
Whole exome sequencing (WES) analysis of transformed small cell lung cancer (SCLC) from lung adenocarcinoma (LUAD)
Introduction
Histologic transformation of EGFR-mutant non-small cell lung cancer (NSCLC) to small cell lung cancer (SCLC) represents a rare mechanism of acquired resistance to epidermal growth factor receptor (EGFR)-targeted tyrosine kinase inhibitors (TKIs), occurring in 3–14% of EGFR-mutant NSCLCs (1). The phenomenon of SCLC transformation in TKI-resistant EGFR-mutant cancers has been identified in small retrospective series and case reports (2-10). The transformation of NSCLC to SCLC has also occasionally been observed in EGFR wild type NSCLC (9,10), raising the question of whether SCLC initially co-exists with NSCLC or rather originates from initial NSCLC.
Several studies have been conducted to elucidate this mechanism. For instance, Niederst et al. (11) demonstrated that while the original EGFR mutation was maintained in transformed cancers, the expression of the EGFR protein was significantly diminished after transformation, thus rendering the transformed tumors unresponsive to EGFR TKI therapy. However, the mechanisms underlying low expression of EGFR in SCLC remain unknown. Oser et al. (12) suggested that, since every transformed SCLC tumor sample in their study had retained its original EGFR-activating mutation, transformed SCLCs are not independent de-novo cancers but a transformed phenotype, which is in contrast with the hypothesis that patients had tumors with combined histology at the time of initial diagnosis, with the SCLC component becoming dominant over time, following treatment with EGFR TKIs. Lee et al. (13) showed that the SCLC clone branched off from the founder clone early, in some cases even before initial clinical cancer diagnosis, and that NSCLCs prone to transformation may show both tumor protein p53 (TP53) and RB1 inactivation at initial diagnosis. Consistently, retrospective analyses have shown that concurrent RB1 and TP53 alterations at baseline define a unique subset of EGFR-mutant NSCLCs at increased risk of SCLC transformation, and poor clinical outcomes (1,14). A recent study reported the association between SCLC transformation and the resistance to ROS TKIs in a patient with ROS1 fusion-positive (ROS1+) lung cancer (15), in which WES was performed in order to define the clonal evolution of the transformed SCLC. However, the tumor tissue from the initial or ROS TKI-resistant LUAD was insufficient for WES, so the WES based clonal evolution analysis of the transformed SCLC from the preceding LUAD was unavailable. According to the findings that while the ROS1 fusion was retained throughout the evolutionary trajectory, its expression was lost, and the ROS1 G2032R resistance mutation previously detected in the ROS TKI-resistant LUAD was not identified in the transformed SCLC, the authors deduced that the transformed SCLC and the preceding LUAD shared a common clonal origin and had an early divergence.
Against this background, we aimed to explore the genomic similarities and differences between initial lung adenocarcinoma (LUAD), the most common subtype of NSCLC and transformed SCLC, and to investigate the origin of transformed SCLC using clonal evolution analysis.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tlcr-20-1278).
Methods
Patient samples and clinical characteristics
Five patients with transformed SCLC from NSCLC who underwent lung biopsy at the Cancer Hospital of the Chinese Academy of Medical Sciences and Peking Union Medical College between November 2013 and January 2019, were selected for this study. The study was approved by the ethics committee of the Cancer Hospital of the Chinese Academy of Medical Sciences (No. 18-102/1680). Written informed consent was obtained from all study participants. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Whole exome sequencing was performed on samples collected from each patient at the time of initial LUAD diagnosis and at the time of transformed SCLC diagnosis (Shanghai Tongshu Biotechnology Co., Ltd, Shanghai, China).
We selected previously reported patients in the Chinese LUAD cohort who did not transform to SCLC until death as controls for this study (16), and finally 16 patients were included. Whole exome sequencing has been performed on the tumor samples from these patients. The basic clinical information was listed in Table S1.
DNA extraction
We performed DNA extraction from serial thick sections cut from tumor tissue samples and control sections. The invasive tumor content was estimated by pathologists, to ensure more than 50% of cells were tumor cells. The DNA was isolated from the FFPE using the DNeasy Blood and Tissue Kit (69504, QIAGEN, Venlo, Netherlands).
