
MicroRNA-30d target TIMP3 induces pituitary tumor cell growth and invasion
Introduction
The annual incidence of pituitary adenoma, one of the most common intracranial tumors, is as high as 16.7%. Approximately 10% of pituitary adenomas behave aggressively and 0.2% metastasize, being classified as pituitary carcinomas, which can lead to long hospital stays and serious endocrine symptoms. Pituitary adenomas are divided into subtypes including prolactin (PRL) adenoma, adrenocorticotropic hormone adenoma (Cushing’s disease), growth hormone (GH) adenoma, thyroid stimulating hormone (TSH) adenoma, gonadotropic hormone adenoma (PGA), and mixed hormone secreting adenoma (1-3). Among them, Cushing’s disease accounts for approximately 14% of all pituitary adenomas and is recognized as the most difficult pituitary adenoma subtype in terms of current diagnosis and treatment. Cushing’s disease can cause extensive damage to various bodily systems and organs, resulting in complicated diagnosis and treatment, and requiring multi-departmental cooperation. Since its discovery and naming nearly 100 years ago, the diagnosis, operative difficulty, and high recurrence rate after operation have persistently posed significant challenges. Therefore, it is necessary to explore the molecular pathogenesis of pituitary adenomas and find effective molecular targets for early clinical diagnosis, as well as follow-up treatment.
MicroRNAs (miRNAs) are important gene expression regulators that consist of approximately 16–29 nucleotides and always bind to the 3'-untranslated regions (3'-UTRs) of target messenger RNAs (mRNAs), causing degradation of mRNAs, and resulting in the prevention of protein translation. This regulatory process plays an important role in the course of the occurrence and development of various diseases (4-6). With the development of sequencing technology, numerous miRNAs have been found. Through studies of leukemia, lung cancer, breast cancer, thyroid cancer, insulinoma, ovarian cancer, and pituitary adenoma, it has been confirmed that miRNAs can regulate cell proliferation, cell differentiation, cell death, and tumorigenesis, with differences in expression observed in the normal group compared to the disease group (7-19). Due to the significant differences in the expression of miRNAs in patients and their various functions, miRNAs are expected to become new targets for the diagnosis and treatment of various diseases.
In our previous studies, TIMP3 mRNA and protein expression were reduced in all types of pituitary adenomas compared to adjacent non-tumorous pituitary tissues. The expression of TIMP3 was significantly correlated with Ki-67 expression and the invasiveness of pituitary adenomas (20). We showed the abnormally high expression of miR-30d in pituitary adenomas by analyzing data in the Gene Expression Omnibus (GEO) database. Through TargetScan Human 7.0 (http://www.targetscan.org/vert_70/), TIMP3 was identified as the direct target of miR-30d. Therefore, we posited that TIMP3 is regulated by miRNA-30d and participates in pituitary tumor growth. However, its potential mechanism has not been fully clarified.
In this article, we aimed to find a new miRNA related to the occurrence and development of pituitary adenomas and attempted to reveal the underlying molecular regulatory mechanism to provide a theoretical basis for its clinical application as a molecular target. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/gs-21-720).
Methods
Participants and tissue samples
Specimens were obtained from patients undergoing surgery in the Department of Neurosurgery of Beijing Union Medical College Hospital. In total, 3 normal pituitary glands were obtained from autopsy patients with no history of endocrine abnormality. Their families signed the informed consent form. None of these cases had a recognized pituitary disease, and their pituitaries exhibited a normal gland appearance. Tissue samples were immediately frozen in liquid nitrogen at the time of surgery and stored at −70 °C until RNA was extracted. A section of the tumor tissue was fixed in 10% formalin, paraffin embedded, and used for immunohistochemistry (IHC). Finally, informed consent for the present study was provided by all participants, and the present study was approved by the Research Ethics Committee of Beijing Tongren Hospital (No. TRECKY2018-012). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and transfection
The mouse pituitary adenoma cell line AtT-20 used in this study was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA; CCL-89™). The cells were seeded in F-12K medium (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and maintained at 37 °C in humidified air with 5% CO2.
