Automatic Lung-RADS™ classification with a natural language processing system
Introduction
Lung cancer is estimated to account for 224,390 new cases and 158,080 deaths in 2016 in the United States (1). The National Lung Screening Trial (NLST) trial showed that CT lung screening (CTLS) can significantly reduce the mortality in a population at high risk for lung cancer (2). As result, the American Cancer Society and U.S. Preventive Services Task Force (USPSTF), together with many professional societies, included a recommendation to screen in their practice guidelines (3,4). It was estimated that 4.9 million Medicare beneficiaries met CTLS criteria in 2014 at an annual cost of approximately $241 per person (5), necessitating a standardized reporting and diagnostic work-up approach (6,7).
Lung-RADS™ (LR) (8) has been shown to increase the positive predictive value of CTLS by a factor of 2.5, without significantly increasing false-negatives compared to the NLST (9,10). Implementing a CTLS reporting system using LR therefore promises to optimize patient outcome while effectively reducing economic burden secondary to unnecessary care escalation. Translation of the imaging findings into the correct LR category is critical for program success. We hypothesize that an automatic algorithm capable of translating image descriptors in a report into the applicable LR category would help the radiologist verify that screening findings have been completely reported and properly classified.
Natural language processing (NLP) is a multi-step process comprised of a computer-based approach analyzing free-form text into a standardized structured format with the help of lexicons and ontologies, to ultimately create standardized and normalized concepts. These concepts in return populate information model knowledge representation of the clinical findings being mined from the text (11,12). NLP of the clinical narrative has been proven to aid clinical decision support by extracting relevant information (13-15). It can assist the radiologist through the life cycle of report development and utilization, starting from real time assistance to reach proper classification of imaging finding and assigning appropriate follow up recommendation, to effectively communicating the results to the primary physician and ultimately populating clinical databases with imaging performance measures to facilitate optimal population management strategies.
Prior applications of NLP algorithms in cancer imaging include the extraction of Bi-RADS® categories from mammography reports (16,17) and the classification of brain tumor progression (17,18). However, few studies have investigated NLP in the reporting of lung malignancies. Reports have demonstrated the ability of a NLP algorithm to analyze radiology reports to identify patients with pulmonary nodules (19) with a sensitivity and specificity of 96% and 86%, respectively. While important, the aim of that study was to detect nodules, but not to further characterize them. Radiologists new to lung cancer screening may benefit from real-time assistance with the use of LR. This approach may help establish an accuracy “floor” for lung screening reporting and as such could represent an important quality control or improvement tool similar to what has been shown for other types of health information technology (20).
For the purpose of this project, we augmented our NLP pipeline with a knowledge representation model of LR (8) to capture all radiologic findings related to lung nodule description in a structured CTLS report. The system suggests the corresponding LR category once the descriptive part of a radiology report is complete. Importantly, the aim of this study was to evaluate the NLP performance of identifying positive exams, rather than a very detailed analysis such as a sub-classification.
Methods
System overview
Our NLP system was developed using Unstructured Information Management Architecture (UIMA) framework (21). UIMA platform supports a pipeline of NLP tasks where each component’s output is used as an input in the consecutive component. Our NLP pipeline starts with pre-processing modules for detecting document structures such as sections, paragraphs, tables, or lists and assigning section Logical Observation Identifiers Names and Codes (LOINC) codes when appropriate (22,23). Once the document structure is detected, subsequent modules detect word tokens and sentence boundaries. Additional modules then define temporality, negation status, and subject at the sentence level. Other modules map text to Systematized Nomenclature of Medicine (SNOMED-CT) (19) concepts such as body parts, disorders, or physiological abnormalities A specialized module detects and normalizes measurements to international standards. The gathered information is aggregated into a clinical information nodule model using a predefined lexicon. This is followed by a rule-based module that maps instances of this information model to the applicable LR categories.
Information models
The lexicon used was made up of SNOMED clinical findings concepts. If a radiology concept was not represented in SNOMED, the RadLex (24) concept (e.g., micronodule) was used to extend SNOMED. Additionally, if the radiologist used lay terminology in the report not explicit in SNOMED-CT, it was added as a synonym to an existing concept when possible or created as needed (e.g., superior margin, lucent-centered, part-solid).
