ISSN: 0973-7510
E-ISSN: 2581-690X
Communication between species from different kingdoms may be as important as intra-kingdom communication. It has recently been confirmed that co-existing bacteria and phytoplankton in aquatic ecosystems do cross-talk. This study examined the signs of possible cross signalling between V. harveyi, one of the predominant bacterial species of the marine ecosystem and a dominant diatom species, S.marinoi, to understand communication over species borders. It is known that V.harveyi employ quorum sensing for cell-to-cell communication, bioluminescence (luxR), and the regulation of the virulence gene (vhp, chiA). Former studies have also shown, this kind of interactions being disrupted by compounds secreted by a few algal species existing in the aquatic ecosystem. We investigated the QS communication by quantifying the expression levels of virulence regulator luxR and virulence factors metalloprotease (vhp) and chitinase (chiA) in four different V. harveyi strains grown in the presence of S. marinoi strain. Results obtained in this study indicate that quorum sensing was activated in strains of V. harveyi analysed but did not regulate the expressions of vhp and chiA virulence factors. This observation suggests that the existence of S. marinoi did not interfere with the QS behaviour of V. harveyi and its interaction with marine diatom; it may be due to the commensalism relationship.
Quorum sensing (QS), diatom-bacteria relationship, Skeletonema, harveyi clade vibrios, virulence gene expression
In aquatic ecosystems, algae and bacteria coexist together, and there are many interactions between them, which are of particular interest to marine ecology.1-4 Such interactions influence the physiology of these organisms, alter ecological and biogeochemical events, and shape the diversity of microbial communities in the ecosystem.5 Quorum sensing (QS) is one such interaction through which density-dependent communication between bacterial cells takes place through the formation, release and acquisition of small signal molecules1,3,6,7 to control the expression of certain genes.1 Vibrio harveyi secretes three different molecules of the quorum sensing [Harveyi Autoinducer 1; Autoinducer 2 and Cholerae Autoinducer 1] that function synergistically in the regulation of gene expression.8-11 With the aid of these QS molecules, bacteria induce changes in downstream gene regulation and control many biological functions beneficial for bacterial fitness and virulence.1,12
Further, QS also modulates gene expression, essential for bacterial cell development, survival and pathogenicity.13,14 The successful colonization and invasion of host by the pathogenic bacteria known to be mediated by QS.13,15-18 Therefore, QS has numerous influences on bacterial physiology and function as well as promotes contact-mediated communication with their symbionts.7
Research on species communication has been focusing primarily on bacteria, with inadequate focus on algae.19 Cross-communication between bacteria and algae has only recently become an area of scientific interest. That inter-kingdom signaling takes place has been confirmed just over the last decade.4,17,20 To date, three forms of inter-kingdom signaling molecules/mechanisms have been identified. Bacterial QS molecule is one such compound involved in cell to cell signalling, similarly, algal pheromones released by diatoms has similar structures and functionality as QS molecules. This similarity may facilitate their use in signalling over species borders.21,22 Many eukaryotes can actively interfere with the QS behaviour of the bacteria by generating compounds that mimic the bacterium’s own signals.2,23,24 Given the potential of these molecules to interfere with QS allows for the biological control of several bacterial pathogens, as there is a correlation between QS and their pathogenesis.25,26
Algae and bacterial biofilms have been found to interact via QS27,28 and marine macro-algae prevent bacterial colonization by disrupting QS.15,29,30 The potential of Delisea pulchra, a marine macroalga, to limit bacterial colonization by releasing halogenated furanones has been extensively explored.15,31,32 The red macro-alga Ahnfeltiopsis flabelliformis has been found to have three new AHL antagonists.33 It inhibits AHL-regulated QS in a dose-dependent manner through a mixture of compounds.34 Likewise, few minor QS-interfering activity has been detected in macro-algae from the families Caulerpaceae, Rhodomelaceae and Galaxauraceae.35 Different marine bacteria have also been reported to produce QS-inhibitory metabolites.36-38
Unlike macro-algae and marine bacteria interaction, little is known about the impact of secondary metabolites generated by micro-algae on bacterial QS. Chlamydomonas reinhardtii is the first micro-algae reported to develop QS-interfering compounds.39,40 Micro-algal interference of QS regulated virulence of many aquaculture pathogens, might constitute a new bio-control strategy for bacterial disease in pisciculture. In a study by Natrah et al30 the effect of different strains of micro-algae, commonly used in aquaculture, from the families Chlorophyceae, Trebouxiophyceae, Eustigmatophyceae, Prymnesiophyceae, Bacillariophyceae and Coscinodiscophyceae was evaluated on QS-regulated gene expression. Different strains of bacteria, including Vibrio harveyi an important pathogen of aquaculture was used to evaluate the ability of these micro-algae to interfere with AHL QS.
