm and nucleus) to produce bioactive molecules. However, few studies have been conducted to understand the interrelationship between these metalloproteinases and how they work together to control cell behavior.
MMP Family
Over the years, the “degradative” activity of MMPs during physiological and pathological processes has led to their association with tissue destruction, due to their unique ability to cleave fibrillar collagens (Sprangers and Everts, 2019). However, “omic” studies and a better understanding of ECM dynamics support a broader role for MMPs in pathological and physiological events (Rodríguez et al., 2010). Several other core matrisome, ECM-associated proteins, and cell surface proteins, cleaved by MMPs, reveal hidden sequences and unblock the diverse cell functions (Butler and Overall, 2009; Deryugina and Quigley, 2010; Bauvois, 2012; Mannello and Medda, 2012).
Extensive reviews focus on MMPs regulation at both intra and extracellular levels and have been extensively reviewed. At the gene transcription level, signals from the ECM (cytokines, growth factors, EMMPRIN/ECM metalloproteinase inducer/CD147, integrins, ECM proteins, cellular stress, morphological changes, among others) significantly impact MMP expression. Mutations and polymorphisms in MMP genes (particularly in promoter regions), together with epigenetic modifications, have been shown to modulate MMP expression. Post-transcriptional regulation includes changes in mRNA stability through microRNAs, decoy RNAs, and degradation pathways. MMPs are targets of several PTMs that are crucial for correct cellular localization (via insertion of the GPI-anchor), intracellular activation of membrane-anchoring MMPs by furins, and insertion of carbohydrates (N and O-glycosylation) (Boyd, 2006; Reuben and Cheung, 2006; Vincenti, 2007; Clark et al., 2008).
In the ECM, soluble proenzymes are activated by other proteinases, mainly active MMPs, via the “cysteine switch mechanism.” The catalytic activity may be inhibited by their endogenous inhibitors in the ECM (TIMPs) or on the cell membrane (RECK). Additionally, their proteolytic activity may be modulated by allosteric control in exosites outside of the catalytic site and also by interactions with proteoglycans and GAGs (Pietraszek-Gremplewicz et al., 2019). For example, MMP-2 interacts with syndecan-2 on the cell surface, which blocks the activation of proMMP-2. In some cases, proenzymes are associated with TIMP (low concentration) for correct activation; for example, the MMP-14/TIMP-2/proMMP-2 ternary complex. MMPs may be associated with other ECM components, such as proteoglycans and GAGs, leading to their specific localization in the cellular perimeter or at ECM sites distant from the cell secretion point (Van den Steen et al., 2001; Hernandez-Barrantes et al., 2002; Ra and Parks, 2007; Gabison et al., 2009; Hadler-Olsen et al., 2011; Piperi and Papavassiliou, 2012; Rietz and Spiers, 2012; Li et al., 2014; Arpino et al., 2015; Karamanos et al., 2019).
Due to their unique ability to cleave fibrillar collagens, collagenases MMP-1, -8, and -13 are the main enzymes for collagen turnover and generate gelatin (classical ¾ and ¼ fragments) (Figure 3D). MMP-2 and -9, along with membrane-anchored MMPs (MMP-14 and MMP-16), may also cleave fibrillar collagen with different affinities, as is the case of collagen I and III, which are preferentially cleaved by MMP-1, whereas collagen II is the preferred substrate for MMP-13. Additionally, stromelysins (MMP-3 and -10) can degrade just collagen III, but not collagen I or II (Sprangers and Everts, 2019). The interaction of the N-terminal site of specific SRLPs (decorin, fibromodulin, and lumican) with collagen fibrils forms a layer that can prevent or delay the collagen cleavage by MMPs (Geng et al., 2006). At the same time, different SRLPs are substrates or inhibitors of MMPs and ADAMTS (Shu et al., 2019; Figure 3C).
ADAMTS Family
N-glycosylation, O-fucosylation, and C-mannosylation are the most frequent post-translational modifications (located at the ancillary domains) to control ADAMTS activity. These modifications control secretion, localization, activation, and catalytic functions. Cleavage of the central thrombospondin type 1 sequence repeat (TSR—probably attached to the cell membrane) regulates both the proteinase activity and the localization of these enzymes. The proenzymes can be activated intracellularly, on the cell surface or within ECM by furin-like pro-protein convertases and then autocatalytic activation. Similarly to ADAMs, ADAMTSs are selectively inhibited by TIMPs, where TIMP-3 is the principal inhibitor (Bekhouche and Colige, 2015; Kelwick et al., 2015; Dancevic et al., 2016).
Although ADAMTS-like homologues lack protease domains, ancillary domains are present and may be involved in the regulation of ADAMTS due to competitive binding to substrates. Furthermore, they may have ADAMTS-independent functions in ECM, cell–EMC, or cell–cell interactions. Recently, an unexpected interaction between LOX-ADAMTSLs was reported, suggesting a role in microfibril assembly (Aviram et al., 2019). Mutations or deletions in ADAMTS have been implicated in many pathologies and syndromes, where they modulate tissue morphogenesis and remodeling, in cancer, inflammation, in the central nervous system, and in cartilage and vascular biology (Kintakas and McCulloch, 2011; Stanton et al., 2011; Lisi et al., 2014; Dubail and Apte, 2015; Hubmacher and Apte, 2015; Kelwick et al., 2015; Rodríguez-Manzaneque et al., 2015; Dancevic et al., 2016; Fu and Kong, 2017; Itoh, 2017; Lemarchant et al., 2017; Yang et al., 2017; Mead and Apte, 2018).
The Roles of MMPs and TIMPs in Palatogenesis
The development of the facial primordia requires remodeling of the ECM, which is mediated in part by MMPs. During embryonic development, the process of morphogenesis involves MMP-mediated changes in the composition of the ECM that further allow for cell migration and differentiation, cell–cell interactions, and tissue resorption. MMPs act on ECM remodeling during palatal shelves orientation and during EMT events for palatal fusion (Brown et al., 2002).
Early studies in mice have provided biological evidence for the roles of MMPs and TIMPs in embryonic development (Gack et al., 1995; Iamaroon et al., 1996; Blavier and DeClerck, 1997). Several MMPs, TIMPs, and RECK mRNA and proteins are expressed, in association with enzymatic activity, throughout the stages of murine palate development. These expressions share the same specific spatial and temporal distribution patterns in areas in which their preferred substrates are also present (Mansell et al., 2000; Morris-Wiman et al., 2000a, b; Blavier et al., 2001; Brown and Nazarali, 2010; de Oliveira Demarchi et al., 2010; Gkantidis et al., 2012).
The secretion of GAGs in the palatal mesenchyme is related to an increase in water content, and the specific accumulation of collagen I in the nasal side of the palatal mesenchyme may be necessary to generate internal forces required for shelf elevation. Furthermore, MMP expression increases in both the medial and oral epithelium before shelf elevation (Morris-Wiman et al., 2000a; de Oliveira Demarchi et al., 2010), as does MMP gelatinolytic activity in the basement membrane and beneath the mesenchyme of the nasopharyngeal epithelial folds that form during palatal shelf reorientation from vertical to horizontal position (Gkantidis et al., 2012; Figure 4A).
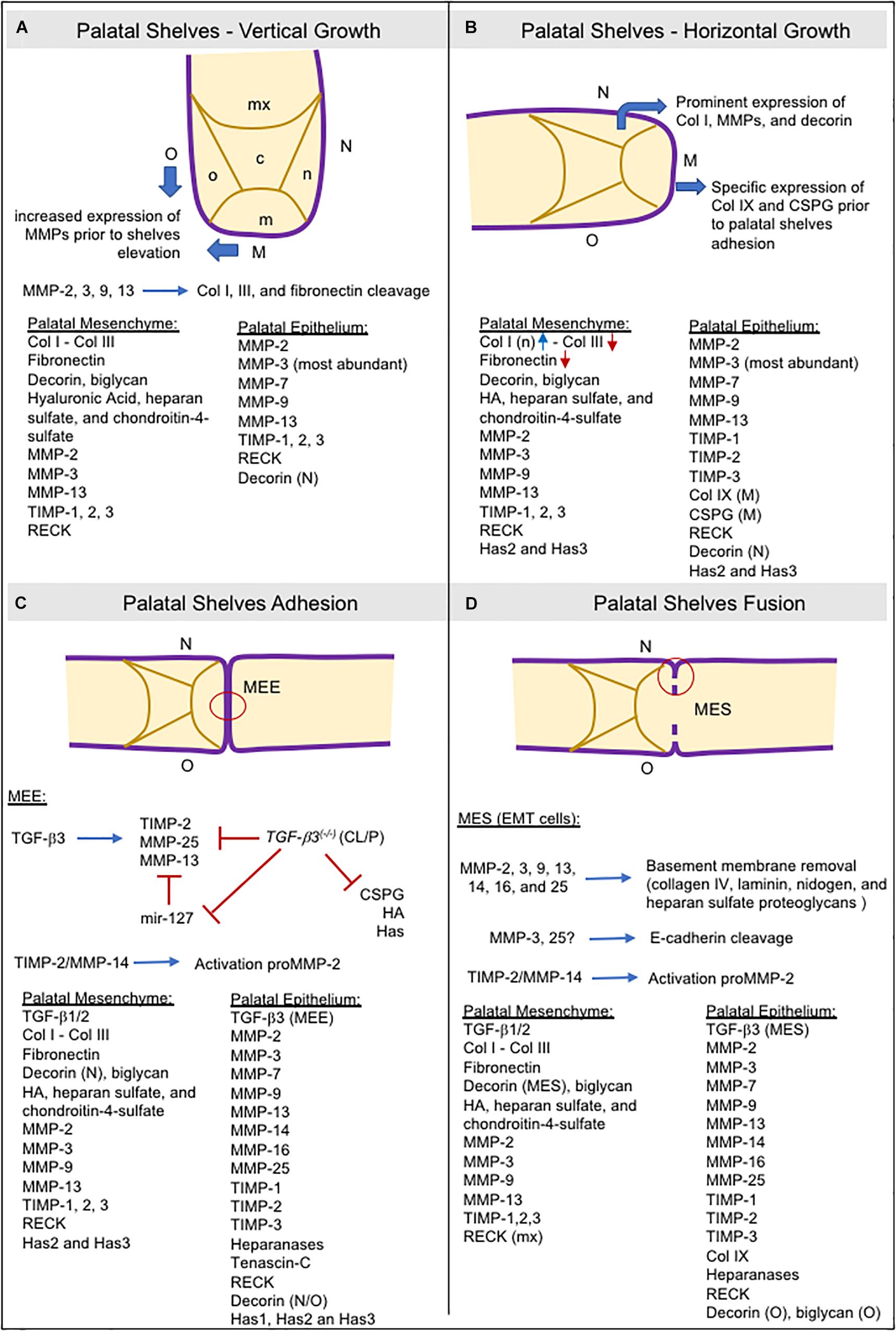
Figure 4. Schematic representation of consecutive steps during secondary palatogenesis and the molecules expressed in palatal mesenchyme ECM and palatal epithelium. Collagens type I and III, fibronectin, GAGs (hyaluronic acid, heparan sulfate, and chondroitin-4-sulfate), proteoglycans (biglycan, decorin), and TGF-β are the most abundant ECM molecules found in provisional ECM during palatogenesis. MMPs are the major protein responsible for ECM remodeling in both the ECM mesenchyme and basement membrane, but also expressing heparanases as well. Expression of TIMPs and RECK occurred, and they are crucial for ECM remodeling balance. (A) Most ECM components are widely present in palatal mesenchyme. MMP expression is high in the oral and medial epithelia before shelve elevation. (B) The increased expression of collagen I and decorin in the nasal region of the palatal mesenchyme may help with palatal elevation. Decorin can bind to collagen I and cause contraction of collagen in vitro. In the medial epithelium, the specific expression of collagen IX and CSPG is vital for adequate palatal shelve adhesion. (C) TGF-β3 modulates the expressions of TIMP-2, MMP-13, MMP-25, and CSPG in the MEE. TGF-β3 knockout mice downregulated these genes. mir-127 is upregulated, leading to repression of MMP-13 expression (Warner et al., 2015). (D) Other MMPs are required for MES disruption and are involved in basement membrane degradation by EMT cells. CSPG: chondroitin sulfate proteoglycans; EMT: epithelial-to-mesenchymal transition; HA: hyaluronic acid; Has: hyaluronan synthases; M: medial site; m: medial region; MEE: medial edge epithelia; MES: medial epithelial seam;mx: maxillary region; n: nasal region; N: nasal site; o: oral region; O: oral site; TGF-β: transforming growth factor beta.
At ED13 and ED14, the secreted inhibitors, TIMP-1 and TIMP-2, display a similar spatial distribution to the MMPs and are widely expressed in the epithelial basement membrane. TIMP-3 is strongly expressed in the palatal epithelium although weakly expressed in the medial mesenchyme (Morris-Wiman et al., 2000a). Of the four TIMPs, TIMP-2 is the most abundant (Mansell et al., 2000), whereas TIMP-3 has different roles than the other TIMPs. Animals lacking the Timp3 gene develop several pathologies associated with increased ECM degradation and loss of tissue integrity due to unregulated MMP, ADAM, and ADAMTS activity (Sahebjam et al., 2007). The anchored-membrane MMP inhibitor, RECK, is also expressed in the mesenchyme (de Oliveira Demarchi et al., 2010). RECK expression has been implicated in tissue integrity since its absence leads to extensive disarrangement of the connective tissue and embryos die in utero before craniofacial development (Oh et al., 2001). Expression of TIMP-3 and RECK in the different sites of the developing palatal epithelium suggests that they function in the maintenance of palatal tissue integrity by regulating epithelial–mesenchymal interactions (Figure 4B).
Matrix metalloproteinases are fundamental for the removal of the basement membrane and are expressed by epithelial cells in the EMT program to detach from MES and migrate to the adjacent mesenchyme to allow palatal fusion (Horejs, 2016). The participation of the membrane-anchored MMPs (MMPs -14, -16, and -25) appear to be crucial in this process (Shi et al., 2008; Brown and Nazarali, 2010). These observations are reinforced by the results of in vitro studies showing that addition of synthetic MMP inhibitors (Blavier et al., 2001), or the silencing of MMP genes (e.g., MMP-25) (Brown and Nazarali, 2010) in palatal cultures can prevent palatal fusion. Furthermore, Tgfβ-3- and Egfr-knockout mice, which display a cleft palate phenotype, have decreased or absent MMP expression in the MEE or MES (Miettinen et al., 1999; Blavier et al., 2001). Heparanase has also been detected in the MEE and MES and co-localized with MMPs -2, -3, and -9 (Hirata et al., 2013). MMP-25 can cleave only collagen IV and, in terms of substrates, displays more similarities to MMP-3 than other MT-MMPs (English et al., 2001). High gelatinolytic activity and laminin expression have also been found in the MEE and MES (Gkantidis et al., 2012). Furthermore, MMP-3 cleaves E-cadherin (Lochter et al., 1997) and MMP-25 is co-localized with E-cadherin in cell–cell junctions (Radichev et al., 2010), a crucial step for epithelial EMT cells to acquire a mesenchymal phenotype. Taken together, the basement membrane and epithelial cell–cell junction degradation require the cooperative proteolytic actions of MMPs and other ECM degrading enzymes in the MEE and MES cells (Figures 4C,D).
TGFβ-3 is expressed explicity in the palatal epithelium, and is co-expressed with MMPs (Blavier et al., 2001; Brown and Nazarali, 2010). In Tgfβ-3 null mice, TIMP-2 and MMP-13 expression in the palatal epithelia were significantly decreased, whereas no changes in expression were noted for MMP-14 (Blavier et al., 2001). Similarly, the incubation of palatal culture with a TGFβ-3-neutralizing antibody decreases MMP-25 expression in the palatal epithelia (Brown and Nazarali, 2010). Collectively, these observations suggest that both MMP-13 and MMP-25 are downstream targets of TGFβ-3 (Figure 4C). MMP-13 is specifically expressed in sites of bone formation in vivo (Fuller and Chambers, 1995; Gack et al., 1995; Mattot et al., 1995; Stahle-Backdahl et al., 1997), and in physiological situations that require rapid and effective remodeling of collagenous ECM.
While MMP and TIMP expressions appear to be critical throughout all stages of palatal development, knockout mice for Timp1, Timp2, Timp3, Timp4, Mmp2, Mmp9, Mmp13, Mmp14, Mmp16, and Mmp25 do not develop cleft palate (Paiva and Granjeiro, 2014; Soria-Valles et al., 2016). Interestingly, combined double knockout of Mmp14 and Mmp16 in mice leads to severe structural and craniofacial defects, including severe dysfunction in palatal shelf formation and cleft palate in 80% of the embryos (Shi et al., 2008). Among the membrane-anchored MMPs, MMP-16 is closely related to MMP-14 in terms of molecular structure and expression patterns in remodeling tissues (Szabova et al., 2005; Shi et al., 2008). While these findings suggest that potential compensatory mechanisms exist to overcome the loss of function of individual MMP genes, they also suggest that the loss of specific MMPs, in combination, may impair embryonic and palate development. Moreover, while individual MMP genes may not contribute a major gene effect to non-syndromic CL/P (NSCLP) susceptibility, they may act as modifiers on the background of other genes.
Cleft LIP And/Or Palate
Orofacial clefts result from the failure of developing embryonic facial and palatal processes to ultimately merge or fuse. A multidisciplinary team including surgical, dental, speech, genetic, and nutrition experts are typically involved in patient care to mitigate the feeding, swallowing, breathing, speech, and hearing complications inherent to the condition. Orofacial clefts can be categorized as syndromic or non-syndromic (also termed isolated), based on the presence of additional structural abnormalities. Over 500 syndromes, including chromosomal abnormalities, have been reported in association with orofacial clefts, comprising 30% of all cleft cases. The remaining 70% of cases are all non-syndromic (i.e., isolated), and may segregate in families or appear in sporadic cases (Mossey et al., 2017).
Non-syndromic orofacial clefts are the most common of congenital craniofacial disabilities, affecting approximately 1 in 700 live births worldwide each year, and have been divided historically into cleft lip, with or without cleft palate (CL/P), and cleft palate only (CP), due to the distinct developmental origins of the lip and the palate (Figure 5). While their reported prevalences vary considerably according to ancestral origin, Asians are the most frequently affected population, with birth prevalence rates as high as 1 in 500 live births, followed by Caucasians with a prevalence rate of about 1 in 1000 live births, and African populations showing the lowest prevalence rates at approximately 1 in 2500 live births (Kadir et al., 2017; Mossey et al., 2017; Ishorst et al., 2018). The presence of CL/P also differs by sex and laterality, with CL/P being more common in males than females at a 2:1 ratio, and CP being more common in females, meanwhile unilateral left CL/P is more common than unilateral right CL/P at a 2:1 ratio.
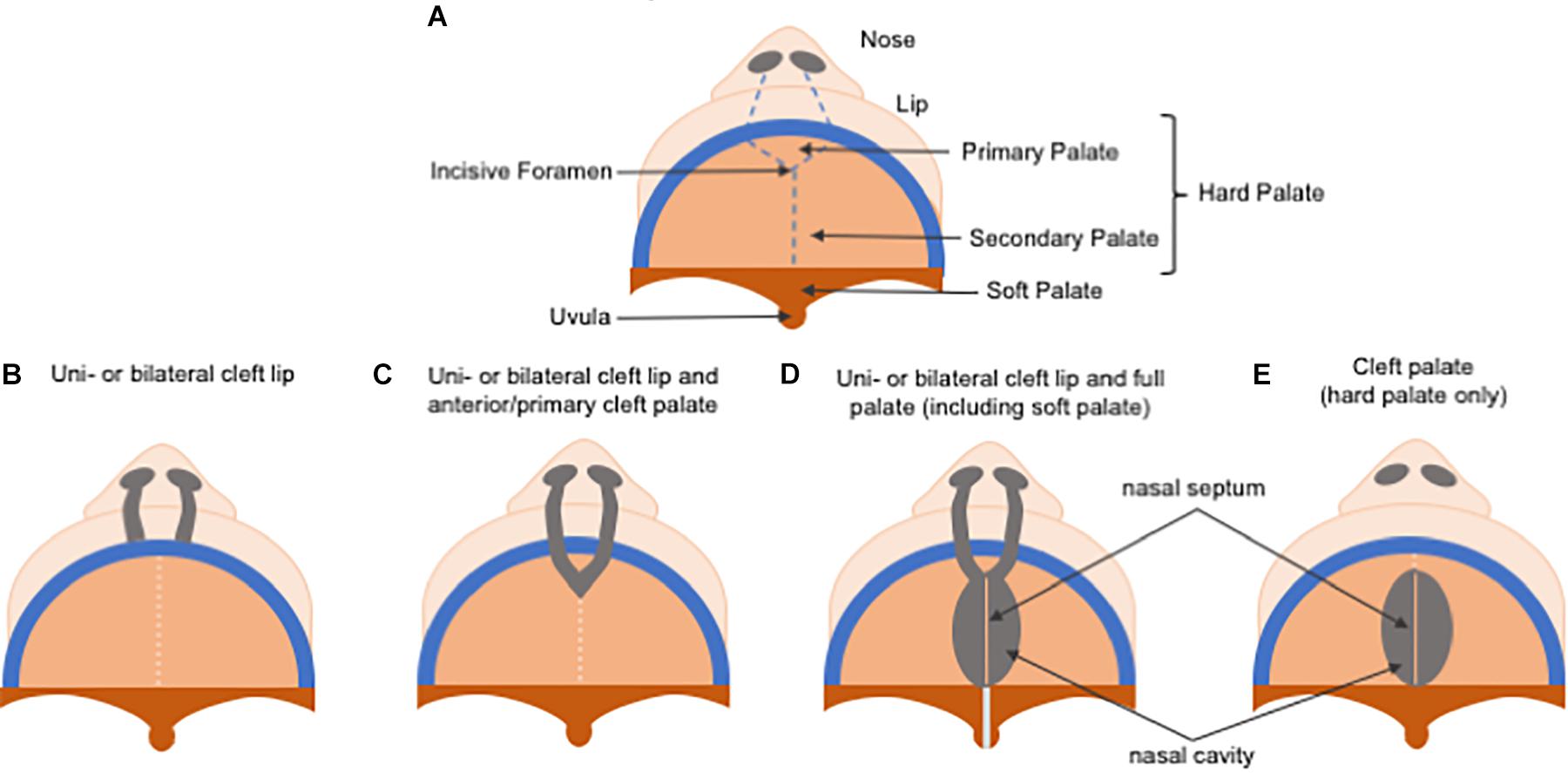
Figure 5. Schematic representation of cleft lip and/or palate classification. (A) Normal palate. (B) In uni or bilateral cleft lip, only lip reconstruction is necessary. (C) In uni or bilateral cleft lip and anterior/primary cleft palate, repair of the alveolar bone repair in the primary hard palate (pre-maxilla) is also required. (D) In uni or bilateral complete cleft lip and palate, treatment is the most challenging due to the need to repair both primary and secondary palate, and the soft palate is affected as well. (E) A cleft palate, which only comprises the secondary palate.
