-
PDF
- Split View
-
Views
-
Cite
Cite
J.J. Loor, A.A. Elolimy, J.C. McCann, Dietary impacts on rumen microbiota in beef and dairy production, Animal Frontiers, Volume 6, Issue 3, July 2016, Pages 22–29, https://doi.org/10.2527/af.2016-0030
Close - Share Icon Share
Abstract
The rumen harbors diverse microorganisms including bacteria, protozoa, fungi, archaea, and viruses. They play a key role in the breakdown and utilization of feedstuff carbohydrate and protein (from forage, grain) through the process of fermentation, resulting in the production of volatile fatty acids (or short-chain fatty acids) and microbial protein.
The volatile fatty acids provide the ruminant with readily available sources of energy and substrates for the synthesis of lipid and glucose by the animal. Microbial protein provides a highly digestible source of amino acids for muscle growth in beef and milk protein in dairy cows. Therefore, rumen microorganisms play an essential role in optimizing nutrient utilization from feed.
Historically, knowledge about the rumen microbial ecology and the nutrition of microorganisms was obtained using classical culture approaches, which by the 1990s, allowed the characterization of at least 22 major bacteria.
Rapid advances in molecular biology and phylogenetic techniques (e.g., multiple sequence alignment) and the rise of “omic” approaches (e.g., high-throughput sequencing) have allowed for understanding the ecology and function of microbial ecosystems in the rumen.
These technical advances have enabled scientists, for the first time, to have a holistic view of the rumen microbiota and enhanced our ability to explore some of the hidden relationships between an “altered” rumen environment and the development of a disorder such as sub-acute ruminal acidosis or milk fat depression (“low fat milk syndrome”).
Here we briefly discuss recent examples demonstrating how age and nutrition can alter the structure, composition, and diversity of the rumen microbiota in beef and dairy production. We also highlight areas where there are knowledge gaps for further research.
Applying an integrative approach, i.e., systems biology, encompassing nutritional management effects on the rumen microbiota, the tissue responses, and the production outcomes will provide added value to nutritionists attempting to optimize further the production of high-quality beef and milk to meet the demands of a growing population worldwide.
Background
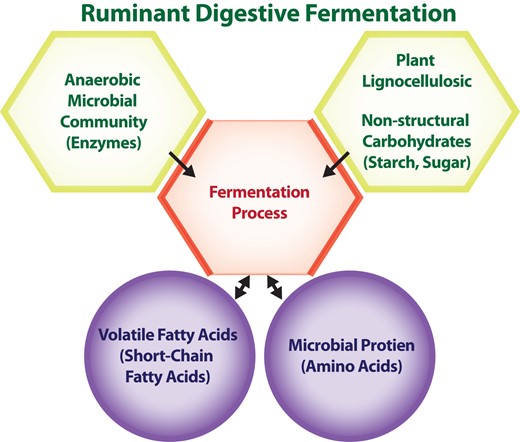
The reticulo-rumen is the largest compartment of the ruminant digestive tract, and it harbors a complex anaerobic microbial community capable of producing a wide array of enzymes, some of which are important for the breakdown of plant lignocellulosic and non-structural carbohydrate (starch, sugars) material through the process of fermentation (Russell and Rychlik, 2001). Important outcomes of microbial fermentation are the production of volatile fatty acids (or short-chain fatty acids) that serve as fuels for the animal's tissues and the synthesis of microbial protein that provides amino acids for the animal to produce high-quality protein for human consumption, i.e., meat and milk. Studies over the last few decades have conclusively demonstrated that the ruminal microbial ecosystem (both of the neonate and mature ruminant) can be altered by nutritional management (McCann et al., 2014b). Therefore, nutrition represents an important tool for manipulating the microbial ecosystem to optimize rumen function while producing high-quality meat and milk for meeting the demands of a growing human population.
The recent development of omic technologies, e.g., sequencing of the 16S and 18S ribosomal RNA gene, metagenomics, and metatranscriptomics, along with bioinformatics tools, have not only enhanced the understanding of the rumen ecology, but also our ability to predict the functional capacity of the microbiota (McCann et al., 2014b). From a production standpoint, it is believed that further gains in knowledge that in the medium-to-long term could lead to practical applications will arise from the integration of taxonomic and functional data with “classical” parameters of rumen digestion, metabolism, and performance (Morgavi et al., 2013). Our aim in this review is to provide recent examples demonstrating how nutrition can alter the structure, composition, and diversity of the rumen microbiota in beef and dairy production. In addition, we attempt to highlight points where there are knowledge gaps for further research.
