Immunologic, genetic, and ecological interplay of factors involved in allergic diseases
- Biomedical Innovations Research for Translational Health Science Laboratory, Department of Biochemistry and Molecular Biology, College of Medicine, University of the Philippines Manila, Manila, Philippines
An allergic or type I hypersensitivity reaction involves a misdirected immune overreaction to innocuous environmental and dietary antigens called allergens. The genetic predisposition to allergic disease, referred to as atopy, can be expressed as a variety of manifestations—e.g., allergic rhinitis, allergic conjunctivitis, atopic dermatitis, allergic asthma, anaphylaxis. Globally, allergic diseases are one the most common types of chronic conditions. Several factors have been identified to contribute to the pathogenesis and progression of the disease, leading to distinctively variable clinical symptoms. The factors which can attenuate or exacerbate allergic reactions can range from genetic heterozygosity, the prominence of various comorbid infections, and other factors such as pollution, climate, and interactions with other organisms and organism-derived products, and the surrounding environment. As a result, the effective prevention and control of allergies remains to be one of the most prominent public health problems. Therefore, to contextualize the current knowledge about allergic reactions, this review paper attempts to synthesize different aspects of an allergic response to describe its significance in the global health scheme. Specifically, the review shall characterize the biomolecular mechanisms of the pathophysiology of the disease based on underlying disease theories and current findings on ecologic interactions and describe prevention and control strategies being utilized. An integrated perspective that considers the underlying genetic, immunologic, and ecologic aspects of the disease would enable the development of more effective and targeted diagnostic tools and therapeutic strategies for the management and control of allergic diseases.
1. Introduction
Adaptive immunity is a complex trait that has persisted in vertebrates across evolutionary history. It consists of humoral and cell-mediated arms, which play distinct yet interconnected roles in the body’s defense against pathogens. Between the two arms of the adaptive immunity, humoral immunity is mediated by different isotypes of immunoglobulins, each of which play a distinctive role against a wide diversity of pathogens. Of the various antibody isotypes, IgE is described as the anti-parasitic antibody isotype that participates in the clearance of helminthic parasites. To mediate this function, IgE binds to high affinity (i.e., FcεRI) and low affinity (i.e., FcεRII or CD23) receptors found on various effector cells (e.g., mast cells, basophils, eosinophils) which stimulate the release of various proinflammatory molecules involved in vasodilation, smooth muscle contraction, and other characteristic manifestations of inflammation (1). Although supposedly beneficial against parasites to prevent further damage to the host, these responses can lead to debilitating outcomes when directed against harmless environmental or dietary antigens known as allergens. Allergens are substances that range from aeroallergens found in the surrounding environment (i.e., pollen), animal detritus and dander, and certain components derived from food products (e.g., shellfish, dairy products, soybeans) which stimulate allergies.
Allergic reactions are described as immediate misdirected immune responses. Such reactions only occur across individuals who are sensitized and exposed to specific allergens. Typically, allergic sensitization or a predisposition to generate an IgE-mediated allergic response (i.e., atopy) occurs as a result of a complex interplay of different genetic factors and environmental exposures. However, studies have identified the involvement of non-IgE immune components in allergic diseases. Interestingly, allergic reactions have been characterized primarily across mammalian species despite the existence of the adaptive immune system across other vertebrates (2). This can be partially attributed to the divergence of the IgG and IgE antibody isotypes from the IgY antibody isotype characteristically found in avian and non-avian reptiles (3). In a previous study by Borges et al., IgY was shown to be involved in mediating antiparasitic immunity across avian species, homologous to the role of IgE in mammalian species (4). However, further investigation revealed that despite its structural resemblance to the IgE antibody isotype, IgY exhibits similar functional characteristics and binding kinetics to the IgG antibody isotype. Differences in the binding sites in the Fc region may be the primary basis for the differences in antibody function, leading to the divergence of the IgG (i.e., Cγ2) and IgE (i.e., Cε3) isotypes (5). Apart from IgY, the role of the IgD isotype in mucosal immunity, immunoactivation, and proinflammation has led many to postulate its potential role in allergic disease progression (6).
Many studies have focused on elucidating the interrelationship of these factors and their role in the progression and severity of allergies. However, the genetic heterogeneity of allergic individuals and the distinctive properties of their immune systems has impeded the full elucidation of the underlying aspects of allergic disease. Moreover, the elusiveness of the allergic response in terms of clinical manifestations and the molecular aspects of the disease stagnated the development of effective and widespread diagnostic and therapeutic strategies.
2. Current theories on allergic diseases
2.1. Allergic march
The development of various types of allergic diseases differs across individuals as a result of different interconnected yet highly variable intrinsic and extrinsic factors. As aforementioned, the innate heterogeneity of these etiologic factors leads to the variation exhibited across allergic individuals. Several studies attempt to characterize the progression of allergic disease development based on the factor of age. Findings suggest that certain diseases tend to predominate and occur at a greater incidence across specific age groups. This theory that describes the temporal trend of allergic disease manifestation is called the “allergic march” (7). Firstly, in the context of atopic dermatitis (AD), it was determined that there exists a causal link between onset of AD and the later manifestation of other allergic diseases. This is attributed to dysfunctionality in the physical barriers of the immune system, which serve as the primary sites of allergic sensitization and colonization of proinflammatory microbiota which are correlated with allergic disease progression. Following this is the induction of type 2 immune responses via effector Th2 cells, which renders hosts susceptible to allergic respiratory responses by upregulating airway hyperresponsiveness to aeroallergens (8). Furthermore, allergic disease progression in the context of aging, beyond the pediatric population, has also been investigated. Recent studies have demonstrated the phenomenon of inflammaging and immunosenescence among the elderly, leading to worsened disease outcomes in the context of acute and chronic inflammatory diseases (9). Moreover, studies indicate that the characteristic hallmarks of cellular aging such as oxidative stress due accumulation of reactive oxygen species, shortening and dysfunctionality of telomeres, and the increased expression of genes associated with aging have been shown to contribute to exacerbation of allergic respiratory diseases, such as allergic rhinitis and bronchial asthma (10–12). These findings suggest that targeted therapies must also consider the temporal progression of the allergy. Administering treatments outside the specific timeframe may result in suboptimal effects to alleviate disease symptoms (13).
2.2. Hygiene hypothesis
On the origin of allergic diseases, many theories have been proposed based on available historical evidence and evolutionary patterns. Firstly, with the advent of modern public health and sanitation practices, many disease-causing pathogens have rapidly declined in prevalence. Of significance in the context of allergic diseases are the parasitic helminths, which are known to elicit IgE-mediated immune responses. Global deworming efforts have been rampant with the aid of widespread treatment strategies and improved diagnostics being a focal agenda of various public health regimes. However, with the eventual decline of helminth populations comes the rise of misdirected immune responses, the allergic reactions. Several studies have demonstrated the immunomodulatory effect of helminth-derived extracts, indicating their possible role in the prevention of allergic diseases (14, 15). This phenomenon was first described as the “hygiene hypothesis,” by the epidemiologist, David Strachan (16, 17).
2.3. Old friends hypothesis
Graham Rook cites that reduced exposure to immunoregulation-inducing microbiota, which persisted across mammalian evolution, leads to poorer control over host inflammatory responses associated with various allergic diseases. He coined this proposed theory as the “Old Friends Hypothesis” to explain the underlying pathogenesis of various chronic inflammatory diseases (18–20). Rook describes the association of chronic inflammatory disorders due to a failure to sustain immunoregulation due to the absence of helminths, non-pathogenic environmental bacteria, and certain gut commensals. These microbes which he referred to as, “Old Friends” drive the expansion of specific populations of immunomodulatory regulatory T cells (Treg) and dendritic cells by the secretion and expression of specific molecules (e.g., IL-10, TGF-β, CRR4) (21–23). These immunomodulatory molecules function by directly acting on immune cells mediating allergic diseases or by downregulating signaling pathways directly involved producing a state of inflammation in the host (24).
2.4. Biota alteration theory
Over time, the advent of new discoveries and the growing attention on the human microbiome, the original concepts of Strachan were reshaped to consider the host ecological interactions with the endogenous microbial symbionts and the surrounding environment (19, 20). One such theory which gained popularity for integrating other inflammatory diseases and the involvement of the host microbiota is the “biota alteration theory” (25).
Based on these theories, several factors are shown to be associated with changes in the host microbiome, leading to various inflammatory diseases—including biota depleting, dirt, and other factors, which affect microbial biodiversity. Biota depleting factors include medical factors such as the use of antibiotics, (i.e., modernization of birthing practices), and clean factors which include improved sanitation and food processing technology, and modern construction. Dirt factors, on the other hand, include increased population density and developed construction practices, which increases the predominance of proinflammatory microbiota.
2.5. Toxin hypothesis
Beyond allergic responses to traditional airborne and dietary allergens, certain iterations of hypersensitivity reactions were historically described as allergies. A theory which illustrates the origin of toxin allergy diseases is the “toxin hypothesis of allergy” which was first described by Margie Profet (26). This theory is based on observations of enzymatic degradation of venom toxin by mast cell-derived proteases, which enhance resistance to venom allergies. The administration of phospholipase A2 (PLA2), a conserved component of bee venom, normally induces a type 2 cell-type response and group 2 innate lymphoid cell activation via the enzymatic cleavage of membrane phospholipids and release of IL-33. As a result of this IgE response to PLA2, protection against anaphylaxis from future challenge can be induced with near-lethal doses of PLA2 (27). Therefore, these findings suggest the possible role of mast cells and IgE-dependent responses in promoting innate and adaptive resistance to venom allergies, forming the basis for immunotherapeutic strategies. However, underlying factors that determine whether a bee venom-induced IgE response leads to pathologic anaphylaxis or protective immunity remains to be poorly elucidated (27–30). The investigation of possible compounds derived from other particles capable of eliciting beneficial immune responses that antagonize the pathologic manifestations of allergic disease can be exploited for the design of novel therapeutics (31, 32).
3. Immunologic basis of allergic diseases
3.1. Pathophysiology
Various components of the immune system have been identified to play essential roles in mediating allergic diseases. The IgE arm of humoral immunity is one of the most prominent drivers of allergic diseases such as allergic rhinitis, allergic asthma, and anaphylaxis. IgE-mediated allergic diseases are classified into three phases—namely the (1) sensitization phase, (2) activation phase, and (3) effector phase (Figure 1). During the sensitization phase, an allergen (i.e., pollen, dust mite dander, food) enters the body via the mucosal surfaces (i.e., respiratory tract, gastrointestinal tract). Upon penetrating the mucosal barrier, an antigen-presenting cell (APC) would take up the allergen, intracellularly degrade it into peptide fragments, and present the peptide onto MHC class II molecules. Phagocytosis of allergen particles are mediated by the presence of protein surfactants (i.e., surfactant protein A, surfactant protein D) which differ in distribution and variety across different types of allergens (33, 34). Upon presenting the allergen peptide to a CD4+ or helper T cell (i.e., Th2), these cells would gain their effector function to stimulate naïve B cells to differentiate into effector IgE plasma cells. Activated plasma cells would then secrete IgE which would subsequently bind to allergens upon re-exposure or challenge during the activation phase. Upon binding of IgE to allergen via the Fab region, effector cells with the corresponding FcεRI or FcεRII receptor would bind to the Fc region of bound IgE thereby activating the effector cell. These effector cells include the mast cells, eosinophils, and basophils. IgE cross-linking was also shown to drive allergic disease progression by impairing monocyte phagocytosis, leading to a pro-inflammatory microenvironment (35). These immune cells carry out various effector functions which are characteristic to the symptoms of different types of allergic diseases via the release of effector molecules (i.e., prostaglandins, leukotrienes, histamines) and the recruitment of other immune cells (i.e., dendritic cells) via cell signaling and chemotaxis to produce a state of inflammation (36, 37). These symptoms include upregulating inflammation, eosinophilia, smooth muscle contraction, excessive mucus secretion, vasodilation, and tissue damage (38). Moreover, various types of alveolar macrophages have also been identified as key regulators of respiratory allergic diseases (39).