The etiology of non-syndromic orofacial clefts is complex, with both genetic and environmental factors contributing to the condition. While the identification of the genes involved in the syndromic forms of clefting has been mostly successful, much remains to be learned about the factors involved in non-syndromic cases. Genetic studies to date, using both family-based and case-control research approaches, have identified several genes and loci that may play a role in susceptibility to oral clefts (Vieira, 2008). Of these, the evidence is particularly strong for MSX1, IRF6, FOXE1, MAFB, WNT3, WNT9B, CRISPLD2, FGFR1, FGF8, BMP4, and the 8q24 chromosome region. Additional variants in numerous other genes (e.g., TGF-β3, TGF-α, MMP3, VAX1, ABCA4, AXIN2) have also been suggested as candidates for oral clefts with population-specific effects (Dixon et al., 2011; Beaty et al., 2016).
ECM Remodeling Enzymes as Candidate Genes for Orofacial Clefts
Matrix metalloproteinases and TIMPs have been considered plausible candidate genes for CLP, based on their expression patterns in craniofacial tissues and their roles in tissue remodeling and morphogenesis during early embryogenesis; however, a functional role for any individual MMP or TIMP in palate development remains unknown (Brown et al., 2002; Verstappen and Von den Hoff, 2006).
In humans, significant associations between polymorphisms in MMP and TIMP genes with NSCLP have been reported (Blanton et al., 2004; Nikopensius et al., 2011; Letra et al., 2012; Figure 6). A large genome scan of multiplex NSCLP families first suggested evidence for linkage within a region on chromosome 16p13.3, in the same location for MMP25 (Blanton et al., 2004). A later large and comprehensive study evaluated 45 polymorphisms spanning all biologically relevant MMP and TIMP genes for their association with NSCLP; significant associations (P < 0.0004) were noted for variants in MMP3 (rs3025058, rs522616) and TIMP2 (rs8179096), when considering CL/P, CP, and all cleft cases combined. Additional nominal associations were also found for variants in MMP16 (rs7828497, P = 0.01) and MMP10 (rs17293607, P = 0.06) in CL/P and all cleft cases. For CP, variants in MMP3, MMP14, and TIMP1 also showed nominal associations (P < 0.05). Of note, certain allele combinations (i.e., haplotypes) involving these MMP and TIMP gene variants were also significantly associated with NSCLP (Letra et al., 2012).
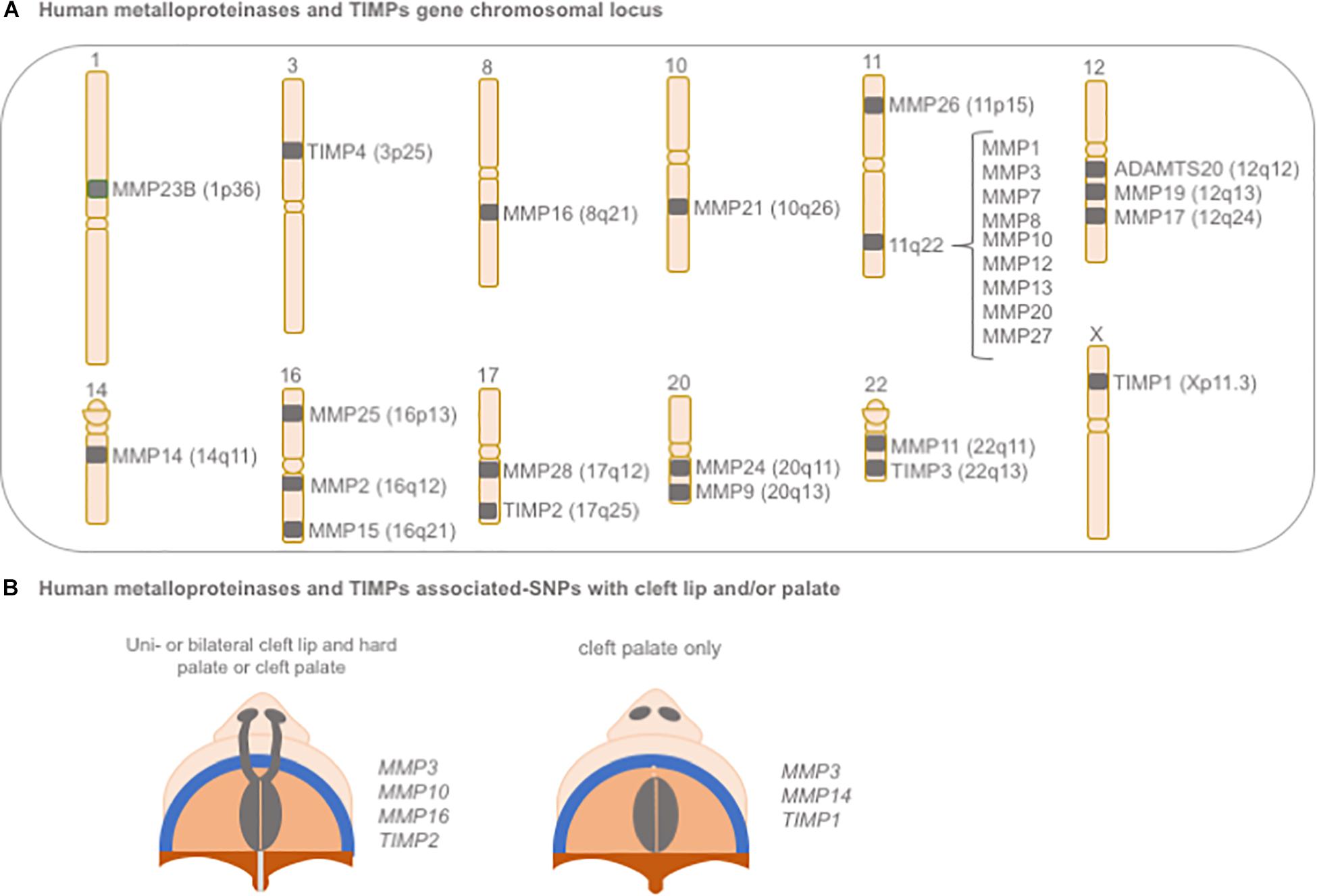
Figure 6. Schematic representation of (A) gene chromosomal locus of human metalloproteinases and TIMPs and (B) their associated-SNPs with cleft lip and/or palate. SNP: single nucleotide polymorphism.
Two polymorphisms, -1171 5A/6A (rs3025058) and -709 A/G (rs522616), located in the MMP3 gene promoter were significantly associated with NSCLP and shown to have functional effects on gene transcription and protein function. The 5A/6A polymorphism consists of a common adenine insertion/deletion polymorphism (5A/6A) at position -1171 of the gene promoter and modulates transcription and local expression of the MMP-3 protein. The 6A allele showed an approximately twofold lower amount of gene product, compared with the 5A allele (Ye et al., 1996), and this difference in promoter activity was attributed to a likely differential binding of the transcriptional repressor to the 6A allele (Mercapide et al., 2003).
A positive regulatory element has also been described for MMP3 -709 A/G, for which significantly higher (∼3.4-fold) promoter activity was found in the presence of allele A. This suggested that allele A can enhance promoter activity, possibly by augmenting transcription factor binding. Furthermore, this variant also appears to be modulated by the concomitant occurrence of the -1171 5A/6A variant. When analyzing the transcriptional effects of haplotypes containing both the -1171 5A/6A and -709 A/G variants, a 1.5-fold decrease in activity was observed for the combination of 5A_A alleles, in comparison with the 5A_G haplotype. Although speculative, this finding may represent a negative feedback loop effect, in an attempt to limit transcription in the presence of the two “high transcription” alleles, 5A and A. In contrast, a fourfold increased activity was found with the 6A_G promoter. The 6A_A haplotype was the least active promoter, suggesting potential gene downregulation with this allelic combination (Letra et al., 2014). Taken together, these observations indicate that the -709 A/G variant may directly regulate MMP3 promoter activity, although its function was shown to the driven by the 1171 5A/6A alleles in the background.
Polymorphisms in the TIMP2 gene have also been associated with NSCLP in different populations. A promoter variant in TIMP2 (-180C/T; rs8179096) was strongly associated with oral clefts in a Brazilian population, whereas additional variants of unknown significance (intronic) were associated with NSCLP in US and Northeastern European populations (Nikopensius et al., 2011; Letra et al., 2012). Furthermore, functional analysis suggests that this variant has distinct allele-dependent effects, with the T allele presenting a 2.5-fold increased promoter activity. Furthermore, both C and T alleles were found to be putative binding sites for NFκB, a key transcription factor involved in the innate immune system. While C and T alleles reduced binding capability when NFκB consensus binding oligo diverges from protein in the same reaction, introduction of a mutant NFκB immunized C and T alleles from binding abolition (Letra et al., 2014). Additional studies are still necessary to unveil the exact mechanisms by which MMPs and TIMPs might contribute to NSCLP; nonetheless, allelic polymorphisms in these genes and their interactions may partly explain the variance in individual susceptibility to oral clefts.
Few studies have described roles for other ECM remodeling or cross-link enzymes during palatogenesis and their potential association with CL/P. ADAMTS-9 and -20 have been shown to participate in versican remodeling during palatogenesis (Enomoto et al., 2010); furthermore, ADAMTS-9 null mice die in utero (Dubail and Apte, 2015). During physiological palate formation, ADAMTS9 expression in the palatal shelves was restricted to microvascular endothelial cells, derived from the mesoderm, whereas CNC-derived mesenchymal cells express ADAMTS20; in contrast, the expressions of ADAMTSs 4 and 5 were not detected during palatogenesis (Enomoto et al., 2010). More recently, a genome-wide association study in both dogs and humans independently identified ADAMTS20 as a potential candidate gene for clefting. A region on chromosome 27 in dogs was found to segregate with a complex phenotype of cleft palate and syndactyly; follow-up whole genome sequencing studies then identified a frameshift mutation in the ADAMTS20 gene as potentially etiologic. Furthermore, four novel risk variants in ADAMTS20 were identified in Guatemalan cases with NSCLP (Wolf et al., 2015).
Additional genes with roles in collagen maturation and other ECM molecules have also been suggested to play a role in NSCLP. The LOXL3 gene located on chromosome 2p13.1 and mutations in this gene have been reported in association with Stickler syndrome (MIM #108300), an autosomal recessive disorder characterized by ocular, auditory, skeletal, and orofacial abnormalities (Alzahrani et al., 2015). In mice, the deletion of Loxl3 causes neonatal mortality due to impaired collagen assembly and cross-linking, as well as spine deformity and cleft palate (Zhang et al., 2015). Furthermore, its ortholog in zebrafish is required for adequate craniofacial morphogenesis, as its loss of function results in abnormal chondrogenesis with micrognathia or agnathia phenotype (van Boxtel et al., 2011). Recently, a missense mutation in LOXL3 (rs17010021; p.Ile615Phe) was reported to contribute to an increased risk of NSCLP (10-fold) in a European population (Khan et al., 2018).
New Approaches to Studying Palatogenesis and Cl/P In Vitro
Studies of both palatogenesis and CL/P are usually carried out in animal models (mouse, chicken, zebrafish, and rat, respectively), especially mice, given the similarity of biological events in these animals to humans (Van Otterloo et al., 2016; Lough et al., 2017). In vitro, the most common method of study employs murine embryonic palatal cell culture and ex vivo palate organ culture (Figure 7A; Abbott, 2019).
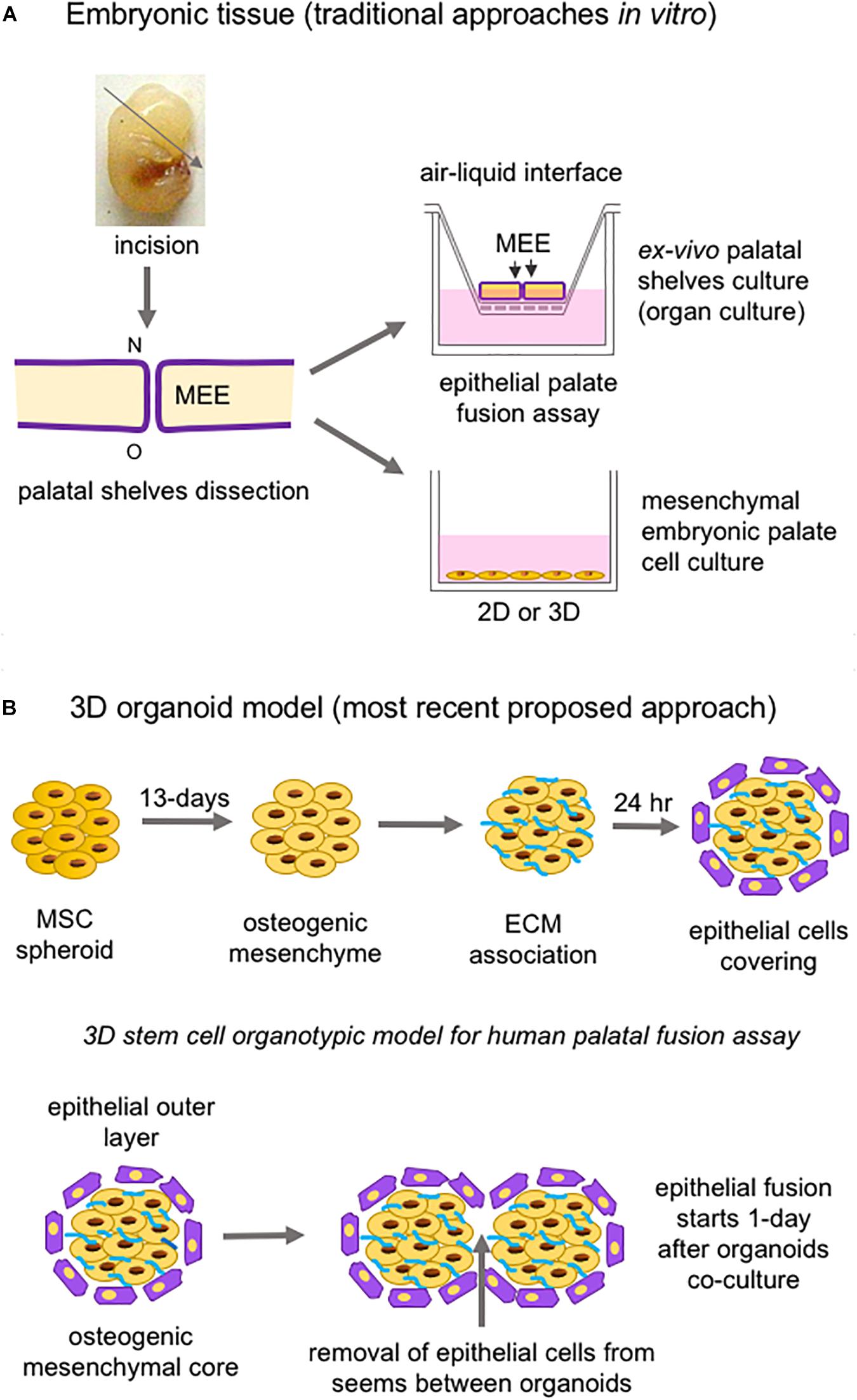
Figure 7. Schematic representation of in vitro approaches to study palatogenesis. (A) The most traditional method is the mouse embryonic palate dissection before palatal shelves fusion; the dissected palate is placed on a membrane insert, and the top part is in an air-liquid interface (organ culture); also, mesenchymal embryonic palate cells are cultivated under adhesion and monolayer on or not embedded into a substrate. (B) The newest proposed 3D organoid model using mesenchymal stem cells and epithelial cells to form spheroids/organoids to mimetize the epithelial fusion. ECM: extracellular matrix; MEE: medial edge epithelia; MSC: mesenchymal stem cell; N: nasal site; o: oral region; O: oral site.
Three-dimensional (3D) cell culture represents a promising approach to better elucidate cell behavior, ECM remodeling, tissue remodeling, and tissue fusion, and ultimately to investigate clinical applications for tissue engineering (Ong et al., 2017). 3D constructs are more similar to the tissue microenvironment than classical 2D cell cultures (plastic-based) as they demonstrate more realistic cell morphology and physiology; furthermore, observation of 3D cultures over time is considered to represent 4D systems (Bissell, 2017). Thus, over the last decade, numerous reviews have illustrated the switch from 2D to 3D cell cultures. Several methods to create 3D structures have been described, such as cell aggregates, spheroids or organoids (Alhaque et al., 2018), seeded onto decellularized matrices (Taylor et al., 2018), and cell sheets (Kim et al., 2019). Among these, cell spheroid is a technology in which cells are cultured in suspension to create a 3D structure using cell–cell interactions and “scaffold-free” strategy (Figure 8B). This technique yields detailed information regarding ECM remodeling through “omics” analysis since the generation, growth, and fusion of cells allow accurate monitoring (Fan et al., 2018; Schnellmann et al., 2018; Wong et al., 2018). Understanding the ECM and its soluble factors are essential to comprehend embryonic development and tissue repair and will contribute to the discovery of new therapeutic tools. Proteomics is an appropriate strategy to characterize ECM components under physiological and pathological conditions. A significant challenge for studying the ECM constitutes its solubilization and protein recovery. An optimized protocol developed by Naba et al. (2017) permitted the digestion of proteins into peptides that could be analyzed by mass spectroscopy; web tools were allowed the annotation and relative quantitation of the ECM proteins. These protocols allow a faster analysis of differentially expressed proteins in the ECM, eliciting the identification of biomarkers and therapeutic targets.
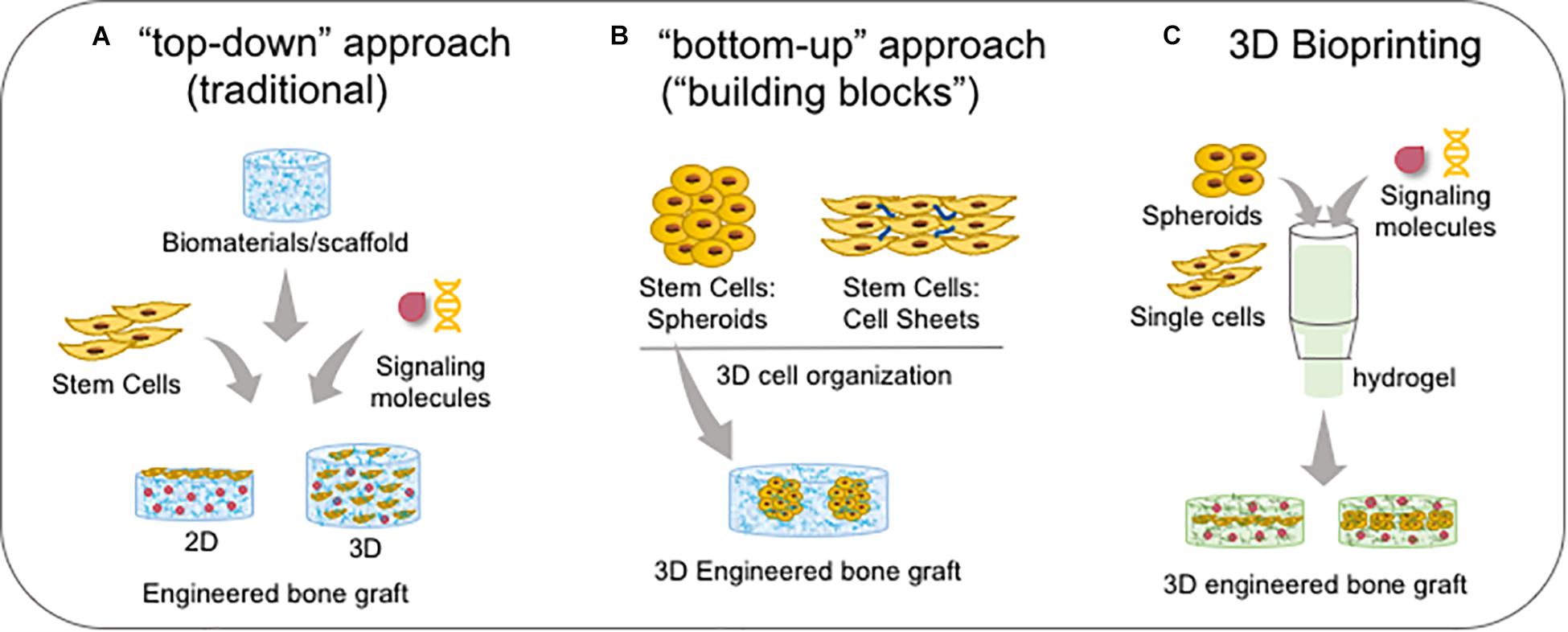
Figure 8. Bone bioengineering based-techniques. (A) Traditional “top-down” approach which stem cells are associated with biomaterials and signalling molecules to create a 2D (when single cells are seeded onto the substrate/biomaterial) or 3D microenvironment (when single cells are seeded into the substrate/biomaterial). (B) The newest “bottom-up” approach which cells are self-assembled in a 3D conformation by cell-cell and cell-ECM interactions as spheroids or cell sheets, forming the functional “building blocks”; spheroids can be embedded into scaffolds/biomaterials. (C) Both sing cells and spheroids and/or signalling molecules are the components of bioink in association with a hydrogel, and then it is bioprinted in a specific 3D engineered bone graft.