For transfection experiments, the AtT-20 cells were cultured in growth medium without antibiotics to 80% confluence and then transfected using a transfection reagent (Lipofectamine 3000, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The miR-30d mimic (cat. no. miR10000245-1-5), the negative control (NC) of the miR-30d mimic (NC miR-30d mimic; cat. no. miR01201-1-5), miR-30d inhibitor (cat. no. miR20000245-1-5), and inhibitor NC (cat. no. miR2N0000001-1-5) were purchased from Guangzhou RiboBio Co. (Guangzhou, China). After incubation for 6 h, the medium was removed and replaced with normal culture medium for 24 h.
Reverse-transcription polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and 2 µg of each sample was reverse transcribed using M-MLV reverse transcriptase (Promega, Madison, WI, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control to show equal loading of the complementary DNA (cDNA) samples. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in an Applied Biosystems Step One Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The Fast SYBR® Green Master Mix was obtained from Applied Biosystems. Data were shown as the relative expression level after normalization to GAPDH expression. Each test was repeated 3 times. The primers for the PCR analysis are listed in Table 1.
Table 1
| Gene symbol | Primer F | Primer R |
|---|---|---|
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| miR-30d | CTCAACTGGTGTCGTGGAG | TCGGCAGGCGGGGATGTTTACA |
| GAPDH | GACAGGATGCAGAAGGAGATTACT | TGATCCACATCTGCTGGAAGGT |
| MMP3 | CCAGGTGTGGAGTTCCTGAT | GGAGTCACCTCTTCCCAGAC |
| MMP9 | GACAAGCTCTTCGGCTTCTG | CAAAGTTCGAGGTGGTAGCG |
| ADAM17 | GCATGGATTCTGCATCGGTT | CCATTCTCTGGTGGTCCAGT |
| TIMP3 | ATGTACCGAGGCTTCACCAA | CTCTCCACGAAGTTGCACAG |
PCR, polymerase chain reaction.
Western blotting
The prepared proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The membranes were immunoblotted with anti-TIMP3 (Abcam, ab39184), anti-MMP3 (Abcam, ab53015), anti-MMP9 (Abcam, ab38898), anti-ADAM17 (Abcam, ab2051), and anti-GAPDH (Proteintech, Rosemont, IL, USA; 10494-1-AP) antibodies overnight at 4 °C and then incubated with IRDye-800-conjugated anti-mouse and anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 30 min at room temperature. The specific proteins were visualized by the Odyssey Infrared Imaging System (Gene Company Limited, Shanghai, China). The expression of GAPDH was used as an internal control to show equal loading of the protein samples. Each test was repeated 3 times.
3-(4,5-diphenyltetrazolium bromide) assay
Each cell group was plated into 96-well plate with 500 cells per well. When the cells adhered to the wall, we used 15 µL 3-(4,5-diphenyltetrazolium bromide) (MTT; 5 mg/mL, Promega Corp., USA) was added to culture the cells for 4 h in incubator with 5% CO2 at 37 °C. Then, 150 µL dimethyl sulfoxide (DMSO) was added and the 96-well plate was shaken for 10 min. A microplate reader (BioTek Instruments, Winooski, VT, USA) was used to measuring the light absorbance at 490 nm, and the cell proliferation in 72 h of each group was also detected. Each test was repeated 3 times.
Transwell invasion assay
The invasion assay was performed using Transwell chambers (Corning Inc., Corning, NY, USA) with polycarbonate membranes (8.0 µm pore size) pre-coated with Matrigel (50 µL; Becton, Dickinson, and Co. Biosciences, Franklin Lakes, NJ, USA). A total of 2×104 cells were suspended in 200 µL fetal bovine serum (FBS)-free Dulbecco’s modified Eagle medium (DMEM) and added to the upper chamber. The lower chamber was filled with 500 µL DMEM containing 10% FBS. Following incubation for 24 h, cells on the lower surface of the membrane were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. Cells in 5 random microscopic fields (magnification, ×20) were counted in triplicate.
Luciferase constructs, site-directed mutagenesis, and luciferase assay
The pmirGLO-TIMP3-WT, the fragment containing the TIMP3 3'-UTR, was ligated into the pmirGLO vector. The pmirGLO-TIMP3-Mutplasmid contained a mutated binding site of miR-30d (TGTAAACA to CAACCCTT). Then, cos-7 cells (2×105/well) were transfected with miR-30d mimics or its respective negative control RNA (RiboBio) in combination with pmirGLO-TIMP3-WT or pmirGLO-TIMP3-Mut. After 48 h, a dual-luciferase assay system (Promega) was used to test luciferase activity.
