
MRI radiomic signature predicts intracranial progression-free survival in patients with brain metastases of ALK-positive non-small cell lung cancer
Introduction
Lung cancer remains the leading cause of cancer-related mortality, with non-small cell lung cancer (NSCLC) accounting for 85–90% of cases (1). Anaplastic lymphoma kinase (ALK) gene rearrangement has been identified as a distinct molecular subtype (3–13%) of NSCLC; the most common alteration is fusion of the ALK and echinoderm microtubule-associated protein-like 4 (EML4) genes (2,3). With the understanding of tumor gene mutations and mechanisms, targeted therapy has become an important treatment for ALK-positive NSCLC, and its efficacy is superior to that of traditional chemotherapy (2,4). Crizotinib is a first-generation ALK inhibitor that exhibits good antitumor properties (5). Despite its success, most patients develop resistance to crizotinib within 1 to 2 years from therapy initiation (6,7), with approximately 50% of patients experiencing progression in the central nervous system (CNS) (8,9), which is an important cause of poor prognosis (10,11). To improve the prognosis of patients with brain metastases and overcome the acquired resistance to crizotinib, several second- and third-generation ALK inhibitors have recently been developed and approved for use in clinical settings (12-14). Ensartinib (X-396, Betta Pharmaceuticals Co. Ltd., Hangzhou, China) is a potent second-generation ALK inhibitor with high activity against a broad range of known crizotinib-resistant ALK mutations and CNS metastases (15,16).
Despite its remarkable success, patients with ALK-positive lung cancer have different responses to ensartinib, particularly those with brain metastases. Prognostic predictions before or early during treatment can potentially aid in patient-outcome assessment, enabling treatment-response evaluation and dynamic therapy adaptation. Various biomarkers for lung cancer, such as circulating tumor cells, circulating tumor cell DNA, and exosome to detect gene mutations, tumor immune-related factors, and inflammatory cytokines (17,18), which to some extent assist in the detection and monitoring of lung cancer, have been determined. However, their concentration in circulating blood is relatively low and requires high detection techniques or the accuracy will be affected. Furthermore, there are few studies on the detection of ALK gene mutations, particularly those evaluating efficacy and prognosis. Clinical response assessment criteria, such as the response evaluation criteria in solid tumors (RECIST), mainly assess the lesion size changes compared to previous examinations (19). Because internal changes in the tumor are usually not considered, this assessment method tends to underestimate the efficacy and has limited prognostic value (20). Therefore, a better method for assessing and predicting the response is still needed. Heterogeneity is a major feature of tumors that varies with time, space, and drug therapy and can reflect tumor changes earlier and more accurately but may not be readily recognized by conventional visual inspection (21).
Radiomics is an emerging field aiming to quantify this heterogeneity by applying a large set of feature extraction algorithms to characterize the tumor phenotype (22-24). It is based on the assumption that the radiographic phenotypes represent underlying pathophysiologies and could thus enable discriminating between disease forms, analyzing the tumor microenvironment, as well as predicting the prognosis and therapeutic response (22). Unlike traditional biopsy-based assays that represent only a sample of the tumor, whole-tumor images reflect the entire tumor burden, providing information on each cancer lesion with a single noninvasive examination. This is of particular importance in targeted therapy, where different lesions can have different microenvironments, potentially leading to heterogeneous response patterns. Previous exploratory studies have aided in tumor diagnosis and pathological typing, gene mutation discrimination, and efficacy and prognosis evaluation (25-29). However, to the best of our knowledge, no studies have explored the relationship between MRI radiomics and risk stratification of intracranial progression in patients with lung cancer that has metastasized to the brain.
The aim of this study was to develop a radiomic signature for risk stratification in patients with crizotinib-resistant, ALK-positive NSCLC treated with ensartinib to predict intracranial progression within 1 year.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-361).
Methods
Patients
This study was done in accordance with the principles of the Declaration of Helsinki (as revised in 2013). The data were collected from September 2017 to July 2019 in a prospective clinical trial (ClinicalTrials.gov identifier: NCT03215693) under approval of the local ethics review board at each participating site after obtaining written informed consent.