Stem cells from many tissue origins, especially MSCs, have been widely employed in 3D constructions due to their effortless isolation from various tissues of the body, potential for differentiation into mesenchymal and non-mesenchymal lineages, and therapeutic use (Han et al., 2019). Cell spheroids to study epithelial palate fusion are available, and strategies employed consist of creating mesenchymal spheroids from human umbilical-derived MSCs undergoing osteogenic differentiation, covered with ECM to mimic basement membrane, which are then seeded with human progenitor epithelial keratinocytes. This 3D organotypic model of human palatal shelves can maintain cell viability for around 4 weeks, expresses alkaline phosphatase, and is responsive to EGF, leading to increased epithelial proliferation and the prevention of epithelial fusion between spheroids (Figure 7B; Wolf et al., 2018). A potential bias relies on the fact that the MSCs used in the spheroid generation do not have the same embryonic origin as the palatal mesenchyme or the epithelial cells.
State-Of-The-Art of Cleft Palate Repair and Palatal Reconstruction
Bone Regeneration
Bone repair is a mechanism in which bone development program is recapitulated to form new intramembranous or endochondral bone at injured sites. Intense ECM bone remodeling by MMPs is required (Paiva and Granjeiro, 2017) as well as the action of several growth factors (TGF-β, BMPs, FGFs, IGFs, PDGFs, and VEGFs), which are implicated in driving osteochondral differentiation, regulation of bone formation, and are responsible for triggering intracellular pathways, such as Wnt (Houschyar et al., 2019). Additionally, other metalloproteinases are important for collagen maturation and mineralization. PPAP-A increases osteoblast proliferation in vitro and bone formation in vivo by augmenting IGF bioavailability upon cleavage of IGFBP-4 (Qin et al., 2006). PPAP-B expression was associated with postnatal skeletal growth, bone mass, and structure due to cleavage of IGFBP-5, the most abundant IGF in bone (Figure 9A; Amiri and Christians, 2015; Christians et al., 2019).
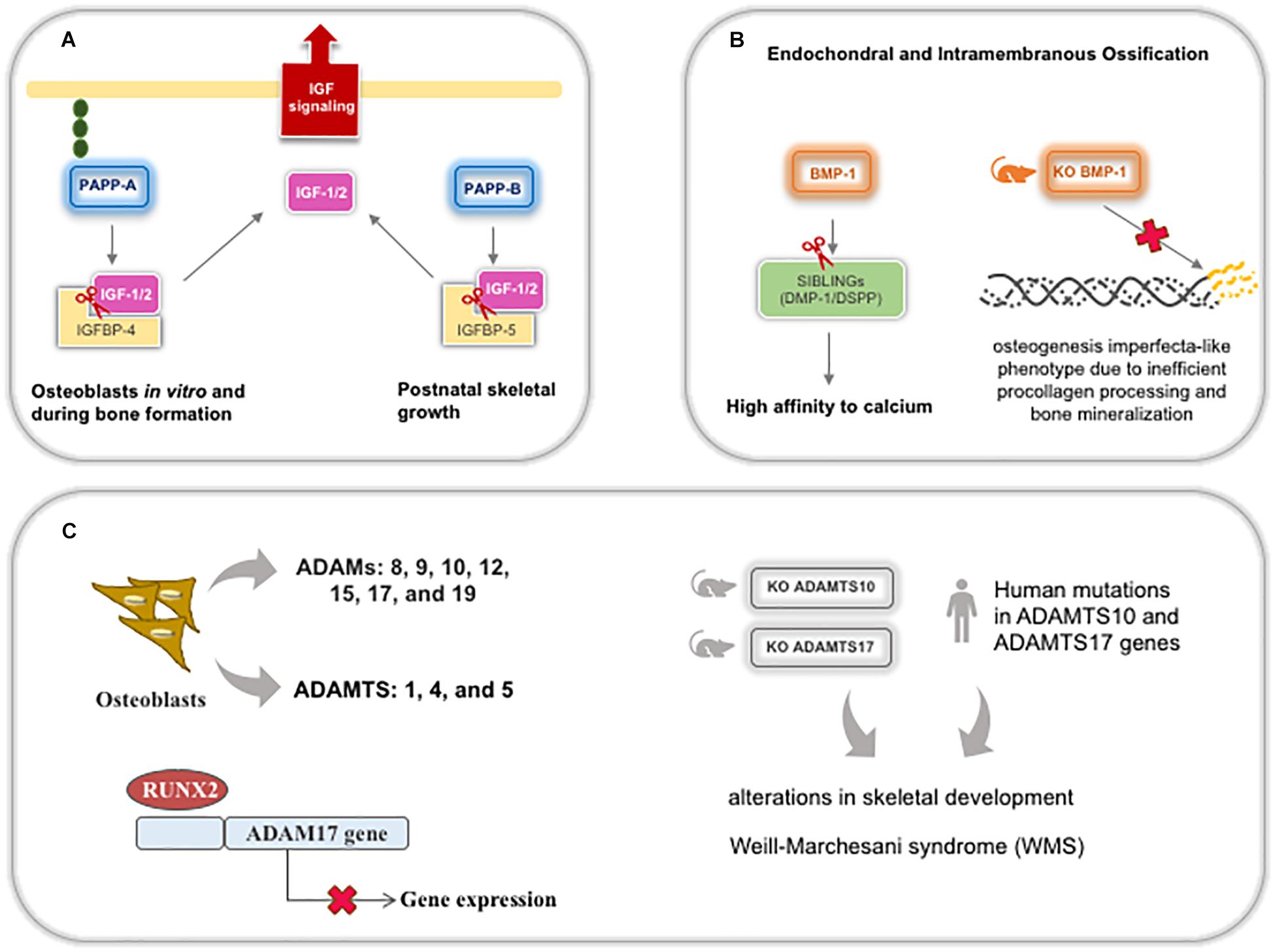
Figure 9. Schematic representation of metalloproteinases (except MMPs) expression and roles during bone development and postnatal skeletal growth. (A) PPAPP-A and B cleavage the complex IGFBP/IGFs to bioavailable IGF for bone cells. (B) BMP-1 is important for SIBLINGs cleavage, specifically DMP-1 and DSPP, leading these glycoproteins to increase their affinity to calcium and contributing for mineralization. (C) Several ADAMs and ADAMTSs are expressed by osteoblasts, being ADAM17 downregulated by RUNX2 during osteoblastic differentiation. Both ADAMTS10 and 17 knockout mice and human mutations display the same bone phenotype recognized in Weill-Marchesani syndrome. ADAM: adisintegrin and metalloproteinase; ADAMTS: adisintegrin and metalloproteinase with thrombospondin motifs; DMP-1: dentin matrix acidic phosphoprotein; DSPP: dentin sialophosphoprotein; IGFBP: insulin-like growth factor binding protein; IGF: insulin-like growth factor; KO: knockout mice; PAPP: pregnancy-associated plasma protein.
In bone and dentin matrices, BTPs can cleave the acidic domain of SIBLINGs (small integrin-binding ligand, N-linked glycoproteins), specifically DMP-1 and DSPP (dentin sialophosphoprotein—generating DPP/dentin phosphoprotein), leading to an increase in the binding affinity of these proteins for calcium, which is necessary for mineralization and improving ECM stiffness (Steiglitz et al., 2004; von Marschall and Fisher, 2010; Tsuchiya et al., 2011). BMP-1 is highly expressed in both endochondral and intramembranous ossification sites during development and contributes to an increase in bone repair (Grgurevic et al., 2011). In addition, BMP-1 expression is modulated by mutations in both alpha procollagen chains (Lindahl et al., 2011) and BMP-1-knockout mice develop an osteogenesis imperfecta-like phenotype due to inefficient procollagen processing and bone mineralization (Figure 9B; Muir et al., 2014). Although meprins also act on procollagen maturation and DSPP cleavage, meprin-β is inhibited in tissues with high concentrations of calcium and is, probably, not crucial in hard tissues since mice knocked out for meprins do not demonstrate alterations in bone and tooth development (Arnold et al., 2017).
Several ADAMs (8, 9, 10, 12, 15, 17, and 19) and ADAMTS (1, 4, and 5) are secreted by osteoblasts and bone tissue. Recently, ADAM17 has been reported to be a target of RUNX2 during osteoblastic differentiation, where ADAM17 is suppressed by RUNX2 (Araya et al., 2018). Human mutations in ADAMTS10 and 17 are associated with related syndromes involved in alterations in skeletal development (Dagoneau et al., 2004; Morales et al., 2009). Recently, knockout mice for both Adamts10 and 17 were shown to develop the recapitulate syndromic phenotype in human (Figure 9C; Mularczyk et al., 2018; Oichi et al., 2019). Reduced hypertrophic zone and increased deposition of fibrillin-2 alter the growth plates during endochondral ossification resulting in adults with short statures. Fibrillins are glycoproteins involved in microfibril formation and elastin deposition. Also, treatment with BMP-2 can rescue terminal chondrocyte differentiation, suggesting that ADAMTS17 is important for ossification through the modulation of BMP signaling (Oichi et al., 2019).
Extracellular matrix cross-linkers also contribute to bone formation and regeneration. In LOX knockout mice, general ECM architecture is profoundly affected in several tissues, and littermates die soon after birth. In bone, early and late osteoblastic differentiation and onset of mineralization are decreased in these mice as well as the gene expression of LOX isoforms (LOXL1-4) and osteoblastic markers (collagen type I, bone sialoprotein, and RUNX2) (Pischon et al., 2004).
The crosstalk between inflammatory cells, MSCs, chondrocytes, osteoblasts, osteoclasts, and osteocytes is necessary (Liu et al., 2018), and the success of bone regeneration results from the balance of the osteoprotegerin/RANK/RANKL axis. Most bone defects regenerate spontaneously; however, extensive bone loss due to trauma or aging-related fractures, metabolic bone disease, and congenital malformations do not regenerate per se, underscoring the search for better drug candidates. Biomaterials, biomolecules, and stem cells have been investigated to support bone repair, overcoming the limitations of autologous and allogeneic grafts (Collignon et al., 2017).
First Generation Palatal Reconstruction: Standard Surgical CL/P Management and Cleft Palate Repair
The treatment of CL/P patients is primarily surgical (plastic surgery for the lip and a bone graft for the palate) followed by orthodontic treatment in multiple stages over the years. The management is dependent on the cleft site, extension, and affected tissues (Farronato et al., 2014). The term “palatal reconstruction” is used to define any intervention able to restore the barrier between the oral and nasal cavities, and physiological functions. As such, several approaches, such as the lip and/or palate closure through plastic surgery, several types of bone graft, or a combination of these have been employed. More recently, the use of bioengineered bone has allowed palatal reconstruction (Figure 10). The decision with regard to the best treatment of choice depends on the type of CL/P and the extension of the tissue loss (Gupta et al., 2012).
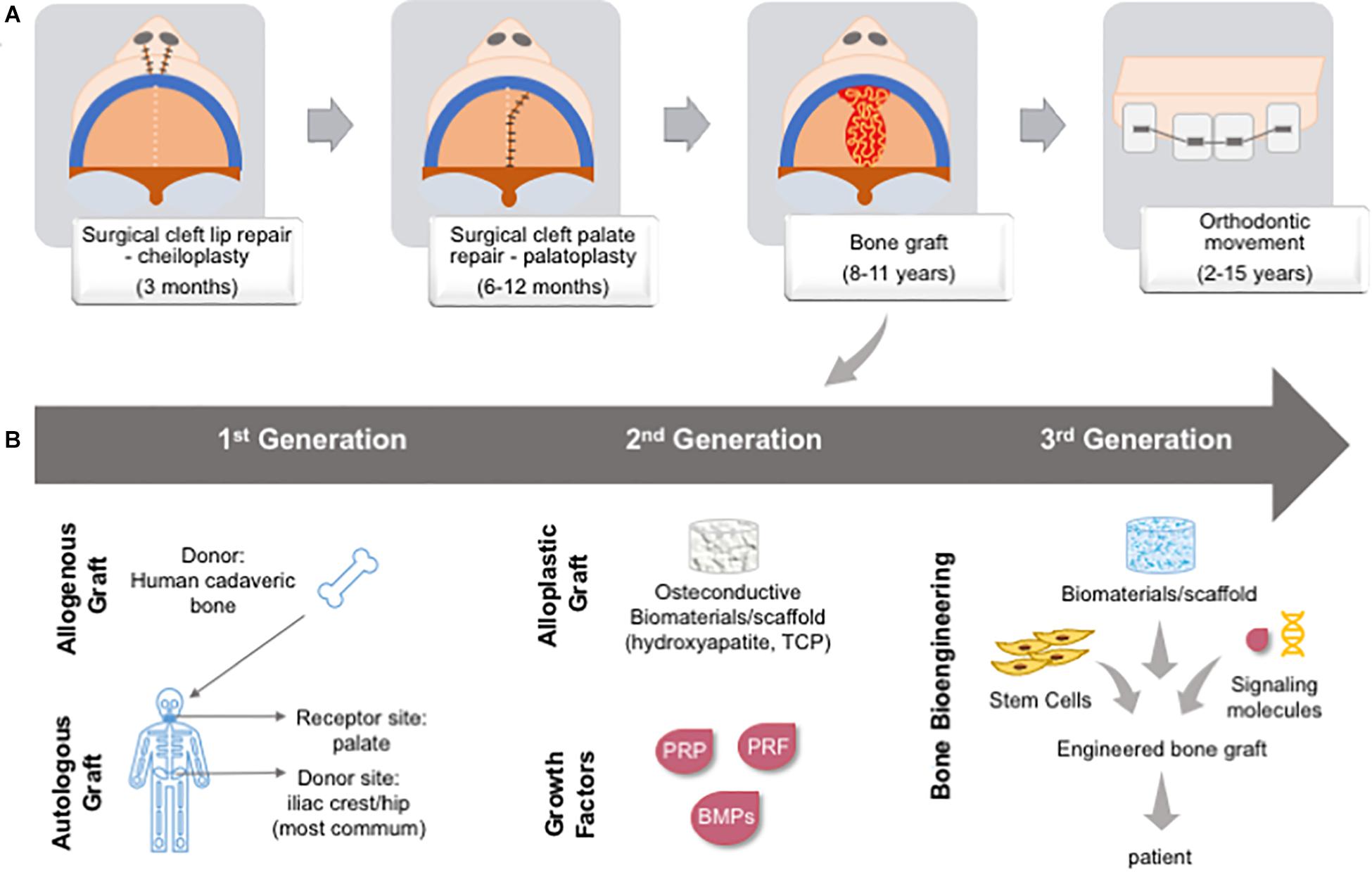
Figure 10. Cleft lip and/or palate patient chronological management over time and types of bone graft. (A) First, patients are submitted to a lip reconstruction (around 3 months); followed by palate reconstruction (between 6–12 months); primary bone graft in the hard palate (between 8–11 years); and, then orthodontic movement (between 2–15 years). (B) Bone graft was developing from gold-standard autologous and allogenous graft (1st generation), the development of osteoconductive biomaterials and recombinant growth factors or natural adjuvants (2nd generation), and bone bioengineering is bringing a new possibility to associate stem cells from patient, biomaterials and signaling molecules to create an in vitro engineered bone graft to be implanted in the patient, being a personalized approach (3rd generation). BMP: bone morphogenetic protein; PRF: platelet-rich fibrin; PRP: platelet-rich plasma; TCP: tricalcium phosphate.
Autogenous bone grafts are the gold standard for alveolar bone and cleft palate repairs. Throughout treatment stages, surgical procedures may require several grafts (e.g., primary, secondary, and tertiary graft). The most common bone donor sites are the anterior iliac crest, proximal tibia, mandibular symphysis, calvaria, and ribs. However, the bone amount and volume available from these sites are restricted, and a most advantageous site is still under debate. The use of autogenous bone graft has many drawbacks related to morbidity at the donor site, and patients may present chronic pain at the site, paresthesia of the thigh, and hypertrophic scar. The loss of the graft due to local inflammation, bone resorption, and the development of oronasal fistulas is also frequently associated with unsuccessful repair (Borba et al., 2014; Tavakolinejad et al., 2014; Shafi et al., 2015).
Second Generation of Palatal Reconstruction: Biomaterials and Growth Factors
Bone graft options available, such as allogeneic bone (Shirani et al., 2017), xenogeneic bone (Thuaksuban et al., 2010), and alloplastic graft (Kumar et al., 2013; Seifeldin, 2016; Sharif et al., 2016), still do not promote effective palatal reconstruction, stimulating the search for better alternatives. To overcome the limitations of autologous and allogeneic bone grafts, studies have investigated the use of the association, or not, of osteoconductive biomaterials, such as hydroxyapatite and tricalcium phosphate, with autologous bone graft or growth factors, such as BMP, which is known to stimulate bone formation and repair (Horch et al., 2006; Weijs et al., 2010; Lazarou et al., 2011; Benlidayi et al., 2012; De Ruiter et al., 2015; Takemaru et al., 2016; Martín-Del-Campo et al., 2019). After the systematic review of 29 studies, just two were used for qualitative and quantitative analysis (Segura-Castillo et al., 2005; Dickinson et al., 2008). However, considering some aspects of randomization, allocation concealment, blinding, outcome data, and reporting, the authors concluded that the two selected articles presented a high risk of bias and that conclusions concerning the benefits of the interventions, compared to traditional iliac grafting, could not be made (Guo et al., 2011).
Growth factors such as BMPs (2, 4, and 7) and “natural adjuvant” platelet concentrates, denominated platelet-rich plasma (PRP) and platelet-rich fibrin (PRF), have been employed in palate reconstruction (Behnia et al., 2012; Seifeldin, 2016). Interestingly, PRF does not affect the expression of the RUNX2, BMP-2, or RANKL, but induces the expression of OPG, leading to the increase of OPG/RANKL, suggesting that PRF could boost osteoblastic differentiation (Sumida et al., 2019).
Clinical studies of alveolar bone repair in CL/P patients tested the association of BMPs or platelet concentrates with the gold standard of autologous bone graft. BMPs make relevant contributions to bone embryogenesis and repair (Salazar et al., 2016) and the inhibition of BMP receptors (Lai et al., 2016), mutations (Sahoo et al., 2011), and gene polymorphism (Savitha et al., 2015) are involved in cleft development. The association of bone graft with human recombinant BMP-2 (hrBMP-2) in vitro and in vivo demonstrated the production of mature bone (Shimakura et al., 2003). Furthermore, clinical studies corroborate that BMPs are, at least, just as efficient as autologous bone graft for the repair of alveolar/palate cleft (Chin et al., 2005; Canan et al., 2012; Ayoub et al., 2016; Hammoudeh et al., 2017). Very recently, 10 years of follow-up evidenced the safe use of BMP-7 for the reconstruction of the alveolar cleft (Ayoub and Gillgrass, 2019).
Evaluating the use of growth factors for the reconstruction of the alveolar cleft, Van Hout group accessed 291 studies using BMP-2, BMP-7, TGF-β, PDGF, IGF, FGF, VEGF, and PRP (van Hout et al., 2011). Only six articles met the authors’ criteria for full analysis, who concluded that BMP-2 improved the quantity of bone formation during the reconstruction of the alveolar cleft, in association with reducing pain at the donor site, a reduction in surgery time, hospital stay, and overall cost. A recent study reported a similar effectiveness of autologous bone and rh-BMP2 during the maxillary reconstruction of cleft lip and palate patients (Scalzone et al., 2019).
A growing field of investigation aims to evaluate the use of cell-based therapies to treat alveolar and palate cleft (Gładysz and Hozyasz, 2015; Brozek et al., 2018) due to the ability of stem cells to differentiate into active osteoblasts that drive bone regeneration. Trabecular bone was enhanced after the treatment of 16 out of 17 alveolar clefts with MSCs together with hrBMP-2, but long-term follow-up studies are still needed (Fallucco and Carstens, 2009).
