A Glimpse Into the Structure and Function of Atypical Type I Chaperonins
- National Centre for Cell Science, Pune, India
Chaperonins are a subclass of molecular chaperones that assist cellular proteins to fold and assemble into their native shape. Much work has been done on Type I chaperonins, which has elucidated their elegant mechanism. Some debate remains about the details in these mechanisms, but nonetheless the roles of these in helping protein folding have been understood in great depth. In this review we discuss the known functions of atypical Type I chaperonins, highlighting evolutionary aspects that might lead chaperonins to perform alternate functions.
Molecular Chaperones
Molecular chaperones comprise of a wide range of proteins playing key roles in cellular homeostasis and are responsible for assisting in protein folding, assembly of multimeric proteins, translocation of proteins within and across cell, degradation of unwanted, or misfolded proteins during normal cellular processes and stabilization of proteins by preventing aggregation and assisting in refolding under stress conditions (Lindquist, 1986; Lindquist and Craig, 1988).
Proteins reported to have chaperone activity were initially discovered as those overexpressed during heat shock and hence were named as the heat shock proteins (Hsp). Apart from heat shock, other stress condition such as carbon, nitrogen, or phosphate limiting conditions were also known to induce molecular chaperones. These proteins are classified according to their molecular weight into five major families: (a) Hsp100 family, (b) Hsp90 family, (c) Hsp70 family, (d) Hsp60 family, and (e) small heat shock protein family (sHsp) (Bohen et al., 1995; Schirmer et al., 1996; Bukau and Horwich, 1998). The chaperones are also classified based on their mode of action into: (a) Foldases, Chaperones that assist refolding of unfolded proteins by using ATP, e.g., Hsp70 and Hsp60, (b) Holdases, Chaperones that bind folding intermediates and prevent aggregation, e.g., sHsp and Hsp40, and (c) Disaggregases, Chaperones which actively disaggregate the harmful protein aggregates, which might lead to their small fragments, e.g., members of AAA + ATPase superfamily and Hsp100. This type of classification holds true with few exceptions (Richter et al., 2010; Kim et al., 2013). Much of our understanding on the mechanisms of chaperone-assisted protein folding has been derived from work on Hsp60 and Hsp70 families of chaperones. This review focuses on Hsp60 class of molecular chaperones, highlighting Hsp60 with atypical structure and function.
Hsp60 Family/Chaperonins
The 60 kDa chaperones form large oligomeric rings, and are also referred to as the chaperonins. Chaperonins can be further sub-classified into two groups on the basis of requirement of co-chaperonins and their cellular location. Type I chaperonins are found in the cytoplasm of prokaryotes and in the mitochondrion and chloroplast of eukaryotes. They require the assistance of the co-chaperonin i.e., Hsp10, which acts as a cap on the ring. The well-studied Type I chaperonin is known as the GroEL-GroES system in Escherichia coli. Its homologs are Cpn60/Cpn20 in chloroplasts, and mtHsp60/mtHsp10 in mitochondrion (Cheng et al., 1989; Hayer-Hartl et al., 1995; Dickson et al., 2000). Type II chaperonins are found in the cytoplasm of eukaryotes and in the archaebacterial micro-organisms. They have an in-built lid and hence do not require co-chaperonins for their function (Ranson et al., 1998). Example of Type II chaperonin includes eukaryotic TriC/CCT machinery (TCP-1 ring complex/chaperonin containing TCP-1 complex), which is made up of 8 subunits and the thermosome in archaebacteria. Contrary to Type I chaperonins, substrate independent capture of Type II chaperonins require the assistance of prefoldin and Hsp70 homologs (Iizuka et al., 2004; Cuéllar et al., 2008). Recently, a third group known as Type III chaperonins was reported which are structurally similar to Type II chaperonins but mechanistically and phylogenetically distinct from both Type I and Type II chaperonins e.g., Carboxydothermus hydrogenoformans chaperonin (Ch-CPN) (Techtmann and Robb, 2010; An et al., 2017; Figure 1). The Type I, II, and III chaperonins are also known as Group I, II, and III chaperonins.
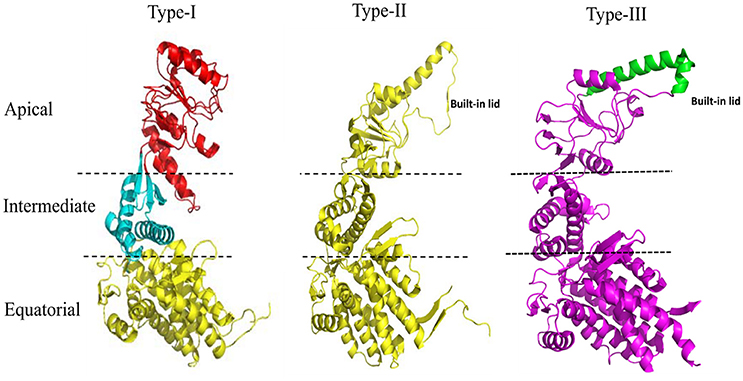
Figure 1. Structural features of the Type I, Type II, and Type III chaperonins. The comparative structure analysis of Type I, Type II, and Type III chaperonins. Structures were downloaded from the RCSB with codes of PDB: 1AON, 3RUW, and 5X9U, respectively. Type I chaperonin is demarcated into Apical, Intermediate, and Equatorial domains, analogous regions of which are shown in Type II and III chaperonins using dotted lines. Type II chaperonin has a characteristic built-in lid in the structure that plays the role of co-chaperonin GroES of Type I chaperonin. Type III chaperonins are structurally similar to Type II chaperonins in having built-in-lid. However, the sequence, structure and function of the lid are distinct in Type II and Type III chaperonins (An et al., 2017). The PyMOL program (PyMOL Molecular Graphics System, version 1.3) was used to generate this figure.