Statistical analysis
Data were expressed as the mean ± standard error (SE), accompanied by the number of experiments performed independently, and were analyzed by the t-test. Differences at P<0.05 were considered statistically significant.
Results
The expression of miR-30d in pituitary adenoma was higher than that in normal pituitary tissues
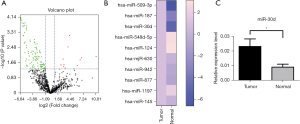
A volcano plot was generated to elucidate the temporal pattern of miRNA changes between Cushing patients and patients with normal pituitary glands, the miRNA expression level changes (Figure 1A). Among them, the expression level of miR-30d increased approximately 4-fold compared to the control, and the related data were plotted in a heat map (Figure 1B). The expression of miR-30d in pituitary adenoma was higher compared to that of normal pituitary tissues (Figure 1C).
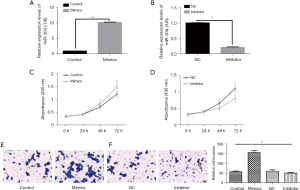
Silencing of miR-30d could inhibit AtT-20 cell proliferation and invasion
To further explore the function of miR-30d in pituitary adenomas, miR-30d mimics and an miR-30d inhibitor were used for additional experiments (Figure 2A,2B). First, an MTT assay was used to detect whether miR-30d could regulate the proliferation of AtT-20 cells. The results showed that there was no significant change in the proliferation of AtT-20 cells regardless of whether miR-30d was overexpressed or silenced within 24 h, compared with the control group. However, after 48 h, overexpression of miR-30d significantly promoted the proliferation of AtT-20 cells, and silencing of miR-30d had the opposite effect (Figure 2C,2D). Subsequently, a Transwell assay was used to detect the effect of miR-30d on the invasive ability of AtT-20 cells. The results showed that when miR-30d was overexpressed for 24 h, the invasive ability of AtT-20 cells was significantly enhanced (Figure 2E), and when miR-30d was silenced, AtT-20 cell invasion was significantly decreased (Figure 2F). These data preliminarily demonstrated that miR-30d could promote pituitary adenoma cell proliferation and invasion.
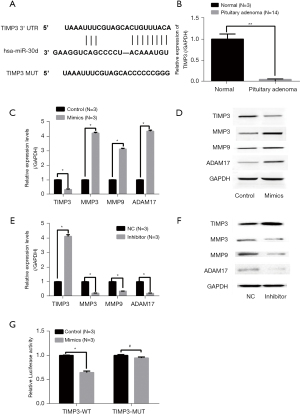
MiR-30d could inhibit TIMP3 expression by binding to the 3'-UTR of TIMP3 mRNA
To explore the molecular mechanism of miR-30d in regulating pituitary adenoma cell invasion, we analyzed potential downstream targets of miR-30d by TargetScan. We found that TIMP3 was a predicted target of miR-30d (Figure 3A). Then, we detected the expression of TIMP3 in normal human pituitary (n=3) and pituitary adenoma (n=14) tissues by real-time RT-PCR. The results confirmed that the expression of TIMP3 in pituitary adenomas was significantly decreased (Figure 3B). To further confirm our prediction, the expression changes in TIMP3 and in genes associated with the TIMP3 pathway were detected in AtT-20 cells overexpressing miR-30d. The real-time RT-PCR and western blot results confirmed that when miR-30d was up-regulated, the expression of TIMP3 and its related genes was inhibited, both at the mRNA and protein levels (Figure 3C,3D). Consistent with previous results, the expression of TIMP3 and its related genes was up-regulated when miR-30d was silenced (Figure 3E,3F). Finally, we analyzed the molecular mechanism of miR-30d in regulating TIMP3, and the luciferase assay results confirmed our predictions that miR-30d regulated the expression of TIMP3 by binding to the 3'-UTR of TIMP3 mRNA (Figure 3G).