Two recent systematic reviews addressed the role of BMP2 (Scalzone et al., 2019) and tissue-engineered bone replacement materials (Kamal et al., 2018) in the efficient treatment of palate and alveolar cleft. Both studies concluded that there was no statistical difference between the autologous bone graft and the alternative materials. The meta-analysis comparing the bone repair with iliac crest bone graft (ICBG) versus BMP-2, acellular dermis matrix membrane, and cranium or rib grafts, indicated that ICBG is still the best choice for treatment (Wu C. et al., 2018). The association of BMP-2 with a collagen sponge provides similar results to those of ICBG, and PRP associated with ICBG increased bone retention for skeletally mature patients. However, better-designed controlled studies are required for long-term analysis of alveolar cleft reconstruction, with follow-ups of greater than 12 months (Seifeldin, 2016; Liang et al., 2018; Wu C. et al., 2018). Furthermore, the development of a consensus for standardized protocols, using multicenter studies, is still needed (de Ladeira and Alonso, 2012).
Third Generation of Palatal Reconstruction: Bone Bioengineering
Tissue bioengineering represents a new therapeutic alternative for palatal reconstruction, associating the use of stem cells, biomaterials/scaffolds, and signaling molecules. Two main approaches are dominant: the “top-down” approach, in which classically cells are seeded on or into biomaterials for recreating a new 3D tissue in vitro; and the “bottom-up” approach, which uses 3D construction techniques (described in the last section) to improve cell secretion of growth factors for tissue regeneration in vivo (Figure 8; Baptista et al., 2018). Furthermore, Good Manufacturing Practice (GMP) and animal-free supplements are crucial for clinical applications. The success of the bioengineered bone graft is directly related to the osteogenic potential of stem cells, biomaterial/scaffold specific properties, and adequate external signaling from growth factors.
Stem cells from different origins and potentialities represent the cutting-edge technology for palatal reconstruction (Zuk, 2008; Gładysz and Hozyasz, 2015). However, few studies of bone bioengineering approaches in animal models of palatal defects have been carried out. A large number of research articles have addressed bone bioengineering approaches for alveolar bone defects created in animal models to reproduce alveolar cleft or mid-palate cleft. However, here we consider only animal models of the mid-palate cleft and the use of human cells. Conejero et al. (2006) demonstrated that rat palatal critical-size defect filled with autogenous engineered graft (fat-derived stem cells previously differentiated into osteoblasts/osteocytes and seeded onto poly-L-lactic acid absorbable scaffolds) could regenerate bone at 6 or 12 weeks after surgery; bone defects filled only with the scaffold or scaffold plus undifferentiated cells had a fibrotic tissue with no bone. Another study used an autogenous multi-layered graft, which was simultaneously bioengineered for palate bone and oral mucosa in a rabbit palatal critical-size defect (Martín-Piedra et al., 2016). Initially, individual cell layers of adipose tissue-derived MSCs were seeded onto fibrin-agarose hydrogels and induced toward osteogenic differentiation; and with fibroblasts and keratinocytes seeded onto fibrin-agarose hydrogels, maintained in epithelial culture medium and under air–liquid conditions. Subsequently, the oral mucosa layer was placed on the top of the osteogenic layer and compressed to induce nanostructured fusion of the mucosal stroma (fibroblasts) and the osteogenic layer. The 3D multi-layered graft was able to integrate with host tissue, and achieved partial bone differentiation; the authors suggested that complex multi-layer constructions could increase the maturation times compared to monolayers in vivo.
Bone-marrow stem cells (BMSCs) are the “gold standard” for several cell- and tissue-based clinical applications. These are, to date, the most studied stem cells and their properties are well known, but it is not clear whether the application of BMSCs or other MSCs in craniofacial bone regeneration requires handling ex vivo and/or pre-differentiation before clinical application (Shanbhag et al., 2019). For alveolar cleft repair in CL/P patients, scaffold-free BMSCs are safe, but this material is not suitable for extensive bone defect repair (Bajestan et al., 2017). The associations of BMSCs with commercial demineralized bone matrix (Osteoset DBM) (Behnia et al., 2009), tricalcium phosphate (Du et al., 2017), platelet-derived growth factor on biphasic hydroxyapatite/tricalcium phosphate (Behnia et al., 2012), or PRF membrane (Mossaad et al., 2019), have been shown to contribute to bone repair. However, the use of BMSC does not reduce the morbidity caused by iliac crest donor site handling even when using minimally invasive techniques. Thus, other sources of MSCs are necessary to eliminate this side effect.
The MSCs found in adult dental tissues display cranial NCC properties (Liu and Cheung, 2016; Niibe et al., 2017; Cui et al., 2018), as these embryonic cells are more similar to palate forming cells than BMSCs. In the oral cavity, human MSCs have been isolated and characterized from odontogenic and non-odontogenic origins, permitting the harvesting of healthy tissues during dental surgical procedures. Thus, a subset of cells displaying MSC properties and osteogenic properties have been described from gingival connective tissue [gingival mesenchymal stem/progenitor cells (GMSCs)] (Yang et al., 2013), oral periosteum of the palate (Caballero et al., 2010; Ceccarelli et al., 2016), the lower and upper vestibule (Ceccarelli et al., 2016), palatal connective tissue (Roman et al., 2013; Páll et al., 2015, Pall et al., 2017), and adipose stem cells from buccal fat pad (Farré-Guasch et al., 2010). Recently, palatal periosteum-derived MSCs cells, cultivated under serum- and xeno-free conditions, and cells were able to retain stem cell properties (Naung et al., 2019). One registered clinical trial was conducted using adipose stem cells from the buccal fat pad (Khojasteh et al., 2017), associated with ICBG, lateral ramus cortical bone plate, and bovine mineral graft, with all groups producing statistically similar results. However, these reports presented a limited source of tissue, and were assessed in specific situations, and therefore more effective cell sources are still required.
Five MSC populations are found in dental tissues: (I) in the dental follicle [dental follicle progenitor stem cells (DFPSCs)]; (II) in the apical papilla [stem cells from apical papilla (SCAPs)]; (III) in the ligament [periodontal ligament stem cells (PDLSCs)]; (IV) in the adult dental pulp [dental pulp stem cells (DPSCs)]; and (V) in the dental pulp of deciduous teeth [stem cells from exfoliated deciduous teeth (SHEDs)] (Baniebrahimi et al., 2018). Of these, SHEDs, which have exfoliative characteristics, are the most easily obtained odontogenic tissue, via a little or non-invasive procedure. The pulp tissue can be obtained during the period of the changing of the child’s teeth, between 5 and 12 years of age, with insignificant ethical implications and provides a suitable alternative for pediatric regenerative medicine (Taguchi et al., 2019). SHEDs display high proliferative capacity, multi-lineage differentiation, secretion of immunomodulatory molecules. DPSCs, similarly to SHEDs, could be an alternative source of cells from teenagers or during adulthood during dental procedures being and are easily harvested from third molars routinely indicated to exodontia (Yamada et al., 2019). Both DPSCs and SHEDs allow cell sheets (Pedroni et al., 2018; Lee et al., 2019) and 3D spheroids cultures (Wang et al., 2010; Xiao et al., 2014). The high regenerative potential of SHEDs and DPSCs could be explained by its particular secretome content, including many types of paracrine soluble molecules and EVs, identified as immunogenic, pro-neurogenic, and pro-angiogenic (Kichenbrand et al., 2018; Marei and El Backly, 2018; Yusof et al., 2018). SHED secretome profile is also modulated during osteogenic differentiation leading to increase angiogenic potential (Mussano et al., 2018).
Concerning bone repair, SHEDs are better for forming new bone in a calvaria critical-size defect model, when compared to other dental MSCs and BMSCs. Recently, human SHEDs seeded onto dense collagen hydrogels, which were primed with FGF-2 and submitted to hypoxia conditions before implantation, improved intramembranous bone formation in an immunodeficient calvaria critical-size bone defect mouse model (Novais et al., 2019). Most of the 56 articles thoroughly analyzed in a systematic review reported good results and the relevance of human DPSCs for bone engineering in animal models or human clinical treatments (Leyendecker Junior et al., 2018). Similarly, a narrative review of 39 studies also concluded that DPSCs and SHEDs were of value for bone tissue regeneration (Cristaldi et al., 2018). Since 2005, many countries have started to collect and store healthy exfoliated teeth, creating biobanks, which are of low cost in comparison to umbilical cord banks (Campanella, 2018). Unfortunately, ex vivo manipulation of stem cells is still required, representing a challenge since this is a high cost and time-consuming procedure.
Little information is available regarding the use of MSCs harvested from CL/P patients. Bueno et al. (2011) isolated and characterized MSCs from orbicularis oris muscle (denominated by the authors as “cleft lip and palate muscle-derived stem cells”), usually discarded during the initial surgery repair (cheiloplasty) of CLP patients. These cells are able to express classical MSC cell surface proteins and differentiate into osteogenic, adipogenic, chondrogenic, and skeletal muscle cell lineages in vitro. The cells, when seeded onto collagen membranes, display the ability to repair bone in an immunocompromised rat critical-size cranial defect model. The analysis of DNA variants affecting the gene expression (cis-eQTLs) of cleft lip and palate muscle-derived stem cells allowed the discovery of a new susceptibility locus for NSCLP was discovered (rs1063588), coincident with the MRPL53 gene (Masotti et al., 2017). Bueno et al. (2011) compared the gene expression profile of SHEDs from healthy donors and CL/P patients and verified that 87 genes presented differential expressions, with more than a half being glycoproteins related to the ECM (collagens, MMPs, integrins, and adhesion proteins). This study showed that MSCs might be a powerful tool for genetic and “omic” studies of CL/P. Later, the same group showed that low power laser therapy could enhance the osteogenic potential of DPSCs from CL/P patients (Pinheiro et al., 2017). These results support the need for the study of MSCs from CL/P patients to better understand cell behavior, ECM secretion, and remodeling, and employ this knowledge to drive new strategies based on bone bioengineering for palatal reconstruction.
Another growing field related to regenerative medicine that could be applied to palate reconstruction is bioprinting. Spheroids are potential building blocks in 3D bioprinting, in a large-scale process for bone and cartilage tissue production. Growing evidence shows that 3D spheroids formed from MSC present increased angiogenic and chemotactic signaling (Costa et al., 2017). 3D bioprinted cryogels, formed from chitosan (CS)/gelatin-based scaffolds for personalized palate reconstruction, have been designed by CL/P computed tomography data (Hixon et al., 2017). A 3D bioprinted bioresorbable scaffold (polycaprolactone—approved by the Korean Ministry of Food and Drug Safety for clinical use) seeded with autologous BMSCs from the iliac crest in the operation room, was implanted in a 10-year-old Korean boy with a history of previously repaired unilateral CL/P presenting a cleft alveolus and an oronasal fistula. At 6 months after transplantation, the new bone formed reached around 45% of the total defect volume, suggesting that this new technology could be a promising alternative (Ahn et al., 2018).
Three-dimensional additive manufacture allows the production of biomaterials/scaffolds used successfully for bone bioengineering have to be biocompatible, specific porosity, chemical and topographical characteristics and surface properties for osteoconduction, biomechanical properties, biodegradability, and radiolucency, to induce osteogenesis (osteoinductivity) and vascular ingrowth. In a new vision, biomaterials may trigger immunological host responses to stimulate tissue regeneration (Franz et al., 2011). As we have previously seen, the provisional matrix is secreted by embryonic palatal cells to drive palatogenesis. Among biopolymers used as biomaterials/scaffolds for bone bioengineering, collagen type I, CS, and HA are the most commonly employed. CS is a polysaccharide chitin-derived present in invertebrate exoskeletons which displays many promising characteristics such as biocompatibility, antibacterial activity, biodegradability, porosity, immunomodulatory properties, promotes cell adhesion, proliferation, migration, and ultimately enhances bone regeneration due to stimulation of osteoblast differentiation and mineralization (Farhadihosseinabadi et al., 2019).
As we have seen previously, LOX and TGs are crucial enzymes for collagen–collagen cross-links. These enzymes have been employed to create cross-links in collagen-based biomaterials (Fortunati et al., 2014; Cai et al., 2017). This modern approach can replace the traditional chemical method employing aldehydes, isocyanates, and carbodiimides. Although this method creates strong stable cross-links, undesirable consequences such as cytotoxicity, calcification, and foreign body response are well known for these chemicals (Sorushanova et al., 2019). Specifically in bone tissue, TG induces the oligomerization of SIBLINSs, and calcium-binding proteins (osteopontin and bone sialoprotein), which may drive mineral nucleation or calcium crystal growth (Forsprecher et al., 2009).
Concluding Remarks
Although several MMPs and their inhibitors TIMPs are expressed during palatogenesis, the cell membrane-anchored MMPs appear to represent the principal pericellular collagenases (MMP-14 and MMP-16) and may be crucial for the development of CL/P. However, recent studies have demonstrated roles for other metalloproteinases, such as ADAMTS in this pathology. There seems to be a crucial compensatory effect between MMPs to ensure that the various stages of palate formation can occur. However, more studies should be carried out on other metalloproteinases to better understand the complexity of ECM remodeling, the generation of bioactive molecules, and the relationship between them.
Cleft therapy still is dependent on bone grafts, mainly of the ICBG, but new approaches have been under evaluation. Administration of BMP-2 and PDGF combined with bone graft is promising, but recent systematic reviews support the need for better-designed randomized controlled clinical trials with long-term follow-up (>12 months). The development of in vitro models using stem cells from CL/P patients may be an exciting approach for further studies of the biology of these cells as well as their potential use in new individualized therapeutic approaches for palatal reconstruction. 3D culture has been used to recapitulate the critical events of development, such as cell–cell interaction, differentiation, growth and cell fusion, which, coupled with “omics” analysis and computational biology, promote considerable advances in tissue remodeling and repair. Additionally, the emerging and exciting field of EVs may provide critical information via “omics” analyses to understand how this component of ECM contributes to the palatogenesis of CL/P.
The development of new biomaterials that simulate a provisional matrix during palatogenesis and provide the controlled release of growth factors may aid to improve new therapeutic approaches based on bone bioengineering. Emerging cell reprogramming or trans-differentiation technologies could provide other unusual sources of therapeutic cells. The former consists of differentiated adult cells, such as fibroblasts, that are forced to overexpress transcription factors that regulate pluripotency generating inducible pluripotent stem cells (iPSCs) displaying the same characteristics as embryonic stem cells (Spyrou et al., 2019). The second approach consists of the direct conversion of a fully differentiated cell type into another one; for example, the conversion of fibroblasts into osteoblasts (Cho and Ryoo, 2018). Furthermore, another possibility is to use EVs released by MSCs as potential cell-free tools for bone regeneration (Praveen Kumar et al., 2019). Finally, we now have a vast knowledge of craniofacial development, CL/P and CL/P treatment, associated with many advances in cell and material engineering technologies. So, multidisciplinary efforts must be made to achieve advances in the quality of life of CL/P patients.
Author Contributions
KP contributed to the design, writing, and financial support of the manuscript. CM, PS, AL, and JG contributed to the design and writing of the manuscript.
Funding
We are grateful for financial support from the São Paulo Research Foundation (FAPESP KBSP fellowship 2017/26813-9).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ABCA4, ATP-binding cassette sub-family A (ABC1) member 4; ADAM, a disintegrin and metalloproteinase; ADAMST, a disintegrin and metalloproteinase with thrombospondin-like motifs; BMP, bone morphogenetic protein; BMSC, bone marrow stem cell; BTP, BMP-1/Tolloid-like proteases; CL/P, cleft and lip palate; CRISPLD2, cysteine-rich secretory protein LCCL domain-containing 2; CSPG, chondroitin sulfate proteoglycans; DFPSC, dental follicle progenitor stem cells; DMP-1, dentin matrix protein-1; DSPP, dentin sialophosphoprotein; DPP, dentin phosphoprotein; DPSC, dental pulp stem cell; ECM, extracellular matrix; ED, embryonic day; EGFR, epidermal growth factor receptor; EMT, epithelial-mesenchymal transition; EV, extracellular vesicles; FGF, fibrobast growth factor; FGFR, fibrobast growth factor receptor; FOXE1, forkhead box E1; GAG, glycosaminoglycans; GDF, growth differentiation factor; GH, growth hormone; GMP, good manufacturing practice; GMSC, gingival mesenchymal stem/progenitor cell; GPI, glycosyl-phosphatidyl-inositol; GW, gestational weeks; HA, hyaluronic acid; Has, hyaluronan synthase; Hh, hedgehog; IGFBPs, insulin-like growth factor binding proteins; IGF, insulin growth factor; IRF6, interferon regulatory factor 6; KO, knockout mice; LOX, lysyl oxidase; LOXL, lysyl oxidase-like; LTBPs, TGF- β binding proteins; MAFB, V-maf musculoaponeurotic fibrosarcoma oncogene homolog B; MEE, medial edge epithelia; MES, medial epithelial seam; MMP, matrix metalloproteinase; MSC, mesenchymal stem cell mTLD, tolloid; MTLL-1, tolloid-like 1; MSX1, Msh homeobox 1; MV, matrix vesicles; NCC, neural crest cells; NFkB, factor nuclear kappa B; NSCLP, nonsyndromic cleft and lip palate; OPG, osteoprotegerin; PAPP, pregnancy-associated plasma protein; PCPE, procollagen C-proteinase enhancer; PDGF, platelet-derived growth factor; PDLSC, periodontal ligament stem cell; PRF, platelet-rich fibrin; PRP, platelet-rich plasma; Rac1, Ras-related C3 botulinum toxin substrate 1; RANK, receptor activator of nuclear factor κ B; RANKL, receptor activator of nuclear factor κ B-ligand; SHED, stem cells from exfoliated deciduous teeth; SCAP, stem cells from apical papilla; SRLP, small rich-leucine proteoglycans; SIBLING, small integrin-binding ligand N-linked glycoprotein); TG, transglutaminase; TGF-α, transforming growth factor alpha; TGF-β, transforming growth factor beta; VAX1, ventral anterior homeobox 1; VEGF, vascular endothelial growth factor.