One hundred and sixty patients with locally advanced or metastatic (stage IIIB/IV) ALK-positive NSCLC from 27 centers across China were enrolled in the clinical trial and we included patients with brain metastases in this study. The inclusion criteria were: (I) age ≥18 years; (II) advanced ALK-positive NSCLC with brain metastases; (III) disease progression during crizotinib treatment, including the progress of brain metastases and the progress of other parts (with a minimum of 7-day washout period); (IV) no previous cranial radiation, and the CNS metastases did not require steroids therapy and were asymptomatic; (V) Eastern Cooperative Oncology Group performance status ≤2; (VI) baseline contrast-enhanced MRI was performed within 4 weeks before ensartinib treatment commencement; and (VII) brain metastases could be measured and delineated. The exclusion criteria were: (I) prior treatment with any ALK inhibitor other than crizotinib; (II) MRI data of poor quality because of motion artifacts or poor contrast injection; and (III) withdrawal from the trial within 51 weeks of follow-up for reasons other than intracranial progression (unacceptable toxicity, withdrawal of consent, tumor progression in other parts of the body).
Tumor assessments were performed using MRI at baseline and every 6 weeks during the first 24 weeks after ensartinib treatment initiation, and every 9 weeks thereafter until occurrence of radiologically documented disease progression by investigators according to RECIST version 1.1. Patients were followed up for at least 51 weeks and divided into progression and non-progression groups according to intracranial progression occurrence within 51 weeks after starting ensartinib treatment. Patients’ demographics were obtained by medical record review.
Image acquisition
Imaging was performed at different sites with different MRI vendors systems and various acquisition parameters, resulting in heterogeneous image data. Four patients were performed on General Electric systems (two at 1.5-T and two at 3.0 T), three, on Siemens systems (at 3.0 T); and 17, on Philips systems (one at 1.5 T and 16 at 3.0 T). All patients underwent axial T1 weighted post-contrast (T1c) scan and T2 weighted (T2) scan; other imaging protocols included non-enhanced T1-weighted, T2-weighted, T2-weighted fluid-attenuated inversion recovery, and diffusion-weighted imaging. T1c images were retrieved for radiomic-based image feature extraction. T1c and T2 images were retrieved for routine MRI feature analysis.
Image analysis
Routine MRI features
Two radiologists with 15- and 4-year’ experience, respectively, in an academic full-service cancer hospital reviewed the MR images for the following parameters: (I) lesion size, defined as the average of the long and the vertical short diameters of the largest cross-sectional area of the lesion on the axial T1c image; (II) location (frontal, parietal, occipital, and temporal lobes; cerebellum, ventricles, brainstem, limbic system, basal ganglia, and others); (III) extent of edema, which was evaluated on T2 images, defined as significant edema (score 2) if the maximum edema thickness was greater than or equal to the lesion diameter, mild (score 1) if the maximum edema thickness was less than the lesion diameter, and no significant edema (score 0); and (IV) range of enhancement, defined as whole (uniform or under-uniform) strengthening or peripheral strengthening. Any disagreement was resolved by consultation.
Region of interest segmentation and radiomic feature extraction
The radiomic workflow is presented in Figure 1. To reduce the effect of slice thickness variation, all images were resampled to voxels of 1×1×1 mm3 (initial dimensions: 1–5 mm in the Z direction) and further intensity standardized by using the Artificial Intelligence Kit software (AK software; GE Healthcare, China). Tumor regions of interest (ROIs) were semi-automatically segmented on the processed axial enhanced MR images using the ITK-SNAP 3.6 software (http://www.iksnap.ong). Brain metastases were first segmented by a radiologist with 4-year’ experience and then reviewed by a radiologist with 15-year’ experience in oncologic imaging. All disagreements were to be resolved by consensus. When consensus could not be reached, the final decision was deferred to a radiologist with more than 30-year’ experience in oncologic imaging. Overall, 396 radiomic features, including the first order histogram (n=42), second order texture (gray level co-occurrence matrix, GLCM, n=144), gray level run-length matrix (n=180), Haralick (n=10), morphological (n=9), and gray level zone size matrix (GLZSM; n=11) features, were extracted from the ROIs using the AK software (Figure S1).