The Symbiosis between Rumen Microbiota and the Host
Rumen microorganisms, termed rumen microbiota, encompass a diverse class diverse class of bacteria, archaea, protozoa, fungi, and viruses. Bacteria compose an estimated 95% of the whole ruminal microbial community (Pitta et al., 2014a). Hence, these microorganisms contribute the most to feedstuff (i.e., forage or forage-grain mixtures) digestion and the conversion of plant materials to volatile fatty acids and microbial protein. Before fermentation can take place, colonization or adhesion by microbiota to feed particles should occur to allow for the formation of enzymatic complexes that break down plant indigestible polymers, e.g., cellulose and hemicellulose, into monomeric and dimeric simple sugars (Russell and Rychlik, 2001). The major end-products of microbial metabolism of feedstuffs are the volatile fatty acids along with microbial protein. The most quantitatively important volatile fatty acids are acetate, propionate, and butyrate, which are used by the animal for fatty acid synthesis, glucose, production of energy (ATP), and ketone bodies. The volatile fatty acids are readily absorbed across the rumen wall and enter the systemic circulation where tissues like the liver, adipose, mammary gland, and muscle could utilize them. Therefore, the volatile fatty acids serve as the main metabolic fuels in tissues of ruminants. The microbial mass that leaves the rumen into the small intestine serves as an important source of amino acids for animal tissues (Storm and Orskov, 1983). In some instances, e.g., at maintenance, microbial protein can supply the animal with 50 to 80% of the amino acid requirements.
Manipulation of Ruminal Fermentation
Sustaining the high levels of productivity in modern dairy and beef cattle cannot be achieved solely with forage. Therefore, nutritionists have long sought methods to enhance beneficial processes (e.g., increase volatile fatty acids production, microbial protein synthesis, and bypass protein), alter inefficient processes such as limit methane and ammonia production, or enhance protection processes in the rumen (e.g., maintain rumen pH using buffers) (Weimer, 1998; Bach et al., 2005; Jeyanathan et al., 2014). As an example, rapidly-fermented grains increase energy (ATP) availability to microbes resulting in volatile fatty acid production; non-protein nitrogen (NPN) inclusion provides rumen microbes with essential nitrogen required for microbial protein synthesis; feeding of heat-treated proteins decrease the ability of microbes to degrade plant amino acids, and hence, those amino acids remain intact and travel directly to the small intestine for absorption; ionophores inhibit gram-positive bacteria that produce hydrogen, ammonia, or lactic acid; and buffers (additives to neutralize acid production; e.g., sodium bicarbonate) counteract the decrease in grain-dependent pH that can be harmful to fiber-degrading microorganisms (Nagaraja et al., 1997; Russell and Rychlik, 2001).
Transfaunation, a common long-practiced medical method used to treat indigestion in ruminants where rumen microorganisms are collected from a healthy donor animal to be transferred to a sick recipient animal to replace the unhealthy microbial ecosystem in the sick recipient with a healthy microbial profile, helps to re-establish a normal rumen fermentation process (DePeters and George, 2014). Classical rumen microbiology work during the 1960s through the 1990s allowed for the discovery and gross characterization of the predominant bacterial species (22 main species; Table 1) that inhabit the rumen, their main nutritional source, and the main fermentation products (see Russell and Rychlik, 2001). Clearly, those studies (mostly using pure cultures of microorganisms) laid the foundation for ruminant nutritionists seeking approaches to optimize feedstuff utilization by the microbes and the animal.
Microbial Methodological Advancements
While rumen function has always been a focal point of cattle nutrition, the growing understanding and interest in ruminal microbiota is directly tied to rapid development in culture-independent techniques. After developments and improvements in polymerase chain reaction (PCR) technology, evaluation of marker genes began being used for microbial community analysis. While the marker genes are present in all members of a population interest, variation in regions of the DNA sequence allows for differentiation among classes of microbes. Observed in all bacterial and archaeal genomes, the 16S rRNA gene is widely used and is supported by the most robust databases. Using primers to amplify a portion of the marker gene facilitated gel-based community “fingerprinting” techniques, which recently have been replaced by high-throughput sequencing the PCR products on “next-generation” DNA sequencers. The increasing ability of these machines to output millions to billions of reads and the capability to multiplex samples with unique barcodes has dramatically reduced the cost and led to the widespread adoption of this technology.
Major characteristics of predominant cultured bacteria in the rumen (Russell and Rychlik, 2001).