The members of the harveyi clade vibrios have been studied for virulence factors necessary for the induction of pathogenesis in susceptible host.41,42 And these virulence factors are under the control of several regulatory systems, including QS and the transmembrane transcriptional regulator ToxR.43,44
Virulence factors and its processes in many pathogens, including Vibrio species, are frequently studied using simplified model systems. Environmental conditions and mechanisms that occur in vivo might be distinct from those that occur in these simplified systems.45 Therefore, it is important to also carry out experiments under conditions that are similar to what the bacteria experience during interaction. So far, most studies reporting on virulence regulation in Vibrio species were performed mainly with macroalgae including green algae. Very few focuses on Vibrio’s interaction with diatoms.30,46 Several groups of diatoms are the major source of atmospheric oxygen responsible for 20-30% of all carbon fixations on the planet. In this study, species of Skeletonema, a globally widespread microalgal genus47,48 was used to study the QS regulation of the virulence gene expression of Harveyi clade vibrios in the presence of a dominant presence phytoplankton species, S. marinoi. The expression level of virulence regulator luxR gene and of the virulence factors metalloprotease (vhp) gene and chitinase (chiA) gene in a group of Harveyi clade vibrios was quantified. QS mediated regulation of selected virulence genes were analyzed in four different strains of V. harveyi (one non-virulent, one strongly virulent and 2 different wild type strains) during co-culturing with axenic cultures of a marine diatom S. marinoi.
The transcription levels of quorum sensing regulator (luxR) and the genes for virulence such as metalloprotease (vhp) and chitinase (chiA) in a group of harveyi clade vibrios (Wild type; BB120 and LMG21363 two QS mutants; QS+ i.e quorum sensing maximally active mutant and QS- i.e quorum sensing inactive mutant) was quantified during co-culturing with axenic cultures of S. marinoi.
Growing S. marinoi and harveyi clade vibrios
Wild type strains BB120 and LMG21363, and quorum sensing mutants JAF 483 (Quorum sensing maximum activity mutant- QS+) and JAF 548 (Quorum sensing inactive mutant- QS-) belonging to the harveyi clade were used in the study. The bacterial strains used in this experiment were obtained from the Laboratory of Aquaculture and Artemia Reference Center, Ghent University, Belgium (Table 1). Single colonies of each bacterial strains were grown in marine broth (Himedia, India) at 28°C under continuous agitation of 150 rpm and cells were subsequently harvested at a concentration of 104 and 107 CFU ml-1.
Table (1):
Wild type and quorum sensing mutants of V. harveyi used in this study.
Strain |
Genotype |
References |
|---|---|---|
BB120 |
Marine isolate from which strains JAF483 and JAF548 are derived |
(Bassler et al)70 |
JAF483 |
Encodes LuxO locked in high cell density conformation (QS+) |
(Freeman and Bassler)71 |
JAF548 |
Encodes LuxO locked in low cell density conformation (QS-) |
(Freeman and Bassler)71 |
LMG21363 |
Isolate from the lymphoid organ of diseased shrimp (P. monodon) juveniles |
(Soto-Rodriguez et al)72 |
S. marinoi, strain HakH from GUMACC (University of Gothenburg Marine Algal Culture Collection) was made axenic following the protocol in65. S. marinoi, and subsequently S. marinoi together with Vibrio, were grown in f/2 medium,66 30 PSU, with a light intensity of 50 μmol photon m-2 s-1 and a 12:12h light dark cycle. The temperature maintained was 26-27°C during the day and 22-23°C during night.
Before starting the experiment, growth curves were established for S. marinoi and the four vibrio strains. Further, to confirm whether there is an impediment of algal growth by bacteria or vice versa was checked in another experiment. BB120, wild type strain of V. harveyi, in different concentrations (102-107 CFU ml-1) was added to a fixed density of S. marinoi. At the time of inoculation, both Vibrio and Skeletonema were in exponential growth phase. After 24 and 48 h of incubation, subsamples were extracted and Skeletonema cell abundance was estimated by using a Sedgewick Rafter chamber and an inverted Axiovert Zeiss microscope. The microscopic counts were compared to the growth curve under controlled conditions. Growth status of bacteria, on the other hand, was assessed by plate count on agar plates.
Specificity of the V. harveyi primers (rpoA, luxR, vhp, chiA) used in the study were checked in order to confirm the absence of primer binding sites in Skeletonema genome by running PCR reactions using Skeletonema DNA. Skeletonema genomic DNA was extracted using the PCI method.67 All the PCR reactions was performed as per the previously described protocol in which BB120 vibrio strain was used as a positive control.67 The amplified amplicons were then electrophoresed on a 1.5% agarose gel, stained with ethidium bromide and the results observed using a gel documentation system.