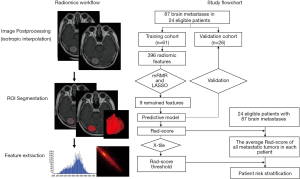
Radiomic feature selection and predictive model construction
All evaluable brain metastases (87 in total) were randomly divided into training and validation datasets at a 7:3 ratio. Multiple metastases in a single patient were assigned either to the training set or to the validation set to prevent bias from cluster correlation.
We used two feature selection methods, the minimum redundancy maximum relevance (mRMR) and least absolute shrinkage and selection operator (LASSO). mRMR was first performed to simultaneously select highly predictive but uncorrelated features based on their ranking by the relevance-redundancy index (30). Next, LASSO was used to select the optimized subset of features and evaluate the corresponding coefficients (31). The predictive model and Rad-score were obtained using 10-fold cross-validation to perform logistic linear regression of the selected features in a linear combination weighted by their respective coefficients and repeated 10 times. Rad-scores were compared between the progression and non-progression groups in both the training and validation datasets using Wilcoxon’s rank-sum test. The prediction model’s performance was assessed by the area under the receiver operator characteristic curve (AUC) in both datasets. The threshold point calculated by maximizing the Youden Index was used to predict each patient’s classification and to construct a confusion matrix, based on which the model’s accuracy, specificity, and sensitivity were calculated. The calibration and Hosmer–Lemeshow tests were performed to assess the goodness of fit of the prediction model. To estimate the clinical utility of the radiomic signature, decision curve analysis was performed by calculating the net benefits for a range of threshold probabilities.
Patient risk stratification
The relationship between the radiomic signature and the measured time to progression was further assessed using Kaplan–Meier analysis. Average Rad-scores for all included brain metastases were calculated for each patient. Patients were divided into high-risk and low-risk groups according to their average Rad-scores and the X-tile-derived threshold (32). Kaplan–Meier curves were constructed for patients in both groups to determine the proportion of patients without disease progression at any given time. The difference in the survival curves was evaluated using the log-rank test.
Statistical analysis
Demographic and regular MRI data were analyzed using the chi-square test or Fisher’s exact test for nominal variables and the Mann–Whitney U test for continuous variables with abnormal distribution. The statistical methods used for radiomic feature extraction and selection, prediction model construction and testing, and patient risk stratification were described in detail in the subsection Image Analysis. All statistical analyses were performed using R (version 3.5.1, http://www.R-project.org). Two-tailed P values <0.05 indicated statistical significance.
Results
Patients and routine MRI Features
From September 2017 to July 2019, 97 patients with brain metastases were enrolled, 40 of whom had measurable intracranial target lesions assessed by the Independent Review Committee based on RECIST 1.1. Of these, 15 were excluded [no enhancement imaging (n=1), tumor progression in other parts of the body within 1 year (n=14)], leaving the data of 24 patients (11 men, 13 women; mean age, 51.25 years; age range, 32–69 years) with 87 brain metastases for analysis (Figure S2). Among the 24 patients, 8 had intracranial progression (1 at 12 weeks after ensartinib treatment, 1 at 18 weeks, 1 at 24 weeks, 2 at 33 weeks, 3 at 42 weeks), and 16 had no intracranial progression within one year.
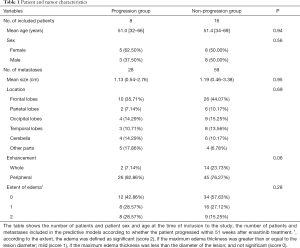
The patients’ demographics and the tumors’ characteristics are presented in Table 1. No significant difference was found between the progression and non-progression groups in demographics or routine MRI features (P=0.06–0.95).
Full table
Predictive model
A total of 396 radiomic features were extracted from each ROI. The most predictive radiomic features with a nonzero coefficient in the LASSO logistic regression model were: Percentile5, Correlation_angle90_offset4, Correlation_angle45_offset4, HaralickCorrelation_AllDirection_offset7, InverseDifferenceMoment_angle90_offset4, LargeAreaEmphasis, ClusterShade_angle90_offset4, Correlation_AllDirection_offset7, and GLCMEntropy_AllDirection_offset1_SD (Figure S3A,B).