Structure-Function of Type I Chaperonins: Prokaryotic Cytosol
E. coli GroEL-GroES
Structural and functional studies on E. coli GroEL have shown that it forms a tetradecameric structure composed of two heptameric rings stacked on each other forming a cavity, which changes its character from being predominantly hydrophobic to hydrophilic upon binding GroES. Substrate protein folding takes place in this cavity with the assistance of co-chaperonin GroES, which is a cap-like heptameric structure (Mande et al., 1996). Each GroES monomer is of 10 kDa size. The GroEL monomer is demarcated into three domains namely apical, intermediate, and equatorial domain. Each monomer is ~57 kDa in size.
There are two models proposed for the GroEL-GroES mediated substrate protein folding. Asymmetric/sequential model, which is accepted widely. In this model the GroEL and GroES are present stoichiometrically in 2:1 ratio (14:7 subunit ratio). In the other model known as the symmetric/simultaneous model, which is based on the recently observed GroEL-GroES complex, both rings of GroEL are capped by co-chaperonin GroES in the stoichiometric ratio of 1:1 i.e., (GroEL-GroES)2, and subunit ratio of 14:14 (Sameshima et al., 2008; Ye and Lorimer, 2013; Fei et al., 2014). Symmetric (GroEL-GroES)2 complex has been observed both in the presence and absence of substrate protein suggesting a transient intermediate state in the folding reaction cycle.
Structure-Function of Type I Chaperonins: Endosymbiotic Organelles
Chloroplast and Mitochondrial Chaperonins
The chloroplast chaperonins are typically referred to as Cpn60 (GroEL homologs) and Cpn10 (GroES homologs). The Cpn60 chaperonins are made up of multiple subunits which are diverged into two related but distinct α and β types (Dickson et al., 2000; Hill and Hemmingsen, 2001). Contrary to bacterial chaperonins, which contain multiple subunits and prefer homo-oligomerization (Ojha et al., 2005; Gould et al., 2007), chloroplast chaperonins form hetero-oligomers with its two types of chaperonin α and β subunits. Heterogeneity also exists in the co-chaperonin structure. Cpn10 is similar to the standard co-chaperonin, forming heptameric single ring of 10 kDa subunits (Koumoto et al., 2001; Sharkia et al., 2003). Cpn20 has two Cpn10-like polypeptide sequences joined in tandem. The purified Cpn20 exists as a tetramer ring-like structure containing 20 kDa subunit. It is fully functional in vitro, helping refolding of denatured protein in presence of both chloroplast Cpn60 and E. coli GroEL (Tang et al., 2006). Moreover, the Chlamydomonas reinhardtii Cpn10 assist GroEL only in presence of Cpn20 (Tsai et al., 2012). Thus, a considerable heterogeneity exists in the oligomeric assembly of chloroplast chaperonins.
The human mitochondrial chaperonin, mtHsp60 is known to have a protein-folding mechanism (mitochondrial protein) distinct from GroEL-GroES system and requires a single heptameric ring to carry out its protein folding function along with its co-chaperonin, mtHsp10 (Viitanen et al., 1992; Nielsen and Cowan, 1998). However, the crystal structure of mitochondrial chaperonin in complex with its co-chaperonin, mtHsp60-mtHsp10 depicts a unique intermediate stage where mtHsp60-mtHsp10 forms a symmetric double-ring football-like structure, (mtHsp60)14 + 2 (mtHsp10)7.
Type I Chaperonins: Non-canonical Features
Multiple Chaperonins Across Species
Analysis of completely sequenced genomes suggest that about 30% of all the genomic sequence data possess multiple copies of gene sequences encoding chaperonins (Lund, 2009; Kumar et al., 2015). Distribution of these multiple chaperonins based on extensive phylogenetic analysis suggest that multiple copies of chaperonin genes exist predominantly in five phyla, namely, (a) phylum Actinobacteria, (b) phylum Firmicutes, (c) phylum Cyanobacteria, (d) phylum Chlamydia, and (e) α-Proteobacteria phylum (Kumar et al., 2015).
Actinobacteria
Actinobacteria are Gram-positive bacteria and possess high G + C content in their genomes, e.g., Mycobacterium tuberculosis, Mycobacterium leprae, and Bifidobacterium longum. These species typically possess two copies of chaperonin genes, with one of the copies being present on an operon-like structure. The other copy of Cpn60 exists as an independent gene without the presence of Cpn10 gene (Rinke de Wit et al., 1992). The actinobacterial chaperonin genes are under the regulatory control of HrcA transcription factor which binds to upstream CIRCE (controlling inverted repeat of chaperone expression) sequence (Duchêne et al., 1994; Grandvalet et al., 1998). In some cases regulation is mediated through HspR transcription factor binding to upstream HAIR (HspR Associated Inverted Repeat) sequence (Barreiro et al., 2004).