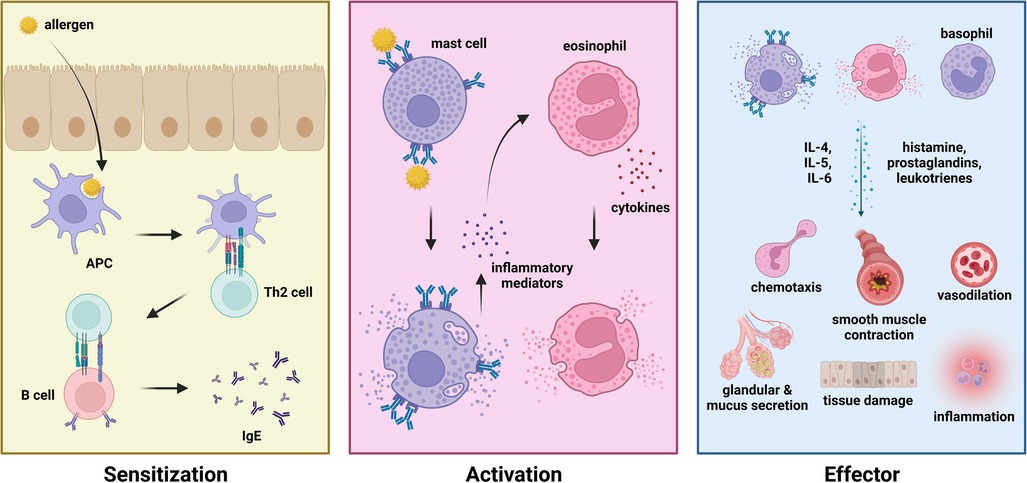
Figure 1. Phases of allergic disease—sensitization (left), activation (middle), and effector (right) phase.
One factor identified to affect the host immune system are sex differences due to hormonal variations. In general, findings suggest that there is increased prevalence of allergic disease across females than in males. One study from Ejima et al. demonstrated the alleviating effect of androgens on airway inflammation, highlighting the role of male sex steroid hormones (i.e., dihydrotestosterone) in suppressing type 2 cytokine production (40). A similar study by Fuseini et al. demonstrated the signal attenuation of house dust mite-induced type 2 and IL-17A inflammatory factors by the action of testosterone (41). Finally, the presence of sex-specific leptin and leptin receptor polymorphisms may also contribute to differences in asthma severity (42). Although the role of estrogen in allergic disease progression has been implicated, the results from such studies are inconclusive on whether the hormone leads to an ameliorative effect by inhibiting inflammasome activation or a pathologic effect by promoting type 2 proinflammatory cytokine production (43, 44).
Despite this, aberrations in the production of IgE leading to pathologically low levels of serum IgE has also been implicated to play a role in allergic disease progression. This condition is described as selective IgE deficiency, and a small number of studies have demonstrated the potential role of deficiency of serum IgE in potentiating various allergic diseases and worsened respiratory function (45). Apart from asthma, this syndrome may also lead to greater risk of other infectious (e.g., upper respiratory tract infection, pneumonia) and non-infectious disease (e.g., bronchiectasis, autoimmune disorders, arthritis), indicating the important role of maintaining a homeostatic level of IgE (46).
3.2. Th1/Th2 interplay
One of the primary mediators of allergic disease is a preferential shift towards Th2 predominant immunity, leading to unregulated inflammation and destructive immune responses. This has been described as the equilibrium model of immunity by Eberl, which highlights the involvement of a dynamic equilibrium between four mutually inhibitory branches of immunity. In the context of allergies, type 2 responses comprise immune cells and other mediators against large parasites (e.g., helminths) and fungi, which are also involved in allergic responses (47). Th1 immunity is typically characterized by beneficial immune responses, leading to better disease outcomes whereas Th2 immunity is associated with worsened disease outcomes, due to the uncontrolled production of proinflammatory cytokines, leading to destructive effects on the host. Although Th1 is known to antagonize Th2 responses, predominance of either Th1 or Th2 immunity was shown to lead to debilitating disease outcomes, such as acute lung pathology, airway hyperreactivity, and chronic inflammation (48). Therefore, it is necessary to devise ways to restore and promote Th1/Th2 balance across patients with allergic diseases.
To improve disease outcomes and control disease progression, several attempts at investigating strategies to regulate Th1/Th2 balance have been done in the context of allergic diseases. This involves the use of the purified carbohydrate, L-arabinose, as a treatment option among patients with wheat allergies, protein S among patients with bronchial asthma, and vitamins such as 1α,25-dihydroxyvitamin D3 against allergic rhinitis (49–51). Immunotherapy has also been identified as a viable strategy in controlling the production of proinflammatory cytokines associated with Th1/Th2 imbalance (52). A better understanding of this complex interplay between the two branches of helper T-cell mediated immunity can help in the development of more robust therapeutics.
4. Genetic factors of allergic diseases
4.1. Genetic predisposition
Several genes have been investigated for their involvement in allergic reactions. However, due to the heterogeneity of the response and how the disease tends to vary from person to person, an exact genetic basis has yet to be fully elucidated for allergic disease predisposition and severity. Several attempts have been made to characterize genes involved in inflammation and maintaining the integrity of the mucosa. Overall, the genes that were identified to be involved with allergic disease severity, progression, and development primarily function in (1) regulating inflammatory responses (i.e., IFN-α, TLR-1, IL-13, IL-4, IL-5, HLA-G, iNOS), (2) maintaining the vascular endothelium and mucosal lining (i.e., FLG, PLAUR, CTNNA3, PDCH1, COL29A1), (3) mediating immune cell function (i.e., IL1RL1, PHF11, H1R, HDC, TSLP, STAT6, RERE, PPP2R3C), and (4) affecting susceptibility to allergic sensitization (i.e., ORMDLR3, CHI3L1) (42, 53–58). Several studies have attempted to characterize the genetic profiles of individuals predisposed to and affected with allergic disease based on their polygenic architecture. Certain loci were identified to be associated with allergic disorders exclusively (e.g., MIIP, CXCR4, SCML4, CYP1B1, ICOS, LINC00824) whereas other pleotropic loci were shown to be associated with both autoimmune and allergic disorders (e.g., PRDM2, G3BP1, HBS1l, POU2AF1) (59). These genes shared a common genetic pathway being involved in inflammation found across different epithelial tissue types (i.e., skin, esophageal, vagina, lung) due to systemic circulation of allergic mediators, indicating the presence of shared genetic components that directly contribute to asthma and allergic disease pathogenesis (60).
Studies on the transcriptomic profiles of atopic individuals revealed the distinctive role of IL-13-associated disease pathways leading to eosinophilic airway inflammation and remodeling, resulting in persistent airflow limitation that is characteristic of allergic asthma. However, gene signatures varied significantly and could be compartmentalized based on function—with expression of genes involved in inflammation being limited to the superficial layer and lumen of the airway whereas genes involved in airway remodeling were limited to endobronchial biopsy samples (61). This distinct pattern of enriched gene signatures was also clearly observable across nasal brushing, sputum, and endobronchial brushing samples. These genes were primarily involved in eosinophilic airway inflammation, mast cell degranulation, and group 3 innate lymphoid cell function, resulting in adult-onset severe asthma (62). Further investigations of the nasal transcriptome revealed other genes involved in airway mucin production (i.e., MUC5AC) and mucus metaplastic transformation of airway epithelium (i.e., FOXA3) causing airway obstruction (63). This compartmentalization of enriched gene signatures presents a novel direction for the development of precision medicine-based therapeutics. However, there still exists numerous gaps in our understanding of the connections pathobiological mechanisms of allergic diseases. Further investigations must elucidate the interactive components involved in allergic diseases, thereby integrating the genetic, immunological, and pathophysiological aspects of allergy.
4.2. Epigenetic basis
The involvement of the epigenome in the pathophysiology of allergic diseases has recently been described and is attributed as one of the major linking factors of allergen and pollutant exposure to disease progression (64–67). Differential methylation of various loci due to exposure to cigarette smoke extract and allergen exposure leading to rapid lung decline, airflow limitation, and overall disease exacerbation (64). DNA methylation of specific genes involved in innate immunity (e.g., IL-1B, IL-6), Th1/Th2 balance (e.g., IL-4, IL-12B, IL-2) and other immune processes (e.g., BDNF, IL-17F, CXCL12, CCR7, RUNX1, CD3E, SERPINE1) have been identified in the progression of peanut allergies (65). Aberrations in DNA methylation have also been shown to be associated with airway epithelial cell dysfunction. Fluctuations in the DNA methylation signatures, due to varied exposure to epigenetic modulatory factors (e.g., traffic-related air pollution, allergen exposure), in genes involved in immune cell function (e.g., CD4+ T cells) and allergic disease pathogenesis (e.g., DUSP22, WTN7B) may also serve as the basis for the heterogeneity of symptoms of asthma and seasonal allergic rhinitis (68, 69). In the context of childhood allergic diseases, nasal DNA methylation signatures in three CpG sites were associated with changes in activated T cell and macrophage populations in the nasal mucosa, leading to allergic inflammation. These DNA methylation signals were also associated with IgE sensitization driving various allergic symptoms (70).
Similarly, the effect of histone acetylation has also been described. Using an IgE-mediated cow’s milk allergy mouse model, reduced percentages of Treg cells were associated with decreased levels of H3 and H4 histone acetylation across specific Treg cell loci. In the same study by Alhamwe et al., decreased histone acetylation was also observed across Th1 loci, preceding the typical reduction of Th1 cell populations associated with allergic responses. This epigenetic mechanism may be involved in a downregulatory mechanism that results in dysregulation of the type 1/type 2 immune response balance (71). On effector cells (e.g., mast cells), a study by Krajewski et al. describes alterations in histone acetylation that regulate the activation of mast cells involved in food allergies. Inhibition of the histone deacetylase with trichostatin A, a broad-spectrum histone deactylase inhibitor, led to changes in the intestinal cytokine profile, decreased expression of FcεRI and prevention of mast cell degranulation (72).
5. Macroecological factors associated with allergic disease
5.1. Exogenous early-life determinants
Several environmental factors have also been identified to lead to an increased risk for developing allergies. A study by Adami et al. confirms the involvement of biota depleting factors in worsening the severity of house dust mite-induced asthma, following administration of antibiotics. By intermittently allowing exposure to antibiotics during the early stages of life, this leads to several debilitating effects on the host—airway eosinophilia, airway hyperreactivity, and reduction in pulmonary Treg cell populations which would lead to a decrease in microbiome diversity and depletion of pro-regulatory species of microbiota (i.e., Lachnospira sp.) (73). Moreover, early life and prenatal exposure to different types of chemical compounds derived from polycarbonate plastics (e.g., bisphenol A) have also been shown to heighten the risk and severity of allergic diseases. Exposure to endocrine-disrupting chemicals has been shown to Th2/Treg cell imbalance, increased levels of IgE, and inflammatory cytokine production. Increased levels mRNA encoding of GATA-3, and decreased levels of mRNAs encoding Foxp3 and Helios due to bisphenol A administration were identified as the primary mediators driving Th2 cell differentiation, leading to exacerbation of allergic asthma (74–76). Interestingly, the interaction of the hormone estrogen and exposure to bisphenol A has also been shown to contribute to increased risk of allergic disease associated with prenatal bisphenol A exposure (74). This pattern was also shown to be present in a study that investigated maternal exposure across different types of bisphenols (i.e., bisphenol F, bisphenol F). The findings of the study revealed a dose and sex-specific effect of bisphenol exposure leading to changes in the mucosal and systemic immune system, leading to impaired immunoregulation and developmental immunotoxicity (76). Overall, these studies implicate the importance of appropriate prenatal and antenatal practices in improving allergic disease outcomes.
5.2. Pollution
Worsening of air quality due to the accumulation of air pollutants and atmospheric particulate matter is one the most prominent factors driving worsened allergic disease outcomes among urban communities. The effect of pollution on allergy pathogenesis manifests in different ways—through the accumulation of reactive oxygen species leading to oxidative stress, enhancement of Th2 responses, upregulation of IgE production, eosinophilia, and impaired mucosal barrier function. Firstly, Jung et al. highlighted in their study the impact of environmental pollutants such as diesel exhaust particles, showing that exposure led to increased levels of the proinflammatory cytokine, IL-17, and worsened disease outcomes (77). Particulate matter and other air pollutants, which are known to carry microbes and viruses from the environment, found in haze are associated with house dust mite allergic sensitization. These atmospheric pollutants are involved in triggering and aggravating cellular inflammatory responses by stimulating sIgE production, contributing to the build-up of oxidative stress, and impairment of mucosal barrier function. (78). A study by Fernandes et al. showed that exposure to smoking and household air pollution also led to worsened disease outcomes across asthmatic adults. Many lifestyle practices such as domestic wood burning, which increased exposure to wood stove smoke, have debilitating effects on lung function, sensitivity to inhaled corticosteroids, and augments airway inflammation, leading to worsened disease outcomes. Across many urban communities, most individuals are subjected to dual exposure to household pollution and smoking, leading to harmful additive effects (79). Therefore, the control of biomass combustion, smoking prevalence, and the production of toxic exhaust fumes are necessary in order to reduce the harmful effects of pollutants.