| Species | Ruminal niche | Fermentation products |
| Fibrobacter succinogenes | CU | S, F, A |
| Ruminococcus albus | CU, HC | A, F, E, H2 |
| Ruminococcus favefaciens | CU, HC | S, F, A, H2 |
| Eubacterium ruminantium | HC, DX, SU | A, F, B, L |
| Ruminobacter amylophilus | ST | S, F, A, E |
| Streptococcus bovis | ST, SU | L, A, F, E |
| Succinomonas amylolytica | ST | S, A, P |
| Prevotella ruminocola, albensis, brevis, and bryantii | ST, PC, XY, SU | S, A, F, P |
| Butyrivibrio fibrisolvens | ST, CU, HC, PC, SU | B, F, A, H2 |
| Selenomonas ruminantium | ST, DX, SU, L, S | L, A, P, B, F, H2 |
| Megasphaera elsdenii | L, SU | P, A, B, Br, H2 |
| Lachnospira multiparus | PC, SU | L, A, F, H2 |
| Succinivibrio dextrinosolvens | PC, DX, SU | S, A, F, L |
| Anaerovibrio lipolytica | GL, SU | A, S, P |
| Peptostreptococcus anaerobius | AA | Br, A |
| Clostridium aminophilum | AA | A, B |
| Clostridium sticklandii | AA | A, Br, B, P |
| Wolinella succinogenes | OA, H2, F | S |
| Methanobrevibacter ruminantium | H2, CO2, F | CH4 |
| Species | Ruminal niche | Fermentation products |
| Fibrobacter succinogenes | CU | S, F, A |
| Ruminococcus albus | CU, HC | A, F, E, H2 |
| Ruminococcus favefaciens | CU, HC | S, F, A, H2 |
| Eubacterium ruminantium | HC, DX, SU | A, F, B, L |
| Ruminobacter amylophilus | ST | S, F, A, E |
| Streptococcus bovis | ST, SU | L, A, F, E |
| Succinomonas amylolytica | ST | S, A, P |
| Prevotella ruminocola, albensis, brevis, and bryantii | ST, PC, XY, SU | S, A, F, P |
| Butyrivibrio fibrisolvens | ST, CU, HC, PC, SU | B, F, A, H2 |
| Selenomonas ruminantium | ST, DX, SU, L, S | L, A, P, B, F, H2 |
| Megasphaera elsdenii | L, SU | P, A, B, Br, H2 |
| Lachnospira multiparus | PC, SU | L, A, F, H2 |
| Succinivibrio dextrinosolvens | PC, DX, SU | S, A, F, L |
| Anaerovibrio lipolytica | GL, SU | A, S, P |
| Peptostreptococcus anaerobius | AA | Br, A |
| Clostridium aminophilum | AA | A, B |
| Clostridium sticklandii | AA | A, Br, B, P |
| Wolinella succinogenes | OA, H2, F | S |
| Methanobrevibacter ruminantium | H2, CO2, F | CH4 |
Abbreviations are as follows: CU, cellulose; HC, hemicellulose; DX, dextrins; SU, sugars; ST, starch; PC, pectin; XY, xylans; L, lactate; S, succinate; GL, glycerol; AA, amino acids; OA, organic acids; H2, hydrogen; F, formate; CO2, carbon dioxide; A, acetate; E, ethanol; B, butyrate; L, lactate; P, propionate; Br, branched-chain volatile fatty acids; and CH4, methane.
Major characteristics of predominant cultured bacteria in the rumen (Russell and Rychlik, 2001).
| Species | Ruminal niche | Fermentation products |
| Fibrobacter succinogenes | CU | S, F, A |
| Ruminococcus albus | CU, HC | A, F, E, H2 |
| Ruminococcus favefaciens | CU, HC | S, F, A, H2 |
| Eubacterium ruminantium | HC, DX, SU | A, F, B, L |
| Ruminobacter amylophilus | ST | S, F, A, E |
| Streptococcus bovis | ST, SU | L, A, F, E |
| Succinomonas amylolytica | ST | S, A, P |
| Prevotella ruminocola, albensis, brevis, and bryantii | ST, PC, XY, SU | S, A, F, P |
| Butyrivibrio fibrisolvens | ST, CU, HC, PC, SU | B, F, A, H2 |
| Selenomonas ruminantium | ST, DX, SU, L, S | L, A, P, B, F, H2 |
| Megasphaera elsdenii | L, SU | P, A, B, Br, H2 |
| Lachnospira multiparus | PC, SU | L, A, F, H2 |
| Succinivibrio dextrinosolvens | PC, DX, SU | S, A, F, L |
| Anaerovibrio lipolytica | GL, SU | A, S, P |
| Peptostreptococcus anaerobius | AA | Br, A |
| Clostridium aminophilum | AA | A, B |
| Clostridium sticklandii | AA | A, Br, B, P |
| Wolinella succinogenes | OA, H2, F | S |
| Methanobrevibacter ruminantium | H2, CO2, F | CH4 |
| Species | Ruminal niche | Fermentation products |
| Fibrobacter succinogenes | CU | S, F, A |
| Ruminococcus albus | CU, HC | A, F, E, H2 |
| Ruminococcus favefaciens | CU, HC | S, F, A, H2 |
| Eubacterium ruminantium | HC, DX, SU | A, F, B, L |
| Ruminobacter amylophilus | ST | S, F, A, E |
| Streptococcus bovis | ST, SU | L, A, F, E |
| Succinomonas amylolytica | ST | S, A, P |
| Prevotella ruminocola, albensis, brevis, and bryantii | ST, PC, XY, SU | S, A, F, P |
| Butyrivibrio fibrisolvens | ST, CU, HC, PC, SU | B, F, A, H2 |
| Selenomonas ruminantium | ST, DX, SU, L, S | L, A, P, B, F, H2 |
| Megasphaera elsdenii | L, SU | P, A, B, Br, H2 |
| Lachnospira multiparus | PC, SU | L, A, F, H2 |
| Succinivibrio dextrinosolvens | PC, DX, SU | S, A, F, L |
| Anaerovibrio lipolytica | GL, SU | A, S, P |
| Peptostreptococcus anaerobius | AA | Br, A |
| Clostridium aminophilum | AA | A, B |
| Clostridium sticklandii | AA | A, Br, B, P |
| Wolinella succinogenes | OA, H2, F | S |
| Methanobrevibacter ruminantium | H2, CO2, F | CH4 |
Abbreviations are as follows: CU, cellulose; HC, hemicellulose; DX, dextrins; SU, sugars; ST, starch; PC, pectin; XY, xylans; L, lactate; S, succinate; GL, glycerol; AA, amino acids; OA, organic acids; H2, hydrogen; F, formate; CO2, carbon dioxide; A, acetate; E, ethanol; B, butyrate; L, lactate; P, propionate; Br, branched-chain volatile fatty acids; and CH4, methane.