References
Abbott, B. D. (2019). Embryonic midfacial palatal organ culture methods in developmental toxicology BT - developmental toxicologys. Methods Mol. Biol. 1965, 93–105. doi: 10.1007/978-1-4939-9182-2_7
Ahn, G., Lee, J.-S., Yun, W.-S., Shim, J.-H., and Lee, U.-L. (2018). Cleft alveolus reconstruction using a three-dimensional printed bioresorbable scaffold with human bone marrow cells. J. Craniofac. Surg. 29, 1880–1883. doi: 10.1097/SCS.0000000000004747
Alhaque, S., Themis, M., and Rashidi, H. (2018). Three-dimensional cell culture: from evolution to revolution. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 373:20170216. doi: 10.1098/rstb.2017.0216
Alzahrani, F., Al Hazzaa, S. A., Tayeb, H., and Alkuraya, F. S. (2015). LOXL3, encoding lysyl oxidase-like 3, is mutated in a family with autosomal recessive stickler syndrome. Hum. Genet. 134, 451–453. doi: 10.1007/s00439-015-1531-z
Amiri, N., and Christians, J. K. (2015). PAPP-A2 expression by osteoblasts is required for normal postnatal growth in mice. Growth Horm. IGF Res. 25, 274–280. doi: 10.1016/j.ghir.2015.09.003
Ansorge, M., and Pompe, T. (2018). Systems for localized release to mimic paracrine cell communication in vitro. J. Control. Release 278, 24–36. doi: 10.1016/j.jconrel.2018.03.028
Araya, H. F., Sepulveda, H., Lizama, C. O., Vega, O. A., Jerez, S., Briceño, P. F., et al. (2018). Expression of the ectodomain-releasing protease ADAM17 is directly regulated by the osteosarcoma and bone-related transcription factor RUNX2. J. Cell. Biochem. 119, 8204–8219. doi: 10.1002/jcb.26832
Arnold, P., Koopmann, L., Peters, F., Birkenfeld, F., Goff, S. V.-L., Damm, T., et al. (2017). Deficiency of the DSPP-cleaving enzymes meprin α and meprin β does not result in dentin malformation in mice. Cell Tissue Res. 367, 351–358. doi: 10.1007/s00441-016-2498-3
Arpino, V., Brock, M., and Gill, S. E. (2015). The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 4, 247–254. doi: 10.1016/j.matbio.2015.03.005
Aviram, R., Zaffryar-Eilot, S., Hubmacher, D., Grunwald, H., Mäki, J. M., Myllyharju, J., et al. (2019). Interactions between lysyl oxidases and ADAMTS proteins suggest a novel crosstalk between two extracellular matrix families. Matrix Biol. 7, 114–125. doi: 10.1016/j.matbio.2018.05.003
Ayoub, A., and Gillgrass, T. (2019). The clinical application of recombinant human bone morphogenetic protein 7 for reconstruction of alveolar cleft: 10 Years’ Follow-Up. J. Oral Maxillofac. Surg. 77, 571–581. doi: 10.1016/j.joms.2018.08.031
Ayoub, A., Roshan, C. P., Gillgrass, T., Naudi, K., and Ray, A. (2016). The clinical application of rhBMP-7 for the reconstruction of alveolar cleft. J. Plast. Reconstr. Aesthet. Surg. 69, 101–107. doi: 10.1016/j.bjps.2015.09.004
Baek, J.-A., Lan, Y., Liu, H., Maltby, K. M., Mishina, Y., and Jiang, R. (2011). Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev. Biol. 350, 520–531. doi: 10.1016/j.ydbio.2010.12.028
Bajestan, M. N., Rajan, A., Edwards, S. P., Aronovich, S., Cevidanes, L. H. S., Polymeri, A., et al. (2017). Stem cell therapy for reconstruction of alveolar cleft and trauma defects in adults: a randomized controlled, clinical trial. Clin. Implant Dent. Relat. Res. 19, 793–801. doi: 10.1111/cid.12506
Baniebrahimi, G., Khanmohammadi, R., and Mir, F. (2018). Teeth-derived stem cells: a source for cell therapy. J. Cell. Physiol. 234, 2426–2435. doi: 10.1002/jcp.27270
Baptista, L. S., Kronemberger, G. S., Côrtes, I., Charelli, L. E., Matsui, R. A. M., Palhares, T. N., et al. (2018). Adult stem cells spheroids to optimize cell colonization in scaffolds for cartilage and bone tissue engineering. Int. J. Mol. Sci. 19:1285. doi: 10.3390/ijms19051285
Barker, T. H., and Engler, A. J. (2017). The provisional matrix: setting the stage for tissue repair outcomes. Matrix Biol. 6, 1–4. doi: 10.1016/j.matbio.2017.04.003
Bauvois, B. (2012). New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim. Biophys. Acta Rev. Cancer 1825, 29–36. doi: 10.1016/j.bbcan.2011.10.001
Beaty, T. H., Marazita, M. L., and Leslie, E. J. (2016). Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Research 5:2800. doi: 10.12688/f1000research.9503.1
Behnia, H., Khojasteh, A., Soleimani, M., Tehranchi, A., and Atashi, A. (2012). Repair of alveolar cleft defect with mesenchymal stem cells and platelet derived growth factors: a preliminary report. J. Cranio Maxillofac. Surg. 40, 2–7. doi: 10.1016/j.jcms.2011.02.003
Behnia, H., Khojasteh, A., Soleimani, M., Tehranchi, A., Khoshzaban, A., Keshel, S. H., et al. (2009). Secondary repair of alveolar clefts using human mesenchymal stem cells. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 108, e1–e6. doi: 10.1016/j.tripleo.2009.03.040
Bekhouche, M., and Colige, A. (2015). The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol. 4, 46–53. doi: 10.1016/j.matbio.2015.04.001
Benkovics, A. H., Skopelitis, D. S., Benkovics, A. H., Husbands, A. Y., and Timmermans, M. C. P. (2017). Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev. Cell 43, 265.e6–273.e6. doi: 10.1016/j.devcel.2017.10.003
Benlidayi, M. E., Tatli, U., Kurkcu, M., Uzel, A., and Oztunc, H. (2012). Comparison of bovine-derived hydroxyapatite and autogenous bone for secondary alveolar bone grafting in patients with alveolar clefts. J. Oral Maxillofac. Surg. 70, e95–e102. doi: 10.1016/j.joms.2011.08.041
Bissell, M. J. (2017). Goodbye flat biology – time for the 3rd and the 4th dimensions. J. Cell Sci. 130, 3–5. doi: 10.1242/jcs.200550
Blanton, S. H., Bertin, T., Serna, M. E., Stal, S., Mulliken, J. B., and Hecht, J. T. (2004). Association of chromosomal regions 3p21.2, 10p13, and 16p13.3 with nonsyndromic cleft lip and palate. Am. J. Med. Genet. 125A, 23–27. doi: 10.1002/ajmg.a.20426
Blavier, L., and DeClerck, Y. A. (1997). Tissue inhibitor of metalloproteinases-2 is expressed in the interstitial matrix in adult mouse organs and during embryonic development. Mol. Biol. Cell 8, 1513–1527. doi: 10.1091/mbc.8.8.1513
Blavier, L., Lazaryev, A., Groffen, J., Heisterkamp, N., DeClerck, Y. A., and Kaartinen, V. (2001). TGF-beta3-induced palatogenesis requires matrix metalloproteinases. Mol. Biol. Cell 12, 1457–1466. doi: 10.1091/mbc.12.5.1457
Bond, J. S. (2019). Proteases: history, discovery, and roles in health and disease. J. Biol. Chem. 294, 1643–1651. doi: 10.1074/jbc.TM118.004156
Borba, A. M., Borges, A. H., Da Silva, C. S. V., Brozoski, M. A., Naclério-Homem, M. D. G., and Miloro, M. (2014). Predictors of complication for alveolar cleft bone graft. Br. J. Oral Maxillofac. Surg. 52, 174–178. doi: 10.1016/j.bjoms.2013.11.001
Boyd, D. D. (2006). Regulation of matrix metalloproteinase gene. Mol. Cell Biochem. 253, 269–285. doi: 10.1002/JCP
Brown, G. D., and Nazarali, A. J. (2010). Matrix metalloproteinase-25 has a functional role in mouse secondary palate development and is a downstream target of TGF-3. BMC Dev. Biol. 10:93. doi: 10.1186/1471-213X-10-93
Brown, N. L., Yarram, S. J., Mansell, J. P., and Sandy, J. R. (2002). Matrix metalloproteinases have a role in palatogenesis. J. Dent. Res. 81, 826–830. doi: 10.1177/154405910208101206
Brozek, R., Kurpisz, M., and Koczorowski, R. (2018). Application of stem cells in dentistry for bone regeneration. J. Physiol. Pharmacol. 69, 23–33. doi: 10.26402/jpp.2018.1.03
Bueno, D. F., Sunaga, D. Y., Kobayashi, G. S., Aguena, M., Raposo-Amaral, C. E., Masotti, C., et al. (2011). Human stem cell cultures from cleft lip/palate patients show enrichment of transcripts involved in extracellular matrix modeling by comparison to controls. Stem Cell Rev. Rep. 7, 446–457. doi: 10.1007/s12015-010-9197-3
Bush, J. O., and Jiang, R. (2012). Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139, 231–243. doi: 10.1242/dev.067082
Butler, G. S., and Overall, C. M. (2009). Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry 48, 10830–10845. doi: 10.1021/bi901656f
Caballero, M., Reed, C. R., Madan, G., and van Aalst, J. A. (2010). Osteoinduction in umbilical cord- and palate periosteum-derived mesenchymal stem cells. Ann. Plast. Surg. 64, 605–609. doi: 10.1097/SAP.0b013e3181ce3929
Cai, L., Xiong, X., Kong, X., and Xie, J. (2017). The role of the lysyl oxidases in tissue repair and remodeling: a concise review. Tissue Eng. Regen. Med. 14, 15–30. doi: 10.1007/s13770-016-0007-0
Campanella, V. (2018). Dental stem cells: current research and future applications. Eur. J. Paediatr. Dent. 19, 257–257. doi: 10.23804/ejpd.2018.19.04.1
Canan, L. W. J., da Silva Freitas, R., Alonso, N., Tanikawa, D. Y. S., Rocha, D. L., and Coelho, J. C. U. (2012). Human bone morphogenetic protein-2 use for maxillary reconstruction in cleft lip and palate patients. J. Craniofac. Surg. 23, 1627–1633. doi: 10.1097/SCS.0b013e31825c75ba
Ceccarelli, G., Graziano, A., Benedetti, L., Imbriani, M., Romano, F., Ferrarotti, F., et al. (2016). Osteogenic potential of human oral-periosteal cells (PCs) isolated from different oral origin: an in vitro study. J. Cell. Physiol. 231, 607–612. doi: 10.1002/jcp.25104
Cerdà-Costa, N., and Gomis-Rüth, F. X. (2014). Architecture and function of metallopeptidase catalytic domains. Protein Sci. 23, 123–144. doi: 10.1002/pro.2400
Chen, G., Li, M. X., Wang, H. X., Hong, J. W., Shen, J. Y., Wang, Q., et al. (2018). Identification of key genes in cleft lip with or without cleft palate regulated by miR-199a-5p. Int. J. Pediatr. Otorhinolaryngol. 111, 128–137. doi: 10.1016/j.ijporl.2018.06.005
Chester, D., and Brown, A. C. (2017). The role of biophysical properties of provisional matrix proteins in wound repair. Matrix Biol. 6, 124–140. doi: 10.1016/j.matbio.2016.08.004
Chin, M., Ng, T., Tom, W. K., and Carstens, M. (2005). Repair of alveolar clefts with recombinant human bone morphogenetic protein (rhBMP-2) in patients with clefts. J. Craniofac. Surg. 16, 778–789. doi: 10.1097/01.scs.0000166802.49021.01
Cho, Y., and Ryoo, H. (2018). Trans-differentiation via epigenetics: a new paradigm in the bone regeneration. J. Bone Metab. 25:9. doi: 10.11005/jbm.2018.25.1.9
Christians, J. K., Amiri, N., Schipilow, J. D., Zhang, S. W., and May-Rashke, K. I. (2019). Pappa2 deletion has sex- and age-specific effects on bone in mice. Growth Horm. IGF Res. 44, 6–10. doi: 10.1016/j.ghir.2018.10.006
Clark, I. M., Swingler, T. E., Sampieri, C. L., and Edwards, D. R. (2008). The regulation of matrix metalloproteinases and their inhibitors. Int. J. Biochem. Cell Biol. 40, 1362–1378. doi: 10.1016/j.biocel.2007.12.006
Cobourne, M. T., and Green, J. B. A. (2012). Hedgehog signalling in development of the secondary palate. Front. Oral Biol. 16, 52–59. doi: 10.1159/000337543
Collignon, A. M., Lesieur, J., Vacher, C., Chaussain, C., and Rochefort, G. Y. (2017). Strategies developed to induce, direct, and potentiate bone healing. Front. Physiol. 8:927. doi: 10.3389/fphys.2017.00927
Conejero, J. A., Lee, J. A., Parrett, B. M., Terry, M., Wear-Maggitti, K., Grant, R. T., et al. (2006). Repair of palatal bone defects using osteogenically differentiated fat-derived stem cells. Plast. Reconstr. Surg. 117, 857–863. doi: 10.1097/01.prs.0000204566.13979.c1
Costa, M. H. G., McDevitt, T. C., Cabral, J. M. S., da Silva, C. L., and Ferreira, F. C. (2017). Tridimensional configurations of human mesenchymal stem/stromal cells to enhance cell paracrine potential towards wound healing processes. J. Biotechnol. 262, 28–39. doi: 10.1016/j.jbiotec.2017.09.020
Cristaldi, M., Mauceri, R., Tomasello, L., Pizzo, G., Pizzolanti, G., Giordano, C., et al. (2018). Dental pulp stem cells for bone tissue engineering: a review of the current literature and a look to the future. Regen. Med. 13, 207–218. doi: 10.2217/rme-2017-0112
Cruz, L., Romero, J. A. A., Iglesia, R. P., and Lopes, M. H. (2018). Extracellular vesicles: decoding a new language for cellular communication in early embryonic development. Front. Cell Dev. Biol. 6:94. doi: 10.3389/fcell.2018.00094
Cui, D., Li, H., Wan, M., Peng, Y., Xu, X., Zhou, X., et al. (2018). The origin and identification of mesenchymal stem cells in teeth: from odontogenic to non-odontogenic. Curr. Stem Cell Res. Ther. 13, 39–45. doi: 10.2174/1574888X12666170913150403
Dagoneau, N., Benoist-Lasselin, C., Huber, C., Faivre, L., Mégarbané, A., Alswaid, A., et al. (2004). ADAMTS10 mutations in autosomal recessive weill-marchesani syndrome. Am. J. Hum. Genet. 75, 801–806. doi: 10.1086/425231
Dancevic, C. M., Mcculloch, D. R., and Ward, A. C. (2016). The ADAMTS hyalectanase family: biological insights from diverse species. Biochem. J. 473, 2011–2022. doi: 10.1042/BCJ20160148
de Ladeira, P. R. S., and Alonso, N. (2012). Protocols in cleft lip and palate treatment: systematic review. Plast. Surg. Int. 2012:562892. doi: 10.1155/2012/562892
de Oliveira Demarchi, A. C. C., Zambuzzi, W. F., Paiva, K. B. S., da Silva-Valenzuela, M. D., Nunes, F. D., de Cássia Sávio Figueira, R., et al. (2010). Development of secondary palate requires strict regulation of ECM remodeling: sequential distribution of RECK, MMP-2, MMP-3, and MMP-9. Cell Tissue Res. 340, 61–69. doi: 10.1007/s00441-010-0931-6
De Ruiter, A., Janssen, N., Van Es, R., Frank, M., Meijer, G., Koole, R., et al. (2015). Micro-structured beta-tricalcium phosphate for repair of the alveolar cleft in cleft lip and palate patients: a pilot study. Cleft Palaten Craniofac. J. 52, 336–340. doi: 10.1597/13-260
Deryugina, E. I., and Quigley, J. P. (2010). Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: contrasting, overlapping and compensatory functions2. Biochim. Biophys. Acta 1803, 103–120. doi: 10.1016/j.bbamcr.2009.09.017
Dickinson, B. P., Ashley, R. K., Wasson, K. L., O’Hara, C., Gabbay, J., Heller, J. B., et al. (2008). Reduced morbidity and improved healing with bone morphogenic protein-2 in older patients with alveolar cleft defects. Plast. Reconstr. Surg. 121, 209–217. doi: 10.1097/01.prs.0000293870.64781.12
Ding, H. L., Hooper, J. E., Batzel, P., Frank Eames, B., Postlethwait, J. H., Artinger, K. B., et al. (2016). MicroRNA profiling during craniofacial development: potential roles for mir23b and mir133b. Front. Physiol. 7:281. doi: 10.3389/fphys.2016.00281
Dixon, M. J., Carette, M. J. M., Moser, B. B., and Ferguson, M. W. J. (1993a). Differentiation of isolated murine embryonic palatal epithelium in culture: exogenous transforming growth factor alpha modulates matrix biosynthesis in defined experimental conditions. Vitr. Cell. Dev. Biol. 29A, 51–61. doi: 10.1007/bf02634371
Dixon, M. J., Foreman, D., Schor, S., William, M., and Ferguson, J. (1993b). Epidermal growth factor and transforming growth factor alpha regulate extracellular matrix production by embryonic mouse palatal mesenchymal cells cultured on a variety of substrata. Roux Arch. Dev. Biol. 203, 140–150. doi: 10.1007/BF00365053
Dixon, M. J., Marazita, M. L., Beaty, T. H., and Murray, J. C. (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 12, 167–178. doi: 10.1038/nrg2933
Du, F., Wu, H., Li, H., Cai, L., Wang, Q., Liu, X., et al. (2017). Bone marrow mononuclear cells combined with beta-tricalcium phosphate granules for alveolar cleft repair: a 12-month clinical study. Sci. Rep. 7:13773. doi: 10.1038/s41598-017-12602-01
Dubail, J., and Apte, S. S. (2015). Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 4, 24–37. doi: 10.1016/j.matbio.2015.03.001
Dupin, E., Calloni, G. W., Coelho-Aguiar, J. M., and Le Douarin, N. M. (2018). The issue of the multipotency of the neural crest cells. Dev. Biol. 444, S47–S59. doi: 10.1016/j.ydbio.2018.03.024
Dworkin, S., Boglev, Y., Owens, H., and Goldie, S. J. (2016). The role of sonic hedgehog in craniofacial patterning, morphogenesis and cranial neural crest survival. J. Dev. Biol. 4:24. doi: 10.3390/jdb4030024
Eberhart, J. K., He, X., Swartz, M. E., Yan, Y. L., Song, H., Boling, T. C., et al. (2008). MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat. Genet. 40, 290–298. doi: 10.1038/ng.82
Eckert, R. L., Kaartinen, M. T., Nurminskaya, M., Belkin, A. M., Colak, G., Johnson, G. V. W., et al. (2014). Transglutaminase regulation of cell function. Physiol. Rev. 94, 383–417. doi: 10.1152/physrev.00019.2013
English, W. R., Velasco, G., Stracke, J. O., Knauper, V., and Murphy, G. (2001). Catalytic activities of membrane-type 6 matrix metalloproteinase (MMP25)117. FEBS Lett. 491, 137–142. doi: 10.1016/s0014-5793(01)02150-0
Enomoto, H., Nelson, C. M., Somerville, R. P. T., Mielke, K., Dixon, L. J., Powell, K., et al. (2010). Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development 137, 4029–4038. doi: 10.1242/dev.050591
Fallucco, M. A., and Carstens, M. H. (2009). Primary reconstruction of alveolar clefts using recombinant human bone morphogenic protein-2: clinical and radiographic outcomes. J. Craniofac. Surg. 20, 1759–1764. doi: 10.1097/SCS.0b013e3181b5d08e
Fan, S. M.-Y., Chen, P.-H., Tseng, Y.-T., Hong, J.-B., Chen, W., Tsai, T.-F., et al. (2018). Preclinical evaluation of melanocyte transplantation by chitosan-based melanocyte spheroid patch to skin prepared by controlled sunburn blistering. J. Biomed. Mater. Res. B Appl. Biomater. 106, 2535–2543. doi: 10.1002/jbm.b.34070
Farhadihosseinabadi, B., Zarebkohan, A., Eftekhary, M., and Heiat, M. (2019). Crosstalk between chitosan and cell signaling pathways. Cell. Mol. Life Sci. 76, 2697–2718. doi: 10.1007/s00018-019-03107-3103
Farré-Guasch, E., Martí-Pagès, C., Hernández-Alfaro, F., Klein-Nulend, J., and Casals, N. (2010). Buccal fat pad, an oral access source of human adipose stem cells with potential for osteochondral tissue engineering: an in vitro study. Tissue Eng. C Methods 16, 1083–1094. doi: 10.1089/ten.tec.2009.0487
Farronato, G., Kairyte, L., Giannini, L., Galbiati, G., and Maspero, C. (2014). How various surgical protocols of the unilateral cleft lip and palate influence the facial growth and possible orthodontic problems? Which is the best timing of lip, palate and alveolus repair? literature review. Stomatologija 16, 53–60.
Fawcett, E. (1906). On the development, ossification, and growth of the palate bone of man. J. Anat. Physiol. 40, 400–406.
Forsprecher, J., Wang, Z., Nelea, V., and Kaartinen, M. T. (2009). Enhanced osteoblast adhesion on transglutaminase 2-crosslinked fibronectin. Amino Acids 36, 747–753. doi: 10.1007/s00726-008-0125-7
Fortunati, D., Chau, D. Y. S., Wang, Z., Collighan, R. J., and Griffin, M. (2014). Cross-linking of collagen I by tissue transglutaminase provides a promising biomaterial for promoting bone healing. Amino Acids 46, 1751–1761. doi: 10.1007/s00726-014-1732-0
Francis-west, P., and Crespo-Enriquez, I. (2016). Vertebrate embryo: craniofacial development. eLS 1–15. doi: 10.1002/9780470015902.a0026602
Franz, S., Rammelt, S., Scharnweber, D., and Simon, J. C. (2011). Immune responses to implants - A review of the implications for the design of immunomodulatory biomaterials. Biomaterials 32, 6692–6709. doi: 10.1016/j.biomaterials.2011.05.078
Fu, Y., and Kong, W. (2017). Cartilage oligomeric matrix protein: matricellular and matricrine signaling in cardiovascular homeostasis and disease. Curr. Vasc. Pharmacol. 15, 186–196. doi: 10.2174/1570161115666170201121232
Fuller, K., and Chambers, T. J. (1995). Localisation of mRNA for collagenase in osteocytic, bone surface and chondrocytic cells but not osteoclasts. J. Cell Sci. 108, 2221–2230.
Funato, N. (2015). Molecular basis of cleft palates in mice. World J. Biol. Chem. 6, 121–138. doi: 10.4331/wjbc.v6.i3.121
Gabison, E. E., Huet, E., Baudouin, C., and Menashi, S. (2009). Direct epithelial-stromal interaction in corneal wound healing: role of EMMPRIN/CD147 in MMPs induction and beyond. Prog. Retin. Eye Res. 28, 19–33. doi: 10.1016/j.preteyeres.2008.11.001
Gack, S., Vallon, R., Schmidt, J., Grigoriadis, A., Tuckermann, J., Schenkel, J., et al. (1995). Expression of interstitial collagenase during skeletal development of the mouse is restricted to osteoblast-like cells and hypertrophic chondrocytes1. Cell Growth Differ. 6, 759–767.