5.3. Global environment
The dispersion of environmental allergens has also been implied to be responsible for variations in the global distribution of allergic disease. Aeroallergens (e.g., pollen, fungal spores) have increased in atmospheric abundance due to urbanization and green architectural practices. The increased public health risk for allergic diseases across developed countries may be attributed to two major driving factors—reduced biochemical diversity of pollen allergens and increased atmospheric pollen counts (80–82). These factors also shape the host microbiome composition, due to micro-ecologic related to lifestyle and the immediate surrounding environment. Seasonal changes in the incidence of allergic symptoms have also been reported, due to fluctuations in atmospheric pollen counts, domestic aeroallergens, and mite allergens across various geographic locations (83). Across temperate countries, there exists a distinct seasonality of different allergic diseases (e.g., allergic rhinitis, asthma, allergic conjunctivitis, atopic dermatitis) due to changes in daily temperature and humidity across different seasons (i.e., spring, summer, autumn, winter) (84). In contrast, among tropical and subtropical countries, a smaller subset of allergen classes typically predominates leading to an increased prevalence of a narrower spectrum of allergic diseases (85). Anthropogenic climate change, leading to increased global temperatures, has led to longer and more widespread pollen seasons, and increased pollen load—resulting in greater exacerbation and negative impacts on the respiratory health of allergic individuals (86, 87). Therefore, strategic design of residential and green spaces must consider the interactions of the type (e.g., temperate, subtropical, tropical) and quality of the ecosystem, its impact of air and soil pollution, and the impact on allergic disease incidence.
6. Microecological factors associated with allergic disease
6.1. Human microbiome
Despite its established significance in the context of other diseases, the microbiome has only recently gained attention in terms of its involvement in allergic disease outcomes. A stable microbiome has generally been shown to be associated with less severe forms of allergic diseases (88, 89). The biota alteration theory attributes change in endogenous microbial communities to inflammatory diseases, of which allergies have been described in detail. Several factors are involved in reshaping the plasticity for healthy state interactions, the stability of the microbial composition, and the adaptability to inflammatory responses (Figure 2). Moreover, there exists a standard microbiota composition that is typically altered in diversity and abundance of specific microbial species leading to pathologic states. The microbial species typically associated with healthy states include Prevotella sp., Lactobacillus spp., and Bacteroides spp. Therefore, the pathology of allergic diseases has been reshaped to recognize the microbiota as a key player in directing the host response and disease outcome (89).
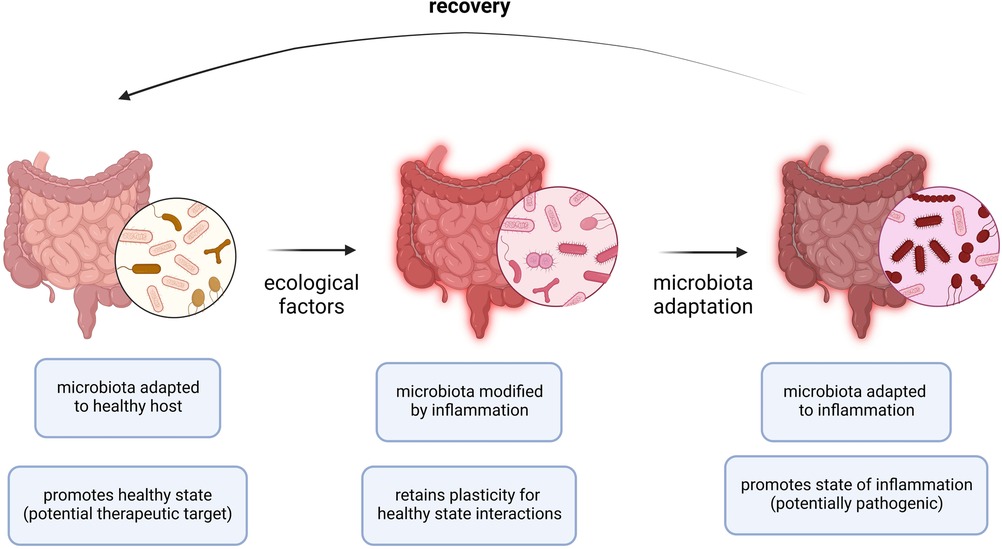
Figure 2. Adaptation of microbiota to allergic inflammation. During allergic inflammation, the microbiota undergoes changes as a result of a complex interplay between internal host and external environmental factors. In a healthy host or microbiota ecosystem, the microbiota is adapted to the healthy host and a healthy state persists. In periods of acute inflammation, ecological factors (i.e., allergen exposure, viral infection) cause the microbiota ecosystem to become inherently unstable and is modified by the immune effector responses while retaining a level of plasticity to preserve interactions occurring in the healthy state. In periods of chronic inflammation (i.e., allergies), the microbiota is adapted to inflammation and the state of inflammation persists. Recovery requires resuscitation from the chronic state of inflammation by restoring the normal endogenous communities comprising the host microbiota.
Recent studies on the human microbiome have attempted to characterize the composition of the gut microbiota in association with the systemic manifestations of various allergic diseases. This would include diseases such as allergic eczema and hives, and more common diseases such as allergic rhinitis. According to Su et al., certain Bacteroides sp., together with Romboustia sp. and Sutterella sp. were associated with the disease eczema. In contrast, species correlated with allergic rhinitis consisted of Clostridia bacteria, Ruminococcaceae bacteria, Lachnospiraceae bacteria, Eubacterium coprostanoligenes, and Atopobium sp., all of which differed in abundance across allergic individuals (88). For skin allergies, Staphylococcus aureus has been shown to affect the severity and progression of skin allergies. Firstly, enterotoxins damage the epithelial lining of the skin, resulting in impairment of the protective barrier. As a result, a state of inflammation would occur and be maintained by the bacteria (90). Furthermore, the intake of specific dietary factors may also serve as adjuvants to allergic disease-associated immune responses, leading to sensitization against certain foodborne allergens. This, in turn, leads to the production of harmful bioactive metabolites that causes disruption of the gut barrier. Thus, the systemic manifestation of allergic diseases (i.e., food allergies) may be attributed to loss of structural integrity of the gut epithelial lining and increased gut permeability to inflammatory mediators (91). Other factors which have been identified to contribute to increased risk for allergic diseases due to changes in host microbiota include cultural factors such as a sedentary lifestyle, and nutritional factors such as deficiency in vitamin D (i.e., lack of exposure to sunlight), iron, and other essential micronutrients (25). Alterations in the host microbiota cause changes in levels of several bioactive metabolites that affect the host immune system, leading to changes in the progression of allergic diseases (92–95). Hence, Meyer et al. suggests vitamin and mineral supplementation to reduce the incidence and severity of food allergies among malnourished children (94). Additionally, Trompette et al. described the association of increased levels of circulating short-chain fatty acids (e.g., propionate) and protection against allergic inflammation in the lung (96). In effect, the importance of proper nutrition to attain a balance of microbiota-derived metabolites and improve overall gut microbiota metabolism is emphasized in allergic diseases. By the perspective posed in the biota alteration theory, the need to elucidate the interactions of the host microbiota and the role various metabolites play in allergic disease progression could help to augment the development of microbiome-targeted therapies.
Apart from the gut microbiome, several studies have attempted to characterize the differences in the microbial composition of other regions of the body, such as the respiratory microbiome. A study by Che et al. attempted to characterize and compare the nasal microbiome of individuals with allergic rhinitis to those who were unaffected. Findings from their study revealed that the presence of species such as Vibrio vulnificus and Acinetobacter baumanni in the nasal microbiome allergic individuals led to an increase in allergic mediators (e.g., GM-CSF, IFNβ, IL-27, IL-1β), leading to allergic rhinitis and asthma (97). Differences in the microbial composition of individuals with respiratory allergies were evident in the nasopharyngeal microbiome, presenting a possible predictor for allergic disease progression in early childhood. Dysbiosis of the nasopharyngeal microbiome was shown to potentiate respiratory allergic responses, with the predominance of Moraxella sp. leading to induced pulmonary epithelial damage and increased proinflammatory cytokine expression (98). The microbiota composition of the upper and lower respiratory tract was also shown to be associated with allergic respiratory tract diseases, in a study by Cui et al. From the findings of their study, bacterial species such as Haemophilus sp., Streptococcus sp., Staphylococcus sp., and Clostridium sp. were shown to be associated with inflammation in the lower respiratory tract, parallel to their effect in the gut microbiome. Additionally, pharyngeal colonization of Haemophilus sp. and Streptococcus sp., together with the upper respiratory tract pathogen Moraxella sp., was identified as a risk factor for acute and aggravated allergic asthma episodes among children (99). Finally, the impact of synergistic infections with respiratory pathogens, such as the influenza A virus and Streptococcus pneumoniae was also implicated to interact with factors associated allergic inflammation to Aspergillus fumigatus. These findings indicate a “triple-disease” burden linking asthma, influenza virus infection, and pneumococcal pneumonia in the context of respiratory health (100). Although underlying immunologic responses and host cell signaling pathways may exist that may explain the complex host-pathogen-microbiome interactions present, these have yet to be fully elucidated. Overall, current studies reinforce the existence of a multifactorial crosstalk between different synergistic microbial factors, which may explain the variations in allergic disease progression.
6.2. Indoor microbiome
The effect of the distribution and abundance of microorganisms (e.g., bacteria, fungi) and their derivatives (e.g., endotoxins, spores) in the indoor microbiome has also been characterized in terms of the immunopathogenesis of asthma and exacerbation of airway inflammation through cytokines mediating type 2 immune cell responses (101–104). Several studies across various geographic regions have attempted to characterize the indoor microbiome across different populations, in relation to allergic disease severity. One study done in Malaysia identified various bacterial (i.e., Sphingobium sp., Rhodomicrobium sp., Shimwellia sp., Solirubrobacter sp., Pleurocapsa sp.) and fungal species (e.g., Torulaspora sp., family Leptosphaeriaceae) with protective roles in the context of respiratory health and asthma severity across tropical areas. Similarly, several species of bacteria (e.g., Izhakiella sp., Robinsoniella sp.) were also identified to be associated with asthma exacerbation. Factors which were identified to shape the indoor microbial composition include building infrastructure, the abundance of common household insect pests, and the presence of molds (105). Two separate studies in different schools across China investigated the association of the indoor microbiome and the severity of various allergic diseases (e.g., rhinoconjunctivitis, asthma, rhinitis, eczema). Bacteria such as Prevotella sp., Lactobacillus iners, and Dolosigranulum sp. were shown to induce a protective effect against rhinitis for preschool children. In contrast, fungi such as Aspergillus subversicolor and bacteria such as Collinsella sp. and Cutibacterium sp. were associated with worsened disease outcomes for asthma, rhinitis, and eczema respectively. These opportunistic pathogenic species are often associated with chronic inflammatory diseases and unregulated immunoactivation due to the production of highly potent virulence factors (e.g., cAMPs, porphyrins, hyaluronate lyase) across different mucosal surfaces of the body. Differences in the microbial composition of urban and rural schools were also shown to be a primary factor in shaping the prevalence of various allergic diseases. For example, species of Brachybacterium was more common across rural areas and was generally associated with improved disease outcomes across high school children with allergic rhinitis. In contrast, potentially pathogenic species of microbes such as Pseudoalteromonas sp., Microbacterium foliorum, a prominent member of the phyllosphere microbiome, and the protist, Neospora caninum, were associated with increased incidence of wheeze, rhinitis, and rhinoconjunctivitis. These microbes were shown to interact with different components of the immune system, leading to Th1/Th2 imbalance and IgE antibody production (106, 107). Findings from a study in the United States revealed that other factors involved in altering fungal allergen load (i.e., Aspergillus sp., Alternaria sp.) include the alpha diversity of bacterial species (e.g., Staphylococcus sp., Porphyromonas sp., Moraxella sp., Sutterella sp., Clostridim sp., family Neisseraceae), window opening, and the presence of pets and flowering plants within the vicinity. From these studies, it was established that the microbial species which are most prevalent in an area are referred to as the “core microbiome” (108). The exact influence of changes in the core microbiome and an individual’s susceptibility to allergic disease due to allergen load and changes in host metabolic and immunologic profiles remains to be fully elucidated.
6.3. Virus respiratory infections
Several types of viral infections have been shown to worsen and exacerbate allergic disease outcomes. This is particularly relevant in cases of respiratory allergies, with rhinoviruses, respiratory syncytial viruses, influenza viruses, and coronaviruses identified as exacerbating agents of allergic diseases. For one, adult asthma exacerbations were associated with viral respiratory infections (VRIs) caused by human rhinovirus, human metapneumovirus, influenza virus, and respiratory syncytial virus (109–115). Other factors involved include seasonal variations in viral infection incidence and the genetic divergence of viruses across populations. As a result, the incidence of VRIs were identified as a predictive factor for adult asthma exacerbations in patients across various seasons (116). Conversely, atopic individuals were shown to be predisposed to more severe viral respiratory diseases. The mechanistic basis for the interactions of VRIs and allergic disease progression may be attributed to several factors that are still being investigated. Current findings, however, suggest that respiratory viruses prevent the development of immune tolerance and enhance allergic sensitization to aeroallergens. This results in increased inflammation and hyperresponsiveness in the respiratory tract. As a result, the airway mucosa becomes significantly more permeable to penetration by allergens, leading to various effector functions characteristic of respiratory allergic diseases (117, 118). Additionally, certain viruses (i.e., rhinoviruses) have also been implicated in persistent Th1/Th2 imbalance among patients with allergic asthma (119). Therefore, understanding the complexity surrounding the relationship of VRIs and allergic diseases must be integrated with the growing incidence of viral pathogens (i.e., SARS-CoV-2).