The radiomic signature was constructed with a Rad-score calculated using the following formula:
Rad-score = –0.196*Correlation_angle90_offset4 + –1.095*Percentile5 + –0.447*Correlation_angle45_offset4 + –0.23*HaralickCorrelation_AllDirection_offset7 + –0.642*InverseDifferenceMoment_angle90_offset4 + 0.14*LargeAreaEmphasis + 0.54*ClusterShade_angle90_offset4 + –0.37*Correlation_AllDirection_offset7 + –0.147*GLCMEntropy_AllDirection_offset1_SD + –0.516.
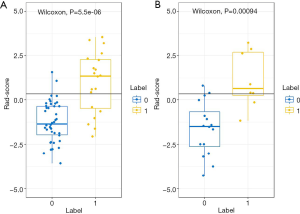
The Rad-scores were significantly higher in the progression than in the non-progression group both in the training and validation datasets (P=5.5e-06 and P=0.00094, respectively; Wilcoxon’s rank-sum test; Figure 2). The contribution of the selected parameters with their regression coefficients for signature construction is presented in the form of a histogram in Figure S4.
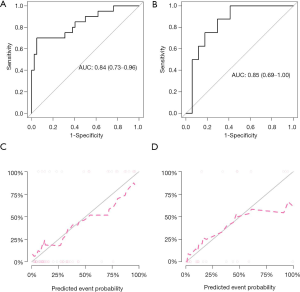
The radiomic prediction model showed favorable predictive efficacy, with AUCs of 0.84 [95% confidence interval (CI): 0.73–0.96; Figure 3A] and 0.85 (95% CI: 0.69–1.00; Figure 3B), accuracies of 0.87 (95% CI: 0.76–0.94) and 0.80 (95% CI: 0.59–0.93), sensitivities of 0.87 and 0.67, and specificities of 0.87 and 0.87 in the training and validation datasets, respectively. The calibration curve and non-significant Hosmer-Lemeshow test showed good calibration both in the training (P=0.50; Figure 3C) and validation (P=0.38; Figure 3D) datasets.
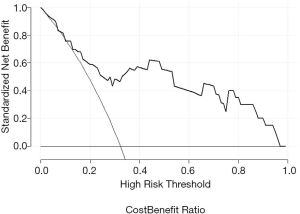
The decision curves in the validation set showed that when the threshold probability was between 0.13 and 0.97, using the radiomic signature added greater net benefit than the assumption of “all patients progress” or “all do not progress” (Figure 4).
Patient risk stratification
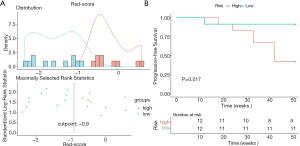
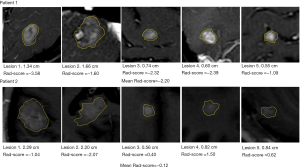
The average Rad-score of all metastatic tumors in each patient was calculated to represent the overall progression risk level of each patient. The Rad-score threshold for dividing patients into high- and low-risk groups was −0.90 (Figure 5A). The Kaplan-Meier curves of the proportion of patients without disease progression were significantly different between the high- and low-risk groups (P=0.017; Figure 5B). Figure 6 shows the brain metastases and corresponding Rad-scores of two patients from different risk groups. Among them, the follow-up images of the patient with both high and low risk lesions are shown in Figure S5.
Discussion
In this study, we performed a retrospective radiomic analysis of clinical trial data (ClinicalTrials.gov, NCT03215693) to develop a noninvasive quantitative prognostic biomarker for risk stratification of intracranial progression based on standard MR images for patients with ALK-positive NSCLC with brain metastases treated with ensartinib. The radiomic model based on the pre-ensartinib treatment images could identify patients at high risk of progression within 51 weeks throughout the treatment course, with AUCs of 0.84 and 0.85 in the training and validation datasets, respectively. We used the Rad-score calculated by the predictive model to quantify the predictive performance of the radiomic features. The higher the Rad-score, the higher the risk of progression within 51 weeks. The Rad-scores were significantly higher in the progression than in the non-progression group in both datasets. The decision curve analysis showed that the radiomic signature added greater net benefit than the assumption of “all patients progress” or “all do not progress” across the majority of reasonable threshold probabilities, indicating the incremental value of the radiomic signature for unified patient monitoring.