Current challenges include different sampling methods (e.g., cannulated vs. gastric tube vs. epimural microbial communities), diverse DNA and RNA extraction methods, DNA- vs. RNA-based comparisons, known biases within the PCR step that may lead to underrepresentation of some bacteria (e.g., Proteobacteria and Actinobacteria are not well represented in the 16S rRNA PCR-based approach compared with metagenomic analysis) as well as varying results from other regions of the 16S rRNA gene (V1/V2 versus V3/V4) for amplicon-based sequencing. Furthermore, the variation in bioinformatics analysis and presentation of the data can make comparisons between studies difficult. It is important to remember that marker gene techniques not only determine the presence of a microbe, but also provide information on its metabolic capability. “Shotgun” metagenomics can be used to determine the genomic capability of all bacteria in a sample while metatranscriptomics evaluates genes that are currently being expressed. Challenges in sample preparation, analysis of bioinformatics, and data interpretation have limited the use of these methods to study rumen microbiota thus far.
Quantitative PCR also has been extensively used in studying ruminal bacteria, methanogens, fungi, and protozoa. This technique focuses on identifying specific, often culturable species, and is a more sensitive tool to determine population changes. Additionally, efforts spearheaded by the Hungate 1000 initiative have sequenced the complete genome of more than 200 cultured microbial strains from the rumen (Creevey et al., 2014). Other techniques are currently available, and their further development and implementation to understand the microbiome within the rumen will improve beef and dairy production.
Dietary Impacts on Rumen Microbiota in Cattle
Dairy calves
The establishment of a microbial ecosystem in the rumen begins early after birth and continues rapidly through weaning to become an “adult-like” rumen microbiome (Jami et al., 2013). The early colonization of the rumen with microbes induces microbial fermentation of dietary complex carbohydrates resulting in the production of volatile fatty acids, which play a vital role in the development and growth of rumen papillae, leading to increase papillae capacity for nutrient absorption and metabolism (Lane and Jesse, 1997). Therefore, an optimal development of the rumen microbiome early in life directly impacts the ability of the calf to become a “fully functioning” ruminant. Interactions among diet, microbiome, and the host are complex but in large part dictate overall metabolic and productive response of the animal.
Various factors can affect the composition of the rumen microbiome in calves including age and nutrition (Tajima et al., 2001). In one of the first studies of its kind, Li et al. (2012) used next-generation sequencing to explore the changes in microbiome composition between 2 and 6 wk of age in dairy calves. The study underscored an age-dependent change in the microbiome. For example, the abundance of Bacteroidetes increased by 1.5-fold from 2 to 6 wk of age. Furthermore, Bacteroides dominated the microbial community at 6 wk while Prevotella was dominant at 2 wk of age. Other subsequent studies revealed that microbiome diversity increases in a sequential fashion with age (Jami et al., 2013; Rey et al., 2014). Such effect is partly due to the gradual introduction of solid feed (e.g., “starter” grain) to the diet of pre-wean calves (Malmuthuge et al., 2013). As a result, volatile fatty acid production in the rumen increases (Laarman et al., 2012) and accelerates rumen epithelium development, which facilitates the weaning transition.
Other research has provided evidence that colostrum can induce quick changes in the microbial composition within the small intestine of the calf. For example, Malmuthuge et al. (2015) used qPCR technology to determine that feeding heat-treated colostrum (60°C, 60 min) to dairy calves increased Bifidobacterium in the small intestine within 12 h after birth. Bifidobacterium is considered a beneficial bacteria that plays a role in enhancing the immune system, improving gut barrier, and reducing enteric pathogens (Fukuda et al., 2011). Heat treatment increases the availability of sialylated oligosaccharides in the colostrum, which can be utilized by Bifidobacterium as an energy source (LoCascio et al., 2007). Heat-treated colostrum also decreased the colonization of the small intestine by Escherichia coli (Malmuthuge et al., 2015), which is a common pathogen associated with diarrhea in calves (Foster and Smith, 2009). A better understanding of the response of the digestive tract microbiota to dietary changes in neonatal calves would not only help optimize rumen development and function but also overall health.