Firmicutes
Firmicutes are Gram-positive bacteria and possess low G + C content in their genomes, e.g., Staphylococcus aureus, Desulfitobacterium dehalogenans, and C. hydrogenoformans. Firmicutes are known to possess both prokaryotic-like Type I chaperonin genes and archael-like chaperonin genes classified as Type III chaperonin. Type I chaperonins are arranged in an operonic arrangement with the co-chaperonin, while Type III chaperonin gene is located in the dnaK operon. Both the Type 1 and Type III chaperonin genes are regulated by HrcA transcription factor (Techtmann and Robb, 2010).
Chlamydiae
Chlamydiae are mostly obligate intracellular pathogens, e.g., Chlamydia trachomatis, Chlamydia pneumonia, and Chlamydia psittaci. Chlamydial species possess three copies of chaperonin genes (McNally and Fares, 2007). Operonic arrangement suggests that only one copy of the chaperonin genes exists along with its co-chaperonin. However, other chaperonin genes are located separately. Regulation of chlamydial chaperonin genes is complex. The first copy of the chaperonin gene is induced by heat shock and regulated by HrcA-CIRCE system. The second copy of the chaperonin gene is induced when Chlamydia are inside monocyte or macrophages (Kol et al., 1999), and the third copy of the chaperonin gene is induced when Chlamydia are in Hep-2 cells (Gérard et al., 2004). Such types of expression and regulation of chaperonin genes suggest life-cycle specific patterns and independent functional roles for them.
α-proteobacteria
Rhizobia, which belong to the α-proteobacteria class, are symbiotic organisms living in association with leguminous plants in the root nodules and are involved in nitrogen fixation, e.g., Bradyrhizobium japonicum, Rhizobium leguminosarum. Rhizobia contain most number of copies of chaperonins. B. japonicum has seven copies of chaperonin genes (Fischer et al., 1993). R. leguminosarum is a well-characterized organism and has three copies of chaperonin genes. Gene arrangement in all these organisms suggests that the three copies of the chaperonin gene form separate operons with their respective co-chaperonin genes (George et al., 2004). One of the chaperonin operons is located on the genomic island that contains genes involved in nitrogen fixation. It is regulated by NiF factors that regulate nitrogen fixation genes (Ogawa and Long, 1995). The second copy of the chaperonin gene is not well-studied and is known to be involved in chaperoning property of several model substrate proteins (George et al., 2004).
Cyanobacteria
Cyanobacteria are largely photosynthetic bacteria, e.g., Synechococcus platensis, Prochlorococcus marinus, and Anabaena variabilis. About 90% of the genomic sequences of the cyanobacterial species contain two copies of chaperonin genes with one of them being arranged on an operon while the other chaperonin gene coded separately. Some cyanobacterial species containing three copies of chaperonin genes, where two of its chaperonin genes being located with respective co-chaperonins in the operon while the third copy of chaperonin genes is independent (Lund, 2009; Kumar et al., 2015). Chaperonin genes existing in the operonic arrangement with their co-chaperonins are essential genes while the ones which exist independent of the co-chaperonin are non-essential (Sato et al., 2008). The two cyanobacterial chaperonin genes are positively regulated by RpoH and negatively regulated by HrcA proteins. Upon heat shock, one of the chaperonin genes is induced rapidly while the other chaperonin gene is gradually induced (Kojima and Nakamoto, 2007; Rajaram and Apte, 2010). The chaperonin gene that is gradually induced on heat shock is known to be directly involved in photosynthesis.
Evolutionary Lineage
As more genomic sequences are becoming available, analysis of chaperonin genes suggests that distribution and frequency of multiple copies of chaperonin genes across phyla and organisms continues to increase (Lund, 2009; Kumar et al., 2015). In order to understand the cause of multiplicity of chaperonin genes is either due to horizontal gene transfer or gene duplication, phylogenetic analysis was carried on GroEL proteins across species, which revealed that the causes of existence of multiple copies of GroELs are non-uniform. In a few cases there is gene duplication event followed by evolutionary selection such as that observed in myxobacterial GroELs, mycobacterial first and second copy of GroEL and few rhizobial GroELs. In the case of the third mycobacterial GroEL homolog, few rhizobial GroELs and methanosarcinal GroELs, horizontal gene transfer occurred (Goyal et al., 2006; Kumar et al., 2015).
It has been proposed earlier in our lab that mycobacterial GroEL has been duplicated and undergone various selective pressures to perform distinctive structural and functional role during the course of evolution (Goyal et al., 2006). Biophysical and biochemical studies on recombinantly purified M. tuberculosis GroELs have shown that GroEL1 and GroEL2 exist as lower oligomeric species contrary to tetradecameric GroEL structure of E. coli (Qamra et al., 2004). The crystal structure of M. tuberculosis GroEL2 in its dimeric form highlighted the presence of distinct residues at the interface region, probably responsible for the change in oligomerization (Figure 2; Qamra and Mande, 2004). Gene shuffling and domain swapping studies on M. tuberculosis GroEL1 suggest that the equatorial domain is responsible for failed oligomerization. The apical domain can withstand large insertions and deletions (Kumar et al., 2009). Around the same time it was shown that GroEL1 has evolved to promiscuously bind nucleic acids (Basu et al., 2009) and oligomerization is facilitated by phosphorylation of serine residues (Kumar et al., 2009). Since GroEL2 is known to be essential chaperonin in Mycobacteria, whereas the oligomeric assembly of GroEL1 is regulated post-translationally, it was reported that tetradecameric assembly and precise inter-domain communication are prerequisite for chaperonin activity (Chilukoti et al., 2015).