Experimental set up
Two density ranges of S. marinoi (1-2 x104 and 4-8 x105 cells ml-1) and two concentrations (104 and 107 CFU ml-1) of V. harveyi were selected for the gene expression study. To each of the two S. marinoi densities, Vibrio strains were added at low and high concentration. All combinations (Table 2) were run in triplicates. Vibrio cells were harvested and washed in sterile distilled water before adding to the S. marinoi cultures and the cell numbers were determined spectrophotometrically (Shimadzu UV-1601, Kyoto, Japan) at 600 nm. Both S. marinoi and V. harveyi cells were grown together for a period of 48 hours and cells were harvested, at different time points, for RNA extraction.
Table (2):
Concentration of S. marinoi and V. harveyi used in this experiment.
Treatment |
S. marinoi concentration (cells ml-1) |
Vibrio concentration (CFU ml-1) |
|---|---|---|
Q1 |
8×105 |
107 |
Q2 |
4×105 |
104 |
Q3 |
2×104 |
107 |
Q4 |
1×104 |
104 |
RNA extraction and Reverse transcription
For quantification of gene expression, bacterial cells were collected at 0, 12, 24, 36 and 48 h after inoculation of different isolates of Vibrio strains to the S. marinoi cultures. Sampled cells were washed with autoclaved seawater and suspended in bacterial RNA protective reagent (Qiagen, Germany) according to the manufacturer’s guidelines. The pellets were stored at -80°C until RNA extraction. Before the RNA extraction, tissue debris were homogenized using Qiashredder (Qiagen, Germany) and RNA was extracted using the commercially available RNA extraction Kit (Qiagen, Germany) according to the producer’s instructions. Extracts were subsequently treated with DNase I, according to the manufacturer’s protocol to remove the contaminating DNA. RNA quantity was checked spectrophotometrically. The extracted RNA was converted to cDNA using the reverse transcriptase (Fermentas International Inc., Canada) as per the standard protocol.
Real-time PCR
Real-time PCR was used to examine the expression levels of luxR, vhp (metalloprotease) and chiA (chitinase) genes. Specific primers were used for these genes (Table 3) at a concentration of 200 nM. The rpoA (RNA polymerase A subunit) gene, which is considered to be a housekeeping gene, was used as a control.68 Real-time PCR was performed in StepOne™ Real-Time PCR System (Applied Biosystems, USA) according to the protocol followed by Ruwandeepika et al.55 Data acquisition was performed at the end of each extension step and analysis was performed using 2-ΔΔCt method.69
Table (3):
Primers used for PCR and real-time PCR amplification.
Gene |
Primer Sequence (5’-3’) |
Amplicon size (bp) |
References |
|---|---|---|---|
luxR |
F: TCAAGATTGCAAAGAGACCTCG R: AGCAAACACTTCAAGAGCGA |
84 |
(Defoirdt et al) 68 |
metalloprotease (vhp) |
F: CTGAACGACGCCCATTATTT R: CGCTGACACATCAAGGCTAA |
201 |
(Ruwandeepika et al)55 |
Chitinase A (chiA) |
F: GGAAGATGGCGTGATTGACT R: GGCATCAATTTCCCAAGAGA |
232 |
(Defoirdt et al)59 |
RNA Polymerase subunit (rpoA) |
F: CGTAGCTGAAGGCAAAGATGA R: AAGCTGGAACATAACCACGA |
197 |
(Defoirdt et al)68 |
Statistical analysis of gene expression data
Three-way ANOVA (SPSS statistics v. 23, IBM) was used to determine whether there was a three-way interaction between the three independent variables: V. harveyi strain, V. harveyi cell concentration and S. marinoi cell concentration in our experiment. The net effect of such an interaction was tested separately on luxR, vhp and chiA gene expression. V. harveyi cell concentration (high vs. low), S. marinoi cell concentration (high vs. low), V. harveyi strains (QS+, QS-, BB120 and LMG21363) were treated as fixed factors. Significance levels were set at p < 0.05. The maximum expression values of the genes in focus were used as response variables in these tests, and before running the analyses; these were checked for homogeneity of variance and normality. Since all the three response variables were found to have heterogeneous variances, attempts were made to transform the data. Data for vhp successfully passed Levene’s test of variance upon log transformation, but luxR and chiA values failed to pass this test even after log transformation or square-root transformation. However, as the interaction output (p-value of the ANOVA) for the gene vhp was found to be similar for both transformed and non-transformed data, we decided to use the ANOVA approach for all three genes using non-transformed data in all analyses so as to avoid any discrepancies in comparisons.
Controls for growth and primer specificity
A control experiment was performed to test if the bacteria impeded S. marinoi growth and vice versa. This experiment confirmed that the microscopic counts of S. marinoi in the presence of Vibrio corresponded to the growth curve without bacteria present, and plate counts of Vibrio wild type strain BB120 in the presence of S. marinoi were similar to the counts in the absence of the diatom. It was, therefore, concluded that neither of the two inhibited each other’s growth.
