Abstract
Aims: We aim to define parameters that affect the safety and long-term transgene expression of attenuated HSV-1 vectors and optimize the expression cassettes to achieve robust and sustained expression in CNS.
Background: Engineered, attenuated Herpes Simplex Virus (HSV) vectors are promising vehicles for gene delivery to the peripheral and central nervous systems. The virus latent promoter (LAP) is commonly used to drive exogenous gene expression; however, parameters affecting the safety and longterm transgene expression of attenuated HSV-1 vectors have not been fully understood.
Objective: The study aimed at using CRISPR-Cas9 system to construct attenuated HSV-1 vectors and examine the influence of transgene cassette construction and insertion site on transgene expression and vector safety.
Methods: In this study, we used a CRISPR-Cas9 system to accurately and efficiently edit attenuated HSV-1 strain 1716, and construct two series of recombinant virus LMR and LMRx with different sets of gene cassettes insertion in Exon1(LAP2) and 2.0 kb intron downstream of LAP, respectively. The transgene expression and viral gene transcriptional kinetics were compared in in vitro cell lines. The reporter gene expression and safety profiles of each vector were further evaluated in mouse hippocampus gene transduction model.
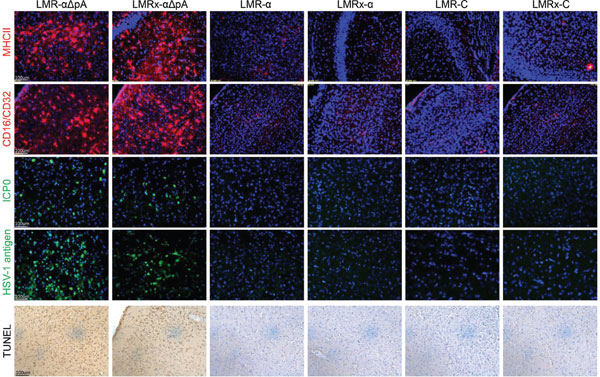
Results: The in vitro cell line analysis indicated that the insertion of a gene expression cassette would disrupt virus gene transcription. Mouse hippocampus transducing analysis suggested that complete expression cassette insertion at 2.0 kb intron could achieve robust and longtime gene expression than the other constructs. Recombinants with gene expression cassettes lacking Poly (A) induced significant neuronal inflammation due to persistent viral antigen expression and microglia activation.
Conclusion: Our results indicated that the integrity of LAT transcripts was not necessary for establishment of long-term latent expression. Exogenous strong promoters (like cBh promoter) could remain active during latency when placed in Exon1 or 2.0 Kb Intron of LAT locus, although their transcriptional activity declined with time. Consistent with previous research, the foreign gene expression would last much longer when the gene cassette was located downstream of Exon1, which suggested a role of LAP2 in maintaining promoter activity during latency. Besides, over-transcription of the downstream part of LAT may induce continuous activation of the attenuated vectors, which suggests an important role of LAT in maintaining viral reactivation potential.
Keywords: HSV-1 vector, CRISPR-Cas9 genome editing, latency-associated transcription, hippocampus gene transduction, long-term transgene expression, recombinant virus.
[http://dx.doi.org/10.1086/521262] [PMID: 17806053]
[PMID: 2824816]
[http://dx.doi.org/10.1126/science.2434993]
[PMID: 9696825]
[http://dx.doi.org/10.1073/pnas.88.3.790] [PMID: 1846963]
[http://dx.doi.org/10.1038/nature07103] [PMID: 18596690]
[http://dx.doi.org/10.1128/JVI.74.12.5604-5618.2000] [PMID: 10823868]
[http://dx.doi.org/10.1006/viro.1993.1632]
[http://dx.doi.org/10.1128/JVI.68.4.2239-2252.1994] [PMID: 8139009]
[http://dx.doi.org/10.1016/j.ymthe.2004.04.004] [PMID: 15233943]
[http://dx.doi.org/10.1128/JVI.74.8.3613-3622.2000] [PMID: 10729137]
[http://dx.doi.org/10.1128/JVI.80.5.2358-2368.2006] [PMID: 16474142]
[http://dx.doi.org/10.1128/mBio.02372-17] [PMID: 29437926]
[http://dx.doi.org/10.1073/pnas.1423556112] [PMID: 25775541]
[http://dx.doi.org/10.1128/JVI.00536-18] [PMID: 29950408]
[http://dx.doi.org/10.1128/JVI.01185-09] [PMID: 19656888]
[http://dx.doi.org/10.1016/j.chom.2014.03.004] [PMID: 24721573]
[http://dx.doi.org/10.1007/s13365-015-0362-y] [PMID: 26069184]
[http://dx.doi.org/10.1126/science.287.5457.1500] [PMID: 10688801]
[http://dx.doi.org/10.1038/srep34531] [PMID: 27713537]
[http://dx.doi.org/10.4049/jimmunol.1203265] [PMID: 23382563]
[http://dx.doi.org/10.1371/journal.ppat.1004090] [PMID: 24788700]
[http://dx.doi.org/10.1099/jgv.0.000012] [PMID: 25502645]
[http://dx.doi.org/10.1007/978-1-4939-1862-1_5]
[http://dx.doi.org/10.1016/j.ymthe.2005.06.478] [PMID: 16122987]
[http://dx.doi.org/10.1038/mtm.2014.59] [PMID: 26052529]
[http://dx.doi.org/10.1128/JVI.71.9.6714-6719.1997] [PMID: 9261395]
[http://dx.doi.org/10.1128/JVI.71.4.3197-3207.1997] [PMID: 9060683]
[http://dx.doi.org/10.1089/vim.2011.0091] [PMID: 22512280]
[http://dx.doi.org/10.1128/JVI.63.7.2893-2900.1989] [PMID: 2542601]
[http://dx.doi.org/10.1016/0042-6822(90)90060-5] [PMID: 2152989]
[http://dx.doi.org/10.1006/viro.1993.1078] [PMID: 8380666]
[http://dx.doi.org/10.1128/JVI.68.12.8045-8055.1994] [PMID: 7966594]
[http://dx.doi.org/10.1371/journal.ppat.1005539] [PMID: 27055281]
[http://dx.doi.org/10.1099/0022-1317-79-3-525] [PMID: 9519831]
[http://dx.doi.org/10.1099/vir.0.056176-0] [PMID: 23907392]
[http://dx.doi.org/10.1038/nn.3161] [PMID: 22837040]