Galloway, J. L., Jones, S. J., Mossey, P. A., and Ellis, I. R. (2013). The control and importance of hyaluronan synthase expression in palatogenesis. Front. Physiol. 4:10. doi: 10.3389/fphys.2013.00010
Gao, S., Moreno, M., Eliason, S., Cao, H., Li, X., Yu, W., et al. (2015). TBX1 protein interactions and microRNA-96-5p regulation controls cell proliferation during craniofacial and dental development: implications for 22q11.2 deletion syndrome. Hum. Mol. Genet. 24, 2330–2348. doi: 10.1093/hmg/ddu750
Garantziotis, S., and Savani, R. C. (2019). Hyaluronan biology: a complex balancing act of structure, function, location and context. Matrix Biol. 78-79, 1–10. doi: 10.1016/j.matbio.2019.02.002
Geng, Y., McQuillan, D., and Roughley, P. J. (2006). SLRP interaction can protect collagen fibrils from cleavage by collagenases. Matrix Biol. 25, 484–491. doi: 10.1016/j.matbio.2006.08.259
Gkantidis, N., Blumer, S., Katsaros, C., Graf, D., and Chiquet, M. (2012). Site-specific expression of gelatinolytic activity during morphogenesis of the secondary palate in the mouse embryo3. PLoS One 7:e47762. doi: 10.1371/journal.pone.0047762
Gładysz, D., and Hozyasz, K. K. (2015). Stem cell regenerative therapy in alveolar cleft reconstruction. Arch. Oral Biol. 60, 1517–1532. doi: 10.1016/j.archoralbio.2015.07.003
Graf, D., Malik, Z., Hayano, S., and Mishina, Y. (2016). Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 27, 129–139. doi: 10.1016/j.cytogfr.2015.11.004
Grassia, V., Lombardi, A., Kawasaki, H., Ferri, C., Perillo, L., Mosca, L., et al. (2018). Salivary microRNAs as new molecular markers in cleft lip and palate: a new frontier in molecular medicine. Oncotarget 9, 18929–18938. doi: 10.18632/oncotarget.24838
Greene, R. M., and Pisano, M. M. (2010). Palate morphogenesis: current understanding and future directions. Birth Defects Res. C. Embryo Today 90, 133–154. doi: 10.1002/bdrc.20180
Grgurevic, L., Macek, B., Mercep, M., Jelic, M., Smoljanovic, T., Erjavec, I., et al. (2011). Bone morphogenetic protein (BMP)1-3 enhances bone repair. Biochem. Biophys. Res. Commun. 408, 25–31. doi: 10.1016/j.bbrc.2011.03.109
Guo, J., Li, C., Zhang, Q., Wu, G., Deacon, S. A., Chen, J., et al. (2011). Secondary bone grafting for alveolar cleft in children with cleft lip or cleft lip and palate. Cochrane Database Syst. Rev. 6:CD008050. doi: 10.1002/14651858.CD008050.pub2
Gupta, V., Hospital, M. S., and Arshad, H. (2012). Palatal reconstruction. Curr. Opin. Otolaryngol. Head Neck Surg. 20, 225–230. doi: 10.1097/MOO.0b013e328355389f
Hadler-Olsen, E., Fadnes, B., Sylte, I., Uhlin-Hansen, L., and Winberg, J.-O. (2011). Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 278, 28–45. doi: 10.1111/j.1742-4658.2010.07920.x
Hammond, N. L., Dixon, J., and Dixon, M. J. (2017). Periderm: life-cycle and function during orofacial and epidermal development. Semin. Cell Dev. Biol. 91, 75–83. doi: 10.1016/j.semcdb.2017.08.021
Hammoudeh, J. A., Fahradyan, A., Gould, D. J., Liang, F., Imahiyerobo, T., Urbinelli, L., et al. (2017). A comparative analysis of recombinant human bone morphogenetic protein-2 with a demineralized bone matrix versus iliac crest bone graft for secondary alveolar bone grafts in patients with cleft lip and palate: review of 501 cases. Plast. Reconstr. Surg. 140, 318e–325e. doi: 10.1097/PRS.0000000000003519
Han, H.-W., Asano, S., and Hsu, S. (2019). Cellular spheroids of mesenchymal stem cells and their perspectives in future healthcare. Appl. Sci. 9:627. doi: 10.3390/app9040627
He, F., and Chen, Y. (2012). Wnt signaling in lip and palate development. Front. Oral Biol. 16, 81–90. doi: 10.1159/000337619
Hernandez-Barrantes, S., Bernardo, M., Toth, M., and Fridman, R. (2002). Regulation of membrane type-matrix metalloproteinases. Semin. Biol. 12, 131–138. doi: 10.1006/scbi.2001.0421
Hill, C. R., Yuasa, M., Schoenecker, J., and Goudy, S. L. (2014). Jagged1 is essential for osteoblast development during maxillary ossification. Bone 62, 10–21. doi: 10.1016/j.bone.2014.01.019
Hill, C., Jacobs, B., Kennedy, L., Rohde, S., Zhou, B., Baldwin, S., et al. (2015). Cranial neural crest deletion of VEGFa causes cleft palate with aberrant vascular and bone development. Cell Tissue Res. 361, 711–722. doi: 10.1007/s00441-015-2150-7
Hirata, A., Katayama, K., Tsuji, T., Natsume, N., Sugahara, T., Koga, Y., et al. (2013). Heparanase localization during palatogenesis in mice. Biomed Res. Int. 2013:760236. doi: 10.1155/2013/760236
Hixon, K. R., Melvin, A. M., Lin, A. Y., Hall, A. F., and Sell, S. A. (2017). Cryogel scaffolds from patient-specific 3D-printed molds for personalized tissue-engineered bone regeneration in pediatric cleft-craniofacial defects. J. Biomater. Appl. 32, 598–611. doi: 10.1177/0885328217734824
Horch, H.-H., Sader, R., Pautke, C., Neff, A., Deppe, H., and Kolk, A. (2006). Synthetic, pure-phase beta-tricalcium phosphate ceramic granules (Cerasorb®) for bone regeneration in the reconstructive surgery of the jaws. Int. J. Oral Maxillofac. Surg. 35, 708–713. doi: 10.1016/j.ijom.2006.03.017
Horejs, C. M. (2016). Basement membrane fragments in the context of the epithelial-to-mesenchymal transition. Eur. J. Cell Biol. 95, 427–440. doi: 10.1016/j.ejcb.2016.06.002
Houschyar, K. S., Tapking, C., Borrelli, M. R., Popp, D., Duscher, D., Maan, Z. N., et al. (2019). Wnt pathway in bone repair and regeneration – what do we know so far. Front. Cell Dev. Biol. 6:170. doi: 10.3389/fcell.2018.00170
Huang, G., Zhang, Y., Kim, B., Ge, G., Annis, D. S., Mosher, D. F., et al. (2009). Fibronectin binds and enhances the activity of bone morphogenetic protein 1. J. Biol. Chem. 284, 25879–25888. doi: 10.1074/jbc.M109.024125
Hubmacher, D., and Apte, S. S. (2015). ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 47, 34–43. doi: 10.1016/j.matbio.2015.05.004
Hynes, R. O., and Naba, A. (2012). Overview of the matrisome–an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 4:a004903. doi: 10.1101/cshperspect.a004903
Iamaroon, A., Wallon, U. M., Overall, C. M., and Diewert, V. M. (1996). Expression of 72-kDa gelatinase (matrix metailloproteinase-2) in the developing mouse craniofacial complex. Arch. Oral Biol. 41, 1109–1119. doi: 10.1016/S0003-9969(96)00097-0
Inomata, H. (2017). Scaling of pattern formations and morphogen gradients. Dev. Growth Differ. 59, 41–51. doi: 10.1111/dgd.12337
Iozzo, R. V., and Schaefer, L. (2015). Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol. 42, 11–55. doi: 10.1016/j.matbio.2015.02.003
Ishorst, N., Francheschelli, P., Böhmer, A. C., Khan, M. F. J., Heilmann-Heimbach, S., Fricker, N., et al. (2018). Nonsyndromic cleft palate: an association study at GWAS candidate loci in a multiethnic sample. Birth Defects Res. 110, 871–882. doi: 10.1002/bdr2.1213
Itoh, Y. (2017). Metalloproteinases in rheumatoid arthritis: potential therapeutic targets to improve current therapies. Prog. Mol. Biol. Transl. Sci. 148, 327–338. doi: 10.1016/bs.pmbts.2017.03.002
Iwata, J., Parada, C., and Chai, Y. (2011). The mechanism of TGF-β signaling during palate development. Oral Dis. 17, 733–744. doi: 10.1111/j.1601-0825.2011.01806.x
Iyyanar, P. P. R., and Nazarali, A. J. (2017). Hoxa2 inhibits bone morphogenetic protein signaling during osteogenic differentiation of the palatal mesenchyme. Front. Physiol. 8:929. doi: 10.3389/fphys.2017.00929
Jankowski, R., and Márquez, S. (2016). Embryology of the nose: the evo-devo concept. World J. Otorhinolaryngol. 6:33. doi: 10.5319/wjo.v6.i2.33
Jiang, R., Bush, J. O., and Lidral, A. C. (2006). Development of the upper lip: morphogenetic and molecular mechanisms. Dev. Dyn. 235, 1152–1166. doi: 10.1002/dvdy.20646
Kadir, A., Mossey, P. A., Orth, M., Blencowe, H., Sowmiya, M., Lawn, J. E., et al. (2017). Systematic review and meta-analysis of the birth prevalence of orofacial clefts in low- and middle-income countries. Cleft Palate Craniofac. J. 54, 571–581. doi: 10.1597/15-221
Kamal, M., Ziyab, A. H., Bartella, A., Mitchell, D., Al-Asfour, A., Hölzle, F., et al. (2018). Volumetric comparison of autogenous bone and tissue-engineered bone replacement materials in alveolar cleft repair: a systematic review and meta-analysis. Br. J. Oral Maxillofac. Surg. 56, 453–462. doi: 10.1016/j.bjoms.2018.05.007
Kantar, R. S., Cammarata, M. J., Rifkin, W. J., Diaz-Siso, J. R., Hamdan, U. S., and Flores, R. L. (2019). Foundation-based cleft care in developing countries. Plast. Reconstr. Surg. 143, 1165–1178. doi: 10.1097/PRS.0000000000005416
Karamanos, N. K., Theocharis, A. D., Neill, T., and Iozzo, R. V. (2019). Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biol. 7, 1–11. doi: 10.1016/j.matbio.2018.08.007
Kelwick, R., Desanlis, I., Wheeler, G. N., and Edwards, D. R. (2015). The ADAMTS (A Disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 16:113. doi: 10.1186/s13059-015-0676-3
Khan, M. F. J., Little, J., Mossey, P. A., Steegers-Theunissen, R. P. M., Bonsi, M., Bassi Andreasi, R., et al. (2018). Association between a common missense variant in LOXL3 gene and the risk of non-syndromic cleft palate. Congenit. Anom. 58, 136–140. doi: 10.1111/cga.12288
Khojasteh, A., Kheiri, L., Behnia, H., Tehranchi, A., Nazeman, P., Nadjmi, N., et al. (2017). Lateral ramus cortical bone plate in alveolar cleft osteoplasty with concomitant use of buccal fat pad derived cells and autogenous bone: phase I clinical trial. Biomed Res. Int. 2017:6560234. doi: 10.1155/2017/6560234
Kichenbrand, C., Velot, E., Menu, P., and Moby, V. (2018). Dental pulp stem cell-derived conditioned medium: an attractive alternative for regenerative therapy. Tissue Eng. B Rev. 25, 78–88. doi: 10.1089/ten.teb.2018.0168
Kim, H., Kim, Y., Park, J., Hwang, N. S., Lee, Y. K., and Hwang, Y. (2019). Recent advances in engineered stem cell-derived cell sheets for tissue regeneration. Polymers 11:209. doi: 10.3390/polym11020209
Kintakas, C., and McCulloch, D. R. (2011). Emerging roles for ADAMTS5 during development and disease. Matrix Biol. 30, 311–317. doi: 10.1016/j.matbio.2011.05.004
Krejci, P., Prochazkova, J., Bryja, V., Kozubik, A., and Wilcox, W. R. (2009). Molecular pathology of the fibroblast growth factor family. Hum. Mutat. 30, 1245–1255. doi: 10.1002/humu.21067
Kumar, P., Vinitha, B., and Fathima, G. (2013). Bone grafts in dentistry. J. Pharm. Bioallied Sci. 5, S125–S127. doi: 10.4103/0975-7406.113312
Lai, Y., Xie, C., Zhang, S., Gan, G., Wu, D., and Chen, W. (2016). Bone morphogenetic protein type I receptor inhibition induces cleft palate associated with micrognathia and cleft lower lip in mice. Birth Defects Res. A Clin. Mol. Teratol. 106, 612–623. doi: 10.1002/bdra.23504
Lan, Y., Qin, C., and Jiang, R. (2019). Requirement of hyaluronan synthase-2 in craniofacial and palate development. J. Dent. Res. 98, 1367–1375. doi: 10.1177/0022034519872478
Lazarou, S. A., Contodimos, G. B., and Gkegkes, I. D. (2011). Correction of alveolar cleft with calcium-based bone substitutes. J. Craniofac. Surg. 22, 854–857. doi: 10.1097/SCS.0b013e31820f7f19
Lee, J.-M., Kim, H.-Y., Park, J.-S., Lee, D.-J., Zhang, S., Green, D. W., et al. (2019). Developing palatal bone using human mesenchymal stem cell and stem cells from exfoliated deciduous teeth cell sheets. J. Tissue Eng. Regen. Med. 13, 319–327. doi: 10.1002/term.2811
Lemarchant, S., Wojciechowski, S., Vivien, D., and Koistinaho, J. (2017). ADAMTS-4 in central nervous system pathologies. J. Neurosci. Res. 95, 1703–1711. doi: 10.1002/jnr.24021
Letra, A., Silva, R. M., Motta, L. G., Blanton, S. H., Hecht, J. T., Granjeirol, J. M., et al. (2012). Association of MMP3 and TIMP2 promoter polymorphisms with nonsyndromic oral clefts. Birth Defects Res. A Clin. Mol. Teratol. 94, 540–548. doi: 10.1002/bdra.23026
Letra, A., Zhao, M., Silva, R. M., Vieira, A. R., and Hecht, J. T. (2014). Functional significance of MMP3 and TIMP2 polymorphisms in cleft lip/palate. J. Dent. Res. 93, 651–656. doi: 10.1177/0022034514534444
Levi, B., Brugman, S., Wong, V. W., Grova, M., Longaker, T., Wan, D. C., et al. (2011). Palatogenesis Engineering, pathways and pathologies. Organogenesis 7, 242–254. doi: 10.4161/org.7.4.17926
Leyendecker Junior, A., Gomes Pinheiro, C. C., Lazzaretti Fernandes, T., and Franco Bueno, D. (2018). The use of human dental pulp stem cells for in vivo bone tissue engineering: a systematic review. J. Tissue Eng. 9:2041731417752766. doi: 10.1177/2041731417752766
Li, C., Lan, Y., and Jiang, R. (2017). Molecular and cellular mechanisms of palate development. J. Dent. Res. 96, 1184–1191. doi: 10.1177/0022034517703580
Li, D., Zhang, H., Ma, L., Han, Y., Xu, M., Wang, Z., et al. (2016). Associations between microRNA binding site SNPs in FGFs and FGFRs and the risk of non-syndromic orofacial cleft. Sci. Rep. 6, 1–8. doi: 10.1038/srep31054
Li, J., Zou, J., Li, Q., Chen, L., Gao, Y., Yan, H., et al. (2016). Assessment of differentially expressed plasma microRNAs in nonsyndromic cleft palate and nonsyndromic cleft lip with cleft palate. Oncotarget 7, 86266–86279. doi: 10.18632/oncotarget.13379
Li, L., Meng, T., Jia, Z., Zhu, G., and Shi, B. (2010). Single nucleotide polymorphism associated with nonsyndromic cleft palate influences the processing of miR-140. Am. J. Med. Genet. A 152, 856–862. doi: 10.1002/ajmg.a.33236
Li, N.-G., Tang, Y.-P., Duan, J.-A., and Shi, Z.-H. (2014). Matrix metalloproteinase inhibitors: a patent review (2011 – 2013). Expert Opin. Ther. Pat. 24, 1039–1052. doi: 10.1517/13543776.2014.937424
Li, R., Chen, Z., Yu, Q., Weng, M., and Chen, Z. (2018). The function and regulatory network of pax9 gene in palate development. J. Dent. Res. 98, 277–287. doi: 10.1177/0022034518811861
Li, Y., Aoki, T., Mori, Y., Ahmad, M., Miyamori, H., Takino, T., et al. (2004). Cleavage of lumican by membrane-type matrix metalloproteinase-1 abrogates this proteoglycan-mediated suppression of tumor cell colony formation in soft agar. Cancer Res. 64, 7058–7064. doi: 10.1158/0008-5472.CAN-04-1038
Liang, F., Leland, H., Jedrzejewski, B., Auslander, A., Maniskas, S., Swanson, J., et al. (2018). Alternatives to autologous bone graft in alveolar cleft reconstruction: the state of alveolar tissue engineering. J. Craniofac. Surg. 29, 584–593. doi: 10.1097/SCS.0000000000004300
Lindahl, K., Barnes, A. M., Fratzl-Zelman, N., Whyte, M. P., Hefferan, T. E., Makareeva, E., et al. (2011). COL1 C-propeptide cleavage site mutations cause high bone mass osteogenesis imperfecta. Hum. Mutat. 32, 598–609. doi: 10.1002/humu.21475
Lisi, S., D’Amore, M., and Sisto, M. (2014). ADAM17 at the interface between inflammation and autoimmunity. Immunol. Lett. 162, 159–169. doi: 10.1016/j.imlet.2014.08.008
Liu, H., Li, D., Zhang, Y., and Li, M. (2018). Inflammation, mesenchymal stem cells and bone regeneration. Histochem. Cell Biol. 149, 393–404. doi: 10.1007/s00418-018-1643-1643
Liu, J. A., and Cheung, M. (2016). Neural crest stem cells and their potential therapeutic applications. Dev. Biol. 419, 199–216. doi: 10.1016/j.ydbio.2016.09.006
Lochter, A., Galosy, S., Muschler, J., Freedman, N., Werb, Z., and Bissell, M. J. (1997). Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J. Cell Biol. 139, 1861–1872. doi: 10.1083/jcb.139.7.1861
López-Otín, C., and Bond, J. S. (2008). Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 283, 30433–30437. doi: 10.1074/jbc.R800035200
Lough, K. J., Byrd, K. M., Spitzer, D. C., and Williams, S. E. (2017). Closing the gap: mouse models to study adhesion in secondary palatogenesis. J. Dent. Res. 96, 1210–1220. doi: 10.1177/0022034517726284
Ma, L., Xu, M., Li, D., Han, Y., Wang, Z., Yuan, H., et al. (2014). A miRNA-binding-site SNP of MSX1 is associated with NSOC susceptibility. J. Dent. Res. 93, 559–564. doi: 10.1177/0022034514527617
Macho, P., Bohac, M., Fedeles, J. Jr., Fekiacova, D., and Fedeles, J. Sr. (2017). Impact of cleft lip and/or palate in children on family quality of life before and after reconstructive surgery. Bratisl. Lek. Listy. 118, 370–373. doi: 10.4149/BLL
Malinowski, M., Pietraszek, K., Perreau, C., Boguslawski, M., Decot, V., Stoltz, J. F., et al. (2012). Effect of lumican on the migration of human mesenchymal stem cells and endothelial progenitor cells: involvement of matrix metalloproteinase-14. PLoS One 7:e50709. doi: 10.1371/journal.pone.0050709
Mammadova, A., Zhou, H., Carels, C. E. L., and Von den Hoff, J. W. (2016). Retinoic acid signalling in the development of the epidermis, the limbs and the secondary palate. Differentiation 92, 326–335. doi: 10.1016/j.diff.2016.05.001
Mannello, F., and Medda, V. (2012). Nuclear localization of matrix metalloproteinases. Prog. Histochem. Cytochem. 47, 27–58. doi: 10.1016/j.proghi.2011.12.002
Mansell, J. P., Kerrigan, J., McGill, J., Bailey, J., TeKoppele, J., and Sandy, J. R. (2000). Temporal changes in collagen composition and metabolism during rodent palatogenesis. Mech. Ageing Dev. 119, 49–62. doi: 10.1016/s0047-6374(00)00168-8
Marei, M. K., and El Backly, R. M. (2018). Dental mesenchymal stem cell-based translational regenerative dentistry: from artificial to biological replacement. Front. Bioeng. Biotechnol. 6:49. doi: 10.3389/fbioe.2018.00049
Martín-Del-Campo, M., Rosales-Ibañez, R., and Rojo, L. (2019). Biomaterials for cleft lip and palate regeneration. Int. J. Mol. Sci. 20:E2176. doi: 10.3390/ijms20092176
Martín-Piedra, M. A., Alaminos, M., Fernández-Valadés-gámez, R., España-López, A., Liceras-Liceras, E., Sánchez-Montesinos, I., et al. (2016). Development of a multilayered palate substitute in rabbits: a histochemical ex vivo and in vivo analysis. Histochem. Cell Biol. 147, 377–388. doi: 10.1007/s00418-016-1489-5
Maruhashi, T., Kii, I., Saito, M., and Kudo, A. (2010). Interaction between periostin and BMP-1 promotes proteolytic activation of lysyl oxidase. J. Biol. Chem. 285, 13294–13303. doi: 10.1074/jbc.M109.088864
Masotti, C., Brito, L. A., Nica, A. C., Ludwig, K. U., Nunes, K., Savastano, C. P., et al. (2017). MRPL53, a new candidate gene for orofacial clefting, identified using an eqtl approach. J. Dent. Res. 97, 33–40. doi: 10.1177/0022034517735805
Massenburg, B. B., Jenny, H. E., Saluja, S., Meara, J. G., Shrime, M. G., and Alonso, N. (2016). Barriers to cleft lip and palate repair around the world. J. Craniofac. Surg. 27, 1741–1745.