EGFR mutation analysis
EGFR mutations were analyzed based on the principle of the amplification refractory mutation system (ARMS). Briefly, resected tumor samples were fixed in 10% neutral buffered formalin and embedded in paraffin wax. Extracted DNA was used for PCR with the Mx3000PtM (Stratagene, La Jolla, USA) using the EGFR 29 Mutations Detection Kit (Amoy Diagnostics, Xiamen, China).
Whole exome sequencing
Targeted capture pulldown and exon-wide libraries were created from native DNA using the xGen® Exome Research Panel (Integrated DNA Technologies, Inc., Skokie, Illinois, US) and TruePrep DNA Library Prep Kit V2 for Illumina (#TD501, Vazyme, Nanjing, China), and paired-end sequence data were generated using Illumina HiSeq machines with an average sequencing depth of 150× for controls and 320× for tumor tissues. The sequence data were aligned to the human reference genome (NCBI build 37) using Burrows-Wheeler Aligner (BWA), and polymerase chain reaction (PCR) duplicates were sorted and removed using sambamba.
Variant calling pipeline
Single nucleotide variants (SNVs), insertions, and deletions were detected using Strelka2 with default parameters. Variants and polymorphisms were annotated using the Ensembl Variant Effect Predictor (VEP). A minimum of 20 reads covering mutated region and 5 reads supporting the variant allele are required for somatic SNV/indel calling. In contrast, sequencing depth need to be ≥20×, and reads supporting the variant <5 at the same site in the normal control sample. Variants with MAF >1% in the ExAC, gnomAD, and esp6500 databases were filtered out as common germline variants.
Tumor mutation burden
The tumor mutation burden (TMB) of a tumor sample is calculated by the number of non-synonymous somatic mutations (single nucleotide variants and small insertions/deletions) per mega-base in coding regions. We defined the non-synonymous somatic mutations as follows: firstly, select the mutations that appeared within the consensus coding sequence region designed by the CDS project; secondly, remove variant with variant count <3, allele frequency <5% or total depth <25; at last, filter out synonymous and splice region mutations, and retain only nonsynonymous mutations (missense, nonsense, insertions and deletions mutations).
Copy number variation analysis
Somatic copy number variations (CNVs) were analyzed using FACETS, and the resulting CNVs were used in further analysis. Recurrent CNVs were identified with GISTIC2.0. Circos were plotted using RCircos.
Intratumoral heterogeneity
Intratumoral heterogeneity (ITH) was evaluated based on two characteristics of ITH, the number of clones forming the tumor (ITH index) and clonal diversity (Shannon diversity index, SDI), according to the methods described previously (17).
Clonal evolution analysis
PyClone was used to cluster subclones inferred from SNVs. With the clustering results used as input, the optimal tree solutions were obtained with the iterative version of citup. Then, the fish plot and phylogenetic tree of each patient were constructed with timescape.
Statistical analysis
All visualizations were generated in the R statistical environment (v3.6.0) using ggpubr, maftools, pheatmap, copynumber and VennDiagram packages. T-test was applied to continuous and normal distribution data; and wilcoxon rank-sum test was applied to discrete variable or non-normal distribution data. P<0.05 was considered as statistical significance.
Data availability
The data that support the findings of this study are openly available from the Sequence Read Archive (SRA) at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA622410. Our scripts are available at https://github.com/Chitanda-Satou/BioinformaticsScripts. All other data are available upon reasonable request.
Results
Clinicopathological characteristics
Five patients who were initially given a diagnosis of advanced LUAD with canonical EGFR-activating mutation and underwent EGFR TKI treatment to achieve a favorable response were selected. The clinical characteristics of these patients are summarized in Table 1. Two patients (patients 1 and 3) were excluded from further analysis. For patient 1, the quality of the WES data was poor due to the degradation of adenocarcinoma DNA; for patient 3, normal tissue was unavailable because of the patient’s death. The clinical history of the three patients included in this study is presented in Figure 1. EGFR mutations were determined through amplification refractory mutation system-polymerase chain reaction (ARMS-PCR). All patients harbored EGFR mutations at the time of LUAD diagnosis and maintained the same EGFR mutation at the time of SCLC diagnosis. For patients 2, 4, and 5, the time to SCLC transformation was 40.9, 16.0, and 56.2 months, respectively, and the overall survival (OS) after transformation was 9.7, 8.5, and >14.5 months, respectively (Table 1).