A specificity test of the PCR primers used for amplifying rpoA, luxR, vhp, chiA genes in Vibrio BB120 genomic DNA and DNA from axenic S. marinoi, as templates for PCR, showed no bands for S. marinoi when PCR products were resolved on an agarose gel. The absence of bands for S. marinoi confirmed the bacterial specificity of these primers.
Gene expression
Relative gene expression varied over the temporal scale of the experiment for all three genes, luxR, vhp and chiA, and in particular for QS+ and the BB120 strains of V. harveyi (Fig. 1-4). Indeed, a few values were exceptionally high (see Fig. 1-2, Q1 and Q2). To compare the effects of the combination of the three different treatment factors (high and low Skeletonema concentration, high and low Vibrio concentration, and strain of Vibrio; QS+, QS-, BB120 and LMG21363) we applied a three-way ANOVA for each of the three genes. Essentially, a three-way ANOVA primarily looks for interactions among the three factors affecting the gene expression. With no interaction, gene expression is to some extent consistent for different levels for at least one of the factors (Vibrio strain), while with a significant three-way interaction the effects are strongly idiosyncratic for the various combinations of concentration levels.
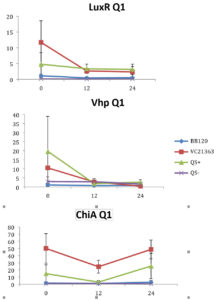
Fig. 1. Relative gene expression levels of luxR, vhp and chiA over a 24h period in treatment with 8×105 cells ml of Skeletonema and Vibrio 107 cells ml-1. Y-axis= Relative expression to RpoA, X-axis= Time elapsed (hr). Error bars represent standard error of mean.
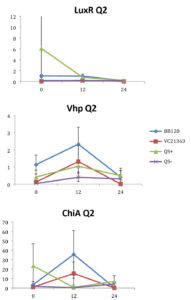
Fig. 2. Relative gene expression levels of luxR, vhp and chiA over a 24h period in treatment with Skeletonema 4×105 cells per ml and Vibrio =104 cells per ml). Y-axis= Relative expression to RpoA, X-axis= Time elapsed (hr). Error bars represent standard error of mean.
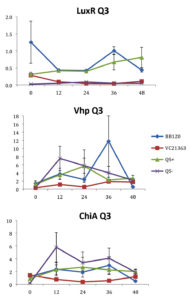
Fig. 3. Relative gene expression levels of luxR, vhp and chiA over a 48h period in treatment Skeletonema 2×104 cells per ml and Vibrio =107 cells per ml. Y-axis= Relative expression to RpoA, X-axis= Time elapsed (hr). Error bars represent standard error of mean.
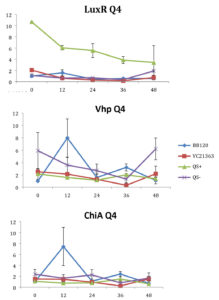
Fig. 4. Relative gene expression levels of luxR, vhp and chiA over a 48h period in treatment Skeletonema marinoi=1×104 cells ml-1, and Vibrio =104 cells ml-1. Y-axis= Relative expression to RpoA, X-axis= Time elapsed (hr). Error bars represent standard error of mean.
We found a statistically significant three-way interaction between the factors V. harveyi strain, V. harveyi concentration and S. marinoi cell concentration for the luxR gene (F(3,23) = 5.483, p = 0.005). This means that the luxR gene expression was statistically different among all the four Vibrio strains, F (3,23) = 7.270, p = 0.001. To disentangle the different effects of the different strains, we continued with pairwise comparisons (Table 4), and found that the expression of luxR was statistically different between the QS- and QS+ strains (p = 0.003) and between the QS- and BB120 strains (p = 0.002), with the QS- strain having lower expression than the other two strains for all combinations of Skeletonema and Vibrio concentrations (Fig. 1-4). However, luxR expression in QS- strain was not found to be statistically different from its expression in strain LMG21362 (p = 0.751). So, luxR expression being higher and statistically different indicates that quorum sensing was initiated in strains QS+ and BB120, though the wild type strain LMG21362 showed relatively low luxR expression. In QS- strain, luxR expression level remained constant.