7. Therapeutic and control strategies for allergic diseases
Within the past decade, several treatment methods have been investigated for their applicability in clinical practice, ranging from the use of pharmaceutical drugs, inhalers, nebulizers, and other immunomodulatory and immunotherapeutic agents (120, 121). Some studies also describe a competition between IgG and IgE antibodies for antigenic sites on specific allergens, with IgG binding leading to desensitization of the host to the specific allergen due to reduced binding sites for IgE. However, the exact mechanistic basis for the competition of IgG and IgE binding and how this can be exploited further to develop new prevention strategies remains poorly elucidated (122, 123). Current findings, however, indicate the involvement of low affinity IgG and its interactions with the inhibitory receptor, FcγRIIb, leading to inhibition of mast cell degranulation (124).
Various public health programs against allergies have been established and implemented globally. While allergen avoidance initially persisted as the primary prevention strategy for allergic diseases, recent findings have suggested the opposite—repeated exposure to allergen can lead to attenuation and possibly eradication of severe disease symptoms. To address the ecological aspect of allergic disease incidence, urban ecosystems must be designed to maximize biodiversity of native, non-allergenic vegetation and grassland species (81). Seasonal variations due to geographic differences must also be considered in the implementation therapeutic strategies (125).
7.1. Pharmaceuticals and biologics
Most pharmaceutical drugs typically target a component involved in mediating the allergic response by disrupting or abrogating its function or inducing a tolerogenic state within the host. For example, leukotriene receptor antagonists which target and suppress cysteinyl leukotrienes that function in eosinophilia, inflammation, and airway hyperresponsiveness associated with viral-induced asthma exacerbation have been identified as effective treatments (126). Antihistamines are another prominent classification of anti-allergy drugs routinely used in clinical practice. This class of pharmaceuticals mainly act by antagonizing the action of smooth muscle cells by stimulating histamine action in the H1-receptors (127). However, a prominent adverse reaction often associated with the use of antihistamines is sedation, which is mainly found in first-generation antihistamines. This is due to low brain uptake when bound to blood proteins such as serum albumin (128). Second-generation antihistamines (e.g., bilastine, loratadine, desloratadine, cetyrisine, levocetirisin) remain as one of the most widely available pharmaceutical drugs for allergic diseases worldwide (129). Many attempts have been made to further characterize the interactions of such widespread pharmaceutical drugs in the context of different types of allergic diseases. A pharmacokinetic study on cetirizine, one of the most widely distributed and cost-effective antihistamine, revealed zwitterionic and lipophilic properties which possible serves as the basis for the differences in antihistamine potency (128). Pharmacogenomic studies revealed the association of certain gene polymorphisms (e.g., CRTH2) and the effective dosage of antihistamine needed to alleviate symptoms of allergic diseases (e.g., chronic urticaria) (130). Investigations on the different molecular properties of the molecule can enable further improvement of the efficacy and potency of pharmaceuticals as treatments for allergic diseases.
In terms of biologics, the humanized recombinant IgG monoclonal antibody, omalizumab (i.e., XOLAIR), has emerged as a prospective therapeutic agent to effectively treat and mitigate IgE-mediated allergic diseases. It functions by targeting IgE to prevent the activation of effector cells which cause the symptoms of allergic disease (131, 132). This causes a competitive binding inhibitory effect, leading to IgE clearance and immune cell inactivation. This is carried out through an exploitation of the intrinsic flexibility of IgE, leading to accelerated dissociation. The presence of specific structural features in IgE, when compared to other antibody isotypes, such as the Cε2 domains and globule-like properties of the Cε3, enable the formation of an allosteric communication pathway to prevent the simultaneous binding of IgE to both of its receptors (i.e., FcεRI, FcεRII). This property would, therefore, prevent allergen-independent activation of mast cells due to crosslinking of FcεRI-bound IgE by CD23. However, the cost of production and resources required limits the widespread distribution and effective implementation of this biologic drug as a treatment for allergic diseases (133). Beyond logistical and economic constraints, there still exists a lack of understanding on the basis for dissociation of allergen specific IgE on mast cells caused by the anti-IgE molecule. Attempts to further circumvent the inherent limitations of omalizumab treatment was done through the development of an omalizumab biobetter antibody with improved stability, binding affinity, and efficacy when compared to the standard omalizumab molecule (134). Despite the pathological role of IgE being implicated by the function of IgE, adverse reactions to omalizumab have also been reported due to the formation of immune complexes between the monoclonal antibody and its target IgE antibody, leading to the manifestation of skin inflammation and anaphylaxis through an IgG receptor-dependent mechanism (i.e., Fcγ) (135).
Ligelizumab, another type of monoclonal anti-IgE antibody, is also being prospected for its enhanced neutralization of free serum IgE, inhibition of IgE binding to FcεRI, basophil activation, IgE production by plasma cells, and prevention of passive systemic anaphylaxis when compared to omalizumab. However, omalizumab remains more potent in terms of inhibiting IgE:CD23 interaction to prevent allergen presentation and other transport processes associated with allergic responses (136). Beyond omalizumab and ligelizumab, several other attempts at developing improved anti-IgE biologics (e.g., UB-221, 8D6, MeDI4212) for treatment of various allergic diseases have also been made but have yet to reach late-stage clinical trials (137–139).
Apart from anti-IgE biologics, Schanin et al. report the use sialic acid-binding immunoglobulin-like lectin immunoregulatory receptor (Siglec)-6 on mast cells as a potential target for therapeutic use. In their study, Siglec-6 monoclonal antibody clones were developed, with the AK04 variant being capable of inducing receptor cluster formation containing inhibitory phosphatase. The epitope-specific agnostic activity of the Siglec-6 mab prevented systemic anaphylaxis with a single dose and reduced overall mast cell activity with chronic dosing (140). Although the therapeutic strategy has shown potential in driving this occurrence, the lack of a mechanistic basis for its mechanism eludes its prospective use in clinical practice.
7.2. Microbiota-targeted therapies
Interestingly, the recognition of the involvement of the microbiome has enabled the investigation of the use of prebiotics and probiotics as attenuating agents against the progression of allergic diseases. However, further investigation is required to elucidate the exact causal mechanism and strategies for improving the efficacy of microbiome-based therapeutic for treatment of allergic diseases (Figure 3) (141). Current findings suggest that allergy attenuation can be achieved by microbiota-dependent upregulation of immunomodulatory Treg cell populations. Turner et al. describes the use of species of Clostridiales to induce the expression of TGF-β1 receptors among Treg cells, leading to regulate allergic disease progression (21). Similarly, Karimi et al. illustrated the potent immunoregulatory capacity of oral treatment with live Lactobacillus reuteri, leading to the attenuation of airway hyperresponsiveness and inflammation (142). Future studies that investigate the immunomodulatory potential of Lactobacillus sp. probiotics must consider the interactions these bacteria have on the amelioration of disease symptoms. The presence of immunosuppressive motifs across the genome sequences of probiotic species of Lactobacillus associated with reduced allergenicity have been identified and could serve as the basis for the selection of probiotic species to be utilized in treatment strategies (143). Additionally, previous investigations suggest prebiotic and probiotic supplementation to promote greater abundance of Prevotella spp. and Bifidobacterium spp., respectively, as microbiota-directed treatment strategies for atopic patients (144, 145). A study by Zhen et al. attempted to elucidate this relationship by correlating the presence of various bacterial enterotypes to the tryptophan metabolic pathways. Findings from the study revealed that the production of indole derivatives was associated with the attenuation of the disease. Indole-3-lactic acid and indole-3-butyric acid inhibited allergic pathogenesis by suppressing the production of IL-4 and IL-5 in macrophages. Indole acetic acid, on the other hand, was shown to be involved in balancing Th17/Treg responses, together with ILA. Plasma components, such as lysozyme C, cystatin-3, and kininogen-1 were also shown to decrease allergic symptom severity (146). Moreover, the consumption of dietary fibers as a prebiotic source may attenuate allergic disease symptoms by the production of short-chain fatty acids which would inhibit the formation of type 2 dendritic cells that would mediate allergic airway inflammation (96).
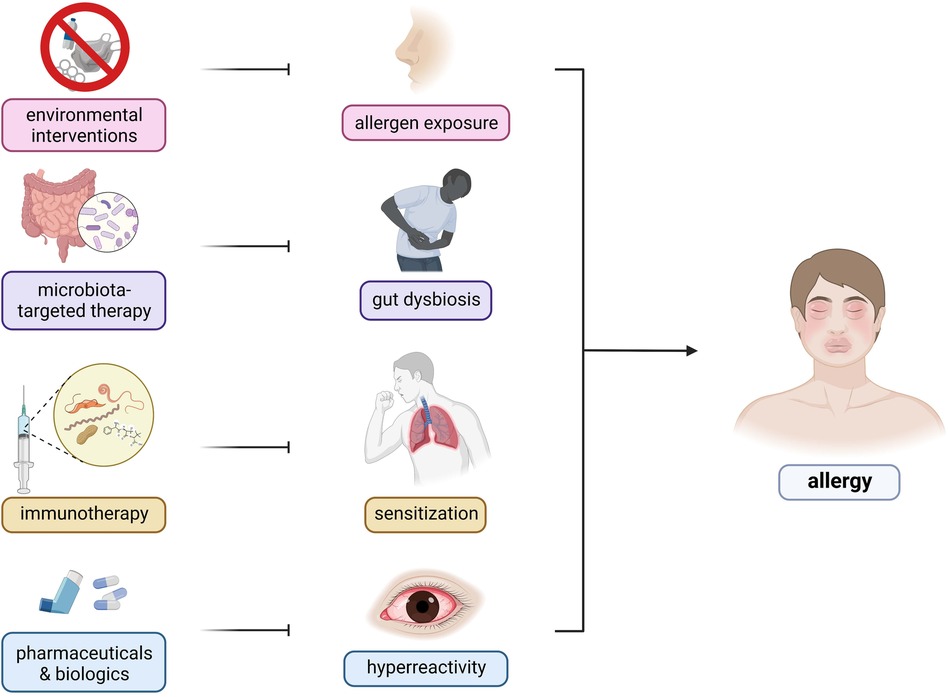
Figure 3. Therapeutic targets and strategies for allergic diseases. There are several therapeutic strategies being developed for the control and mitigation of allergic diseases—(1) environmental interventions, (2) microbiota-targeted therapy, (3) immunotherapy, and (4) pharmaceuticals and biologics. Environmental interactions include public health programs involving infrastructure development and changes to lifestyle factors. Microbiota-based therapies consist of prebiotic, probiotic, and synbiotic use to restore the host microbiome to reduce predisposition to inflammation. Immunotherapies involve repeated exposure to gradually increasing dosages of allergens to ameliorate the host immune response. Pharmaceuticals include traditional oral antihistamines, inhalers, and nebulizers to target specific components of the host immune response whereas biologics include monoclonal antibodies targeting IgE and other components of the allergic response.
7.3. Immunotherapy
The emergence of immunotherapies as effective treatment strategies to various types of allergic diseases can be attributed to several factors impacting several aspects of the immune system. In a study by Scadding et al., patients treated with immunotherapies experienced less severe symptoms, had better respiratory function, and reduced levels of nasal fluid concentrations of IL-4, IL-9 and eotaxin, a potent eosinophil chemoattractant, after challenge with grass pollen (147). Immunotherapies have been tested against milk allergies via oral administration, for hymenopteran venom allergies, grass pollen allergies, and even house dust mite allergies (147–149). Jung et al. demonstrates in their study the viability of two different types of sublingual immunotherapy as a treatment strategy for patients with dust mite-induced allergic rhinitis, each with a distinct set of benefits (150). Similar findings were also reported by Matsumoto et al., wherein long-term immunotherapy with house dust extracts led to amelioration pulmonary function across asthmatic patients (148). Thus, the growing success of such studies highlights the potential of immunotherapies as viable preventive measures for severe allergic disease symptoms. However, safety concerns remain a primary roadblock to the effective implementation of immunotherapies in clinical practices. Additionally, adjuvant therapies using chitin, a major component of various allergy-causing organisms (e.g., house dust mites, crustaceans, fungi), may be utilized to improve the efficacy of therapeutics that attenuate the Th2 response (101). Current studies have attempted to compare different components of immunotherapy extracts that would produce the most desirable clinical outcomes. For example, a study by Du et al. attempted to compare the potency of house dust extracts from house dust mite allergen extracts. Findings from their study revealed greater allergenic potential of house dust mite allergen extracts, implicating its overall preferability over standard house dust extracts (151). The ability to exactly determine at what point and at what dosage re-exposure to an allergen would induce a beneficial, long-lasting, protective effect rather than a life-threatening, anaphylactic shock remains a major gap in current research and necessitates further investigation. The standardization of allergen immunotherapy extracts may improve the therapeutic efficacy birch, ragweed, dog hair, and Alternaria allergies, as highlighted by Due et al. (152).