Full table
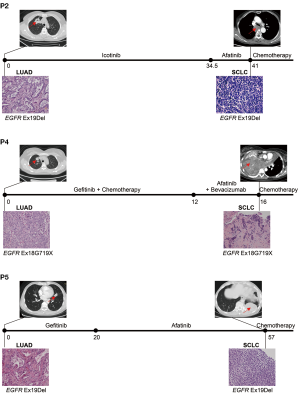
Mutational landscape of initial LUAD and transformed SCLC
Tumor samples obtained at the time of initial LUAD diagnosis and at the time of transformed SCLC diagnosis for each patient were subjected to WES with an average sequencing depth of 320×. Matched DNA from normal lung tissue was used as germline DNA control. The number of nonsynonymous somatic mutations per sample ranged from 106 to 468. The distribution of substitutions showed a strong preference for C>T transitions that is consistent with cytidine deamination.
After SCLC transformation, the mutation spectrum changed, with a decrease in the median proportion of C>T, from 55.8% to 33.3%, and an increase in the median proportion of C>A, from 12.7% to 25.0% (Figure 2A). The mutation spectrum of LUAD with or without SCLC transformation were different (Figure S1A). After SCLC transformation, TMB in patient 2 decreased slightly, but TMB in patient 4 and 5 remained unchanged. The TMB of LUAD and SCLC was 9.7 and 6.1 in patient 2, 1.5 and 1.7 in patient 4, and 1.3 and 1.7 mutations per Mb in patient 5, respectively (Figure 2B). The median TMB of LUAD with SCLC transformation (1.5 mutations/Mb) was comparable to LUAD without SCLC transformation (1.3 mutations/Mb, Table S2). Eleven known lung cancer driver genes (18) were detected in patients with SCLC transformation, among which TP53 and EGFR had a high mutation frequency (Figure 2C). Fifteen known lung cancer driver genes were detected in LUAD without SCLC transformation (Figure S1B). Only two driver genes, TP53 and EGFR, were shared by LUAD with or without SCLC transformation.
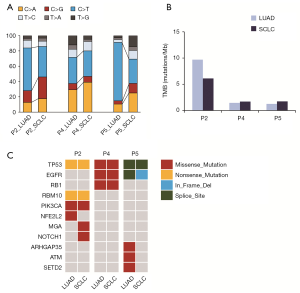
Next, we investigated the most frequently mutated genes in this cohort. As shown in Figure S2, no specific mutations were observed in the transformed SCLC samples. Examination of the alterations in key pathways of typical LUAD (19) (Figure S3A) and de novo SCLC (20) (Figure S3B) revealed that the genes involved in these pathways were rarely mutated except for several recurrently mutated genes (including TP53 and EGFR); however, CNVs were ubiquitous in these genes.
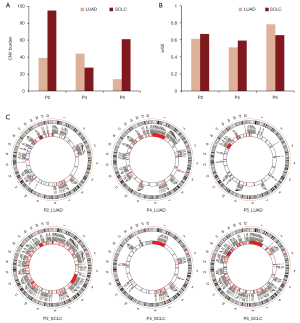
CNV profiles of initial LUAD and transformed SCLC
Compared with that of initial LUAD, the CNV burden of transformed SCLC was significantly elevated in patient 2 (39.0 vs. 94.9) and patient 5 (14.0 vs. 61.1), but it was slightly decreased in patient 4 (44.1 vs. 27.6) (Figure 3A). The median CNV burden of transformed SCLC was higher than that of initial LUAD (39.0 vs. 61.1, Wilcoxon P=0.4). However, the weighted genome instability index (wGII) (21) was comparable between initial LUAD and transformed SCLC (Figure 3B). As shown in Figure 3C, in patients 2 and 5, CNVs were widespread throughout the entire genome of transformed SCLC compared with the initial LUAD, but fewer CNVs were observed in patient 4. Known lung cancer driver genes affected by CNVs are presented in Figure S4. Compared with LUAD without SCLC transformation, the median CNV burden of initial LUAD with SCLC transformation was higher (39.0 vs. 21.5, Wilcoxon P=0.36, Table S2).