Table (4):
Pairwise comparison (p-value) of gene expression (luxR, vhp, chiA) between different Vibrio strains (BB120, LMG21363, QS+, QS-).
| Gene | Strain | p-value | |
|---|---|---|---|
| luxR | QS- | BB120 | 0.002 |
| LMG21363 | 0.751 | ||
| QS+ | 0.003 | ||
| vhp | QS+ | BB120 | 0.080 |
| LMG21363 | 0.783 | ||
| QS- | 0.493 | ||
| chiA | QS+ | BB120 | 0.073 |
| LMG21363 | 0.563 | ||
| QS- | 0.844 | ||
There was no clear difference in the expression of gene vhp, in the Vibrio strains. That is, we found no statistically significant three-way interaction between V. harveyi strains, V. harveyi concentration and S. marinoi cell concentration for this gene (F (3,23) = 2.305, p = 0.104). Thus, Vibrio strains had no significant effect on the vhp gene’s expression level, F (3,23) = 2.382, p = 0.096. This was further confirmed when pairwise comparisons were applied (Table 4), as these showed that in strain QS+ gene vhp did not express differently from strains QS- (p = 0.493), BB120 (p = 0.080) or LMG21363 (p = 0.783). Similarly, the expression levels of gene chiA did not differ significantly among the four Vibrio strains, as shown by the non-significant three-way interaction between V. harveyi strain, V. harveyi concentration and S. marinoi cell concentration for this gene F (3,24) = 1.760, p = 0.182). The different Vibrio strains showed no significant effect on the expression of chiA, F (3,24) = 1.743, p = 0.185, and this was also obvious from the pairwise comparisons (Table 4) showing that in QS+ strain, gene chiA did not express differently than in QS- strains (p = 0.844), BB120 (p = 0.073) or VC21363 (p = 0.563). This suggests that even though quorum sensing was activated in strains QS+, BB120, and LMG21363, it did not change the expression levels of vhp and chiA in the presence of the algae.
According to previous studies (in vivo and in vitro), luxR gene is activated in the presence of sufficient amount of signal molecules49-51 and the activation of luxR gene is an indication of the activation of quorum sensing in Harveyi clade. In V. harveyi, luxR directly activates the lux operon,52 whereas majority of other QS-regulated genes and some virulence factors appear to be indirectly controlled by luxR.53 Metalloprotease (vhp) and chitinase (chiA) genes are two such virulence factors. QS has been shown to regulate metalloprotease expression in V. harveyi positively,54,55 V. cholerae and other marine vibrios as well.56–58 QS regulation of chitinase has been seen in V. harveyi59 with a negative regulation on chitinase A gene expression.
In our study, gene luxR was activated in all the strains. However, the expression in the QS maximally activated (QS+) and wild type strain (BB120) was higher and significantly different from its expression in the QS- strain. This confirmed the activation of quorum sensing in these strains, inferring that S. marinoi did not produce any QS interfering signals. LuxR gene expression in strain LMG21363 (wild type), however, was not seen to be significantly different from its expression in strain QS-. There is a possibility of time-based lag between the presence of both autoinducers and the luxR gene expression of in LMG21363. As suggested by Mok et al,54 an extended presence of both autoinducers is required to stimulate the expression of bioluminescence in this pathogen. So, despite QS being initiated in strain LMG21363, the luxR signal wasn’t strong enough to be significantly different from its expression in QS inactivated strain (QS-).
In vitro and in vivo studies of V. harveyi show that while metalloprotease (vhp) is positively regulated by luxR,51,54 chitinase gene (chiA) is negatively regulated.59 As per our observations, even though QS was activated in strains QS+ and BB120, it did not regulate the expression levels of vhp and chiA. The expression of these genes was not significantly different in these strains compared to quorum sensing inhibited strain QS-. One possible explanation for this result is that these virulence factors were not expressed as they would be as part of Vibrio species’s pathogenesis or defense mechanism. However, this response of V. harveyi might be specific for this particular S. marinoi strain. The process of interaction of V. harveyi is still not fully understood and may well be different in different strains or populations of S. marinoi as the response of the V. harveyi strain has been shown to be host-strain specific.60 vhp gene response in V. harveyi has been studied only during the infection of brine shrimp larvae.51,55 Studies on V. harveyi clade infection of crustaceans, molluscs and fish have investigated responses at physiological level.61-63 The expression levels of genes coding for virulence factors and/or QS regulated genes among these organisms is yet to be studied.
Moreover, different genes may be involved and expressed at different stages of the infection. So, the virulence genes included in our experiment may not have been activated until after 48 hours, unlike the V. harveyi-brine shrimp system where 48 hours of infection observed a consequent virulence gene response.51 Running our experiment longer than 48 hours might have seen regulation by luxR in a different manner at some later point. However, based on the gene expression recorded during the first 48 hours of exposure, with different concentration combinations of Vibrio and Skeletonema, it seems that the Vibrio strains BB120, LMG21363 and JAF483 do not use this S. marinoi strain in a potential host pathogen relationship, but rather form a in commensalism relationship. This would eliminate the possibility of using S. marinoi as a potential bio-control agent against aquaculture pathogens. Another possibility is that the luxR transcriptional regulator levels were not sufficiently high to allow expression of the virulence factors.50 This, perhaps, can be the case as the expression of the quorum sensing-regulated genes is proportional to the levels of the signal molecules.64
Indeed, we may be looking for the needle in the haystack, as the quorum sensing regulation is fundamentally complex, involving different virulence factors with tentatively specific roles, and involving an essentially unknown time-dimension in relation to the stage of the infection and production and activation of virulence factors. Finally, whether or not different virulence genes are responsible for infection or interaction with different hosts remains yet another challenging question.