The peripartum or ‘transition’ period in dairy cows
The “transition” period in dairy cows is defined as the stage between 3 wk before parturition through the first 3 wk postpartum. This physiological stage is characterized by dramatic physiologic and metabolic adaptations, and it is now well established that the cow is at the highest risk of succumbing to health disorder during this time (Loor et al., 2013a). From a rumen microbiota standpoint, nutrition-induced changes during the transition period could play a vital role in providing optimal amounts of volatile fatty acids, propionate in particular, to help alleviate the demands of the mammary gland for glucose and energy. The shortfall in glucose availability after parturition is one key driver of ketosis, a potentially deadly metabolic disorder (Bobe et al., 2004). Because the diversity, richness, and composition of ruminal microbiota are diet specific (McCann et al., 2014b), nutritionists can manipulate the proportions of forage and grain fed and, for example, increase fibrolytic bacteria or amylolytic bacteria.
A common approach to manage high-producing pregnant dairy cows during the last 3 wk of the non-lactating period is to shift from a high-forage, low-energy diet to a high-grain, high-energy diet (Roche et al., 2013) to enhance the population of microorganisms capable of fermenting starch and sugars (non-structural carbohydrates), and hence, produce more volatile fatty acids and propionate. Such an approach also could enhance ruminal epithelium development and potentially increase the volatile fatty acids absorptive capacity after parturition when cows will normally take a week or longer to reach appropriate levels of feed intake.
A recent study detected increases after parturition in abundance of Lactobacillus spp., Streptococcus bovis, Megasphaera elsdenii, Ruminobacter amylophilus, Prevotella ruminicola, and Selenomonas ruminantium (Minuti et al., 2015). A decrease in fibrolytic bacteria such as Ruminococcus flavefaciens, Fibrobacter succinogenes, and Butyrivibrio fibrisolvens also was detected after parturition. Such responses were primarily driven by the feeding of a higher-grain, higher-energy diet after parturition, which is a common management practice in the field. Similar results were reported by others utilizing PCR or sequencing approaches (Fernando et al., 2010; Wang et al., 2012) and agree with the characteristics of these predominant bacteria identified in vitro (Russell and Rychlik, 2001).
In addition to changes in bacterial populations, Kumar et al. (2015) and Lima et al. (2015) reported a decrease in fungi after parturition when a high-grain, high-energy diet was fed and suggested that fungi may play an important role in fiber degradation. Lima et al. (2015) revealed that Litostomatea, a ruminal protozoal taxa, was more abundant with the high-energy diet but the biologic role of this change could not be discerned.
Subacute ruminal acidosis
The onset of subacute ruminal acidosis is common and somewhat difficult to diagnose in the field, but it causes substantial economic losses to dairy farmers because it decreases milk production, increases premature culling, and also could result in death if not detected in a timely fashion. Although shifting postpartum dairy cows to a high-grain, high-energy diet is a widely used feeding strategy in the dairy industry, grain-based diets can reduce rumen pH and dramatically alter the microbial ecosystem, which often leads to onset of subacute ruminal acidosis and, if uncontrolled, could lead to death (Nagaraja and Titgemeyer, 2007). Onset of subacute ruminal acidosis, which is not associated with accumulation of lactic acid, is often diagnosed when ruminal pH after feeding remains below 5.5 for a period of 4 h or more after feeding (Garrett et al. (1999).
Khafipour et al. (2009) and Mao et al. (2013) investigated the shift in the rumen microbiota associated with subacute ruminal acidosis incidence in lactating cows fed high-starch diets. Common to both studies was the marked reduction in the diversity of the rumen microbiota during subacute ruminal acidosis. Analyses revealed that 14 bacterial phyla (a taxonomic rank), including 5,664 species within 155 genera (a taxonomic rank), were affected during the onset of subacute ruminal acidosis. At the genus level, subacute ruminal acidosis increased numbers of microorganisms involved in starch fermentation such as Ruminococcus, Bifidobacterium, and Atopobium. In contrast, the growth of fibrolytic microorganisms such as Prevotella, Treponema, Anaeroplasma, Papillibacter, and Acinetobacter decreased in large part because of their intolerance to low rumen pH. In general, the low rumen pH associated with high-grain diets increases gram-positive bacteria such as the Firmicutes phylum members, which can readily ferment starch and sugars, while it reduces gram-negative bacteria such as the Bacteroidetes phylum.