In patients with crizotinib-resistant ALK-positive NSCLC, despite the initial positive effect of ensartinib, progression-free survival (PFS) varies widely and is difficult to predict by conventional imaging features. Using the Rad-score, high- and low-risk patients were successfully distinguished, with significant differences in the 51-week PFS. In routine image evaluation and previous radiomic studies, greater attention had been paid to larger lesions; however, these were not always representative of the patient’s overall status. For example, in one case of multiple metastases (the bottom row of Figure 6), two large lesions had a Rad-score lower than −0.9, representing low risk, but the average Rad-score for all lesions was higher than 0.9, and the patient was classified in the high-risk group. Follow-up data confirmed that this patient required early intervention (PFS =24 weeks), attesting for the need of a comprehensive analysis of the histological characteristics of multiple lesions in patients. It was reported that the median intracranial PFS of ALK-positive lung cancer treated with an ALK-inhibitor was 11.9 months (7), and according to the follow-up plan of this clinical trial, the closest follow-up time was 51 weeks; therefore, we selected PFS at 51 weeks as the evaluation cut-off point. Compared with the long-term overall survival outcome, PFS is an endpoint that avoids extended follow-up and enables earlier adjustment of therapy (33). The radiomic signature has successfully identified high-risk patients with poor survival outcomes who need further intensive monitoring and timely improvement of treatment regimens, such as radiation.
Previous studies have shown that the average entropy and uniformity of multiple metastatic tumors in patients with renal cancer can predict targeted therapy efficacy and assess the PFS (20). In addition to the first-order histogram features, we comprehensively analyzed the high-order radiomic features. Multiple feature combinations can better reflect the complex heterogeneity of tumors, which will undoubtedly improve the accuracy of prediction.
Among the nine best performing radiomic features, Percentile5 is a histogram feature, Correlation_angle90_offset4, Correlation_angle45_offset4, HaralickCorrelation_AllDirection_offset7, InverseDifferenceMoment_angle90_offset4, ClusterShade_angle90_offset4, Correlation_AllDirection_offset7, and GLCMEntropy_AllDirection_offset1_SD are GLCM features, and LargeAreaEmphasis is a GLZSM feature. The first-order histogram feature is mainly based on the statistics of the lesion area grayscale or luminance information, followed by the overall distribution of the lesion area grayscale information. The GLCM and GLZSM features are texture parameters that describe the complexity, degree of change, and texture thickness of the lesion microstructure. Most of these radiomic features are high-order texture features that reflect image heterogeneity and were similar to those in other proposed signatures for NSCLC (34-36). According to the radiomic hypothesis, intratumoral heterogeneity assessed through imaging could reflect genomic heterogeneity, which would indicate worse prognosis, as tumors with greater genomic heterogeneity are more likely to develop resistance to treatment and to progress (22,37). As shown in the current study, the identified signature could predict the survival outcomes, supporting the idea that radiomic signatures have the ability to evaluate the intratumoral heterogeneity in a noninvasive manner and are thus associated with patient prognosis.
The combination of clinical and radiomic features may have greater clinical significance (38,39). Unfortunately, in our study, the clinical and conventional imaging features were not found to be statistically significantly associated with intracranial progression at 51 weeks for patients with ALK-positive NSCLC, which may be largely attributed to the limited size of the study population.
The reason we analyzed the T1c MR images was that a previous study found that enhanced images contain the most useful information and enhanced scanning was routinely performed for patients with brain metastases without contraindication (40).
The existing radiomic-based analyses have focused on molecular, histologic, and prognosis-related classifications using imaging biomarkers of primary tumors. To the best of our knowledge, this study was the first to use MR-based radiomic features to predict the intracranial efficacy of ALK-targeted therapy in patients with ALK-positive NSCLC. Moreover, we used prospectively acquired high-quality data from a clinical trial, which had predefined criteria for patient enrollment, treatment, and follow-up. As this was a multicenter trial, the MRI protocols and equipment varied across institutions, further highlighting the potential generalizability of our proposed model. Another advantage of radiomic analysis is that it imposes no additional burden on patients because it is an additional postprocessing step of standard radiology images obtained for response assessment.