Our study shows how bacterial communication functions in the presence of a diatom. It adds on to the existing knowledge on inter-kingdom signaling, highlighting the quorum sensing regulation in V. harveyi clade in the presence of a diatom species. It paves the way for future studies investigating the communication and signaling between bacteria and diatoms.
ACKNOWLEDGMENTS
We dedicate this article to the Principal Investigator of the project, Late Dr. Anna Godhe, University of Gothenburg, Goteborg, for her scientific contribution and dedicated unparalleled leadership. The Indian PI (IK) and the Sri Lankan PI (HADR) and all authors are grateful to the Swedish Research Council (VR 2016-05655) for the award of the project to the Swedish Principal Investigator, Late Dr. Anna Godhe.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
GK applied the methodology and carried out the experiments. BKK and TD performed the data analysis and did the validation. TD did the interpretation work. BKK performed experiment supervision and wrote the first draft. HDR performed formal analysis and edited the draft. INK conceptualized and administered the project, performed final review and edited the manuscript. All listed authors have read and approved the final manuscript for publication.
FUNDING
This study was supported by grants from the Swedish Research Council (VR 2016-05655).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
The datasets generated during the current study are available from the corresponding author on reasonable request.
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319-346.
Crossref - Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2009;6(2):111-120.
Crossref - Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng. 2008;10:145-167.
Crossref - Amin SA, Hmelo LR, van Tol HM, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522(7554):98-101.
Crossref - Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55(1):165-199.
Crossref - Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35(1):439-468.
Crossref - Bassler BL. Small Talk : Cell-to-Cell Communication in Bacteria. Cell. 2002;109(4):421-424.
Crossref - Cao J, Meighens EA. Purification and Structural Identification of an Autoinducer for the Luminescence System of Vibrio harueyi. J Biol Chem. 1989;264(36):21670-21676.
Crossref - Chen X, Schauder S, Potier N, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415(6871):545-549.
Crossref - Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450(7171):883-886.
Crossref - Boyer M, Wisniewski-Dye F. Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol. 2009;70(1):1-19.
Crossref - Mangwani N, Dash HR, Chauhan A, Das S. Bacterial Quorum Sensing : Functional Features and Potential Applications in Biotechnology. J Mol Microbiol Biotechnol. 2012;22(4):215-227.
Crossref - de Kievit TR, Iglewski BH. Bacterial Quorum Sensing in Pathogenic Relationships. Infect Immun. 2000;68(9):4839-4849.
Crossref - Williams P, Camara M, Hardman A, et al. Quorum sensing and the population-dependent control of virulence. Philos Trans R Soc Lond B Biol Sci. 2000;355(1397):667-680.
Crossref - Givskov M, de Nys R, Manefield M, et al. Eukaryotic Interference with Homoserine Lactone-Mediated Prokaryotic Signalling. J Bacteriol. 1996;178(22):6618-6622.
Crossref - Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1999;96(5):2408-2413.
Crossref - Visick KL, Ruby EG. The emergent properties of quorum sensing: consequences to bacteria of autoinducer signaling in their natural environment. Cell-cell signaling in bacteria. ASM Press, Washington, DC. 1999;333-352.
- Loh J, Pierson EA, Pierson III LS, Stacey G, Chatterjee A. Quorum sensing in plant-associated bacteria. Curr Opin Plant Biol. 2002;5(4):285-290.
Crossref - Amin SA, Parker MS, Armbrust EV. Interactions between Diatoms and Bacteria. Microbiol Mol Biol Rev. 2012;76(3):667-684.
Crossref - Steinberg PD, Rice SA, Campbell AH, Mcdougald D, Harder T. Interfaces Between Bacterial and Eukaryotic ‘” Neuroecology “‘. Integr Comp Biol. 2011;51(5):794-806.
Crossref - Zhou J, Lao Y-M, Cai Z-H. Draft Genome Sequence of Providencia sneebia Strain ST1 , a Quorum Sensing Bacterium Associated with Marine Microalgae. J Genomics. 2016;4:10-12.
Crossref - Zhou J, Lyu Y, Richlen ML, Anderson DM, Cai Z. Quorum Sensing Is a Language of Chemical Signals and Plays an Ecological Role in Algal-Bacterial Interactions. Crit Rev Plant Sci. 2016;35(2):81-105.