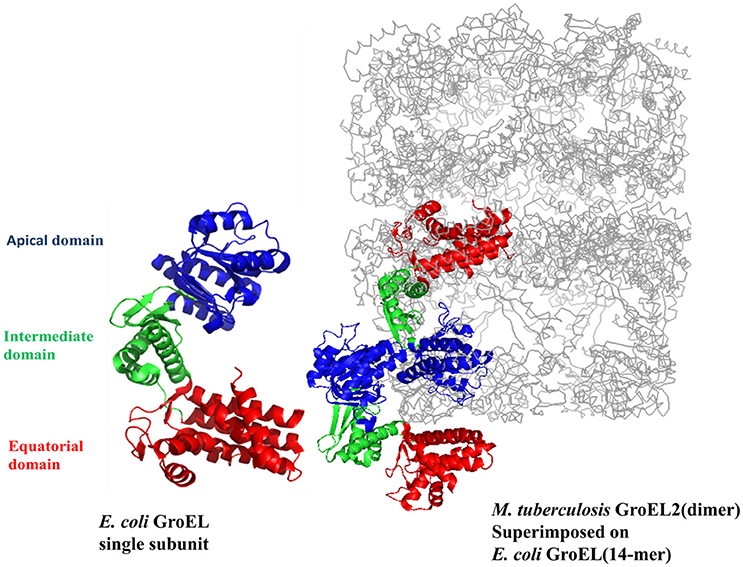
Figure 2. The crystal structure of M. tuberculosis GroEL2 superimposed on E. coli GroEL-ES structure. The structure of M. tuberculosis GroEL2 (PDB ID:1SJP) shows lower oligomeric status (dimer). Colored in blue, green, and red are the Apical, Intermediate and Equatorial domain, respectively. Compared to E. coli GroEL (PDB ID: 1AON) shown in gray color, the inter-subunit interaction is mediated through Apical domain in M. tuberculosis GroEL2 structure whereas inter-subunit interaction is through Equatorial domain in E. coli GroEL. Single subunit of M. tuberculosis GroEL2 is aligned to E. coli GroES bound GroEL ring representing asymmetric model. GroES structure has been removed for simplicity. A single subunit of E. coli GroEL has been shown with the same color-coded domains compared to M. tuberculosis GroEL2 for comparative analysis. The PyMOL program (PyMOL Molecular Graphics System, version 1.3) was used to generate this figure.
Functional Diversity
It is important to examine whether the presence of multiple copies of chaperonins are responsible for behaving as canonical chaperonins or they have diverged to carry out novel functions. It is also important to note whether these multiple chaperonins act on common substrates or on distinct pool of substrates. GroELs are highly conserved across different species and it has been shown that homologs of chaperonins from other bacteria are able to function in E. coli suggesting overlapping of substrate proteins and common mechanism of GroEL function. The interactions of substrate proteins with GroEL are hydrophobic in nature, so conformational change mediated exposure of the apical and the equatorial domains in the cavity plays a key role in substrate recognition and assists protein folding. Binding of substrate proteins to GroEL is through α/β domains of proteins with no sequence similarity (Kerner et al., 2005; Kumar and Mande, 2011) and further studies suggest that GroEL selectively binds globular substrates rather than extended polypeptides (Robinson et al., 1994; Goldberg et al., 1997). Multiple copies of chaperonins in an organism have also been reported to have evolved to carry out novel functions. GroEL homolog in an insect symbiont, Xenorhabdus nematophila has been shown to be toxic to insects which is mediated through binding to alpha-chitin. Mutational analysis on these GroEL homologs suggests that the amino acid critical for this kind of activity is distinct from the essential chaperonin (Joshi et al., 2008). In M. tuberculosis, GroEL2 acts as a generalist chaperonin (Hu et al., 2008) while GroEL1 is reported to be associated with nucleoids (Basu et al., 2009). Thus, it is apparent that gene duplication of groEL genes has led to the functional diversity of chaperonins and/or distinct substrate spectrum for intracellular protein folding.
Post-translational Modifications/Biofilm Formation
Post-translational modifications in proteins are employed by organisms to modulate their physiological processes and adapt to constantly changing environment (Bernal et al., 2014). Chaperonins have been reported to be post-translationally modified in certain organisms, and this modification has been reported to gain/loss of their function. For example, fractionation of M. tuberculosis cell lysate has shown that tetradecameric form of GroEL1 is attained only upon phosphorylation at serine residues (Kumar et al., 2009). Similarly in another report it has been shown that phosphorylation occurs at threonine residues (Canova et al., 2009). Both of these observations suggest that oligomerization of GroEL1 is a result of post-translational modification.