There were also limitations to this study. First, because of the low incidence of ALK rearrangements in NSCLC, although the study was a national multicenter study, only a small number of patients (and lesions) were included. Random noise in small datasets can often be mistakenly interpreted as meaningful (a problem known as overfitting); consequently, the model may not perform as well in independent datasets. Second, the non-progression group (and the low risk group) had twice the data samples of the progression group (and the high-risk group) and the number of brain metastases varied among patients, which may have caused statistical analysis bias, affecting the predictive capability and accuracy of the model. Third, the overall tumor burden of the patients (primary lung cancer and extracranial metastases), was not considered, which may affect the prognosis. We are also studying the relationship between primary lung cancer and systemic metastases and patient prognosis, which may be reported in subsequent articles. Fourth, changes in radiomic characteristics from one time point to the next (delta radiomics), such as changes before and after treatment, could dynamically respond to changes in tumor heterogeneity and might have a higher predictive value than extracted from a single time point (41,42). However, due to the small number of cases, and some patients did not undergo enhanced scan during follow-up, we only analyzed the baseline radiomic features before treatment. We will further analyze the relationship between delta radiomic features and prognosis when more patients are included. Fifth, we did not include genomic characteristics and circulating tumor markers in the analysis, although this is undoubtedly a medical hotspot to promote the progress of tumor diagnosis.
Conclusions
This study showed that radiomics based on MRI had prognostic value for PFS and progression and allowed pretreatment risk stratification in patients with ALK-positive NSCLC receiving ensartinib treatment, which allowed the choice of follow-up and treatment to be tailored to each patient’s individual risk profile. Prospective validation in a large and diverse population is needed to acquire high-level evidence for the radiomic signature’s clinical application.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (No. 2017YFC1308700), the Chinese Academy of Medical Sciences Initiative for Innovative Medicine Program (No. 2017-I2M-1-005).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-361
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-361
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-361
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-361). LD and LM are employees of Betta Pharmaceuticals. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was done in accordance with the principles of the Declaration of Helsinki (as revised in 2013). The data were collected from September 2017 to July 2019 in a prospective clinical trial (ClinicalTrials.gov identifier: NCT03215693) under approval of the local ethics review board at each participating site after obtaining written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Marsilje TH, Pei W, Chen B, et al. Synthesis, structure-activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulfonyl)phenyl)pyrimidine-2,4-diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J Med Chem 2013;56:5675-90. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Johung KL, Yeh N, Desai NB, et al. Extended Survival and Prognostic Factors for Patients With ALK-Rearranged Non-Small-Cell Lung Cancer and Brain Metastasis. J Clin Oncol 2016;34:123-9. [Crossref] [PubMed]
- Costa DB, Kobayashi S, Pandya SS, et al. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J Clin Oncol 2011;29:e443-5. [Crossref] [PubMed]
- Castellanos EH, Horn L. Re-evaluating progression in an era of progress: A review of first-and second-line treatment options in anaplastic lymphoma kinase-positive non-small cell lung cancer. Oncologist 2016;21:755-61. [Crossref] [PubMed]
- Gállego Pérez-Larraya J, Hildebrand J. Brain metastases. Handb Clin Neurol 2014;121:1143-57. [Crossref] [PubMed]
- Costa DB, Shaw AT, Ou SH, et al. Clinical Experience With Crizotinib in Patients With Advanced ALK-Rearranged Non-Small-Cell Lung Cancer and Brain Metastases. J Clin Oncol 2015;33:1881-8. [Crossref] [PubMed]