Mattot, V., Raes, M. B., Henriet, P., Eeckhout, Y., Stehelin, D., Vandenbunder, B., et al. (1995). Expression of interstitial collagenase is restricted to skeletal tissue during mouse embryogenesis. J. Cell Sci. 108(Pt 2), 529–535.
Mead, T. J., and Apte, S. S. (2018). ADAMTS proteins in human disorders. Matrix Biol. 7, 225–239. doi: 10.1016/j.matbio.2018.06.002
Mercapide, J., Lopez De Cicco, R., Castresana, J. S., and Klein-Szanto, A. J. P. (2003). Stromelysin-1/matrix metalloproteinase-3 (MMP-3) expression accounts for invasive properties of human astrocytoma cell lines. Int. J. Cancer 106, 676–682. doi: 10.1002/ijc.11286
Miettinen, P. J., Chin, J. R., Shum, L., Slavkin, H. C., Shuler, C. F., Derynck, R., et al. (1999). Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat. Genet. 22, 69–73. doi: 10.1038/8773
Mimura, T., Han, K. Y., Onguchi, T., Chang, J. H., Kim, T. I., Kojima, T., et al. (2009). MT1-MMP-Mediated cleavage of decorin in corneal angiogenesis. J. Vasc. Res. 46, 541–550. doi: 10.1159/000226222
Morales, J., Al-Sharif, L., Khalil, D. S., Shinwari, J. M. A., Bavi, P., Al-Mahrouqi, R. A., et al. (2009). Homozygous mutations in ADAMTS10 and ADAMTS17 Cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am. J. Hum. Genet. 85, 558–568. doi: 10.1016/j.ajhg.2009.09.011
Morris-Wiman, J., Burch, H., and Basco, E. (2000a). Temporospatial distribution of matrix metalloproteinase and tissue inhibitors of matrix metalloproteinases during murine secondary palate morphogenesis. Anat. Embryol. 202, 129–141. doi: 10.1007/s004290000098
Morris-Wiman, J., Du, Y., and Brinkley, L. (2000b). Occurrence and temporal variation in matrix metalloproteinases and their inhibitors during murine secondary palatal morphogenesis. J. Craniofac. Genet. Dev. Biol. 19, 201–212.
Mossaad, A., Badry, T., El Abdelrahaman, M., Abdelazim, A., Ghanem, W., Hassan, S., et al. (2019). Alveolar cleft reconstruction using different grafting techniques. Open Access Maced. J. Med. Sci. 7, 1369–1373. doi: 10.3889/oamjms.2019.236
Mossey, P. A., Little, J., Steegers-Theunissen, R., Molloy, A., Peterlin, B., Shaw, W. C., et al. (2017). Genetic interactions in nonsyndromic orofacial clefts in Europe—EUROCRAN study. Cleft Palate Craniofac. J. 54, 623–630. doi: 10.1597/16-037
Muir, A. M., Ren, Y., Butz, D. H., Davis, N. A., Blank, R. D., Birk, D. E., et al. (2014). Induced ablation of Bmp1 and Tll1 produces osteogenesis imperfecta in mice. Hum. Mol. Genet. 23, 3085–3101. doi: 10.1093/hmg/ddu013
Mularczyk, E. J., Singh, M., Godwin, A. R. F., Galli, F., Humphreys, N., Adamson, A. D., et al. (2018). ADAMTS10-mediated tissue disruption in Weill–Marchesani syndrome. Hum. Mol. Genet. 27, 3675–3687. doi: 10.1093/hmg/ddy276
Mussano, F., Genova, T., Petrillo, S., Roato, I., Ferracini, R., and Munaron, L. (2018). Osteogenic differentiation modulates the cytokine, chemokine, and growth factor profile of ASCs and SHED. Int. J. Mol. Sci. 19:1454. doi: 10.3390/ijms19051454
Naba, A., Pearce, O. M. T., Del Rosario, A., Ma, D., Ding, H., Rajeeve, V., et al. (2017). Characterization of the extracellular matrix of normal and diseased tissues using proteomics. J. Proteome Res. 16, 3083–3091. doi: 10.1021/acs.jproteome.7b00191
Nakajima, A. F., Shuler, C., Gulka, A. O. D., and Hanai, J.-I. (2018). TGF-β signaling and the epithelial-mesenchymal transition during palatal fusion. Int. J. Mol. Sci. 19:E3638. doi: 10.3390/ijms19113638
Nassif, A., Senussi, I., Meary, F., Loiodice, S., Hotton, D., Robert, B., et al. (2014). Msx1 role in craniofacial bone morphogenesis. Bone 66, 96–104. doi: 10.1016/j.bone.2014.06.003
Naung, N. Y., Duncan, W., Silva, R., and Coates, D. (2019). Localization and characterization of human palatal periosteum stem cells in serum-free, xeno-free medium for clinical use. Eur. J. Oral Sci. 127, 99–111. doi: 10.1111/eos.12603
Nawshad, A., LaGamba, D., and Hay, E. D. (2004). Transforming growth factor β (TGFβ) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT). Arch. Oral Biol. 49, 675–689. doi: 10.1016/j.archoralbio.2004.05.007
Neill, T., Painter, H., Buraschi, S., Owens, R. T., Lisanti, M. P., Schaefer, L., et al. (2012). Decorin antagonizes the angiogenic network: concurrent inhibition of met, hypoxia inducible factor 1α, vascular endothelial growth factor A, and induction of thrombospondin-1 and tiMP3. J. Biol. Chem. 287, 5492–5506. doi: 10.1074/jbc.M111.283499
Nelson, E. R., Levi, B., Sorkin, M., James, A. W., Liu, K. J., Quarto, N., et al. (2011). Role of GSK-3β in the osteogenic differentiation of palatal mesenchyme. PLoS One 6:e25847. doi: 10.1371/journal.pone.0025847
Nie, X., Luukko, K., and Kettunen, P. (2006). FGF signalling in craniofacial development and developmental disorders. Oral Dis. 12, 102–111. doi: 10.1111/j.1601-0825.2005.01176.x
Niewiarowska, J., Brézillon, S., Sacewicz-Hofman, I., Bednarek, R., Maquart, F.-X., Malinowski, M., et al. (2011). Lumican inhibits angiogenesis by interfering with α2β1 receptor activity and downregulating MMP-14 expression. Thromb. Res. 128, 452–457. doi: 10.1016/j.thromres.2011.06.011
Niibe, K., Zhang, M., Nakazawa, K., Morikawa, S., Nakagawa, T., Matsuzaki, Y., et al. (2017). The potential of enriched mesenchymal stem cells with neural crest cell phenotypes as a cell source for regenerative dentistry. Jpn. Dent. Sci. Rev. 53, 25–33. doi: 10.1016/j.jdsr.2016.09.001
Nikopensius, T., Kempa, I., Ambrozaitytë, L., Jagomägi, T., Saag, M., Matulevièienë, A., et al. (2011). Variation in FGF1, FOXE1, and TIMP2genes is associated with nonsyndromic cleft lip with or without cleft palate. Birth Defects Res. A Clin. Mol. Teratol. 91, 218–225. doi: 10.1002/bdra.20791
Novais, A., Lesieur, J., Sadoine, J., Slimani, L., Baroukh, B., Saubaméa, B., et al. (2019). Priming dental pulp stem cells from human exfoliated deciduous teeth with fibroblast growth factor-2 enhances mineralization within tissue-engineered constructs implanted in craniofacial bone defects. Stem Cells Transl. Med. 8, 844–857.
Oh, J., Takahashi, R., Kondo, S., Mizoguchi, A., Adachi, E., Sasahara, R. M., et al. (2001). The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis175. Cell 107, 789–800. doi: 10.1016/s0092-8674(01)00597-9
Oichi, T., Taniguchi, Y., Soma, K., Oshima, Y., Yano, F., Mori, Y., et al. (2019). Adamts17 is involved in skeletogenesis through modulation of BMP-Smad1/5/8 pathway. Cell. Mol. Life Sci. 76, 4795–4809. doi: 10.1007/s00018-019-03188-3180
Okano, J., Udagawa, J., and Shiota, K. (2014). Roles of retinoic acid signaling in normal and abnormal development of the palate and tongue. Congenit. Anom. 54, 69–76. doi: 10.1111/cga.12049
Ong, C. S., Yesantharao, P., Huang, C. Y., Mattson, G., Boktor, J., Fukunishi, T., et al. (2017). 3D bioprinting using stem cells. Pediatr. Res. 83:223. doi: 10.1038/pr.2017.252
Paiva, K. B. S., and Granjeiro, J. M. (2014). Bone tissue remodeling and development: focus on matrix metalloproteinase functions. Arch. Biochem. Biophys. 561, 74–87. doi: 10.1016/j.abb.2014.07.034
Paiva, K. B. S., and Granjeiro, J. M. (2017). Matrix metalloproteinases in bone resorption, remodeling, and repair. Prog. Mol. Biol. Transl. Sci. 148, 203–303. doi: 10.1016/bs.pmbts.2017.05.001
Pall, E., Cenariu, M., Kasaj, A., Florea, A., Soancã, A., Roman, A., et al. (2017). New insights into the cellular makeup and progenitor potential of palatal connective tissues. Microsc. Res. Tech. 80, 1270–1282. doi: 10.1002/jemt.22925
Páll, E., Florea, A., Soriţău, O., Cenariu, M., Petruţiu, A. S., and Roman, A. (2015). Comparative assessment of oral mesenchymal stem cells isolated from healthy and diseased tissues. Microsc. Microanal. 21, 1249–1263. doi: 10.1017/S1431927615014749
Pan, A., Chang, L., Nguyen, A., and James, A. (2013). A review of hedgehog signaling in cranial bone development. Front. Physiol. 4:61. doi: 10.3389/fphys.2013.00061
Pan, Y., Li, D., Lou, S., Zhang, C., Du, Y., Jiang, H., et al. (2018). A functional polymorphism in the pre-miR-146a gene is associated with the risk of nonsyndromic orofacial cleft. Hum. Mutat. 39, 742–750. doi: 10.1002/humu.23415
Parada, C., and Chai, Y. (2012). Roles of BMP signaling pathway in lip and palate development. Front. Oral Biol. 16, 60–70. doi: 10.1159/000337617
Pauws, E., and Stanier, P. (2007). FGF signalling and SUMO modification: new players in the aetiology of cleft lip and/or palate. Trends Genet. 23, 631–640. doi: 10.1016/j.tig.2007.09.002
Pedroni, A. C. F., Sarra, G., de Oliveira, N. K., Moreira, M. S., Deboni, M. C. Z., and Marques, M. M. (2018). Cell sheets of human dental pulp stem cells for future application in bone replacement. Clin. Oral Investig. 23, 2713–2721. doi: 10.1007/s00784-018-2630-8
Pietraszek, K., Chatron-Colliet, A., Brézillon, S., Perreau, C., Jakubiak-Augustyn, A., Krotkiewski, H., et al. (2014). Lumican: a new inhibitor of matrix metalloproteinase-14 activity. FEBS Lett. 588, 4319–4324. doi: 10.1016/j.febslet.2014.09.040
Pietraszek-Gremplewicz, K., Karamanou, K., Niang, A., Dauchez, M., Belloy, N., Maquart, F. X., et al. (2019). Small leucine-rich proteoglycans and matrix metalloproteinase-14: key partners? Matrix Biol. 7, 271–285. doi: 10.1016/j.matbio.2017.12.006
Pinheiro, C. C. G., de Pinho, M. C., Aranha, A. C., Fregnani, E., and Bueno, D. F. (2017). Low power laser therapy: a strategy to promote the osteogenic differentiation of deciduous dental pulp stem cells from cleft lip and palate patients. Tissue Eng. Part A 24, 569–575. doi: 10.1089/ten.tea.2017.0115
Piperi, C., and Papavassiliou, A. G. (2012). Molecular mechanisms regulating matrix metalloproteinases. Curr. Top. Med. Chem. 12, 1095–1112. doi: 10.2174/1568026611208011095
Pischon, N., Darbois, L. M., Palamakumbura, A. H., Kessler, E., and Trackman, P. C. (2004). Regulation of collagen deposition and lysyl oxidase by tumor necrosis factor-α in osteoblasts. J. Biol. Chem. 279, 30060–30065. doi: 10.1074/jbc.M404208200
Pla, P., and Monsoro-Burq, A. H. (2018). The neural border: induction, specification and maturation of the territory that generates neural crest cells. Dev. Biol. 444, S36–S46. doi: 10.1016/j.ydbio.2018.05.018
Praveen Kumar, L., Kandoi, S., Misra, R., Vijayalakshmi, S., and Verma, R. S. (2019). The mesenchymal stem cell secretome: a new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev. 46, 1–9. doi: 10.1016/j.cytogfr.2019.04.002
Prochazkova, M., Prochazka, J., Marangoni, P., and Klein, O. D. (2018). Bones, glands, ears and more: the multiple roles of FGF10 in craniofacial development. Front. Genet. 9:542. doi: 10.3389/fgene.2018.00542
Qin, X., Wergedal, J. E., Rehage, M., Tran, K., Newton, J., Lam, P., et al. (2006). Pregnancy-associated plasma protein-A increases osteoblast proliferation in vitro and bone formation in vivo. Endocrinology 147, 5653–5661. doi: 10.1210/en.2006-1055
Ra, H. J., and Parks, W. C. (2007). Control of matrix metalloproteinase catalytic activity. Matrix Biol. 26, 587–596. doi: 10.1016/j.matbio.2007.07.001
Radichev, I. A., Remacle, A. G., Shiryaev, S. A., Purves, A. N., Johnson, S. L., Pellecchia, M., et al. (2010). Biochemical characterization of the cellular glycosylphosphatidylinositol-linked membrane type-6 matrix metalloproteinase. J. Biol. Chem. 285, 16076–16086. doi: 10.1074/jbc.M110.107094
Raghunathan, R., Sethi, M. K., Klein, J. A., and Zaia, J. (2019). Proteomics, glycomics, and glycoproteomics of matrisome molecules. Mol. Cell. Proteomics 18, 2138–2148. doi: 10.1074/mcp.R119.001543
Ray, H. J., and Niswander, L. (2012). Mechanisms of tissue fusion during development. Development 139, 1701–1711. doi: 10.1242/dev.068338
Reiss, K., and Bhakdi, S. (2017). The plasma membrane: penultimate regulator of ADAM sheddase function. Biochim. Biophys. Acta Mol. Cell Res. 1864, 2082–2087. doi: 10.1016/j.bbamcr.2017.06.006
Reuben, P. M., and Cheung, H. S. (2006). Research service & geriatric research, education and clinical center, veterans administration Medical Center, Miami, Florida 33125, Department of Biomedical Engineering, University of Miami, Coral Gables, Florida 33146 and Department of Medicine, University of Miami School of Medicine, Miami, Florida 33101. Front. Biosci. 11, 1199–1215.
Reynolds, K., Kumari, P., Sepulveda Rincon, L., Gu, R., Ji, Y., Kumar, S., et al. (2019). Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis. Model. Mech. 12:dmm037051. doi: 10.1242/dmm.037051
Ricard-Blum, S., and Vallet, S. D. (2016). Proteases decode the extracellular matrix cryptome. Biochimie 122, 300–313. doi: 10.1016/j.biochi.2015.09.016
Ricard-Blum, S., and Vallet, S. D. (2019). Fragments generated upon extracellular matrix remodeling: biological regulators and potential drugs. Matrix Biol. 7, 170–189. doi: 10.1016/j.matbio.2017.11.005
Rietz, A., and Spiers, J. (2012). The relationship between the MMP system, adrenoceptors and phosphoprotein phosphatases. Br. J. Pharmacol. 166, 1225–1243. doi: 10.1111/j.1476-5381.2012.01917.x
Riou, J.-F., Umbhauer, M., Shi, D. L., and Boucaut, J.-C. (1992). Tenascin: a potential modulator of cell-extracellular matrix interactions during vertebrate embryogenesis. Biol. Cell 75, 1–9. doi: 10.1016/0248-4900(92)90118-K
Rodríguez, D., Morrison, C. J., and Overall, C. M. (2010). Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim. Biophys. Acta Mol. Cell Res. 1803, 39–54. doi: 10.1016/j.bbamcr.2009.09.015
Rodríguez-Manzaneque, J. C., Fernández-Rodríguez, R., Rodríguez-Baena, F. J., and Iruela-Arispe, M. L. (2015). ADAMTS proteases in vascular biology. Matrix Biol. 4, 38–45. doi: 10.1016/j.matbio.2015.02.004
Rodriguez-Pascual, F., and Rosell-Garcia, T. (2018). Lysyl oxidases: functions and disorders. J. Glaucoma 27(Suppl. 1), S15–S19. doi: 10.1097/IJG.0000000000000910
Roman, A., Şoanca, A., Florea, A., and Páll, E. (2013). In vitro characterization of multipotent mesenchymal stromal cells isolated from palatal subepithelial tissue grafts. Microsc. Microanal. 19, 370–380. doi: 10.1017/S143192761201433X
Rosell-García, T., Paradela, A., Bravo, G., Dupont, L., Bekhouche, M., Colige, A., et al. (2019). Differential cleavage of lysyl oxidase by the metalloproteinases BMP1 and ADAMTS2/14 regulates collagen binding through a tyrosine sulfate domain. J. Biol. Chem. 294:11087. doi: 10.1074/jbc.RA119.007806
Rothstein, M., Bhattacharya, D., and Simoes-Costa, M. (2018). The molecular basis of neural crest axial identity. Dev. Biol. 444, S170–S180. doi: 10.1016/j.ydbio.2018.07.026
Sahebjam, S., Khokha, R., and Mort, J. S. (2007). Increased collagen and aggrecan degradation with age in the joints of Timp3-/- mice. Arthritis Rheum. 56, 905–909. doi: 10.1002/art.22427
Sahoo, T., Theisen, A., Sanchez-Lara, P. A., Marble, M., Schweitzer, D. N., Torchia, B. S., et al. (2011). Microdeletion 20p12.3 involving BMP2 contributes to syndromic forms of cleft palate. Am. J. Med. Genet. A 155A, 1646–1653. doi: 10.1002/ajmg.a.34063
Salazar, V. S., Ohte, S., Capelo, L. P., Gamer, L., and Rosen, V. (2016). Specification of osteoblast cell fate by canonical Wnt signaling requires Bmp2. Development 143, 4352–4367. doi: 10.1242/dev.136879
Sanderson, R. D., Bandari, S. K., and Vlodavsky, I. (2019). Proteases and glycosidases on the surface of exosomes: newly discovered mechanisms for extracellular remodeling. Matrix Biol. 7, 160–169. doi: 10.1016/j.matbio.2017.10.007
Savitha, S., Sharma, S. M., Veena, S., and Rekha, R. (2015). Single nucleotide polymorphism of bone morphogenetic protein 4 gene: a risk factor of non-syndromic cleft lip with or without palate. Indian J. Plast. Surg. 48, 159–164. doi: 10.4103/0970-0358.163053
Savoca, M. P., Tonoli, E., Atobatele, A. G., and Verderio, E. A. M. (2018). Biocatalysis by transglutaminases: a review of biotechnological applications. Micromachines 9, 9–11. doi: 10.3390/mi9110562
Scalzone, A., Flores-Mir, C., Carozza, D., d’Apuzzo, F., Grassia, V., and Perillo, L. (2019). Secondary alveolar bone grafting using autologous versus alloplastic material in the treatment of cleft lip and palate patients: systematic review and meta-analysis. Prog. Orthod. 20:6. doi: 10.1186/s40510-018-0252-y
Scherrer, K. (2018). Primary transcripts: from the discovery of RNA processing to current concepts of gene expression - Review. Exp. Cell Res. 373, 1–33. doi: 10.1016/j.yexcr.2018.09.011
Schnellmann, R., Sack, R., Hess, D., Annis, D. S., Mosher, D. F., Apte, S. S., et al. (2018). A selective extracellular matrix proteomics approach identifies fibronectin proteolysis by a disintegrin-like and metalloprotease domain with thrombospondin type 1 motifs (ADAMTS16) and its impact on spheroid morphogenesis. Mol. Cell. Proteomics 17, 1410–1425. doi: 10.1074/mcp.RA118.000676
Schoen, C., Aschrafi, A., Thonissen, M., Poelmans, G., Von den Hoff, J. W., and Carels, C. E. L. (2017). MicroRNAs in palatogenesis and cleft palate. Front. Physiol. 8:165. doi: 10.3389/fphys.2017.00165
Schoen, C., Glennon, J. C., Abghari, S., Bloemen, M., Aschrafi, A., Carels, C. E. L., et al. (2018). Differential microRNA expression in cultured palatal fibroblasts from infants with cleft palate and controls. Eur. J. Orthod. 40, 90–96. doi: 10.1093/ejo/cjx034
Schwarzbauer, J. E., and DeSimone, D. W. (2011). Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 3:a005041. doi: 10.1101/cshperspect.a005041
Seelan, R. S., Mukhopadhyay, P., Warner, D. R., Appana, S. N., Brock, G. N., Pisano, M. M., et al. (2014). Methylated MicroRNA genes of the developing murine palate. MicroRNA 3, 160–173. doi: 10.2174/2211536604666150131125805
Segura-Castillo, J. L., Aguirre-Camacho, H., González-Ojeda, A., and Michel-Perez, J. (2005). Reduction of bone resorption by the application of fibrin glue in the reconstruction of the alveolar cleft. J. Craniofac. Surg. 16, 105–112. doi: 10.1097/00001665-200501000-00020
Seifeldin, S. A. (2016). Is alveolar cleft reconstruction still controversial? (Review of literature). Saudi Dent. J. 28, 3–11. doi: 10.1016/j.sdentj.2015.01.006
Shafi, A., Ah-kee, E., and Khan, A. (2015). Cleft lip and palate and alveolar bone grafting in the united kingdom: a brief overview for non-specialists. West Indian Med. J. 65, 185–188. doi: 10.7727/wimj.2015.191
Shanbhag, S., Suliman, S., Pandis, N., Stavropoulos, A., Sanz, M., and Mustafa, K. (2019). Cell therapy for orofacial bone regeneration: a systematic review and meta-analysis. J. Clin. Periodontol. 46(Suppl. 21), 162–182. doi: 10.1111/jcpe.13049
Sharif, F., Ur Rehman, I., Muhammad, N., and MacNeil, S. (2016). Dental materials for cleft palate repair. Mater. Sci. Eng. C Mater. Biol. Appl. 61, 1018–1028. doi: 10.1016/j.msec.2015.12.019
Sharpe, P. M., Brunet, C. L., Foreman, D. M., and Ferguson, M. W. J. (1993). Localisation of acidic and basic fibroblast growth factors during mouse palate development and their effects on mouse palate mesenchyme cells in vitro. Roux’s Arch. Dev. Biol. 202, 132–143. doi: 10.1007/BF00365303
Shaw, W. (2004). Global strategies to reduce the health care burden of craniofacial anomalies: report of who meetings on international collaborative research on craniofacial anomalies. Cleft Palate Craniofac. J. 41, 238–243. doi: 10.1597/03-214.1
Shi, J., Son, M.-Y., Yamada, S., Szabova, L., Kahan, S., Chrysovergis, K., et al. (2008). Membrane-type MMPs enable extracellular matrix permissiveness and mesenchymal cell proliferation during embryogenesis. Dev. Biol. 313, 196–209. doi: 10.1016/j.ydbio.2007.10.017
Shimakura, Y., Yamzaki, Y., and Uchinuma, E. (2003). Experimental study on bone formation potential of cryopreserved human bone marrow mesenchymal cell/hydroxyapatite complex in the presence of recombinant human bone morphogenetic protein-2. J. Craniofac. Surg. 14, 108–116. doi: 10.1097/00001665-200301000-00021
Shirani, G., Abbasi, A. J., Mohebbi, S. Z., and Moharrami, M. (2017). Comparison between autogenous iliac bone and freeze-dried bone allograft for repair of alveolar clefts in the presence of plasma rich in growth factors: a randomized clinical trial. J. Cranio Maxillofac. Surg. 45, 1698–1703. doi: 10.1016/j.jcms.2017.08.001
Shu, C. C., Flannery, C. R., Little, C. B., and Melrose, J. (2019). Catabolism of fibromodulin in developmental rudiment and pathologic articular cartilage demonstrates novel roles for MMP-13 and ADAMTS-4 in C-terminal processing of SLRPs. Int. J. Mol. Sci. 20:E579. doi: 10.3390/ijms20030579
Smith, T., Lozanoff, S., Iyyanar, P., and Nazarali, A. J. (2013). Molecular signaling along the anterior–posterior axis of early palate development. Front. Physiol. 3:488. doi: 10.3389/fphys.2012.00488
Snyder-Warwick, A. K., and Perlyn, C. A. (2012). Coordinated events: FGF signaling and other related pathways in palatogenesis. J. Craniofac. Surg. 23, 397–400. doi: 10.1097/SCS.0b013e318240fed7
Soria-Valles, C., Gutiérrez-Fernández, A., Osorio, F. G., Carrero, D., Ferrando, A. A., Colado, E., et al. (2016). MMP-25 metalloprotease regulates innate immune response through NF-κB signaling. J. Immunol. 197, 296–302. doi: 10.4049/jimmunol.1600094
Sorushanova, A., Delgado, L. M., Wu, Z., Shologu, N., Kshirsagar, A., Raghunath, R., et al. (2019). The collagen suprafamily: from biosynthesis to advanced biomaterial development. Adv. Mater. 31, 1–39. doi: 10.1002/adma.201801651
Sprangers, S., and Everts, V. (2019). Molecular pathways of cell-mediated degradation of fibrillar collagen. Matrix Biol. 7, 190–200. doi: 10.1016/j.matbio.2017.11.008
Spyrou, J., Gardner, D. K., and Harvey, A. J. (2019). Metabolism is a key regulator of induced pluripotent stem cell reprogramming. Stem Cells Int. 2019:7360121. doi: 10.1155/2019/7360121
Stahle-Backdahl, M., Sandstedt, B., Bruce, K., Lindahl, A., Jimenez, M. G., Vega, J. A., et al. (1997). Collagenase-3 (MMP-13) is expressed during human fetal ossification and re-expressed in postnatal bone remodeling and in rheumatoid arthritis. Lab. Invest. 76, 717–728.