Many pathogens evade innate immune response and become resistant to antibiotics by forming biofilms on epithelial cells (Hall-Stoodley and Stoodley, 2005). The role of GroEL in biofilm formation has been elucidated in a few organisms. For example, GroEL1 mutant of M. smegmatis fails to form biofilm. Mechanistic studies revealed that M. smegmatis GroEL1 interacts with the KasA enzyme, which is critical for mycolic acid biosynthesis involved in biofilm formation (Ojha et al., 2005). Interestingly, it has been recently reported that GroEL in pathogenic strain B. anthracis gets phosphorylated and thereby modulates biofilm formation. These findings highlight that phosphorylation of GroEL has functional implications (Arora et al., 2017). Acetylation is another post-translational modification associated with E. coli and M. tuberculosis chaperonins, however a functional role has not yet been ascribed to this modification (Liu et al., 2014). Similarly, mitochondrial co-chaperonin (mtHsp10) undergoes acetyl modification and controls folding of mitochondrial proteins under excess nutrient condition (Lu et al., 2015).
C-Terminal Diversity
Various studies highlight the importance of the C-terminal residues of GroEL in the overall functioning of the chaperonin (Tang et al., 2006; Chen et al., 2013). In cases pertaining to multiple copies of chaperonins, they have distinct pattern of C-terminal residues. While the C-terminus of GroEL (from E. coli) has a 13 residue motif (GGM)4M, GroEL homologs from other organisms (which contain multiple copies of chaperonins) have distinct C-terminal motifs, such as:
a) Histidine-rich C-terminal, e.g., Mycobacteria (Colaco and MacDougall, 2014)
b) Pattern-less C-terminus, e.g., Rhizobia (George et al., 2004)
c) Similar (GGM)4M repeats, e.g., Myxobacteria (Wang et al., 2013)
d) Lack of GGM-like tail, e.g., Methanosarcina (Figueiredo et al., 2004)
It is clearly seen that many chaperonin paralogs in different organisms have GGM-like C-terminus. A wide range of genomic organization is seen in these chaperonins. Moreover, differences are also seen in their co-expression with co-chaperonin and essentiality of their function. Thus, these paralogs are perplexingly observed to be either essential or non-essential, co-expressed with their co-chaperonin or not co-expressed, and possibly function as housekeeping chaperonins. On the other hand chaperonins not possessing the GGM-like C-terminus have possibly evolved to carry out novel functions (Ojha et al., 2005; Wang et al., 2013; Figure 3).
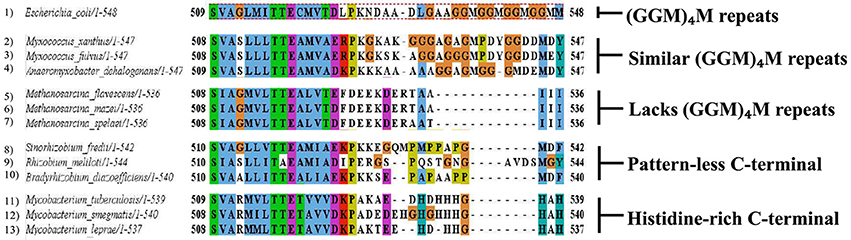
Figure 3. Multiple chaperonins in bacteria displaying diversity at C-terminal. Sequence alignment highlighting C-terminal regions of the representative bacterial GroEL homologs with the E. coli GroEL. The last C-terminal residues of selected multiple GroELs in different bacteria show divergence from the canonical (GGM)4M motif of the E. coli GroEL shown in dotted red box. Sequences were retrieved from www.uniprot.org and aligned in MEGA6 using MUSCLE algorithm (www.megasoftware.net). Formatting of aligned sequences were done in Jalview alignment viewer (www.jalview.org). Residues in the alignment follow the default Clustal color scheme of Jalview.
Concluding Remarks
Type I chaperonins are important by virtue of their role in intracellular protein folding. GroEL-GroES system in bacteria helps folding of about 10–15% of cytosolic proteins. Various structures of GroEL solved in apo-form, nucleotide-bound form as well as in complex with co-chaperonin GroES attempt to explain the role of these chaperonins in protein folding (Saibil et al., 2013). The existence of multiple chaperonins and their role in varied functions hints evolutionary pressure toward adapting to different environmental conditions. The structure of M. tuberculosis GroEL2 highlights lower oligomeric state and more exposed hydrophobic surfaces, probably to increase substrate pool and energy conservation (Qamra and Mande, 2004; Qamra et al., 2004; Kumar and Mande, 2011). Owing to the presence of Histidine-rich C-terminal in multiple chaperonins, these have been proposed to help in alternate biological functions. M. smegmatis GroEL1 binding to iron may help in biofilm formation (Ojha et al., 2005). Survival defect of M. tuberculosis groEL1 knockout strain under low aeration condition might help in oxygen sensing by directly binding to metals or help certain metalloproteins in folding (Sharma et al., 2016). The structure of other homologous chaperonin proteins will probably answer the myriad of questions associated with the novel functions of chaperonin homologs.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MYA thanks the financial support for Senior Research Fellowship of the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India. The authors would like to gratefully acknowledge the financial support for work in the SCM laboratory through grants BT/PR15450/COE/34/46/2016 and BT/PR3260/BRB/10/967/2011 of the DBT.