Clonal evolution analysis
Initial LUAD and transformed SCLC were found to share a small proportion of nonsynonymous mutations, and there were over 60% unique mutations (Figure S5). Four genes, TP53, mucin-17 (MUC17), EGFR, and pumilio RNA binding family member 1 (PUM1), were frequently mutated in both initial LUAD and transformed SCLC (Figure S2). The intratumoral heterogeneity (ITH) index (Figure 4A) and Shannon diversity index (SDI) (Figure 4B) of transformed SCLC were higher than those of initial LUAD. Histograms of CCF distribution also showed transformed SCLC to have a higher ITH index (Figure S6).
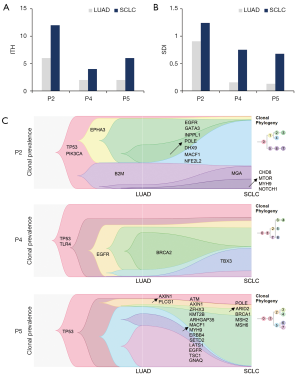
The clonal evolution analysis showed that the clonal components of initial LUAD and transformed SCLC were different. In patient 2, the LUAD was mainly composed of clones 2, 3, and 5, while the SCLC was mainly composed of clones 4, 6, and 7 (Figure 4C). The LUAD of patient 4 was mainly composed of clones 0 and 4, while the SCLC was mainly composed of clones 0, 3, and 6 (Figure 4C). In patient 5, the LUAD was mainly composed of clones 6 and 7, and the SCLC was mainly composed of clones 0 and 4 (Figure 4C). As shown, the major clones in the transformed SCLC did not originate from the major clones in the LUAD; rather, they originated from subclones generated before diagnosis. In all the three cases, mutations in TP53 occurred early in the tumorigenesis, and mutations in EGFR occurred relatively late. Other driver genes detected in the clones varied between the patients. These results suggest that LUAD and transformed SCLC originate from the same clone but the transformed SCLC clones do not originate directly from the major clones of the initial LUAD.
CNV burden is associated with the prognosis of patients with transformed SCLC
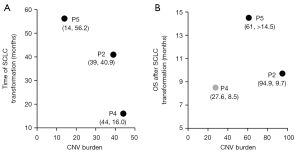
For the three patients (patient 5, 2, 4) in this study, the transformation time from LUAD to SCLC was 56.2, 40.9, and 16 months, and the CNV burden of LUAD was 14.0, 39.0, and 44.1, respectively (Table 1, Figure 5A). Together with the result that showed higher CNV burden of initial LUAD with SCLC transformation than LUAD without SCLC transformation (Table S2), these results suggested that the higher the CNV burden, the shorter the time to transformation into SCLC. The OS of the patient 5, 2 and 4 after SCLC transformation was >14.5, 9.7, and 8.5 months, with a CNV burden of SCLC of 61, 94.9, and 27.6, respectively (Table 1, Figure 5B). Because of the small sample size (N=3) whether CNV burden of transformed SCLC is prognostic remains to be determined.
Discussion
Transformation from LUAD into SCLC is a rare event in patients with lung cancer, and occurs in approximately 3–7% of EGFR-mutant NSCLC treated with TKIs. In this study we investigated the genomic correlates of SCLC transformation from 3 cases of EGFR-mutant LUAD. Although limited by the small sample size, our analysis provided some important insights into the genetic characteristics of transformed SCLC from initial LUAD.