Other prospective targeted therapies are also being explored for their capacity as immunotherapeutic strategies for allergic diseases. In the context of cow’s milk allergy, oral immunotherapy alleviated allergic symptoms and reduced levels of cow’s milk-specific IgE. However, adverse reactions were observed in several individuals on oral immunotherapy (153). As a potential alternative, the holoprotein form of the lipocalin beta-lactoglobulin (holo-BLG) protein is being investigated for its immunotherapeutic potential. Despite being a component of cow’s milk, which has been shown to induce allergies as well, this protein was identified as a novel component in inducing a farm protective effect against pollen allergies among farmers by promoting regulatory cell function and downregulating antigen presentation to effector cells. Prophylactic treatment with the protein results in disruption of specific IgE production and attenuated the type 2 response (154). Although not readily used or available for the clinical treatment of cow’s milk allergy, these findings on holo-BLG represent a myriad of potential directions for immunotherapeutic development for allergic disease mitigation.
An interesting prospect that could be undertaken in future research on immunotherapy potency is the combination of immunotherapeutic extract use with administration of biologics and other readily available treatments. A study by Bożek and colleagues showed that a combination of an allergen vaccine with the biologic, omalizumab, significantly increased the efficacy of allergen immunotherapy by reducing the incidence of asthma exacerbation and the daily dose of inhaled corticosteroids required by patients with house dust mite-driven asthma (155). Together, these findings highlight the importance of an integrated approach to provide complete amelioration of quality-of-life and improve overall disease outcomes for allergic individuals.
8. Discussion
The past century has been a revolutionary period for immunology, with new findings reshaping our understanding of the fundamental aspects of the immune system. From the discovery of underlying molecular pathways and mechanisms to the development of novel theories on the evolution of the immune system, these breakthroughs culminate in the current state of our global health scheme. This is especially relevant in the context of allergic diseases, which are regarded as one of the leading chronic inflammatory diseases globally.
To better understand the interrelationships of the various factors that underlie the pathogenesis and progression of allergic diseases, this review has adopted an integrative approach to correlate the historical, immunologic, and genetic aspects of the disease. Accordingly, a framework for these factors which utilized Tinbergen’s four questions is proposed (Figure 4) (156). In terms of evolutionary history, the ancestral allergic response is postulated to primarily be a form of antiparasitic immunity utilized by mammalian species, mediated primarily by the IgE immune responses. A primitive form of antiparasitic immunity is also exhibited in other vertebrates such as avian and non-avian reptiles mediated by the IgY antibody (4–6). However, the survival value for IgE responses has been largely masked with the advent of global deworming efforts, improved sanitation, and changes in global environmental conditions (16, 25, 157). Several theories postulating the basis of the continued persistence of the IgE responses, leading into allergic diseases, have been made in relation to the interactions of the immune system with host genetic, micro-ecologic, and macro-ecologic factors (18, 25). However, the concurrent evolution of these responses resulted in the reliance on a state of complete immunologic, genetic, and ecologic homeostasis as evidenced by the emergence of selective IgE deficiency syndromes that occur as a result of dysregulated immune function (45, 46). These observations highlight the importance of attenuating excessive, uncontrolled immune reactions and effector functions by restoring a proper balance (e.g., Th1/Th2, microbiome composition) in various aspects involved in allergic diseases. Therapeutics and control strategies at both the individual and macroscale level must consider this complex interplay of immunologic, genetic, and ecologic factors in order to better achieve desirable health outcomes without severely affecting other aspects.
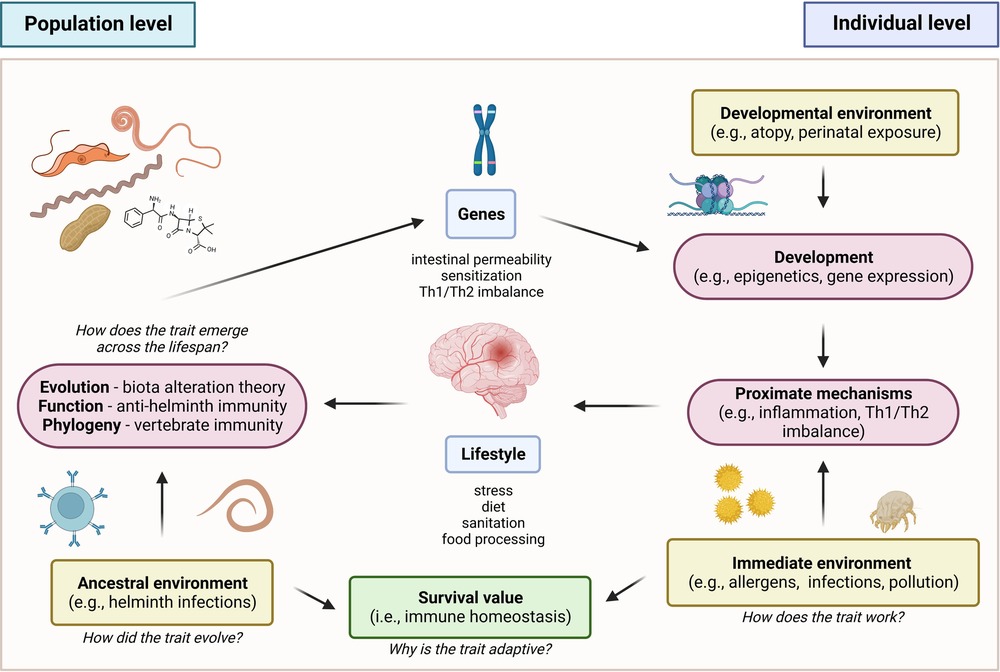
Figure 4. Proposed framework for allergic disease based on tinbergen’s four questions (156). In the context of evolutionary history, the ancestral allergic responses are postulated to be a form of antiparasitic immunity that persisted across vertebrates. In terms of development, the emergence of global de-worming efforts and improved sanitation practices resulted in changes in the persistence and primary function of the trait, masking its original function against helminthic parasites. This resulted in a mechanism involving misdirected responses against innocuous allergens. However, the trait persisted due to its conferred survival value for immune homeostasis.
In the pursuit of more effective diagnostic, preventive, and therapeutic strategies, a better understanding of the interplay of these factors is essential. Reshaping current perspectives about allergic diseases would help in the development of effective approaches in the global health scheme, to remedy the public health burden posed by such chronic conditions.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Acknowledgments
The authors would like to thank the kind support of the members of the Biomedical Innovations Research for Translational Health Science (BIRTHS) Laboratory, Department of Biochemistry, College of Medicine, and the members of the MD-PhD (Molecular Medicine) Program of the University of the Philippines Manila.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Acevedo N, Sánchez J, Zakzuk J, Bornacelly A, Quiróz C, Alvarez Á, et al. Particular characteristics of allergic symptoms in tropical environments: follow up to 24 months in the FRAAT birth cohort study. BMC Pulm Med. (2012) 12:1–15. doi: 10.1186/1471-2466-12-13
2. Daschner A, González Fernández J. Allergy in an evolutionary framework. J Mol Evol. (2020) 88:66–76. doi: 10.1007/s00239-019-09895-3
3. Nett-Mettler CS. Allergies in birds. Veterinary Allergy. (2014) 1:422–7. doi: 10.1002/9781118738818.ch60
4. Borges IP, Silva MF, Santiago FM, de Faria LS, Júnior ÁF, da Silva RJ, et al. Antiparasitic effects induced by polyclonal IgY antibodies anti-phospholipase A2 from bothrops pauloensis venom. Int J Biol Macromol. (2018) 112:333–42. doi: 10.1016/j.ijbiomac.2018.01.178
5. Taylor AI, Gould HJ, Sutton BJ, Calvert RA. Avian IgY binds to a monocyte receptor with IgG-like kinetics despite an IgE-like structure. J Biol Chem. (2008) 283:16384–90. doi: 10.1074/jbc.M801321200
6. Li L, Wang T, Sun Y, Cheng G, Yang H, Wei Z, et al. Extensive diversification of IgD-, IgY-, and truncated IgY(ΔFc)-encoding genes in the red-eared turtle (trachemys scripta elegans). J Immunol. (2012) 189:3995–4004. doi: 10.4049/jimmunol.1200188
7. Goksör E, Loid P, Alm B, Åberg N, Wennergren G. The allergic march comprises the coexistence of related patterns of allergic disease not just the progressive development of one disease. Acta Paediatr (2016) 105:1472–9. doi: 10.1111/apa.13515
8. Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. (2014) 05:1–8. doi: 10.4172/2155-9899.1000202
9. Fulop T, Larbi A, Pawelec G, Khalil A, Cohen AA, Hirokawa K, et al. Immunology of aging: the birth of inflammaging. Clin Rev Allergy Immunol. (2021) 64(2):109–22. doi: 10.1007/s12016-021-08899-6
10. Piñeiro-Hermida S, Martínez P, Blasco MA. Short and dysfunctional telomeres protect from allergen-induced airway inflammation. Aging Cell. (2021) 20:1–19. doi: 10.1111/acel.13352
11. Celik M, Tuncer A, Soyer OU, Saçkesen C, Tanju Besler H, Kalayci O. Oxidative stress in the airways of children with asthma and allergic rhinitis. Pediatr Allergy Immunol. (2012) 23:556–61. doi: 10.1111/j.1399-3038.2012.01294.x
12. McGeachie MJ, Sordillo JE, Dahlin A, Wang AL, Lutz SM, Tantisira KG, et al. Expression of SMARCD1 interacts with age in association with asthma control on inhaled corticosteroid therapy. Respir Res. (2020) 21:1–8. doi: 10.1186/s12931-020-1295-4
13. Jimenez J, Paller AS. The atopic march and its prevention. Ann Allergy Asthma Immunol (2021) 127:289–90. doi: 10.1016/j.anai.2021.04.021
14. Sun S, Li H, Yuan Y, Wang L, He W, Xie H, et al. Preventive and therapeutic effects of trichinella spiralis adult extracts on allergic inflammation in an experimental asthma mouse model. Parasit Vectors. (2019) 12:1–10. doi: 10.1186/s13071-019-3561-1
15. Pascoal VF, Da Cunha AA, Morassutti AL, Antunes GL, Da Silveira KA, Silveira JS, et al. Immunomodulatory effect of different extracts from angiostrongylus cantonensis on airway inflammation in an allergic asthma model. Parasitol Res. (2020) 119:3719–28. doi: 10.1007/s00436-020-06884-0/Published
16. Parker W, Sarafian JT, Broverman SA, Laman JD. Between a hygiene rock and a hygienic hard place. Evol Med Public Health. (2021) 9:120–30. doi: 10.1093/emph/eoab006
17. Perkin MR, Strachan DP. The hygiene hypothesis for allergy—conception and evolution. Front Allergy. (2022) 3:1–17. doi: 10.3389/falgy.2022.1051368
18. Frew JW. The hygiene hypothesis, old friends, and new genes. Front Immunol. (2019) 10:1–3. doi: 10.3389/fimmu.2019.00388
19. Rook GAW. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: darwinian medicine and the “hygiene” or “old friends” hypothesis. Clin Exp Immunol. (2010) 160(1):70–9. doi: 10.1111/j.1365-2249.2010.04133.x
20. Rook GAW, Lowry CA, Raison CL. Microbial “old friends”, immunoregulation and stress resilience. Evol Med Public Health (2013) 2013:46–64. doi: 10.1093/emph/eot004
21. Turner JA, Stephen-Victor E, Wang S, Rivas MN, Abdel-Gadir A, Harb H, et al. Regulatory T cell-derived TGF-β1 controls multiple checkpoints governing allergy and autoimmunity. Immunity. (2020) 53:1202–1214.e6. doi: 10.1016/j.immuni.2020.10.002
22. Faustino L, da Fonseca DM, Takenaka MC, Mirotti L, Florsheim EB, Guereschi MG, et al. Regulatory T cells migrate to airways via CCR4 and attenuate the severity of airway allergic inflammation. J Immunol. (2013) 190:2614–21. doi: 10.4049/jimmunol.1202354
23. Coomes SM, Kannan Y, Pelly VS, Entwistle LJ, Guidi R, Perez-Lloret J, et al. CD4 + Th2 Cells are directly regulated by IL-10 during allergic airway inflammation. Mucosal Immunol. (2017) 10:150–61. doi: 10.1038/mi.2016.47