In addition to nodule and organ description, specific lexicon concepts were developed to support the reasoning layer in the engine and extend SNOMED to represent all LR categories. Concepts were developed to recognize key verbiage such as ‘evidence of lung infection’ and presence of lymph nodes (e.g., concepts related to pneumonia, bronchitis, multi-focal tree-in-bud abnormality, etc.) Other concepts were added to capture status of nodule and whether it was ‘new’ or not (e.g., new, stable, growing, etc.)
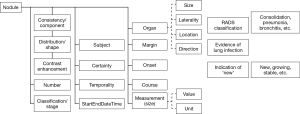
A high-level knowledge representation of a ‘nodule’ is shown in Figure 1; it includes clinical and radiological attributes—values needed to describe any radiologic nodule including consistency, distribution, number, margin, course and measurement, etc. The ‘organ’ attribute has its own knowledge representation of site, laterality and direction, etc. Separating the ‘organ’ in its own model allows the re-capture of ‘nodule’ in different organs that exist in same radiology report. It is imperative to exclude non-pulmonary nodules such as ‘subcutaneous nodules’, ‘lymph nodes’ or ‘thyroid nodules’ before the algorithm can make use of the other attributes to predict the proper LR classification.
Algorithm reasoning
The NLP pipeline through pre-processing first identifies the ‘Findings’ section of the radiologist report and then assigns each nodule mentioned within this section its own information model. The nodule is only marked as a ‘Baseline’ nodule if no reference to previous observation and/or size is made in the report. Figure 2 shows how the pipeline would populate the model from a nodule description “Nodule (Image 179, Series 4) 22 mm × 32 mm ground-glass, ill-defined, parenchymal nodule right lower lobe previously 22 mm × 32 mm”. The concept nodule attribute-value pairs detected are: site = lower lobe of lung, laterality = right, size that is normalized to unit = cm and value =2.2 and 3.2, shape = ill-defined and course = unchanged. The nodule is marked as ‘Baseline’ nodule if no reference to previous observation and/or size was specified in the report. Finally, through a series of rule-based reasoning steps, all nodules are assigned to their specific categories and then compared to each other; the nodule with the highest category is selected. For example, if the previous nodule is the only one mentioned in the report, a ‘Lung-RADS Suggestion’ (Figure 3) would be triggered to indicate that LR category value equals ‘Lung-RADS 2’ and the reason equals ‘Unchanged ground-glass nodule, greater than or equal to 20 mm’.
If the report doesn’t contain sufficient information to populate the critical values in the nodule model, for example the size of nodule is missing, and then an ‘alert’ is triggered to indicate the algorithm could not find sufficient data to calculate a LR category. The goal was to only assign a category if sufficient information was available.
CT imaging
The CTLS protocol and image analysis have been previously described (25). In brief, examinations were performed on ≥64-row multidetector CT scanners [LightSpeed VCT and Discovery VCT (GE Medical Systems, Milwaukee, Wisconsin); Somatom Definition (Siemens AG, Erlangen, Germany); iCT (Philips Medical Systems, Andover, Massachusetts)] at 100 kV and 30 to 100 mA depending on the scanner and the availability of iterative reconstruction software, which can reconstruct diagnostic quality images from very low doses. Axial images were obtained at 1.25 to 1.5 cm thickness with 50% overlap. Images were reconstructed using both, a soft tissue and lung algorithm. Axial maximum-intensity projections (16 mm × 2.5 mm) and coronal and sagittal multiplanar reformatted images were reconstructed and used for interpretation.
Lung screening database and patient selection
This was a retrospective, single-center study approved by the institutional review board (IRB). We reviewed imaging results for consecutive individuals undergoing clinical CTLS at our institution from April 28, 2014 through August 25, 2015. The NLP algorithm was not used when these examinations were initially reported as part of routine clinical care. To qualify for screening, patients had to satisfy the NCCN Guidelines®, Lung Cancer Screening Version 1.2012 high-risk criteria for lung cancer, be asymptomatic, have a physician order for CTLS, be free of lung cancer for ≥5 years, and have no known metastatic disease.
Institutional policy dictates that exams with findings concerning for infection or inflammation are assigned an overall assessment of ‘2i’ (‘i’ designating infection/inflammation). As there are no guidelines for how to handle such cases in LR, we combined categories ‘2’ and ‘2i’.
All included exams were divided into two groups: group 1; training set: comprised of 3 sets of lung screening reports for algorithm training purpose (500, 120 and 383 reports respectively), and group 2; test set: a set of 498 radiology reports.