C>T transitions were predominant in all three patients included in our analysis, all of whom were never-smokers. This finding is consistent with a previous study in which C>T transitions were found to be predominant in never-smokers and former light smokers with lung cancer, whereas a predominance of C>A transversions was observed in smokers (22). C>T transitions are resulted from the deamination of cytosine to uracil. A recent study suggested that activation-induced cytidine deaminase hypermutation could serve as an early predictor of SCLC transformation (14). In our study, C>T transitions were decreased in transformed SCLC, while C>A transversions were increased. Cancers with enrichment of C>T transitions have been reported to exhibit low levels of chromosomal alterations, and cancers with a preponderance of C>A transversions have been observed to have significantly more copy number gains (23), which is consistent with our findings. We also observed a significantly higher CNV burden in transformed SCLC than in initial LUAD in patients with increased C>A transversions and decreased C>T transitions. However, compared with that of initial LUAD, the wGII of transformed SCLC did not change, which allowed us to conclude that genome instability was not the cause of the increase in CNVs. Moreover, patients 2 and 5 had more DDR gene alterations in their transformed SCLC than patient 4 whose CNV burden did not increase after transformation to SCLC, which indicates that incorrect repair of DNA damage may be the source of increased CNVs (24). It is not clear whether the hypothetical mechanism of SCLC transformation for non-smokers is applicable to smokers.
The origin of SCLC subclones is a question still in need of investigation. Several studies have suggested that EGFR-mutant lung cancers that develop in alveolar type II cells might have the potential to transform into SCLC (25-27). Lee et al. suggested that EGFR TKI-resistant SCLCs may branch out early from the LUAD clones, even before EGFR TKI treatment (13). Our results were consistent with the findings of Lee et al., suggesting that SCLC diverges early from LUAD. We also found TP53 mutation to be an early event in both LUAD and SCLC, while EGFR mutation occurred later.
In previous studies, deactivation of TP53 and RB1 has been reported to occur in the majority of transformed SCLCs (13). In our study, all three cases harbored TP53 mutations in both LUAD and SCLC, and one of the cases harbored RB1 mutations. Moreover, CNV loss of RB1 occurred in all but one of six samples. However, accumulating experimental evidence has demonstrated that while dual inactivation of RB and p53 is essential for SCLC lineage transformation in EGFR-mutant LUAD, it is not sufficient to induce this transformation alone, which suggests that additional factors are required (11). A retrospective study described the clinical outcomes of patients with EGFR-mutant transformed SCLCs. Among 67 patients, all maintained their founder EGFR mutation, and 15 of 19 with prior EGFR T790M positivity were T790 wild-type at transformation. Other recurrent mutations identified in SCLC samples were TP53 (79%, 38/48), RB1 (58%, 18/31), and Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) (27%, 14/52) (1). MYC amplification and PIK3CA mutation have been proposed to potentially cooperate with RB/p53 loss to facilitate transformation, and specific epigenetic regulators may also provide the appropriate context for lineage reprogramming to occur (13). Park et al. (28) reported that loss of RB and inactivation of p53 combined with expression of myristoylated AKT1 and overexpression of MYC and B-cell lymphoma 2 (BCL2), leads to the development of aggressive SCLC. In our study, in transformed SCLCs, 100% [6/6] of samples had TP53 mutations, 33.3% [2/6] of samples had RB1 mutations, and 33.3% [3/6] of samples had PIK3CA mutations. We attempted to explore the genetic features of transformed SCLC, but no common mutations were found among the 3 cases. Our failure to determine the genetic features of transformed SCLC may mainly be attributed to the number of patients in this study, which was too small to identify common features.
The results of previous studies indicate that transformed SCLC takes on many features of de novo SCLC, including inactivation of the tumor suppressors RB and p53 in nearly all cases, similar gene expression profiles, including reduced or absent EGFR expression, and heightened sensitivity to BCL-2 family inhibition (11,29). However, a systematic review showed that current treatment strategies derived from de novo SCLC seemed to be largely inefficacious for transformed SCLC (30). Another previous study showed that few de novo SCLCs harbored EGFR mutations (12). From these observations, transformed SCLC may be genetically different from de novo SCLC.
Furthermore, we found that the CNV burden of initial LUAD with SCLC transformation was higher than LUAD without SCLC transformation. There was an association between CNV burden and the prognosis of patients with transformed SCLC. We observed that the higher the CNV burden in the initial LUAD, the shorter the transformation time to SCLC, and the higher CNV burden in the transformed SCLC, the shorter the OS after SCLC transformation. Nevertheless, the value of CNV burden in predicting SCLC transformation has not been studied. In future studies, we will employ transcriptome and epigenome analysis to elucidate the causal relations between increased CNV burden and SCLC transformation.