Stanier, P., and Pauws, E. (2012). Development of the lip and palate: fgf signalling. Front. Oral Biol. 16, 71–80. doi: 10.1159/000337618
Stanton, H., Melrose, J., Little, C. B., and Fosang, A. J. (2011). Proteoglycan degradation by the ADAMTS family of proteinases. Biochim. Biophys. Acta Mol. Basis Dis. 1812, 1616–1629. doi: 10.1016/j.bbadis.2011.08.009
Steiglitz, B. M., Ayala, M., Narayanan, K., George, A., and Greenspan, D. S. (2004). Bone morphogenetic protein-1/tolloid-like proteinases process dentin matrix protein-1. J. Biol. Chem. 279, 980–986. doi: 10.1074/jbc.M310179200
Stöcker, W., Grams, F., Baumann, U., Reinemer, P., Gomis-Rüth, F. X., McKay, D. B., et al. (1995). The metzincins–topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 4, 823–840. doi: 10.1002/pro.5560040502
Sumida, R., Maeda, T., Kawahara, I., Yusa, J., and Kato, Y. (2019). Platelet-rich fibrin increases the osteoprotegerin/receptor activator of nuclear factor-κB ligand ratio in osteoblasts. Exp. Ther. Med. 18, 358–365. doi: 10.3892/etm.2019.7560
Sundell, A. L., Törnhage, C.-J., and Marcusson, A. (2017). A comparison of health-related quality of life in 5- and 10-year-old Swedish children with and without cleft lip and/or palate. Int. J. Paediatr. Dent. 27, 238–246. doi: 10.1111/ipd.12253
Suzuki, A., Abdallah, N., Gajera, M., Jun, G., Jia, P., Zhao, Z., et al. (2018). Genes and microRNAs associated with mouse cleft palate: a systematic review and bioinformatics analysis. Mech. Dev. 150, 21–27. doi: 10.1016/j.mod.2018.02.003
Szabova, L., Yamada, S. S., Birkedal-Hansen, H., and Holmbeck, K. (2005). Expression pattern of four membrane-type matrix metalloproteinases in the normal and diseased mouse mammary gland. J. Cell Physiol. 205, 123–132. doi: 10.1002/jcp.20385
Taguchi, T., Yanagi, Y., Yoshimaru, K., Zhang, X.-Y., Matsuura, T., Nakayama, K., et al. (2019). Regenerative medicine using stem cells from human exfoliated deciduous teeth (SHED): a promising new treatment in pediatric surgery. Surg. Today 49, 316–322. doi: 10.1007/s00595-019-01783-z
Takemaru, M., Sakamoto, Y., Sakamoto, T., and Kishi, K. (2016). Assessment of bioabsorbable hydroxyapatite for secondary bone grafting in unilateral alveolar cleft. J. Plast. Reconstr. Aesthet. Surg. 69, 493–496. doi: 10.1016/j.bjps.2015.10.040
Tang, M., Wang, Y., Han, S., Guo, S., and Wang, D. (2013). Transforming growth factor-beta3 gene polymorphisms and nonsyndromic cleft lip and palate risk: a meta-analysis. Genet. Test. Mol. Biomarkers 17, 881–889. doi: 10.1089/gtmb.2013.0334
Tang, Q., Li, L., Jin, C., Lee, J.-M., and Jung, H.-S. (2015). Role of region-distinctive expression of Rac1 in regulating fibronectin arrangement during palatal shelf elevation. Cell Tissue Res. 361, 857–868. doi: 10.1007/s00441-015-2169-2169
Tang, Q., Li, L., Lee, M.-J., Ge, Q., Lee, J.-M., and Jung, H.-S. (2016). Novel insights into a retinoic-acid-induced cleft palate based on Rac1 regulation of the fibronectin arrangement. Cell Tissue Res. 363, 713–722. doi: 10.1007/s00441-015-2271-z
Tarr, J., Lambi, A., Bradley, J., Barbe, M., and Popoff, S. (2018). Development of normal and cleft palate: a central role for connective tissue growth factor (CTGF)/CCN2. J. Dev. Biol. 6:18. doi: 10.3390/jdb6030018
Tavakolinejad, S., Ebrahimzadeh Bidskan, A., Ashraf, H., and Hamidi Alamdari, D. (2014). A glance at methods for cleft palate repair. Iran. Red Crescent Med. J. 16:e15393. doi: 10.5812/ircmj.15393
Taylor, D. A., Sampaio, L. C., Ferdous, Z., Gobin, A. S., and Taite, L. J. (2018). Decellularized matrices in regenerative medicine. Acta Biomater. 74, 74–89. doi: 10.1016/j.actbio.2018.04.044
Thompson, J., Mendoza, F., Tan, E., Bertol, J. W., Gaggar, A. S., Jun, G., et al. (2019). A cleft lip and palate gene, Irf6, is involved in osteoblast differentiation of craniofacial bone. Dev. Dyn. 248, 221–232. doi: 10.1002/dvdy.13
Thuaksuban, N., Nuntanaranont, T., and Pripatnanont, P. (2010). A comparison of autogenous bone graft combined with deproteinized bovine bone and autogenous bone graft alone for treatment of alveolar cleft. Int. J. Oral Maxillofac. Surg. 39, 1175–1180. doi: 10.1016/j.ijom.2010.07.008
Tsuchiya, S., Simmer, J. P., Hu, J. C. C., Richardson, A. S., Yamakoshi, F., and Yamakoshi, Y. (2011). Astacin proteases cleave dentin sialophosphoprotein (Dspp) to generate dentin phosphoprotein (Dpp). J. Bone Miner. Res. 26, 220–228. doi: 10.1002/jbmr.202
Tweedie, A. R. (1910). Specimen of the face and mouth of a female infant, showing arrest of development of the right half of the tongue, combined with a cleft of the soft palate and a palatolingual fold. Proc. R. Soc. Med. 3, 99–102. doi: 10.1177/003591571000300673
van Boxtel, A. L., Gansner, J. M., Hakvoort, H. W. J., Snell, H., Legler, J., and Gitlin, J. D. (2011). Lysyl oxidase-like 3b is critical for cartilage maturation during zebrafish craniofacial development. Matrix Biol. 30, 178–187. doi: 10.1016/j.matbio.2010.12.002
Van den Steen, P. E., Opdenakker, G., Wormald, M. R., Dwek, R. A., and Rudd, P. M. (2001). Matrix remodelling enzymes, the protease cascade and glycosylation. Biochim. Biophys. Acta 1528, 61–73. doi: 10.1016/s0304-4165(01)00190-8
van Hout, W. M. M. T., Mink van der Molen, A. B., Breugem, C. C., Koole, R., and Van Cann, E. M. (2011). Reconstruction of the alveolar cleft: can growth factor-aided tissue engineering replace autologous bone grafting? a literature review and systematic review of results obtained with bone morphogenetic protein-2. Clin. Oral Investig. 15, 297–303. doi: 10.1007/s00784-011-0547-546
Van Otterloo, E., Williams, T., and Artinger, K. B. (2016). The old and new face of craniofacial research: how animal models inform human craniofacial genetic and clinical data. Dev. Biol. 415, 171–187. doi: 10.1016/j.ydbio.2016.01.017
Verstappen, J., and Von den Hoff, J. W. (2006). Tissue inhibitors of metalloproteinases (TIMPs): their biological functions and involvement in oral disease. J. Dent Res. 85, 1074–1084. doi: 10.1177/154405910608501202
Vieira, A. R. (2008). Unraveling human cleft lip and palate research. J. Dent. Res. 87, 119–125. doi: 10.1177/154405910808700202
Vincenti, M. P. (2007). Signal transduction and cell-type specific regulation of matrix metalloproteinase gene expression: can MMPs be good for you? J. Cell Physiol. 213, 355–364. doi: 10.1002/JCP
von Marschall, Z., and Fisher, L. W. (2010). Decorin is processed by three isoforms of bone morphogenetic protein-1 (BMP1). Biochem. Biophys. Res. Commun. 391, 1374–1378. doi: 10.1016/j.bbrc.2009.12.067
Wang, J., Bai, Y., Li, H., Greene, S. B., Klysik, E., Yu, W., et al. (2013). MicroRNA-17-92, a direct Ap-2α transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 9:e1003785. doi: 10.1371/journal.pgen.1003785
Wang, J., Wang, X., Sun, Z., Wang, X., Yang, H., Shi, S., et al. (2010). Stem cells from human-exfoliated deciduous teeth can differentiate into dopaminergic neuron-like cells. Stem Cells Dev. 19, 1375–1383. doi: 10.1089/scd.2009.0258
Wang, S., Sun, C., Meng, Y., Zhang, B., Wang, X., Su, Y., et al. (2017). A pilot study: screening target miRNAs in tissue of nonsyndromic cleft lip with or without cleft palate. Exp. Ther. Med. 13, 2570–2576. doi: 10.3892/etm.2017.4248
Warner, D., Ding, J., Mukhopadhyay, P., Brock, G., Smolenkova, I., Seelan, R., et al. (2015). Temporal expression of mirnas in laser capture microdissected palate medial edge epithelium from tgfβ3-/- mouse fetuses. MicroRNA 4, 64–71. doi: 10.2174/2211536604666150710125743
Weijs, W. L. J., Siebers, T. J. H., Kuijpers-Jagtman, A. M., Bergé, S. J., Meijer, G. J., and Borstlap, W. A. (2010). Early secondary closure of alveolar clefts with mandibular symphyseal bone grafts and β-tri calcium phosphate (β-TCP). Int. J. Oral Maxillofac. Surg. 39, 424–429. doi: 10.1016/j.ijom.2010.02.004
Weng, M., Chen, Z., Xiao, Q., Li, R., and Chen, Z. (2018). A review of FGF signaling in palate development. Biomed. Pharmacother. 103, 240–247. doi: 10.1016/j.biopha.2018.04.026
Weston, J. A., and Thiery, J. P. (2015). Pentimento: neural crest and the origin of mesectoderm. Dev. Biol. 401, 37–61. doi: 10.1016/j.ydbio.2014.12.035
Whitehead, A. L. (1902). The influence of nasal and nasopharyngeal obstruction upon the development of the teeth and palate. Br. Med. J. 1, 949–951. doi: 10.1136/bmj.1.2155.949
Wight, T. N. (2017). Provisional matrix: a role for versican and hyaluronan. Matrix Biol. 6, 38–56. doi: 10.1016/j.matbio.2016.12.001
Wolf, C. J., Belair, D. G., Becker, C. M., Das, K. P., Schmid, J. E., and Abbott, B. D. (2018). Development of an organotypic stem cell model for the study of human embryonic palatal fusion. Birth Defects Res. 110, 1322–1334. doi: 10.1002/bdr2.1394
Wolf, Z. T., Brand, H. A., Shaffer, J. R., Leslie, E. J., Arzi, B., Willet, C. E., et al. (2015). Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 11:e1005059. doi: 10.1371/journal.pgen.1005059
Wong, M. K., Shawky, S. A., Aryasomayajula, A., Green, M. A., Ewart, T., Selvaganapathy, P. R., et al. (2018). Extracellular matrix surface regulates self-assembly of three-dimensional placental trophoblast spheroids. PLoS One 13:e0199632. doi: 10.1371/journal.pone.0199632
Wu, C., Pan, W., Feng, C., Su, Z., Duan, Z., Zheng, Q., et al. (2018). Grafting materials for alveolar cleft reconstruction: a systematic review and best-evidence synthesis. Int. J. Oral Maxillofac. Surg. 47, 345–356. doi: 10.1016/j.ijom.2017.08.003
Wu, M., Li, J., Engleka, K. A., Zhou, B., Lu, M. M., Plotkin, J. B., et al. (2008). Persistent expression of Pax3 in the neural crest causes cleft palate and defective osteogenesis in mice. J. Clin. Invest. 118, 2076–2087. doi: 10.1172/JCI33715
Wu, N., Yan, J., Han, T., Zou, J., and Shen, W. (2018). Integrated assessment of differentially expressed plasma microRNAs in subtypes of nonsyndromic orofacial clefts. Medicine 97:e11224. doi: 10.1097/MD.0000000000011224
Xavier, G. M., Seppala, M., Barrell, W., Birjandi, A. A., Geoghegan, F., and Cobourne, M. T. (2016). Hedgehog receptor function during craniofacial development. Dev. Biol. 415, 198–215. doi: 10.1016/j.ydbio.2016.02.009
Xiao, L., Kumazawa, Y., and Okamura, H. (2014). Cell death, cavitation and spontaneous multi-differentiation of dental pulp stem cells-derived spheroids in vitro: a journey to survival and organogenesis. Biol. Cell 106, 405–419. doi: 10.1111/boc.201400024
Xu, J., Huang, Z., Wang, W., Tan, X., Li, H., Zhang, Y., et al. (2018). FGF8 signaling alters the osteogenic cell fate in the hard palate. J. Dent. Res. 97, 589–596. doi: 10.1177/0022034517750141
Xu, M., Ma, L., Lou, S., Du, Y., Yin, X., Zhang, C., et al. (2018). Genetic variants of microRNA processing genes and risk of non-syndromic orofacial clefts. Oral Dis. 24, 422–428. doi: 10.1111/odi.12741
Yamada, Y., Nakamura-Yamada, S., Kusano, K., and Baba, S. (2019). Clinical potential and current progress of dental pulp stem cells for various systemic diseases in regenerative medicine: a concise review. Int. J. Mol. Sci. 20:1132. doi: 10.3390/ijms20051132
Yang, C. Y., Chanalaris, A., and Troeberg, L. (2017). ADAMTS and ADAM metalloproteinases in osteoarthritis – looking beyond the ‘usual suspects.’. Osteoarthr. Cartil. 25, 1000–1009. doi: 10.1016/j.joca.2017.02.791
Yang, H., Gao, L.-N., An, Y., Hu, C.-H., Jin, F., Zhou, J., et al. (2013). Comparison of mesenchymal stem cells derived from gingival tissue and periodontal ligament in different incubation conditions. Biomaterials 34, 7033–7047. doi: 10.1016/j.biomaterials.2013.05.025
Ye, S., Eriksson, P., Hamsten, A., Kurkinen, M., Humphries, S. E., and Henney, A. M. (1996). Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression. J. Biol. Chem. 271, 13055–13060. doi: 10.1074/jbc.271.22.13055
Yusof, M. F. H., Zahari, W., Hashim, S. N. M., Osman, Z. F., Chandra, H., Kannan, T. P., et al. (2018). Angiogenic and osteogenic potentials of dental stem cells in bone tissue engineering. J. Oral Biol. Craniofac. Res. 8, 48–53. doi: 10.1016/j.jobcr.2017.10.003
Zhang, J., Yang, R., Liu, Z., Hou, C., Zong, W., Zhang, A., et al. (2015). Loss of lysyl oxidase-like 3 causes cleft palate and spinal deformity in mice. Hum. Mol. Genet. 24, 6174–6185. doi: 10.1093/hmg/ddv333
Zhang, S., Wang, C., Xie, C., Lai, Y., Wu, D., Gan, G., et al. (2017). Disruption of hedgehog signaling by vismodegib leads to cleft palate and delayed osteogenesis in experimental design. J. Craniofac. Surg. 28, 1607–1614. doi: 10.1097/SCS.0000000000003790
Keywords: palatogenesis, extracellular matrix, extracellular matrix remodeling, metalloproteinases, cleft lip/palate, palatal reconstruction, tissue bioengineering, biomaterials
Citation: Paiva KBS, Maas CS, Santos PM, Granjeiro JM and Letra A (2019) Extracellular Matrix Composition and Remodeling: Current Perspectives on Secondary Palate Formation, Cleft Lip/Palate, and Palatal Reconstruction. Front. Cell Dev. Biol. 7:340. doi: 10.3389/fcell.2019.00340
Received: 27 February 2019; Accepted: 29 November 2019;
Published: 13 December 2019.
Edited by:
Charles D. Little, University of Kansas Medical Center, United StatesReviewed by:
Lidija Radenovic, University of Belgrade, SerbiaZhizhan Gu, Dana-Farber Cancer Institute, United States
Copyright © 2019 Paiva, Maas, Santos, Granjeiro and Letra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katiúcia Batista Silva Paiva, katipaiva@usp.br
 Katiúcia Batista Silva Paiva
Katiúcia Batista Silva Paiva Clara Soeiro Maas
Clara Soeiro Maas Pâmella Monique dos Santos
Pâmella Monique dos Santos José Mauro Granjeiro
José Mauro Granjeiro Ariadne Letra
Ariadne Letra