Training set: an initial set of 500 documents was used to build the lexicon and train the NLP pipeline for document processing. The documents were changed to proper HL7-CDA (Clinical Document Architecture) format (26), and all appropriate concepts were captured into their proper attribute-value position. The second set of 120 radiology documents was used to develop the reasoning layer. The third set was used to train the engine and optimize some of the rules.
For the purpose of this study, we considered the LR category in the radiologist report as reference standard for both groups. All reading radiologists were board certified in radiology, fulfilled USPSTF eligibility criteria, and had at least 2 years interpreting CTLS exams (27). All results were reviewed and discussed with a radiologist who had >4 years of experience in clinical CTLS interpretation.
Results
Patient characteristics
A total of 498 consecutive patients scanned between June 2015 and August 2015 were included for the final assessment of the trained, optimized algorithm.
Of the 498 patients, 267 (53.6%) were male, 255 (51.2%) were actively smoking, and 123 (24.7%) were NCCN group 2 and required an additional risk factor to qualify for screening. The average patient was 63.8 years old with a 48.9 pack-year smoking history. Among former smokers, the average quit time was 10.5 years. There were 135 (27.1%) baseline lung screening exams, 284 (57.0%) routine annual follow-up screening exams for which at least one prior lung screening exam was available, and 79 (15.9%) interval follow-up exams for previously detected pulmonary nodules. A total of 432 (86.7%) findings were classified as benign (LR 1 or 2) by the radiologist. Of these, 106 were category 1. Sixty-six (13.3%) findings were classified as category 3 or 4. Of these, 36 were category 3 (Table 1).
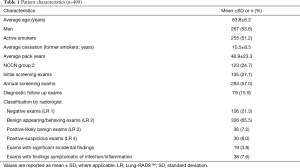
Full table
In 38 (7.6%) reports, findings were designated LR 2 with the modifier ‘i’, indicating that these findings were suspicious for infection or inflammation.
Of the 498 included cases, 27 (5.4%) were found to have findings designated ‘insufficient’ by the algorithm and no LR category could be assigned based on available descriptors in the language (see Table S1 for a detailed case-by-case). Those cases were excluded from the further analysis.
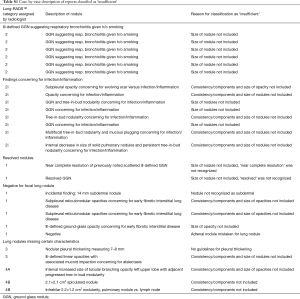
Full table
Identifying suspicious cases (LR 4)
The NLP algorithm was able to identify suspicious nodules (LR 4) with an overall sensitivity of 74.1% and an overall specificity of 98.6% (Table 2). The distinction between non-suspicious (LR 1–3) and suspicious findings (LR 4) differed between the NLP algorithm and the assessment by the radiologist in 13 (2.8%) cases (Table 3).
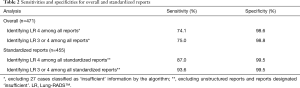
Full table

Full table
In 5 of the 13 cases, misclassification was due to “special findings” for which currently no specific ACR LR guidelines exist: In three patients, the exams had findings concerning for infection or inflammation. Institutional policy dictated these cases be assigned a final overall assessment of ‘2i’ (‘i’ designating infection/inflammation). The NLP algorithm classified these findings as LR 4. In two cases, the nodules did not meet criteria for growth on follow-up as defined by LR (≥1.5 mm). However, the radiologist suspected that growth below the LR threshold was present. Institutional policy dictated that such cases be classified as LR 3 or LR 4 depending on the duration of time since to the prior exam.
In 3 of the 13 cases, there were imaging findings that increased the suspicion of malignancy. None of these were described using the standard language in the structured templates as they showed a high degree of complexity. All of these cases were designated LR 4X by the reading radiologist.
In 5 of the 13 cases, the NLP algorithm misidentified information given in the report: in two cases, the exam was not identified as a follow-up imaging exam. Consequently, one case was classified as LR 3 instead of 4B and one as LR 4A instead of 2. In three cases, the report did not specify the size of the solid component of a part-solid nodule. As a result, the NLP algorithm failed to identify those three cases as ‘insufficient’ and classified one case as LR 3 instead of 4A, one as LR 4A instead of 2, and one as LR 2 instead of 4A.
Table 2 summarizes the NLP performance after the exclusion of unstructured reports where non-standardized language was used and cases for which there are currently no specific LR guidelines.