Crossref - Kjelleberg S, Steinberg P, Givskov M, Gram L, Manefield M, De Nys R. Do marine natural products interfere with prokaryotic AHL regulatory systems?. Aquatic Microbial Ecology 1997;13(1):85-93.
Crossref - Bauer WD, Teplitski M. Can plants manipulate bacterial quorum sensing ? Functional Plant Biology. 2001;28(9:913-921.
Crossref - Finch RG, Pritchard DI, Bycroft BW, Williams P, Stewart GS. Quorum sensing: a novel target for anti-infective therapy. J Antimicrob Chemother. 1998;42(5):569-571.
Crossref - Donabedian H. Quorum Sensing and its Relevance to Infectious Diseases. J Infect. 2003;46(4):207-214.
Crossref - Joint I, Tait K, Wheeler G. Cross-kingdom signalling : exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos Trans R Soc Lond B Biol Sci. 2007;362(1483):1223-1233.
Crossref - Rivas MO, Vargas P, Riquelme CE. Interactions of Botryococcus braunii Cultures with Bacterial Biofilms. Microb Ecol. 2010;60(3):628-635.
Crossref - Dobretsov S, Teplitski M, Paul V. Mini-review : quorum sensing in the marine environment and its relationship to biofouling. Biofouling. 2009;25(5):413-427.
Crossref - Natrah FMI,Kenmegne MM, Wiyoto W, Sorgeloos P, Bossier P, Defoirdt T. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture. 2011;317(1-4):53-57.
Crossref - Dworjanyn SA, De Nys R, Steinberg PD. Localisation and surface quantification of secondary metabolites in the red alga Delisea pulchra. Marine Biology. 1999;133(4):727-736.
Crossref - Rasmussen TB, Givskov M. Quorum sensing inhibitors : a bargain of effects. Microbiology. 2006;152(4):895-904.
Crossref - Kim JS, Kim YH, Seo YW, Park S. Quorum sensing inhibitors from the red alga, Ahnfeltiopsis flabelliformis. Biotechnology and Bioprocess Engineering. 2007;12(3):308-311.
Crossref - Liu HB, Koh KP, Kim JS, Seo Y, Park S. The effects of Betonicine, Floridoside , and Isethionic Acid from the red alga Ahnfeltiopsis flabelliformis on quorum-sensing activity. Biotechnology and Bioprocess Engineering. 2008;13(4):458-463.
Crossref - Skindersoe ME, Ettinger-Epstein P, Rasmussen TB, Bjarnsholt T, de Nys R, Givskov M. Quorum sensing antagonism from marine organisms. Marine Biotechnology. 2008;10(1):56-63.
Crossref - Kanagasabhapathy M, Yamazaki G, Ishida A, Sasaki H, Nagata S. Presence of quorum-sensing inhibitor-like compounds from bacteria isolated from the brown alga Colpomenia sinuosa. Lett Appl Microbiol. 2009;49(5):573-579.
Crossref - Dobretsov S, Teplitski M,Alagely A, Gunasekera SP, Paul VJ. Malyngolide from the cyanobacterium Lyngbya majuscula interferes with quorum sensing circuitry. Environ Microbiol Rep. 2010;2(6):739-744.
Crossref - Teasdale ME, Donovan KA, Forschner-dancause SR, Rowley DC. Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Marine Biotechnol. 2011;13(4):722-732.
Crossref - Teplitski M, Chen H, Rajamani S, et al. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 2015;134(1):137-146.
Crossref - Rajamani S, Bauer WD, Robinson JB, et al. The vitamin riboflavin and its derivative lumichrome activate the lasr bacterial quorum-sensing receptor. Mol Plant-microbe Interact. 2008;21(9):1184-1192.
Crossref - Zhang X, Austin B. Haemolysins in Vibrio species. J Appl Microbiol. 2005;98(5):1011-1019.
Crossref - Ruwandeepika HAD, Defoirdt T, Bhowmick PP, Shekar M, Bossier P, Karunasagar I. Presence of typical and atypical virulence genes in vibrio isolates belonging to the Harveyi clade. J Appl Microbiol. 2010;109(3):888-899.
Crossref - Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator toxr is a fansmembrane dna binding protein. Cell. 1987;48(2):271-279.
Crossref - Milton DL. Quorum sensing in vibrios : Complexity for diversification. Int J Med Microbiol. 2006;296(2-3):61-71.
Crossref - Fux CA, Shirtliff M, Stoodley P, Costerton JW. Can laboratory reference strains mirror ‘ real-world ‘ pathogenesis ?. Trends in Microbiol. 2005;13(2):58-63.
Crossref - Molina-cardenas CA, Sanchez-saavedra MP. Inhibitory effect of benthic diatom species on three aquaculture pathogenic vibrios. Algal Res. 2017;27:131-139.