24. Xu W, Lan Q, Chen M, Chen H, Zhu N, Zhou X, et al. Adoptive transfer of induced-treg cells effectively attenuates murine airway allergic inflammation. PLoS One. (2012) 7:1–10. doi: 10.1371/journal.pone.0040314
25. Villeneuve C, Kou HH, Eckermann H, Palkar A, Anderson LG, McKenney EA, et al. Evolution of the hygiene hypothesis into biota alteration theory: what are the paradigms and where are the clinical applications? Microbes Infect. (2018) 20:147–55. doi: 10.1016/j.micinf.2017.11.001
26. Profet M. The functon of allergy: immunological defense against toxins. Q Rev Biol. (1991) 66:23–62. doi: 10.1086/417049
27. Palm NW, Rosenstein RK, Yu S, Schenten DD, Florsheim E, Medzhitov R. Bee venom phospholipase A2 induces a primary type 2 response that is dependent on the receptor ST2 and confers protective immunity. Immunity. (2013) 39:976–85. doi: 10.1016/j.immuni.2013.10.006
28. Glaeser A, Müller C, Bode S. Anaphylactic reactions in the build-up phase of rush immunotherapy for bee venom allergy in pediatric patients: a single-center experience. Clin Mol Allergy. (2022) 20:1–6. doi: 10.1186/s12948-022-00170-3
29. Stock R, Fischer T, Aẞmus K, Zoeller N, Ackermann H, Kaufmann R, et al. Safety and tolerability of venom immunotherapy: evaluation of 581 rush- and ultra-rush induction protocols (safety of rush and ultra-rush venom immunotherapy). World Allergy Organ J (2021) 14:1–13. doi: 10.1016/j.waojou.2020.100496
30. Kołaczek A, Skorupa D, Antczak-Marczak M, Kuna P, Kupczyk M. Safety and efficacy of venom immunotherapy: a real life study. Postepy Dermatol Alergol. (2017) 34:159–67. doi: 10.5114/ada.2017.67082
31. Tsai M, Starkl P, Marichal T, Galli SJ. Testing the “toxin hypothesis of allergy”: mast cells, IgE, and innate and acquired immune responses to venoms. Curr Opin Immunol. (2015) 36:80–7. doi: 10.1016/j.coi.2015.07.001
32. Pritchard A. The evolution of IgE-mediated type I hypersensitivity and its immunological value. Allergy. (2022) 76:1024–40. doi: 10.1111/all.14570
33. Erpenbeck VJ, Malherbe DC, Sommer S, Schmiedl A, Steinhilber W, Ghio AJ, et al. Surfactant protein D increases phagocytosis and aggregation of pollen-allergen starch granules. Am J Physiol Lung Cell Mol Physiol. (2005) 288:692–8. doi: 10.1152/ajplung
34. Ding J, Umstead TM, Floros J, Phelps DS. Factors affecting SP-A-mediated phagocytosis in human monocytic cell lines. Respir Med. (2004) 98:637–50. doi: 10.1016/j.rmed.2003.12.018
35. Pyle DM, Yang VS, Gruchalla RS, Farrar JD, Gill MA. Ige cross-linking critically impairs human monocyte function by blocking phagocytosis. J Allergy Clin Immunol. (2013) 131:491–500. doi: 10.1016/j.jaci.2012.11.037
36. Lauzon-Joset JF, Marsolais D, Langlois A, Bissonnette EY. Dysregulation of alveolar macrophages unleashes dendritic cell-mediated mechanisms of allergic airway inflammation. Mucosal Immunol. (2014) 7:155–64. doi: 10.1038/mi.2013.34
37. Lukacs NW, Standiford TJ, Chensue SW, Kunkel RG, Strieter RM, Kunkel SL. C-C chemokine-induced eosinophil chemotaxis during allergic airway inflammation. J Leukoc Biol. (1996) 60:573–8. doi: 10.1002/jlb.60.5.573
38. Kang JW, Lee YH, Kang MJ, Lee HJ, Oh R, Min HJ, et al. Synergistic mucus secretion by histamine and IL-4 through TMEM16A in airway epithelium. Am J Physiol Lung Cell Mol Physiol. (2017) 313:466–76. doi: 10.1152/ajplung.00103.2017.-Histamine
39. Kulkarni N, Kantar A, Costella S, Ragazzo V, Piacentini G, Boner A, et al. Macrophage phagocytosis and allergen avoidance in children with asthma. Front Pediatr. (2018) 6:1–8. doi: 10.3389/fped.2018.00206
40. Ejima A, Abe S, Shimba A, Sato S, Uehata T, Tani-ichi S, et al. Androgens alleviate allergic airway inflammation by suppressing cytokine production in Th2 cells. J Immunol. (2022) 209:1083–94. doi: 10.4049/jimmunol.2200294
41. Fuseini H, Yung JA, Cephus JY, Zhang J, Goleniewska K, Polosukhin VV, et al. Testosterone decreases house dust Mite–induced type 2 and IL-17A–mediated airway inflammation. J Immunol. (2018) 201:1843–54. doi: 10.4049/jimmunol.1800293
42. Ziyab AH, Mukherjee N, Kurukulaaratchy RJ, Zhang H, Ewart S, Arshad H, et al. Leptin receptor gene polymorphisms and sex modify the association between Acetaminophen use and asthma among young adults: results from two observational studies. Respir Res. (2018) 19:1–9. doi: 10.1186/s12931-018-0892-y
43. Cheng C, Wu H, Wang M, Wang L, Zou H, Li S, et al. Estrogen ameliorates allergic airway inflammation by regulating activation of NLRP3 in mice. Biosci Rep. (2019) 39:1–12. doi: 10.1042/BSR20181117
44. Cai Y, Zhou J, Webb DC. Estrogen stimulates Th2 cytokine production and regulates the compartmentalisation of eosinophils during allergen challenge in a mouse model of asthma. Int Arch Allergy Immunol. (2012) 158:252–60. doi: 10.1159/000331437
45. Unsworth DJ, Virgo PF, Lock RJ. Immunoglobulin E deficiency: a forgotten clue pointing to possible immunodeficiency? Ann Clin Biochem. (2011) 48:459–61. doi: 10.1258/acb.2011.011052
46. Picado C, de Landazuri I O, Vlagea A, Bobolea I, Arismendi E, Amaro R, et al. Spectrum of disease manifestations in patients with selective immunoglobulin E deficiency. J Clin Med. (2021) 10:1–15. doi: 10.3390/jcm10184160
48. Hansen G, Berry G, DeKruyff RH, Umetsu DT. Allergen-specific Th1 cells fail to counterbalance Th2 cell-induced airway hyperreactivity but cause severe airway inflammation. J Clin Invest. (1999) 103:175–83. doi: 10.1172/JCI5155
49. Wang Y, Sun J, Xue L, Liu J, Nie C, Fan M, et al. L -Arabinose attenuates gliadin-induced food allergy via regulation of Th1/Th2 balance and upregulation of regulatory T cells in mice. J Agric Food Chem. (2021) 69:3638–46. doi: 10.1021/acs.jafc.0c07167
50. Keles E, Ozkara S, Ilhan N, Gungor H, Karlidag T, Yalcin S. The relationship between Th1/Th2 balance and 1α, 25-dihydroxyvitamin D3 in patients with allergic rhinitis. Turk Arch Otolaryngol. (2016) 53:139–43. doi: 10.5152/tao.2015.1187
51. Asayama K, Kobayashi T, D’Alessandro-Gabazza CN, Toda M, Yasuma T, Fujimoto H, et al. Protein S protects against allergic bronchial asthma by modulating Th1/Th2 balance. Allergy. (2020) 75:2267–78. doi: 10.1111/all.14261
52. Zhu Y, Yu J, Zhu XH, Yuan JS, Dai MN, Bao YW, et al. Experimental observation of the effect of immunotherapy on CD4+ T cells and Th1/Th2 cytokines in mice with allergic rhinitis. Sci Rep (2023) 13:5273. doi: 10.1038/s41598-023-32507-6
53. Yang CA, Chiang BL. Toll-like receptor 1 N248S polymorphism affects T helper 1 cytokine production and is associated with serum immunoglobulin E levels in Taiwanese allergic patients. J Microbiol Immunol Infect (2017) 50:112–7. doi: 10.1016/j.jmii.2015.01.004
54. Sadanaga Y, Ishikawa T, Yasueda H, Okudaira H, Nishimura Y. HLA Class II association with type I allergy to house dust mite and Japanese cedar pollen in Japanese subjects. Allergol Int. (1998) 47:285–91. doi: 10.1046/j.1440-1592.1998.00105.x
55. Ranjbar M, Matloubi M, Assarehzadegan M-A, Fallahpour M, Sadeghi F, Soleyman-Jahi S, et al. Association between two single nucleotide polymorphisms of thymic stromal lymphopoietin (TSLP) gene and asthma in Iranian population. Iran J Allergy Asthma Immunol. (2020) 19:362–72. doi: 10.18502/ijaai.v19i4.4111
56. Luukkonen TM, Kiiski V, Ahola M, Mandelin J, Virtanen H, Pöyhönen M, et al. The value of flg null mutations in predicting treatment response in atopic dermatitis: an observational study in Finnish patients. Acta Derm Venereol. (2017) 97:456–63. doi: 10.2340/00015555-2578
57. Sato S, Wang X, Saito J, Fukuhara A, Uematsu M, Suzuki Y, et al. Exhaled nitric oxide and inducible nitric oxide synthase gene polymorphism in Japanese asthmatics. Allergol Int. (2016) 65:300–5. doi: 10.1016/j.alit.2016.02.007
58. Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. (2017) 49:1752–7. doi: 10.1038/ng.3985
59. Shirai Y, Nakanishi Y, Suzuki A, Konaka H, Nishikawa R, Sonehara K, et al. Multi-Trait and cross-population genome-wide association studies across autoimmune and allergic diseases identify shared and distinct genetic component. Ann Rheum Dis. (2022) 81:1301–12. doi: 10.1136/annrheumdis-2022-222460
60. Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, et al. A genome-wide cross-trait analysis from UK biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. (2018) 50:857–64. doi: 10.1038/s41588-018-0121-0
61. Hekking PP, Loza MJ, Pavlidis S, de Meulder B, Lefaudeux D, Baribaud F, et al. Transcriptomic gene signatures associated with persistent airflow limitation in patients with severe asthma. Eur Respir J. (2017) 50:857–64. doi: 10.1183/13993003.02298-2016
62. Hekking PP, Loza MJ, Pavlidis S, de Meulder B, Lefaudeux D, Baribaud F, et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J Allergy Clin Immunol. (2018) 141:1280–90. doi: 10.1016/j.jaci.2017.06.037
63. Sajuthi SP, Everman JL, Jackson ND, Saef B, Rios CL, Moore CM, et al. Nasal airway transcriptome-wide association study of asthma reveals genetically driven mucus pathobiology. Nat Commun. (2022) 13:1–17. doi: 10.1038/s41467-022-28973-7
64. Chen YC, Tsai YH, Wang CC, Liu SF, Chen TW, Fang WF, et al. Epigenome-wide association study on asthma and chronic obstructive pulmonary disease overlap reveals aberrant DNA methylations related to clinical phenotypes. Sci Rep. (2021) 11:1–14. doi: 10.1038/s41598-021-83185-1
65. Zhou X, Han X, Lyu SC, Bunning B, Kost L, Chang I, et al. Targeted DNA methylation profiling reveals epigenetic signatures in peanut allergy. JCI Insight. (2021) 6:1–15. doi: 10.1172/jci.insight.143058
66. Glass K, Thibault D, Guo F, Mitchel JA, Pham B, Qiu W, et al. Integrative epigenomic analysis in differentiated human primary bronchial epithelial cells exposed to cigarette smoke. Sci Rep. (2018) 8:1–11. doi: 10.1038/s41598-018-30781-3
67. Seiler CL, Song JUM, Kotandeniya D, Chen J, Kono TJY, Han Q, et al. Inhalation exposure to cigarette smoke and inflammatory agents induces epigenetic changes in the lung. Sci Rep. (2020) 10:1–15. doi: 10.1038/s41598-020-67502-8
68. Clifford RL, Patel J, MacIsaac JL, McEwen LM, Johnson SR, Shaw D, et al. Airway epithelial cell isolation techniques affect DNA methylation profiles with consequences for analysis of asthma related perturbations to DNA methylation. Sci Rep. (2019) 9:1–12. doi: 10.1038/s41598-019-50873-y
69. Nestor CE, Barrenäs F, Wang H, Lentini A, Zhang H, Bruhn S, et al. DNA Methylation changes separate allergic patients from healthy controls and may reflect altered CD4+ T-cell population structure. PLoS Genet. (2014) 10:1–8. doi: 10.1371/journal.pgen.1004059
70. van Breugel M, Qi C, Xu Z, Pedersen CET, Petoukhov I, Vonk JM, et al. Nasal DNA methylation at three CpG sites predicts childhood allergic disease. Nat Commun. (2022) 13:1–13. doi: 10.1038/s41467-022-35088-6