Identifying positive cases (LR 3 or 4)
The NLP algorithm was able to identify positive nodules (LR 3 or 4) with an overall sensitivity of 75.0% and an overall specificity of 98.8% (Table 2). The distinction between positive (LR 3 or 4) and negative/benign (LR 1 or 2) findings differed between the NLP algorithm and the assessment by the radiologist in 20 (4.2%) cases (Table 3).
In 12 of the 20 cases, the difference in classification was due to the presence of “special findings” for which currently there are no specific ACR LR guidelines. In three cases, imaging findings that increased the suspicion of malignancy cases lead to a LR 4X classification by the reading radiologist. In 5 of the 20 cases, the NLP algorithm misidentified information given in the report. A detailed description is available in the online-only supplement.
Table 2 summarizes the NLP performance after the exclusion of unstructured reports where non-standardized language was used and cases for which there are currently no specific LR guidelines.
Discussion
NLP has been shown to be able to extract relevant information and help clinical decision making (13-15). In our study on NLP in the setting of CTLS exams, we found that an NLP algorithm can detect lung nodule characteristics and identify suspicious nodules (LR 4) with an overall sensitivity and specificity among all exams of 74.1% and 98.6%, respectively. To our knowledge, this is the first study to show the feasibility to classify CTLS exams using NLP.
A mismatch between the NLP algorithm and the expert radiologist in our study resulted from either true misclassification by the algorithm or from findings for which currently no ACR LR guidelines exist. The latter encompassed primarily enlarged lymph nodes and findings suspicious for infection or inflammation. In addition, this included cases where the radiologist suspected growth of the nodule compared to prior exam, but which did not meet ACR LR criteria for growth. These mismatched cases highlight gaps for the ACR LR committee to address in future revisions of the LR system. The NLP algorithm can be updated to account for such changes.
Two major challenges that resulted in true misclassifications by the algorithm were the identification of nodules for which prior imaging exams were available and the identification of measurements. Notably, it was not possible to simply refer to the comparison section in the report to determine if a prior exam is available as patients often had thoracic or abdominal CTs performed for other indications. As these exams might only capture part of the lung parenchyma, it cannot be inferred that a nodule present on a screening exam could have been present on the prior exam. We therefore programmed the algorithm to determine if certain linguistic concepts such as ‘new’ or ‘stable’ were mentioned in the findings section together with the nodule. However, misclassifications can arise if those concepts are not recognized by the algorithm as being directly related to a specific nodule. Accessibility to patient record and previous studies can help the algorithm to overcome this.
The extraction of measurements is a well-known challenge in NLP and has been addressed previously (28). Despite the great number of measurements in each report, we primarily saw misclassifications only if several measurements for one nodule were present—as is the case with part-solid nodules. In these cases, more extensive descriptions by the radiologist might be necessary to describe the findings as the consistency and the size of the nodules can change from exam to exam. A different report structure with those more extensive descriptions might have limited the NLP algorithm’s ability to correctly recognize how measurements are related. More training data would be needed and probably a more consistent and standardized description of the whole versus parts of nodule might help resolve this issue.
Our results have to be interpreted in context with the limitations of the study design. Most importantly, the reports at our institution used for this study are highly structured and descriptors are highly standardized. Our radiologists use a defined set of approximately 40 language macros and some free text to generate their reports. The accuracy of the algorithm decreased if non-standardized language was used to describe more complex findings as in LR 4X cases. The generalizability of this algorithm from our single center study to other organizations is unknown. We make our report language building blocks available to interest other institutions free of charge and one can hypothesize that usage of this very language by other centers would set them up for successful implementation of this assistive algorithm. However, the NLP algorithm in its current form and as used for this research is proprietary and available through MModal. Further, the reading radiologists have either interpreted thousands of clinical CTLS exams or undergone an extensive internal credentialing/training process beyond the USPSTF requirements (6).
Correct LR classification is not only paramount for patient care, but participating programs also need to upload certain required structured data elements into a CMS-approved registry to qualify for payment. The long term viability of this important national screening program hinges on its performance in clinical practice. Any tool that can help to raise the quality bar of lung screening interpretation and speed the learning curve of radiologists should therefore be of value.
Conclusions
In conclusion, the algorithm may be able to assist the radiologist in real time by not only suggesting the appropriate LR category, but also by identifying reports that might contain insufficient data about a nodule to assign the correct LR class. Consequently, it could prompt review and improvement of the report in such situations.