References
An, Y. J., Rowland, S. E., Na, J. H., Spigolon, D., Hong, S. K., Yoon, Y. J., et al. (2017). Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium. Nat. Commun. 8:827. doi: 10.1038/s41467-017-00980-z
Arora, G., Sajid, A., Virmani, R., Singhal, A., Kumar, C. M. S., Dhasmana, N., et al. (2017). Ser/Thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. NPJ Biofilms Microbiomes 3:7. doi: 10.1038/s41522-017-0015-4
Barreiro, C., González-Lavado, E., Pátek, M., and Martín, J. F. (2004). Transcriptional analysis of the groES-groEL1, groEL2, and dnaK genes in Corynebacterium glutamicum: characterization of heat shock-induced promoters. J. Bacteriol. 186, 4813–4817. doi: 10.1128/JB.186.14.4813-4817.2004
Basu, D., Khare, G., Singh, S., Tyagi, A., Khosla, S., and Mande, S. C. (2009). A novel nucleoid-associated protein of Mycobacterium tuberculosis is a sequence homolog of GroEL. Nucleic Acids Res. 37, 4944–4954. doi: 10.1093/nar/gkp502
Bernal, V., Castaño-Cerezo, S., Gallego-Jara, J., Écija-Conesa, A., de Diego, T., Iborra, J. L., et al. (2014). Regulation of bacterial physiology by lysine acetylation of proteins. N. Biotechnol. 31, 586–595. doi: 10.1016/j.nbt.2014.03.002
Bohen, S. P., Kralli, A., and Yamamoto, K. R. (1995). Hold ‘em and fold' em: chaperones and signal transduction. Science 268, 1303–1304.
Bukau, B., and Horwich, A. L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366. doi: 10.1016/S0092-8674(00)80928-9
Canova, M. J., Kremer, L., and Molle, V. (2009). The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J. Bacteriol. 191, 2876–2883. doi: 10.1128/JB.01569-08
Chen, D. H., Madan, D., Weaver, J., Lin, Z., Schröder, G. F., Chiu, W., et al. (2013). Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell 153, 1354–1365. doi: 10.1016/j.cell.2013.04.052
Cheng, M. Y., Hartl, F. U., Martin, J., Pollock, R. A., Kalousek, F., Neupert, W., et al. (1989). Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337, 620–625. doi: 10.1038/337620a0
Chilukoti, N., Kumar, C. M., and Mande, S. C. (2015). GroEL2 of Mycobacterium tuberculosis reveals the importance of structural pliability in chaperonin function. J. Bacteriol. 198, 486–497. doi: 10.1128/JB.00844-15
Colaco, C. A., and MacDougall, A. (2014). Mycobacterial chaperonins: the tail wags the dog. FEMS Microbiol. Lett. 350, 20–24. doi: 10.1111/1574-6968.12276
Cuéllar, J., Martín-Benito, J., Scheres, S. H., Sousa, R., Moro, F., López-Viñas, E., et al. (2008). The structure of CCT-Hsc70 NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nat. Struct. Mol. Biol. 15, 858–864. doi: 10.1038/nsmb.1464
Dickson, R., Weiss, C., Howard, R. J., Alldrick, S. P., Ellis, R. J., Lorimer, G., et al. (2000). Reconstitution of higher plant chloroplast chaperonin 60 tetradecamers active in protein folding. J. Biol. Chem. 275, 11829–11835. doi: 10.1074/jbc.275.16.11829
Duchêne, A. M., Thompson, C. J., and Mazodier, P. (1994). Transcriptional analysis of groEL genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 245, 61–68. doi: 10.1007/BF00279751
Fei, X., Ye, X., LaRonde, N. A., and Lorimer, G. H. (2014). Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc. Natl. Acad. Sci. U.S.A. 111, 12775–12780. doi: 10.1073/pnas.1412922111
Figueiredo, L., Klunker, D., Ang, D., Naylor, D. J., Kerner, M. J., Georgopoulos, C., et al. (2004). Functional characterization of an archaeal GroEL/GroES chaperonin system: significance of substrate encapsulation. J. Biol. Chem. 279, 1090–1099. doi: 10.1074/jbc.M310914200
Fischer, H. M., Babst, M., Kaspar, T., Acuña, G., Arigoni, F., and Hennecke, H. (1993). One member of a gro-ESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12, 2901–2912.
George, R., Kelly, S. M., Price, N. C., Erbse, A., Fisher, M., and Lund, P. A. (2004). Three GroEL homologues from Rhizobium leguminosarum have distinct in vitro properties. Biochem. Biophys. Res. Commun. 324, 822–828. doi: 10.1016/j.bbrc.2004.09.140
Gérard, H. C., Whittum-Hudson, J. A., Schumacher, H. R., and Hudson, A. P. (2004). Differential expression of three Chlamydia trachomatis hsp60-encoding genes in active vs. persistent infections. Microb. Pathog. 36, 35–39. doi: 10.1016/j.micpath.2003.08.005
Goldberg, M. S., Zhang, J., Sondek, S., Matthews, C. R., Fox, R. O., and Horwich, A. L. (1997). Native-like structure of a protein-folding intermediate bound to the chaperonin GroEL. Proc. Natl. Acad. Sci. U.S.A. 94, 1080–1085. doi: 10.1073/pnas.94.4.1080
Gould, P. S., Burgar, H. R., and Lund, P. A. (2007). Homologous cpn60 genes in Rhizobium leguminosarum are not functionally equivalent. Cell Stress Chaperones 12, 123–131. doi: 10.1379/CSC-227R.1
Goyal, K., Qamra, R., and Mande, S. C. (2006). Multiple gene duplication and rapid evolution in the groEL gene: functional implications. J. Mol. Evol. 63, 781–787. doi: 10.1007/s00239-006-0037-7
Grandvalet, C., Rapoport, G., and Mazodier, P. (1998). hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 180, 5129–5134.