The greater loss of gram-negative bacteria, due to cellular death and lysing with the low pH, can lead to the release of “endotoxin,” a large molecule consisting of lipid and a polysaccharide (e.g., lipopolysaccharide or LPS) into the ruminal fluid (Nagaraja and Titgemeyer, 2007). In addition, lowering rumen pH for extended periods of time because of volatile fatty acid production could damage the epithelial tight junctions of the rumen wall, allowing for transport of endotoxin to peripheral blood and stimulation of inflammation. In the short term, an uncontrolled inflammatory response reduces feed intake and compromises liver function, both of which are detrimental to the animal (Loor et al., 2013a).
Milk fat depression
The lactating mammary gland of dairy cows is a “lipid-synthesizing machine,” and it relies primarily on the volatile fatty acids acetate and butyrate as substrates for “lipogenesis” (making a 16-carbon fatty acid from the 2-carbon acetate). The overall process of making milk fat includes not only the availability of acetate and butyrate, but also a number of enzymes that work in a coordinated fashion to synthesize fatty acids and use them to make “fat,” i.e., fatty acids attached to a glycerol backbone (“triacylglycerol”) (Bionaz and Loor, 2008). Milk fat synthesis also relies on the availability of dietary and rumen-derived fatty acids that are absorbed in the small intestine during digestion. Reducing fat yield could represent an economic loss to dairy producers because milk fat content is one important component that influences milk price.
Milk fat depression occurs for a number of reasons, e.g., heat stress and dietary forage chopped too fine, but elucidating fundamental biological mechanisms underlying milk fat depression from the standpoint of rumen microbiota and diet interactions would be helpful to nutritionists. Perhaps the best-studied model of nutrition-induced milk fat depression is that involving rumen-derived “trans fatty acids” (Davis and Brown, 1970), i.e., unsaturated fatty acids with a trans configuration around a double bond on the carbon chain. High-grain, low-forage diets alone or supplemented with polyunsaturated fatty acids (e.g., 18:2, 18:3) or with ionophores (lipid-soluble compounds that eliminate gram-positive bacteria) often induce milk fat depression, also known as the “low-fat milk syndrome.” This condition is characterized by a reduction in milk fat yield by 50% or more during early lactation with no effect on milk yield or other milk components.
Knowledge about the metabolism of unsaturated fatty acids in the rumen dates to 1957 when Shorland et al. (1957) demonstrated the conversion of linoleic (18-carbons, 2 double bonds; 18:2) and linolenic (18-carbons, 3 double bonds; 18:3) acid to saturated fatty acids (18:0 primarily) using whole-rumen contents. Subsequent research during the 1960s and 1970s utilizing pure cultures of various species of microorganisms elucidated the microbial enzymes involved as well as the intermediates and end-products of the process (Harfoot, 1978). More recently (2000s), advances in chromatography to better identify biohydrogenation intermediates along with modern microbial genetics and molecular phylogenetic techniques for identifying and classifying microorganisms based on gene sequences (small-subunit rRNA) have substantially advanced knowledge of the role and contribution of specific microbial species in the process of biohydrogenation (Jenkins et al., 2008).
Recent work using molecular technologies has been useful in determining with greater certainty the changes in the microbial ecology that are associated with milk fat depression. For example, feeding high-starch diets not only increases amylolytic bacteria, but also the abundance of Megasphaera elsdenii, which ferments lactate and sugars (Table 1). Weimer et al. (2010) reported that high abundance of Megasphaera elsdenii in the rumen microbiota was strongly correlated with milk fat depression. More recently, Rico et al. (2015) confirmed the relationship between Megasphaera elsdenii and diet-induced milk fat depression as well as with Selenomonas ruminantium (a lactate utilizer) and Streptococcus bovis (starch and sugars fermenter). That study also detected a decrease in fungi and ciliate protozoa abundance with milk fat depression. From the standpoint of protozoa, the data support the hypothesis of Lima et al. (2015) that these microorganisms might play an important role in fiber degradation in the rumen.
Dietary effects on rumen microbiota in beef production
Cattle diets vary more than other livestock species in their nutrient composition. They range from almost all low-quality forage diets to nearly all grain concentrates depending on the production system and location. These dietary changes completely alter the substrates available to the rumen microbiota and, in turn, may lead to effects on microbial community composition, function, and fermentation end-products (Petri et al., 2013). Dietary transition in growing beef cattle occurs when animals move from one segment of the industry to another with the typical pattern being from cow-calf to stocker and from stocker to feedlot.
The shift from a medium-quality forage to winter wheat pastures is often seen in the southern US after the calf is weaned. Dietary increases in energy and protein facilitate greater animal performance but require the rumen microbiome to adjust accordingly. These effects were first observed by evaluating rumen bacteria when transitioning from a bermudagrass hay diet to wheat pasture (Pitta et al., 2010). The 16S rRNA sequencing indicated Prevotella and Rikenella were the most abundant genera observed, but the shift to wheat pasture increased relative abundance of Prevotella while it decreased Rikenella. The genus Succiniclasticum also increased on the wheat pasture diet and coincided with an expected greater propionate production. Bacterial community diversity decreased on wheat pasture as is generally observed with greater dietary energy. Among the rumen epimural microbes, Petri et al. (2013) reported that a high-grain diet increased the abundance of Atopobium, Desulfocurvus, Fervidicola, Lactobacillus, and Olsenella. Further work has revealed that increased phyla Bacteroidetes relative to Firmicutes in the liquid fraction may be linked to greater biofilm formation for cattle grazing wheat pasture, which causes frothy bloat (Pitta et al., 2014b).