71. Alhamwe BA, Meulenbroek LAPM, Veening-Griffioen DH, Wehkamp TMD, Alhamdan F, Miethe S, et al. Decreased histone acetylation levels at Th1 and regulatory loci after induction of food allergy. Nutrients. (2020) 12:1–17. doi: 10.3390/nu12103193
72. Krajewski D, Kaczenski E, Rovatti J, Polukort S, Thompson C, Dollard C, et al. Epigenetic regulation via altered histone acetylation results in suppression of mast cell function and mast cell-mediated food allergic responses. Front Immunol. (2018) 9:1–15. doi: 10.3389/fimmu.2018.02414
73. Adami AJ, Bracken SJ, Guernsey LA, Rafti E, Maas KR, Graf J, et al. Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatr Res. (2018) 84:426–34. doi: 10.1038/s41390-018-0031-y
74. Zhou A, Chang H, Huo W, Zhang B, Hu J, Xia W, et al. Prenatal exposure to bisphenol A and risk of allergic diseases in early life. Pediatr Res. (2017) 81:851–6. doi: 10.1038/pr.2017.20
75. Wang Y, Cao Z, Zhao H, Ren Y, Hao L, Gu Z. Bisphenol A exacerbates allergic inflammation in an ovalbumin-induced mouse model of allergic rhinitis. J Immunol Res. (2020) 2020:1–9. doi: 10.1155/2020/7573103
76. Malaisé Y, Lencina C, Cartier C, Olier M, Ménard S, Guzylack-Piriou L. Bisphenol A, S or F mother’s dermal impregnation impairs offspring immune responses in a dose and sex-specific manner in mice. Sci Rep. (2021) 11:1–11. doi: 10.1038/s41598-021-81231-6
77. Jung HJ, Ko YK, Shim WS, Kim HJ, Kim DY, Rhee CS, et al. Diesel exhaust particles increase nasal symptoms and IL-17A in house dust mite-induced allergic mice. Sci Rep. (2021) 11:1–9. doi: 10.1038/s41598-021-94673-9
78. Ye Q, Zhang T, Mao JH. Haze facilitates sensitization to house dust mites in children. Environ Geochem Health. (2020) 42:2195–203. doi: 10.1007/s10653-019-00481-6
79. Fernandes AGO, De Souza-Machado C, Pinheiro GP, De Oliva ST, Mota RCL, De Lima VB, et al. Dual exposure to smoking and household air pollution is associated with an increased risk of severe asthma in adults in Brazil. Clin Transl Allergy. (2018) 8:1–11. doi: 10.1186/s13601-018-0235-6
80. Tegart LJ, Johnston FH, Arriagada NB, Workman A, Dickinson JL, Green BJ, et al. ‘Pollen potency’: the relationship between atmospheric pollen counts and allergen exposure. Aerobiologia (Bologna) (2021) 37:825–41. doi: 10.1007/s10453-021-09726-3
81. Bernard-Verdier M, Seitz B, Buchholz S, Kowarik I, Mejía SL, Jeschke JM. Grassland allergenicity increases with urbanisation and plant invasions. Ambio. (2022) 51:2261–77. doi: 10.1007/s13280
82. Parmes E, Pesce G, Sabel CE, Baldacci S, Bono R, Brescianini S, et al. Influence of residential land cover on childhood allergic and respiratory symptoms and diseases: evidence from 9 European cohorts. Environ Res. (2020) 183:1–10. doi: 10.1016/j.envres.2019.108953
83. Eberlein B, Gulyas AF, Schultz K, Lecheler J, Flögel S, Wolfmeyer C, et al. Domestic allergens and endotoxin in three hospitals offering in-patient rehabilitation for allergic diseases in the alpine mountain climate of bavaria—the AURA study. Int J Hyg Environ Health. (2009) 212:21–6. doi: 10.1016/j.ijheh.2007.09.003
84. Lee YC, Ju HJ, Kwon JW, Bae JM. Seasonality of allergic diseases: real-world evidence from a nationwide population-based study. Immun Inflamm Dis. (2020) 8:360–2. doi: 10.1002/iid3.316
85. Andiappan AK, Puan KJ, Lee B, Nardin A, Poidinger M, Connolly J, et al. Allergic airway diseases in a tropical urban environment are driven by dominant mono-specific sensitization against house dust mites. Allergy. (2014) 69:501–9. doi: 10.1111/all.12364
86. Anderegg WRL, Abatzoglou JT, Anderegg LDL, Bielory L, Kinney PL, Ziska L. Anthropogenic climate change is worsening north American pollen seasons. Environ Sci (Ruse) (2021) 118:e124–31. doi: 10.1073/pnas.2013284118/-/DCSupplemental
87. Ziska LH, Makra L, Harry SK, Bruffaerts N, Hendrickx M, Coates F, et al. Temperature-related changes in airborne allergenic pollen abundance and seasonality across the northern hemisphere: a retrospective data analysis. Lancet Planet Health. (2019) 3:e124–31. doi: 10.1016/S2542-5196(19)30015-4
88. Su YJ, Luo SD, Hsu CY, Kuo HC. Differences in gut microbiota between allergic rhinitis, atopic dermatitis, and skin urticaria A pilot study. Medicine (United States) (2021) 100. doi: 10.1097/MD.0000000000025091
89. De Filippis F, Paparo L, Nocerino R, Della Gatta G, Carucci L, Russo R, et al. Specific gut microbiome signatures and the associated pro-inflamatory functions are linked to pediatric allergy and acquisition of immune tolerance. Nat Commun. (2021) 12. doi: 10.1038/s41467-021-26266-z
90. Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with atopic dermatitis colonized with Staphylococcus aureus have a distinct phenotype and endotype. J Invest Dermatol. (2018) 138:2224–33. doi: 10.1016/j.jid.2018.03.1517
91. Drønen EK, Namork E, Dirven H, Nygaard UC. Suspected gut barrier disruptors and development of food allergy: adjuvant effects and early immune responses. Front Allergy. (2022) 3. doi: 10.3389/falgy.2022.1029125
92. Solidoro P, Bellocchia M, Aredano I, Mattei A, Pivetta E, Patrucco F, et al. Asthmatic patients with vitamin D deficiency have decreased exacerbations after vitamin replacement. Nutrients. (2017) 9. doi: 10.3390/nu9111234
93. Nowak S, Wang H, Schmidt B, Jarvinen KM. Vitamin D and iron status in children with food allergy. Ann Allergy Asthma Immunol. (2021) 127:57–63. doi: 10.1016/j.anai.2021.02.027
94. Meyer R, De Koker C, Dziubak R, Skrapac AK, Godwin H, Reeve K, et al. A practical approach to vitamin and mineral supplementation in food allergic children. Clin Transl Allergy. (2015) 5. doi: 10.1186/s13601-015-0054-y
95. Sahoyama Y, Hamazato F, Shiozawa M, Nakagawa T, Suda W, Ogata Y, et al. Multiple nutritional and gut microbial factors associated with allergic rhinitis: the hitachi health study. Sci Rep. (2022) 12. doi: 10.1038/s41598-022-07398-8
96. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
97. Che Y, Wang N, Ma Q, Liu J, Xu Z, Li Q, et al. Microbial characterization of the nasal cavity in patients with allergic rhinitis and non-allergic rhinitis. Front Cell Infect Microbiol. (2023) 13. doi: 10.3389/fcimb.2023.1166389
98. Tsai MH, Shih HJ, Su KW, Liao SL, Hua MC, Yao TC, et al. Nasopharyngeal microbial profiles associated with the risk of airway allergies in early childhood. J Microbiol Immunol Infect. (2022) 55:777–85. doi: 10.1016/j.jmii.2022.01.006
99. Cui J, Zhang Y, Zhao H, Sun X, Chen Z, Zhang Q, et al. The relationship between lower respiratory tract microbiome and allergic respiratory tract diseases in children. Front Microbiol. (2021) 12. doi: 10.3389/fmicb.2021.630345
100. LeMessurier KS, Iverson AR, Chang TC, Palipane M, Vogel P, Rosch JW, et al. Allergic inflammation alters the lung microbiome and hinders synergistic co-infection with H1N1 influenza virus and Streptococcus pneumoniae in C57BL/6 mice. Sci Rep. (2019) 9. doi: 10.1038/s41598-019-55712-8
101. Arae K, Morita H, Unno H, Motomura K, Toyama S, Okada N, et al. Chitin promotes antigen-specific Th2 cell-mediated murine asthma through induction of IL-33-mediated IL-1β production by DCs. Sci Rep. (2018) 8. doi: 10.1038/s41598-018-30259-2
102. Reeder KM, Dunaway CW, Blackburn JP, Yu Z, Matalon S, Hastie AT, et al. The common γ-chain cytokine IL-7 promotes immunopathogenesis during fungal asthma. Mucosal Immunol. (2018) 11:1352–62. doi: 10.1038/s41385-018-0028-1
103. Hanashiro J, Muraosa Y, Toyotome T, Hirose K, Watanabe A, Kamei K. Schizophyllum commune induces IL-17-mediated neutrophilic airway inflammation in OVA-induced asthma model mice. Sci Rep. (2019) 9. doi: 10.1038/s41598-019-55836-x
104. Arae K, Ikutani M, Horiguchi K, Yamaguchi S, Okada Y, Sugiyama H, et al. Interleukin-33 and thymic stromal lymphopoietin, but not interleukin-25, are crucial for development of airway eosinophilia induced by chitin. Sci Rep. (2021) 11. doi: 10.1038/s41598-021-85277-4
105. Fu X, Norbäck D, Yuan Q, Li Y, Zhu X, Hashim JH, et al. Indoor microbiome, environmental characteristics and asthma among junior high school students in johor bahru, Malaysia. Environ Int. (2020) 138. doi: 10.1016/j.envint.2020.105664
106. Sun Y, Meng Y, Ou Z, Li Y, Zhang M, Chen Y, et al. Indoor microbiome, air pollutants and asthma, rhinitis and eczema in preschool children—a repeated cross-sectional study. Environ Int. (2022) 161. doi: 10.1016/j.envint.2022.107137
107. Fu X, Ou Z, Zhang M, Meng Y, Li Y, Wen J, et al. Indoor bacterial, fungal and viral species and functional genes in urban and rural schools in shanxi province, China–association with asthma, rhinitis and rhinoconjunctivitis in high school students. Microbiome. (2021) 9. doi: 10.1186/s40168-021-01091-0
108. Richardson M, Gottel N, Gilbert JA, Gordon J, Gandhi P, Reboulet R, et al. Concurrent measurement of microbiome and allergens in the air of bedrooms of allergy disease patients in the Chicago area. Microbiome. (2019) 7. doi: 10.1186/s40168-019-0695-5
109. Bjerregaard A, Laing IA, Poulsen N, Backer V, Sverrild A, Fally M, et al. Characteristics associated with clinical severity and inflammatory phenotype of naturally occurring virus-induced exacerbations of asthma in adults. Respir Med. (2017) 123:34–41. doi: 10.1016/j.rmed.2016.12.010
110. Xiang WQ, Li L, Wang BH, Ali AF, Li W. Profiles and predictive value of cytokines in children with human metapneumovirus pneumonia. Virol J. (2022) 19. doi: 10.1186/s12985-022-01949-1
111. Schwarze J, Hamelmann E, Bradley KL, Takeda K, Gelfand EW. Respiratory syncytial virus infection results in airway hyperresponsiveness and enhanced airway sensitization to allergen. J Clin Invest. (1997) 100:226–33. doi: 10.1172/JCI119516
112. Siegle JS, Hansbro N, Herbert C, Rosenberg HF, Domachowske JB, Asquith KL, et al. Early-life viral infection and allergen exposure interact to induce an asthmatic phenotype in mice. Respir Res. (2010) 11(1):14. doi: 10.1186/1465-9921-11-14
113. Suzuki S, Suzuki Y, Yamamoto N, Matsumoto Y, Shirai A, Okubo T. Influenza A virus infection increases IgE production and airway responsiveness in aerosolized antigen-exposed mice. J Allergy Clin Immunol. (1998) 102:732–40. doi: 10.1016/S0091-6749(98)70012-0
114. Park J-S, Kim Y-H, Kwon E, Callaway Z, Fujisawa T, Kim C-K. Comparison of nasal cytokine profiles of human metapneumovirus and respiratory syncytial virus. Asia Pac Allergy. (2017) 7:206–12. doi: 10.5415/apallergy.2017.7.4.206
115. Schauer U, Hoffjan S, Bittscheidt J, Köchling A, Hemmis S, Bongartz S, et al. RSV Bronchiolitis and risk of wheeze and allergic sensitisation in the first year of life. Eur Respir J. (2002) 20:1277–83. doi: 10.1183/09031936.02.00019902