Supplementary
Methods
Model development
We divided the Lung-RADS™ categorization task into two specific tasks. The first task was to find an annotation of ‘lung nodule’. The second task was to classify the resulting annotations into the proper category.
Annotation of lung nodule: this comprised multiple steps: (I) identification of radiology document type of CT lung screen using the title; (II) identification of findings and impression sections by transforming the radiology report into CDA level III and mapping the sections into the appropriate LOINC codes; (III) the annotation parser then compiles all needed attribute-values of a nodule site, certainty and negation, temporality, nodule components and consistency, nodule measurement and solid component measurement; (IV) define the nodule course as baseline, new, stable; (V) annotation of concepts denoting lung infection.
Classification of the report also undergoes multiple steps: (I) based on the annotation output of nodules size and components the each individual nodule is classified into a Lung-RADS category, if information is insufficient for classification it is marked as such; (II) then the lung category is adjusted based on the course (new vs. pre-existing nodule); (III) finally the highest Lung-RADS category is selected and if data is insufficient, no category is assigned.
Results
Identifying positive cases (LR 3 or 4)
The NLP algorithm was able to identify positive nodules (LR 3 or 4) with an overall sensitivity of 75.0% and an overall specificity of 98.8% (Table 2). The distinction between positive (LR 3 or 4) and negative/benign (LR 1 or 2) findings differed between the NLP algorithm and the assessment by the radiologist in 20 (4.2%) cases (Table 3).
In 12 of the 20 cases, the difference in classification was due to the presence of “special findings” for which currently there are no specific ACR LR guidelines. In four patients, enlarged lymph nodes in the mediastinum and/or hilum were present in the absence of a positive pulmonary nodule. Institutional policy dictated that such lymph nodes without documented stability are to be assigned a final overall assessment of LR 3 (1–1.5 cm minimum dimension) or LR 4 (>1.5 cm in minimum dimension, growing, or with suspicious features such as internal necrosis). The NLP algorithm misclassified all those cases as LR 2.
In three patients, the exams had findings concerning for infection or inflammation. Institutional policy dictated these cases be assigned a final overall assessment of ‘2i’ (‘i’ designating infection/inflammation). The NLP algorithm misclassified these findings as LR 4.
In four cases, the nodules did not meet criteria for growth on follow-up as defined by LR (≥1.5 mm). However, the radiologist suspected growth below the threshold was present. Institutional policy dictated such cases be classified as LR 2, LR 3 or LR 4 depending on the duration of time since to the prior exam.
In one case, a ground-glass nodule <20 mm was present that was increasing in size. As the reading radiologist considered the growth to be more than “slow”, the nodule was designated LR 3.
In 3 of the 20 cases, there were imaging findings that increased the suspicion of malignancy. None of these were described using the standard language in the structured templates as they showed a high degree of complexity. All of these cases were designated LR 4X by the reading radiologist.
In 5 of the 20 cases, the NLP algorithm misidentified information given in the report.
In three cases, the exam was not identified as a follow-up imaging exam. Consequently, one case was classified as LR 4A instead of 2. In the other two cases, new <20 mm ground-glass nodules were incorrectly designated LR 2 instead of LR 3 as they were not recognized as new nodules.
In two cases, the report did not specify the size of the solid component of a part-solid nodule. As a result, the NLP algorithm failed to identify those three cases as ‘insufficient’ and classified one as LR 4A instead of 2 and one as LR 2 instead of 4A.
NLP algorithm performance during training
The first training set was used to build the ontology without an evaluation of the algorithm performance. Overall, in training sets 2 and 3 the algorithm incorrectly classified 3.7% and 12.0% of reports, respectively. The sensitivity/specificity to identify suspicious cases (LR 4) was 93.3%/99.4% and 50.0%/98.9%. The sensitivity/specificity to identify positive cases (LR 3 and 4) was 82.4%/98.8% and 53.6%/98.5%.
Acknowledgements
The authors wish to thank Szymon Letowski, the lead engineer of the NLP algorithm.