Beef cattle eating hay (source: ellisia/stock.adobe.com).
Calves enter feedlots immediately after weaning or the stocker phase as calf-feds or yearlings, respectively. Although cattle in feedlots may be susceptible to stressful conditions aside from diet-related issues (e.g., mixing, regrouping, antibiotic treatments, or vaccination), which may alter the composition and function of the microbiome, to our knowledge, no attention has been placed in exploring the dynamics of the microbiome to such potentially adverse effects. The initial receiving diet is highly palatable and often contains high-quality forages such as alfalfa to stimulate intake. The receiving diet is then blended with a predominantly concentrate-based finishing diet at incremental steps until cattle are completely on the finishing diet. This progression allows cattle time to physiologically adjust to dietary changes in terms of rumen microbial and epithelial function while reducing occurrence of nutritional disorders such as acidosis. While many experiments have described differences between forage and concentrate diets on rumen microbiota, Fernando et al. (2010) was the first to describe this adaptation using a DNA-based evaluation of ruminal bacteria. Greater concentrate inclusion corresponded to increased Selenomonas ruminantium and Megasphaera elsdenii. While both species are capable of utilizing lactate (Table 1; Russell and Baldwin, 1978; Counotte et al., 1981) that can be produced by rapid starch fermentation, S. ruminantium also can produce propionate (Table 1), which increases with greater concentrate inclusion.
Ruminal production of propionate is an important fermentation end-product as it is the main gluconeogenic precursor in ruminants. Furthermore, two fibrolytic bacteria, Fibrobacer succinogenes and Butyrivibrio fibrisolvens, decreased 45 to 15-fold, respectively, in the final finishing diet compared with the pre-transition control (Fernando et al., 2010). More recent results suggest that the composition of the receiving diet also can affect the rate on transition by the rumen bacteria to the finishing diet (Anderson et al., 2016). Researchers compared the use of a complete commercial starter diet that is high in corn gluten feed and low in forage (RAMP) with an alfalfa- and corn-based control as both were blended with increasing amounts of a similar finishing diet. The rumen bacterial community for cattle on RAMP shifted after the second and third step diet to more readily adapt to the final finishing diet than cattle on the control (Anderson et al., 2016). Multiple operational taxonomic units (OTUs), a quantitative measure of species, classified at the family level as Prevotellaceae and Paraprevotellaceae, were important adaptations to dietary changes as some OTUs increased while others decreased. These findings reinforce the importance of the need for understanding the various roles of Prevotella in the rumen and its relevance to dietary changes (Stevenson and Weimer, 2007).
While the majority of rumen microbiota research focuses on growing beef cattle or high-producing dairy cattle, much less attention is given to moderate-to-low-quality forage diets consumed by the 30 million beef cows in the US. The diets are high in fibrous plant material that highlights the signature functions of rumen microbiota. A variety of nutritional strategies or supplements are commonly implemented to meet the cow's maintenance requirements and vary by region, season, and stocking intensity. Considering the dearth of research in this area, it is difficult to understand the importance of the ruminal microbiome composition beyond the main energetic contribution of fermentation end-products.
Protein is often supplemented in low amounts to cattle consuming low-quality forage to improve ruminal fermentation. Recent research compared a novel protein source, postextraction algal residue, with an isonitrogenous cottonseed meal control as well as an unsupplemented treatment (McCann et al., 2014a). The most significant impact on the rumen microbiome occurred in the liquid fraction as increasing algal coproduct inclusion resulted in greater phyla Firmicutes and corresponding families Ruminococcaceae, Lachnospiraceae, and Clostridiaceae. While the isonitrogenous cottonseed meal and algal coproduct diets led to similar levels of forage intake and utilization, the bacterial community for the cottonseed meal treatment was most similar to the unsupplemented control. These findings suggest bacterial community composition alone may not predict well the utilization of low-quality forages because ruminal fermentation is a complex process requiring interactions among different microbial groups.