116. Saraya T, Kimura H, Kurai D, Ishii H, Takizawa H. The molecular epidemiology of respiratory viruses associated with asthma attacks. Medicine (United States) (2017) 96. doi: 10.1097/MD.0000000000008204
117. Schwarze J, Gelfand EW. Respiratory viral infections as promoters of allergic sensitization and asthma in animal models. Eur Respir J. (2002) 19:341–9. doi: 10.1183/09031936.02.00254302
118. Edwards MR, Strong K, Cameron A, Walton RP, Jackson DJ, Johnston SL. Viral infections in allergy and immunology: how allergic inflammation influences viral infections and illness. J Allergy Clin Immunol. (2017) 140:909–20. doi: 10.1016/j.jaci.2017.07.025
119. Muehling LM, Heymann PW, Wright PW, Eccles JD, Agrawal R, Carper HT, et al. Human TH1 and TH2 cells targeting rhinovirus and allergen coordinately promote allergic asthma. J Allergy Clin Immunol. (2020) 146:555–70. doi: 10.1016/j.jaci.2020.03.037
120. Prescott S, Nowak-Wgrzyn A. Strategies to prevent or reduce allergic disease. Ann Nutr Metab. (2011) 59:28–42. doi: 10.1159/000334150
121. Arshad SH. Primary prevention of asthma and allergy. J Allergy Clin Immunol. (2005) 116:3–14. doi: 10.1016/j.jaci.2005.03.043
122. Giallongo A, Oreste U, Cocchiara R, Ruffilli A, Geraci D. Ige and IgG antibodies compete for antigenic determinants of Parietaria officinalis allergen. Mol Immunol. (1980) 17:1019–24. doi: 10.1016/0161-5890(80)90095-4
123. Kim K-E, Rosenberg A, Roberts S, Blumenthals MN. The affinity of allergen specific IgE and the competition between IgE and IgG for the allergen amb a V sensitive individuals. Mol Immunol. (1996) 33:873–80. doi: 10.1016/0161-5890(96)84613-X
124. Zha L, Leoratti FMS, He L, Mohsen MO, Cragg M, Storni F, et al. An unexpected protective role of low-affinity allergen-specific IgG through the inhibitory receptor FcγRIIb. J Allergy Clin Immunol. (2018) 142:1529–1536.e6. doi: 10.1016/j.jaci.2017.09.054
125. Larenas-Linnemann D, Michels A, Dinger H, Shah-Hosseini K, Mösges R, Arias-Cruz A, et al. Allergen sensitization linked to climate and age, not to intermittent-persistent rhinitis in a cross-sectional cohort study in the (sub)tropics. Clin Transl Allergy. (2014) 4. doi: 10.1186/2045-7022-4-20
126. Ujino M, Sugimoto N, Koizumi Y, Ro S, Kojima Y, Asae KH, et al. Leukotriene receptor antagonist attenuated airway inflammation and hyperresponsiveness in a double-stranded RNA-induced asthma exacerbation model. Allergol Int. (2017) 66:S21–6. doi: 10.1016/j.alit.2017.05.004
127. Sher N, Siddiqui FA, Hasan N, Shafi N, Zubair A, Mirza AZ. Simultaneous determination of antihistamine anti-allergic drugs, cetirizine, domperidone, chlorphenamine maleate, loratadine, meclizine and buclizine in pharmaceutical formulations, human serum and pharmacokinetics application. Anal Methods. (2014) 6:2704–14. doi: 10.1039/c3ay41698d
128. Pagliara A, Testa B, Carrupt P-A, Jolliet P, Morin C, Morin D, et al. Molecular properties and pharmacokinetic behavior of cetirizine, a zwitterionic H 1-receptor antagonist. J Med Chem. (1998) 41:853–63. doi: 10.1021/jm9704311
129. Kardas G, Panek M, Kuna P, Cieszyński J, Kardas P. Primary non-adherence to antihistamines—conclusions from E-prescription pilot data in Poland. Front Pharmacol. (2020) 11:1–8. doi: 10.3389/fphar.2020.00783
130. Palikhe NS, Kim SH, Ye YM, Hur GY, Cho BY, Park HS. Association of CRTH2 gene polymorphisms with the required dose of antihistamines in patients with chronic urticaria. Pharmacogenomics. (2009) 10:375–83. doi: 10.2217/14622416.10.3.375
131. Al-Ahmad M, Arifhodzic N, Nurkic J, Maher A, Rodriguez-Bouza T, Al-Ahmed N, et al. “Real-life” efficacy and safety aspects of 4-year omalizumab treatment for asthma. Med Princ Pract. (2018) 27:260–6. doi: 10.1159/000487482
132. Molimard M, Mala L, Bourdeix I, Le Gros V. Observational study in severe asthmatic patients after discontinuation of omalizumab for good asthma control. Respir Med. (2014) 108:571–6. doi: 10.1016/j.rmed.2014.02.003
133. Davies AM, Allan EG, Keeble AH, Delgado J, Cossins BP, Mitropoulou AN, et al. Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab. J Biol Chem. (2017) 292:9975–87. doi: 10.1074/jbc.M117.776476
134. Liu P, Pan Z, Gu C, Cao X, Liu X, Zhang J, et al. An omalizumab biobetter antibody with improved stability and efficacy for the treatment of allergic diseases. Front Immunol. (2020) 11:1–15. doi: 10.3389/fimmu.2020.596908
135. Balbino B, Herviou P, Godon O, Stackowicz J, Goff OR, Iannascoli B, et al. The anti-IgE mAb omalizumab induces adverse reactions by engaging fcγ receptors. J Clin Invest. (2020) 130:1330–5. doi: 10.1172/JCI129697
136. Gasser P, Tarchevskaya SS, Guntern P, Brigger D, Ruppli R, Zbären N, et al. The mechanistic and functional profile of the therapeutic anti-IgE antibody ligelizumab differs from omalizumab. Nat Commun. (2020) 11:1–14. doi: 10.1038/s41467-019-13815-w
137. Chen JB, Ramadani F, Pang MOY, Beavil RL, Holdom MD, Mitropoulou AN, et al. Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody. Sci Rep. (2018) 8:1–13. doi: 10.1038/s41598-018-29664-4
138. Kuo BS, Li CH, Chen JB, Shiung YY, Chu CY, Lee CH, et al. IgE-neutralizing UB-221 mAb, distinct from omalizumab and ligelizumab, exhibits CD23-mediated IgE downregulation and relieves urticaria symptoms. J Clin Invest. (2022) 132:1–16. doi: 10.1172/JCI157765
139. Cohen ES, Dobson CL, Kac¨k H, Wang B, Sims DA, Lloyd CO, et al. A novel IgE-neutralizing antibody for the treatment of severe uncontrolled asthma. MAbs. (2014) 6:756–64. doi: 10.4161/mabs.28394
140. Schanin J, Korver W, Brock EC, Leung J, Benet Z, Luu T, et al. Discovery of an agonistic siglec-6 antibody that inhibits and reduces human mast cells. Commun Biol. (2022) 5:1–13. doi: 10.1038/s42003-022-04207-w
141. Fujiwara D, Wakabayashi H, Watanabe H, Nishida S, Iino H. A double-blind trial of Lactobacillus paracasei strain KW3110 administration for immunomodulation in patients with pollen allergy. Allergol Int (2005) 54:143–9. doi: 10.2332/allergolint.54.143
142. Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med. (2009) 179:186–93. doi: 10.1164/rccm.200806-951OC
143. Mazhary Z, Allahyari Fard N, Minuchehr Z, Javanshir N. Package of anti-allergic probiotic Lactobacillus by focusing on the regulatory role of immunosuppressive motifs in allergy. Inform Med Unlocked. (2020) 18:1–11. doi: 10.1016/j.imu.2019.100280
144. Laigaard A, Krych L, Zachariassen LF, Ellegaard-Jensen L, Nielsen DS, Hansen AK, et al. Dietary prebiotics promote intestinal Prevotella in association with a low-responding phenotype in a murine oxazolone-induced model of atopic dermatitis. Sci Rep. (2020) 10:1–11. doi: 10.1038/s41598-020-78404-0
145. Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut. (2002) 51:51–5. doi: 10.1136/gut.51.1.51
146. Zhen J, Zhao P, Li Y, Cai Y, Yu W, Wang W, et al. The multiomics analyses of gut Microbiota, urine metabolome and plasma proteome revealed significant changes in allergy featured with indole derivatives of tryptophan. J Asthma Allergy. (2022) 15:117–31. doi: 10.2147/JAA.S334752
147. Scadding GW, Eifan AO, Lao-Araya M, Penagos M, Poon SY, Steveling E, et al. Effect of grass pollen immunotherapy on clinical and local immune response to nasal allergen challenge. Allergy. (2015) 70:689–96. doi: 10.1111/all.12608
148. Matsumoto T, Kimoto K, Muraoka N, Miike T. Long-term house dust immunotherapy improves pulmonary functions in children and adolescents with bronchial asthma. Allergol Int. (1998) 47:213–8. doi: 10.2332/allergolint.47.213
149. Ruëff F, Przybilla B, Biló MB, Müller U, Scheipl F, Seitz MJ, et al. Clinical effectiveness of hymenoptera venom immunotherapy: a prospective observational multicenter study of the European academy of allergology and clinical immunology interest group on insect venom hypersensitivity. PLoS One. (2013) 8:1–8. doi: 10.1371/journal.pone.0063233
150. Jung JH, Kang TK, Kang IG, Kim ST. Comparison of sublingual immunotherapy in patients with allergic rhinitis sensitive to house dust mites in Korea. Ear Nose Throat J. (2021) 100:505S–12S. doi: 10.1177/0145561319882593
151. Du W, Fukano C, Yonemoto M, Matsuoka T, Masuyama K, Ohashi-Doi K. Comparison of the allergenic potency of house dust extract and house dust mite allergen extract for subcutaneous allergen immunotherapy. Biol Pharm Bull. (2019) 42:601–6. doi: 10.1248/bpb.b18-00710
152. Dua B, Park J, Kim H. Comparison of weight per volume and protein nitrogen units in non-standardized allergen extracts: implications for prescribing subcutaneous immunotherapy. Allergy Asthma Clin Immunol (2021) 17:1–10. doi: 10.1186/s13223-021-00588-5
153. Maeda M, Imai T, Ishikawa R, Nakamura T, Kamiya T, Kimura A, et al. Effect of oral immunotherapy in children with milk allergy: the ORIMA study. Allergol Int. (2021) 70:223–8. doi: 10.1016/j.alit.2020.09.011
154. Afify SM, Pali-Schöll I, Hufnagl K, Hofstetter G, El-Bassuoni MAR, Roth-Walter F, et al. Bovine holo-Beta-lactoglobulin cross-protects against pollen allergies in an innate manner in BALB/c mice: potential model for the farm effect. Front Immunol. (2021) 12:1–14. doi: 10.3389/fimmu.2021.611474
155. Bożek A, Fischer A, Bogacz-Piaseczynska A, Canonica GW. Adding a biologic to allergen immunotherapy increases treatment efficacy. ERJ Open Res. (2023) 9(2):00639–2022. doi: 10.1183/23120541.00639-2022
156. Bergman TJ, Beehner JC. Leveling with tinbergen: four levels simplified to causes and consequences. Evol Anthropol. (2022) 31:12–9. doi: 10.1002/evan.21931
157. Marín-Cassinello A, Vega-Hernández MC, Lumbreras-Lacarra B, De Arriba-Méndez S, Pellegrini-Belinchón J. Prevalence of symptoms, severity and diagnosis of asthma in adolescents in the province of salamanca, Spain: global asthma network (GAN) phase I. Allergol Immunopathol (Madr) (2021) 49:106–12. doi: 10.15586/aei.v49i5.438
Keywords: allergy, immunology, genetic, ecology, microbiome, helminths, sensitization, IgE
Citation: Falcon RMG and Caoili SEC (2023) Immunologic, genetic, and ecological interplay of factors involved in allergic diseases. Front. Allergy 4:1215616. doi: 10.3389/falgy.2023.1215616
Received: 2 May 2023; Accepted: 24 July 2023;
Published: 3 August 2023.
Edited by:
Spyridon Megremis, University of Leicester, United KingdomReviewed by:
Hontian Wang, Capital Medical University, ChinaClaudia Alessandri, Centri Associati Allergologia Molecolare, Italy
© 2023 Falcon and Caoili. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robbi Miguel G. Falcon rgfalcon@up.edu.ph
 Robbi Miguel G. Falcon
Robbi Miguel G. Falcon Salvador Eugenio C. Caoili
Salvador Eugenio C. Caoili