In conclusion, we demonstrated that the transformed SCLC did not evolve directly from the initial LUAD but branched off from LUAD early. The CNV burden was associated with the time to SCLC transformation and with the OS of patients after SCLC transformation.
Acknowledgments
We thank all participating subjects for their kind cooperation in this study and appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: This work was sponsored by Shanghai Tongshu Biotechnology Co., Ltd.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1278
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1278
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1278). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethics committee of the Cancer Hospital of the Chinese Academy of Medical Sciences (No. 18-102/1680). Written informed consent was provided by all participants. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Marcoux N, Gettinger SN, O'Kane G, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol 2019;37:278-85. [Crossref] [PubMed]
- Zakowski MF, Ladanyi M, Kris MG. Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213-5. [Crossref] [PubMed]
- Morinaga R, Okamoto I, Furuta K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer 2007;58:411-3. [Crossref] [PubMed]
- Popat S, Wotherspoon A, Nutting CM, et al. Transformation to "high grade" neuroendocrine carcinoma as an acquired drug resistance mechanism in EGFR-mutant lung adenocarcinoma. Lung Cancer 2013;80:1-4. [Crossref] [PubMed]
- Watanabe S, Sone T, Matsui T, et al. Transformation to small-cell lung cancer following treatment with EGFR tyrosine kinase inhibitors in a patient with lung adenocarcinoma. Lung Cancer 2013;82:370-2. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Norkowski E, Ghigna MR, Lacroix L, et al. Small-cell carcinoma in the setting of pulmonary adenocarcinoma: new insights in the era of molecular pathology. J Thorac Oncol 2013;8:1265-71. [Crossref] [PubMed]
- Ahmed T, Vial MR, Ost D, et al. Non-small cell lung cancer transdifferentiation into small cell lung cancer: A case series. Lung Cancer 2018;122:220-3. [Crossref] [PubMed]
- Ferrer L, Giaj Levra M, Brevet M, et al. A Brief Report of Transformation From NSCLC to SCLC: Molecular and Therapeutic Characteristics. J Thorac Oncol 2019;14:130-4. [Crossref] [PubMed]
- Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377. [Crossref] [PubMed]
- Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. [Crossref] [PubMed]
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Lin JJ, Langenbucher A, Gupta P, et al. Small cell transformation of ROS1 fusion-positive lung cancer resistant to ROS1 inhibition. NPJ Precis Oncol 2020;4:21. [Crossref] [PubMed]
- Gu W, Wang N, Gu W, et al. Molecular gene mutation profiles, TMB and the impact of prognosis in Caucasians and east Asian patients with lung adenocarcinoma. Transl Lung Cancer Res 2020;9:629-38. [Crossref] [PubMed]
- Wolf Y, Bartok O, Patkar S, et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell 2019;179:219-235.e21. [Crossref] [PubMed]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;173:371-385.e18. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Endesfelder D, Burrell R, Kanu N, et al. Chromosomal instability selects gene copy-number variants encoding core regulators of proliferation in ER+ breast cancer. Cancer Res 2014;74:4853-63. [Crossref] [PubMed]
- Fumagalli C, Guerini-Rocco E, Barberis M. Broad-based genomic sequencing in advanced non-small cell lung cancer in the dock. Transl Lung Cancer Res 2019;8:S360-3. [Crossref] [PubMed]
- McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15-26. [Crossref] [PubMed]
- Hastings PJ, Lupski JR, Rosenberg SM, et al. Mechanisms of change in gene copy number. Nat Rev Genet 2009;10:551-64. [Crossref] [PubMed]
- Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011;19:754-64. [Crossref] [PubMed]
- Mainardi S, Mijimolle N, Francoz S, et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:255-60. [Crossref] [PubMed]
- Sutherland KD, Song JY, Kwon MC, et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:4952-7. [Crossref] [PubMed]
- Park JW, Lee JK, Sheu KM, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 2018;362:91-5. [Crossref] [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Roca E, Gurizzan C, Amoroso V, et al. Outcome of patients with lung adenocarcinoma with transformation to small-cell lung cancer following tyrosine kinase inhibitors treatment: A systematic review and pooled analysis. Cancer Treat Rev 2017;59:117-22. [Crossref] [PubMed]