- Kim DW, Tiseo M, Ahn MJ, et al. Brigatinib in Patients With Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase II Trial. J Clin Oncol 2017;35:2490-8. [Crossref] [PubMed]
- Delmonte A, Burgio MA, Verlicchi A, et al. New generation anaplastic lymphoma kinase inhibitors. Transl Lung Cancer Res 2019;8:S280-9. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Yang Y, Zhou J, Zhou J, et al. Efficacy, safety, and biomarker analysis of ensartinib in crizotinib-resistant, ALK-positive non-small-cell lung cancer: a multicentre, phase 2 trial. Lancet Respir Med 2020;8:45-53. [Crossref] [PubMed]
- Horn L, Infante JR, Reckamp KL, et al. Ensartinib (X-396) in ALK-Positive Non-Small Cell Lung Cancer: Results from a First-in-Human Phase I/II, Multicenter Study. Clin Cancer Res 2018;24:2771-9. [Crossref] [PubMed]
- Ma W, Gilligan BM, Yuan J, et al. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol 2016;9:47. [Crossref] [PubMed]
- Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Goh V, Ganeshan B, Nathan P, et al. Assessment of response to tyrosine kinase inhibitors in metastatic renal cell cancer: CT texture as a predictive biomarker. Radiology 2011;261:165-71. [Crossref] [PubMed]
- Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338-45. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Wang H, Ou Q, Li D, et al. Genes associated with increased brain metastasis risk in non-small cell lung cancer: Comprehensive genomic profiling of 61 resected brain metastases versus primary non-small cell lung cancer (Guangdong Association Study of Thoracic Oncology 1036). Cancer 2019;125:3535-44. [Crossref] [PubMed]
- Grossmann P, Narayan V, Chang K, et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol 2017;19:1688-97. [Crossref] [PubMed]
- Bulens P, Couwenberg A, Intven M, et al. Predicting the tumor response to chemoradiotherapy for rectal cancer: Model development and external validation using MRI radiomics. Radiother Oncol 2020;142:246-52. [Crossref] [PubMed]
- Bhatia A, Birger M, Veeraraghavan H, et al. MRI radiomic features are associated with survival in melanoma brain metastases treated with immune checkpoint inhibitors. Neuro Oncol 2019;21:1578-86. [Crossref] [PubMed]
- Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol 2019;30:998-1004. [Crossref] [PubMed]
- Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 2005;3:185-205. [Crossref] [PubMed]
- Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B (Methodol) 1996;58:267-88. [Crossref]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Mauguen A, Pignon JP, Burdett S, et al. Surrogate endpoints for overall survival in chemotherapy and radiotherapy trials in operable and locally advanced lung cancer: a re-analysis of meta-analyses of individual patients’ data. Lancet Oncol 2013;14:619-26. [Crossref] [PubMed]
- Huang L, Chen J, Hu W, et al. Assessment of a Radiomic Signature Developed in a General NSCLC Cohort for Predicting Overall Survival of ALK-Positive Patients With Different Treatment Types. Clin Lung Cancer 2019;20:e638-51. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non–small-cell lung cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Fried DV, Tucker SL, Zhou S, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non–small-cell lung cancer. Int J Radiat Oncol Biol Phys 2014;90:834-42. [Crossref] [PubMed]
- Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010;467:1109-13. [Crossref] [PubMed]
- Kniep HC, Madesta F, Schneider T, et al. Radiomics of Brain MRI: Utility in Prediction of Metastatic Tumor Type. Radiology 2019;290:479-87. [Crossref] [PubMed]
- Yu W, Tang C, Hobbs BP, et al. Development and Validation of a Predictive Radiomics Model for Clinical Outcomes in Stage I Non-small Cell Lung Cancer. Int J Radiat Oncol Biol Phys 2018;102:1090-7. [Crossref] [PubMed]
- Zhang Z, Yang J, Ho A, et al. A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images. Eur Radiol 2018;28:2255-63. [Crossref] [PubMed]
- Jeong J, Wang L, Ji B, et al. Machine-learning based classification of glioblastoma using delta-radiomic features derived from dynamic susceptibility contrast enhanced magnetic resonance images: Introduction. Quant Imaging Med Surg 2019;9:1201-13. [Crossref] [PubMed]
- Nardone V, Reginelli A, Guida C, et al. Delta-radiomics increases multicentre reproducibility: a phantom study. Med Oncol 2020;37:38. [Crossref] [PubMed]