Crossref - Kooistra WHCF, Sarno D, Balzano S, Gu H, Andersen RA, Zingone A. Global diversity and biogeography of skeletonema species (Bacillariophyta ). Protist. 2008;159(2):177-193.
Crossref - De Vargas C, Audic S, Henry N, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348(6237):121605.
Crossref - Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes & Development. 2007;21(2):221-233.
Crossref - Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi : lessons learned from in vivo work. The ISME Journal. 2008;2(1):19-26.
Crossref - Ruwandeepika HAD, Bhowmick PP, Karunasagar I, Bossier P, Defoirdt T. Quorum sensing regulation of virulence gene expression in Vibrio harveyi in vitro and in vivo during infection of gnotobiotic brine shrimp larvae. Environ Microbiol Rep. 2011;3(5):597-602.
Crossref - Swartzman E, Silverman M, Meighenl EA. The luxR gene product of vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol. 1992;174(22):7490-7493.
Crossref - Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes & Development. 2006;20(19):2754-2767.
Crossref - Mok KC, Wingreen NS, Bassler BL. Vibrio harveyi quorum sensing : a coincidence detector for two autoinducers controls gene expression. The EMBO Journal. 2003;22(4):870-881.
Crossref - Ruwandeepika HAD, Defoirdt T, Bhowmick PP, Karunasagar I, Karunasagar I, Bossier P. In vitro and in vivo expression of virulence genes in Vibrio isolates belonging to the Harveyi clade in relation to their virulence towards gnotobiotic brine shrimp (Artemia franciscana). Environ Microbiol. 2011;13(2):506-517.
Crossref - Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA / protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26(5):1023-1034.
Crossref - Shao C, Hor L. Regulation of Metalloprotease Gene Expression in Vibrio vulnificus by a Vibrio harveyi LuxR Homologue. J Bacteriol. 2001;183(4):1369-1375.
Crossref - Croxatto A, Pride J,Hardman A,Williams P,Camara M, Milton DL. A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol Microbiol. 2004;52(6):1677-1689.
Crossref - Defoirdt T, Ruwandeepika HAD, Karunasagar I, Boon N, Bossier P. Quorum sensing negatively regulates chitinase in Vibrio harveyi. Environ Microbiol Rep. 2010;2(1):44-49.
Crossref - Ruwandeepika HAD, Jayaweera TSP, Bhowmick PP, Karunasagar I, Bossier P, Defoirdt T. Pathogenesis, virulence factors and virulence regulation of vibrios belonging to the Harveyi clade. Rev Aquac. 2012;4(2):59-74.
Crossref - Nicolas JL, Basuyaux O, Mazurie J, Thebault A. Vibrio carchariae, a pathogen of the abalone Haliotis tuberculata. Diseases of Aquatic Organisms. 2002;50(1):35-43.
Crossref - Liu P, Lin J, Chuang W, Lee K. Isolation and characterization of pathogenic Vibrio harveyi ( V. carchariae) from the farmed marine cobia fish Rachycentron canadum L. with gastroenteritis syndrome. World J Microbiol Biotechnol. 2004;20(5):495-499.
Crossref - Martin GG, Rubin N, Swanson E. Vibrio parahaemolyticus and V. harveyi cause detachment of the epithelium from the midgut trunk of the penaeid shrimp Sicyonia ingentis. Diseases of Aquatic Organisms. 2004;60(1):21-29.
Crossref - Henke JM, Bassler BL. Three Parallel Quorum-Sensing Systems regulate Gene Expression in Vibrio harveyi. J Bacteriol. 2004;186(20):6902-6914.
Crossref - Nagai S, Imai I, Manabe T. A simple and quick technique for establishing axenic cultures of the centric diatom Coscinodiscus wailesii Gran. Journal of Plankton Research. 1998;20(7):1417-1420.
Crossref - Robert RLG. Culture of phytoplankton for feeding marine invertebrates. Journal of Plankton Research. 1975;20(7):29-60.
Crossref - Ausubel FM. Current Protocols in Molecular Biology Current Protocols in Molecular Biology. 1987;1 and 2. ISBN: 0471625949
- Defoirdt T, Miyamoto CM, Wood TK, et al. The natural furanone (5Z) -4-bromo-5- (bromomethylene) -3-butyl-2 (5H) -furanone disrupts quorum sensing-regulated gene expression in Vibrio harveyi by decreasing the DNA-binding activity of the transcriptional regulator protein luxR. Environ Microbiol. 2007;9(10):2486-2495.
Crossref - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402-408.
Crossref - Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum- sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179(12):4043-4045.
Crossref - Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31(2):665-677.
Crossref - Soto-Rodriguez SA, Roque A, Lizarraga-Partida ML, Guerra-Flores AL, Gomez-Gil B. Virulence of luminous vibrios to Artemia franciscana nauplii. Diseases of Aquatic Organisms. 2003;53(3):231-240.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.