Feed efficiency
The ability of cattle to convert dietary nutrients to retail end-products such as milk and meat is the basis for various measures of efficiency. Animals that maintain similar levels of performance while consuming less feed are classified as efficient with a lower numerical residual feed intake (RFI). While many factors may contribute to efficiency differences within a population, some data do support a role for rumen microbiota in mediating observed phenotypical differences in efficiency (Jewell et al., 2015). Some of the earliest research in this area using fingerprinting techniques suggested more efficient cattle had greater levels of butyrate production and differences in bacterial communities compared with the least efficient animals (Guan et al., 2008). While sequencing of efficiency-associated denaturing gel electrophoresis bands revealed multiple bacteria were associated with efficient and inefficient cattle, qPCR confirmed Eubacterium rectale had the strongest relationship with inefficient cattle on a high-energy diet (Hernandez-Sanabria et al., 2012). Others have observed Prevotella to be associated with inefficient (high RFI) cattle under various experimental conditions (Carberry et al., 2012; McCann et al., 2014c).
Greater sequencing depth per sample allowed a recent study to detect many associations with efficiency using different classifications based on average daily gain and feed intake (Myer et al., 2015). Most significantly affected bacteria such as Dialister and Acidaminoccous were linked with differences in average daily gain, which were greater on cattle with higher average daily gain. It was noteworthy that a Prevotella-identified OTU was much greater in animals with high average daily gain and low feed intake (Myer et al., 2015). Considering that some studies have detected Prevotella at more than 30% of all bacteria in the rumen, it is not surprising that Prevotella-identified OTUs have been positively and negatively associated with more efficient animals. Cattle classified as more efficient on a concentrate-based diet may not be similarly classified within the same contemporary group on a high-forage diet (Durunna et al., 2011; Carberry et al., 2012). Therefore, microbiota associated with efficiency differences are likely dependent on diet. Knowing that multiple factors contribute to efficiency differences observed with a population suggests that the relative importance of these factors will change depending on genetics, diet, and management of the evaluated contemporary group.
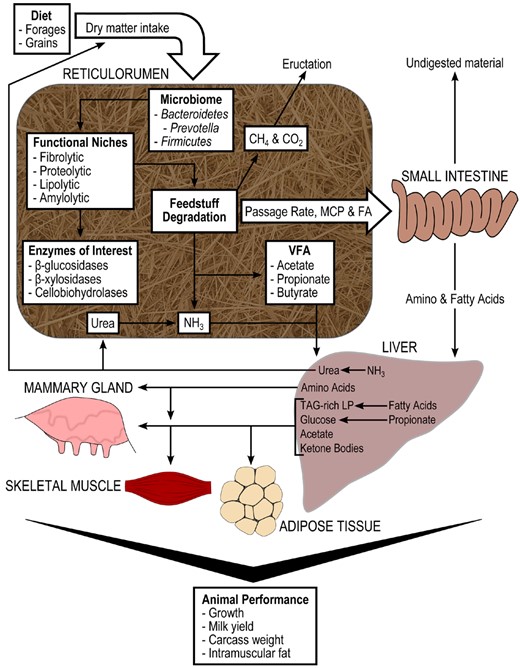
Diet-microbiota-host interactions in cattle (McCann et al., 2014b).
Integration in Ruminant Nutrition: the Future Direction
The concept of integration in ruminant nutrition and physiology in an effort to apply “systems biology” concepts and tools has been the subject of increasing interest during the last few years. The review by Loor et al. (2013b) outlined and discussed the major milestones that have been accomplished by applying systems concepts in dairy cows. The main goal when applying systems biology concepts is to integrate information at various levels of biological regulation (e.g., gene to mRNA, mRNA to protein) as a means to arrive at a holistic view of how animals function. The applicability of high-throughput technologies (e.g., sequencing, proteomics, and metabolomics) to discern functional biological networks has been demonstrated through work in humans and model organisms. Ruminants are obviously complex organisms, and ample evidence indicates that information gathering and integration are far better than continuing with the reductionist approach (Loor et al., 2013b). Both McNamara (2012) and Loor et al. (2013b, 2015) argue that another important goal of systems biology is to discover new properties that may arise from examining the interactions between components of a system, e.g., nutrition and the tissue genome. The same can be envisioned when the rumen microbiome is specifically placed as one of the key components of ruminant systems (see McCann et al., 2014b; also Figure 1).
Despite the abundance of knowledge about many specific rumen microorganisms and their function (e.g., Russell and Rychlik, 2001), our holistic understanding of the ruminal microbiome and how it cooperatively functions to degrade a wide array of substrates is still limited in many regards. Despite high-throughput methods allowing researchers to further define microbial community occupants, their metabolic potential, and what they are actively doing, we are just beginning to learn how to use these data to better understand the ruminant system. A systems biology approach will facilitate integration of the data further defining the host–microbiome relationship and allowing researchers to address pertinent questions facing ruminants.
The knowledge generated to date in livestock nutrition by utilizing a more holistic approach to dietary management underscores the validity of molecular technologies in helping build diets tailored to create a desired outcome at the whole-animal level (Loor et al., 2015). Lack of available data still limit our ability to take into account when formulating diets any specific effects of nutrients on the microbiome and the tissue response. An important undertaking for the field of ruminant nutrition in the medium to long term will be the generation of such kind of data for inclusion into diet formulation systems.



Literature Cited