Footnote
Conflicts of Interest: BJ McKee reports financial activities from Covidien/Medtronic and Grail, Inc., outside the submitted work. SM Regis reports personal fees from Covidien/Medtronic, outside the submitted work. AB McKee reports personal fees from Covidien/Medtronic and Grail, Inc., outside the submitted work. G El Saadawi reports personal fees from MModal, during the conduct of the study, and outside the submitted work. C Wald reports personal fees from Philips HealthTech for advisory board function, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The study was institution review board (IRB) approved with a waiver of individual patient consent.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Smith RA, Andrews K, Brooks D, et al. Cancer screening in the United States, 2016: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 2016;66:96-114. [Crossref] [PubMed]
- Moyer VA. U.S. Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160:330-8. [PubMed]
- Pyenson BS, Henschke CI, Yankelevitz DF, et al. Offering lung cancer screening to high-risk medicare beneficiaries saves lives and is cost-effective: an actuarial analysis. Am Health Drug Benefits 2014;7:272-82. [PubMed]
- McKee BJ, McKee AB, Kitts AB, et al. Low-dose computed tomography screening for lung cancer in a clinical setting: essential elements of a screening program. J Thorac Imaging 2015;30:115-29. [Crossref] [PubMed]
- Mulshine JL, D'Amico TA. Issues with implementing a high-quality lung cancer screening program. CA Cancer J Clin 2014;64:352-63. [Crossref] [PubMed]
- Lung CT Screening Reporting and Data System (Lung-RADS). Available online: http://www.acr.org/Quality-Safety/Resources/LungRADS
- McKee BJ, Regis SM, McKee AB, et al. Performance of ACR Lung-RADS in a clinical CT lung screening program. J Am Coll Radiol 2015;12:273-6. [Crossref] [PubMed]
- Pinsky PF, Gierada DS, Black W, et al. Performance of Lung-RADS in the National Lung Screening Trial: a retrospective assessment. Ann Intern Med 2015;162:485-91. [Crossref] [PubMed]
- Kimia AA, Savova G, Landschaft A, et al. An Introduction to Natural Language Processing: How You Can Get More From Those Electronic Notes You Are Generating. Pediatr Emerg Care 2015;31:536-41. [Crossref] [PubMed]
- Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161-74. [Crossref] [PubMed]
- Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp 1997.829-33. [PubMed]
- Nassif H, Woods R, Burnside E, et al. Information Extraction for Clinical Data Mining: A Mammography Case Study. Proc IEEE Int Conf Data Min 2009.37-42. [PubMed]
- Raja K, Jonnalagadda SR. Natural language processing and data mining for clinical text. Healthcare Data Anal 2015;36:219.
- Mamlin BW, Heinze DT, McDonald CJ. Automated extraction and normalization of findings from cancer-related free-text radiology reports. AMIA Annu Symp Proc 2003.420-4. [PubMed]
- Cai T, Giannopoulos AA, Yu S, et al. Natural Language Processing Technologies in Radiology Research and Clinical Applications. Radiographics 2016;36:176-91. [Crossref] [PubMed]
- Cheng LT, Zheng J, Savova GK, et al. Discerning tumor status from unstructured MRI reports--completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging 2010;23:119-32. [Crossref] [PubMed]
- Danforth KN, Early MI, Ngan S, et al. Automated identification of patients with pulmonary nodules in an integrated health system using administrative health plan data, radiology reports, and natural language processing. J Thorac Oncol 2012;7:1257-62. [Crossref] [PubMed]
- Chaudhry B, Wang J, Wu S, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med 2006;144:742-52. [Crossref] [PubMed]
- Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Natural Language Engineering 2004;10:327-348. [Crossref]
- The international standard for identifying health measurements, observations, and documents. Available online: http://loinc.org/
- Chen ES, Melton GB, Engelstad ME, et al. Standardizing Clinical Document Names Using the HL7/LOINC Document Ontology and LOINC Codes. AMIA Annu Symp Proc 2010;2010:101-5. [PubMed]
- Radiological Society of North America (RSNA) RadLex. Available online: http://www.rsna.org/RadLex.aspx
- McKee BJ, Regis SM, McKee AB, et al. Performance of ACR Lung-RADS in a Clinical CT Lung Screening Program. J Am Coll Radiol 2016;13:R25-9. [Crossref] [PubMed]
- Health Level Seven Clinical Document Architecture (HL7 CDA). Available online: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=7
- Decision Memo for Screening for Lung Cancer with Low Dose Computed Tomography (LDCT) (CAG-00439N). Available online: https://www.cms.gov/medicare-coverage-database/details/nca-decision-memo.aspx?NCAId=274
- Sevenster M, Buurman J, Liu P, et al. Natural Language Processing Techniques for Extracting and Categorizing Finding Measurements in Narrative Radiology Reports. Appl Clin Inform 2015;6:600-110. [Crossref] [PubMed]