Hall-Stoodley, L., and Stoodley, P. (2005). Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13, 7–10. doi: 10.1016/j.tim.2004.11.004
Hayer-Hartl, M. K., Martin, J., and Hartl, F. U. (1995). Asymmetrical interaction of GroEL and GroES in the ATPase cycle of assisted protein folding. Science 269, 836–841. doi: 10.1126/science.7638601
Hill, J. E., and Hemmingsen, S. M. (2001). Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 6, 190–200. doi: 10.1379/1466-1268(2001)006<0190:ATTIAI>2.0.CO;2
Hu, Y., Henderson, B., Lund, P. A., Tormay, P., Ahmed, M. T., Gurcha, S. S., et al. (2008). A Mycobacterium tuberculosis mutant lacking the groEL homologue cpn60.1 is viable but fails to induce an inflammatory response in animal models of infection. Infect. Immun. 76, 1535–1546. doi: 10.1128/IAI.01078-07
Iizuka, R., So, S., Inobe, T., Yoshida, T., Zako, T., Kuwajima, K., et al. (2004). Role of the helical protrusion in the conformational change and molecular chaperone activity of the archaeal group II chaperonin. J. Biol. Chem. 279, 18834–18839. doi: 10.1074/jbc.M400839200
Joshi, M. C., Sharma, A., Kant, S., Birah, A., Gupta, G. P., Khan, S. R., et al. (2008). An insecticidal GroEL protein with chitin binding activity from Xenorhabdus nematophila. J. Biol. Chem. 283, 28287–28296. doi: 10.1074/jbc.M804416200
Kerner, M. J., Naylor, D. J., Ishihama, Y., Maier, T., Chang, H. C., Stines, A. P., et al. (2005). Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220. doi: 10.1016/j.cell.2005.05.028
Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M., and Hartl, F. U. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355. doi: 10.1146/annurev-biochem-060208-092442
Kojima, K., and Nakamoto, H. (2007). A novel light- and heat-responsive regulation of the groE transcription in the absence of HrcA or CIRCE in cyanobacteria. FEBS Lett. 581, 1871–1880. doi: 10.1016/j.febslet.2007.03.084
Kol, A., Bourcier, T., Lichtman, A. H., and Libby, P. (1999). Chlamydial and human heat shock protein 60s activate human vascular endothelium, smooth muscle cells, and macrophages. J. Clin. Invest. 103, 571–577. doi: 10.1172/JCI5310
Koumoto, Y., Shimada, T., Kondo, M., Hara-Nishimura, I., and Nishimura, M. (2001). Chloroplasts have a novel Cpn10 in addition to Cpn20 as co-chaperonins in Arabidopsis thaliana. J. Biol. Chem. 276, 29688–29694. doi: 10.1074/jbc.M102330200
Kumar, C. M., Khare, G., Srikanth, C. V., Tyagi, A. K., Sardesai, A. A., and Mande, S. C. (2009). Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation-mediated oligomerization. J. Bacteriol. 191, 6525–6538. doi: 10.1128/JB.00652-09
Kumar, S., and Mande, S. (2011). Protein chaperones and non-protein substrates: on substrate promiscuity of GroEL. Curr. Sci. 100, 1646–1653.
Kumar, C. M., Mande, S. C., and Mahajan, G. (2015). Multiple chaperonins in bacteria—novel functions and non-canonical behaviors. Cell Stress Chaperones 20, 555–574. doi: 10.1007/s12192-015-0598-8
Lindquist, S. (1986). The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191. doi: 10.1146/annurev.bi.55.070186.005443
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Liu, F., Yang, M., Wang, X., Yang, S., Gu, J., Zhou, J., et al. (2014). Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell. Proteomics 13, 3352–3366. doi: 10.1074/mcp.M114.041962
Lu, Z., Chen, Y., Aponte, A. M., Battaglia, V., Gucek, M., and Sack, M. N. (2015). Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. J. Biol. Chem. 290, 2466–2476. doi: 10.1074/jbc.M114.606228
Lund, P. A. (2009). Multiple chaperonins in bacteria–why so many? FEMS Microbiol. Rev. 33, 785–800. doi: 10.1111/j.1574-6976.2009.00178.x
Mande, S. C., Mehra, V., Bloom, B. R., and Hol, W. G. (1996). Structure of the heat shock protein chaperonin-10 of Mycobacterium leprae. Science 271, 203–207. doi: 10.1126/science.271.5246.203
McNally, D., and Fares, M. A. (2007). In silico identification of functional divergence between the multiple groEL gene paralogs in Chlamydiae. BMC Evol. Biol. 7:81. doi: 10.1186/1471-2148-7-81
Nielsen, K. L., and Cowan, N. J. (1998). A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol. Cell 2, 93–99. doi: 10.1016/S1097-2765(00)80117-3
Ogawa, J., and Long, S. R. (1995). The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 9, 714–729. doi: 10.1101/gad.9.6.714
Ojha, A., Anand, M., Bhatt, A., Kremer, L., Jacobs, W. R., and Hatfull, G. F. (2005). GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123, 861–873. doi: 10.1016/j.cell.2005.09.012
Qamra, R., and Mande, S. C. (2004). Crystal structure of the 65-kilodalton heat shock protein, chaperonin 60.2, of Mycobacterium tuberculosis. J. Bacteriol. 186, 8105–8113. doi: 10.1128/JB.186.23.8105-8113.2004
Qamra, R., Srinivas, V., and Mande, S. C. (2004). Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. J. Mol. Biol. 342, 605–617. doi: 10.1016/j.jmb.2004.07.066
Rajaram, H., and Apte, S. K. (2010). Differential regulation of groESL operon expression in response to heat and light in Anabaena. Arch. Microbiol. 192, 729–738. doi: 10.1007/s00203-010-0601-9
Ranson, N. A., White, H. E., and Saibil, H. R. (1998). Chaperonins. Biochem. J. 333(Pt 2), 233–242. doi: 10.1042/bj3330233
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. doi: 10.1016/j.molcel.2010.10.006
Rinke de Wit, T. F., Bekelie, S., Osland, A., Miko, T. L., Hermans, P. W., van Soolingen, D., et al. (1992). Mycobacteria contain two groEL genes: the second Mycobacterium leprae groEL gene is arranged in an operon with groES. Mol. Microbiol. 6, 1995–2007. doi: 10.1111/j.1365-2958.1992.tb01372.x
Robinson, C. V., Gross, M., Eyles, S. J., Ewbank, J. J., Mayhew, M., Hartl, F. U., et al. (1994). Conformation of GroEL-bound alpha-lactalbumin probed by mass spectrometry. Nature 372, 646–651. doi: 10.1038/372646a0
Saibil, H. R., Fenton, W. A., Clare, D. K., and Horwich, A. L. (2013). Structure and allostery of the chaperonin GroEL. J. Mol. Biol. 425, 1476–1487. doi: 10.1016/j.jmb.2012.11.028
Sameshima, T., Ueno, T., Iizuka, R., Ishii, N., Terada, N., Okabe, K., et al. (2008). Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J. Biol. Chem. 283, 23765–23773. doi: 10.1074/jbc.M802541200
Sato, S., Ikeuchi, M., and Nakamoto, H. (2008). Expression and function of a groEL paralog in the thermophilic cyanobacterium Thermosynechococcus elongatus under heat and cold stress. FEBS Lett. 582, 3389–3395. doi: 10.1016/j.febslet.2008.08.034
Schirmer, E. C., Glover, J. R., Singer, M. A., and Lindquist, S. (1996). HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21, 289–296. doi: 10.1016/S0968-0004(96)10038-4
Sharkia, R., Bonshtien, A. L., Mizrahi, I., Weiss, C., Niv, A., Lustig, A., et al. (2003). On the oligomeric state of chloroplast chaperonin 10 and chaperonin 20. Biochim. Biophys. Acta 1651, 76–84. doi: 10.1016/S1570-9639(03)00237-1
Sharma, A., Rustad, T., Mahajan, G., Kumar, A., Rao, K. V., Banerjee, S., et al. (2016). Towards understanding the biological function of the unusual chaperonin Cpn60.1 (GroEL1) of Mycobacterium tuberculosis. Tuberc. Edinb. Scotl. 97, 137–146. doi: 10.1016/j.tube.2015.11.003
Tang, Y. C., Chang, H. C., Roeben, A., Wischnewski, D., Wischnewski, N., Kerner, M. J., et al. (2006). Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914. doi: 10.1016/j.cell.2006.04.027
Techtmann, S. M., and Robb, F. T. (2010). Archaeal-like chaperonins in bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 20269–20274. doi: 10.1073/pnas.1004783107
Tsai, Y. C., Mueller-Cajar, O., Saschenbrecker, S., Hartl, F. U., and Hayer-Hartl, M. (2012). Chaperonin cofactors, Cpn10 and Cpn20, of green algae and plants function as hetero-oligomeric ring complexes. J. Biol. Chem. 287, 20471–20481. doi: 10.1074/jbc.M112.365411
Viitanen, P. V., Lorimer, G. H., Seetharam, R., Gupta, R. S., Oppenheim, J., Thomas, J. O., et al. (1992). Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J. Biol. Chem. 267, 695–698.
Wang, Y., Zhang, W. Y., Zhang, Z., Li, J., Li, Z. F., Tan, Z. G., et al. (2013). Mechanisms involved in the functional divergence of duplicated GroEL chaperonins in Myxococcus xanthus DK1622. PLoS Genet. 9:e1003306. doi: 10.1371/journal.pgen.1003306
Keywords: Type I chaperonins, GroEL, GroES, Mycobacterium tuberculosis, protein folding, gene duplication
Citation: Ansari MY and Mande SC (2018) A Glimpse Into the Structure and Function of Atypical Type I Chaperonins. Front. Mol. Biosci. 5:31. doi: 10.3389/fmolb.2018.00031
Received: 04 January 2018; Accepted: 21 March 2018;
Published: 11 April 2018.
Edited by:
Abdussalam Azem, Tel Aviv University, IsraelReviewed by:
David A. Dougan, La Trobe University, AustraliaAnat Ben-Zvi, Ben-Gurion University of the Negev, Israel
Copyright © 2018 Ansari and Mande. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shekhar C. Mande, shekhar@nccs.res.in; director@nccs.res.in
 Mohammed Y. Ansari
Mohammed Y. Ansari Shekhar C. Mande
Shekhar C. Mande