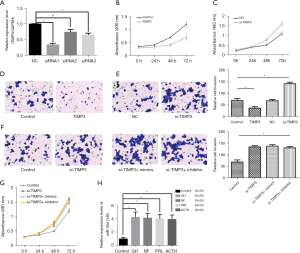
MiR-30d could promote AtT-20 cell proliferation and invasion via TIMP3
As the function of TIMP3 in pituitary adenoma has remained unclear, we examined the effect of TIMP3 on the proliferation and invasion of AtT-20 cells. The results showed that TIMP3 could effectively inhibit the proliferation and invasion of AtT-20 cells (Figure 4A-4E). Then, in AtT-20 cells with silenced TIMP3, miR-30d was overexpressed or silenced to detect cell proliferation and invasion. The results confirmed that miR-30d could not regulate the proliferation (Figure 4F) or invasion (Figure 4G) of AtT-20 cells with TIMP3 silencing. Finally, we also examined the peripheral blood of patients with other subtypes of pituitary adenomas and found that the expression of miR-30d increased to varying degrees compared with that in normal participants (Figure 4H).
Discussion
The correlation between pituitary adenoma and miRNAs was first studied in 2005. In 10 growth hormone (GH)-secreting and 10 prolactin (PRL)-secreting adenoma patients, the expression levels of miR-15a and miR-16-l were significantly down-regulated (63% and 73%, respectively) compared with those in normal pituitary tissues and were negatively correlated with the diameters of the tumors (7). Subsequently, 29 miRNAs were identified and considered as biomarkers of histological classification for pituitary adenomas. For example, the miR-23 family was only highly expressed in GH and PRL adenomas, while the miR-30 family was highly expressed in gonadotropic adenomas, and miR-24-2 was only expressed at low levels in nonfunctional adenomas. Moreover, the expression profile of miR-140 could differentiate between nonfunctional microadenomas and large adenomas (9). However, there are still many controversies regarding whether miRNA-30d is an oncogene or a tumor suppressor gene in the tumorigenesis and development. In previous studies, miRNA-30d was shown to promote aggressive tumor progression by targeting lactate dehydrogenase A (LDHA) in gallbladder carcinoma (21), and that it can contribute to angiogenesis and tumor growth via the MYPT1/c-JUN/VEGFA pathway in prostate cancer (22). However, in other reports, results showed that up-regulated miRNA-30d can attenuate the effects of overexpressing POU3F3 on cancer cell proliferation, migration, and invasion in non-small cell lung cancer (23). Also, miR‑30d functions as a tumor suppressor against tumor cell development in colorectal cancer (24). These results indicate that the occurrence and development of tumors is regulated by a variety of genes, and miRNA, as a micro-regulatory link in the signaling pathway, may play different functions in different types of benign and malignant tumors. Furthermore, an increasing number of studies have shown that there are a large number of miRNAs in the serum of both healthy people and patients. It is worth noting that miRNAs are stable in human peripheral blood. Based on this, significant differences in the expression of miRNAs in the serum of patients with malignant or benign tumors and healthy controls can be used as specific tumor markers (25-27). In this study, we found abnormal expression of miR-30d in pituitary adenoma based on high-throughput sequencing data. Then, the expression of miR-30d in the peripheral blood of Cushing patients and patients with other subtypes of pituitary adenomas was detected by qRT-PCR. The results showed that the expression of miR-30d in the peripheral blood of patients with different subtypes of pituitary adenomas was significantly increased compared with that in normal controls. This further demonstrates the importance of miR-30d in the occurrence and development of pituitary adenomas.
We also explored the molecular regulatory mechanism of miR-30d in pituitary adenoma in vitro and identified a new pathway in which miR-30d could inhibit the migration and invasion of pituitary adenomas via TIMP3. Matrix metalloproteinases (MMPs) are a family of proteases that require zinc ions as co-factors, and can degrade the extracellular matrix in normal physiological processes, such as embryonic development, tissue and bone remodeling, and angiogenesis. Human tissue metalloproteinase inhibitor 3 (TIMP3) is a natural MMP inhibitor that has been shown to play an important role in many tumorigenesis processes. Previous studies have shown that TIMP3 expression is low in breast cancer, colorectal cancer, and prostate cancer, and the decrease in its expression level has been related to the poor prognosis of tumors (28-30). Moreover, high expression of TIMP3 could inhibit angiogenesis in colorectal cancer (31). These reports confirmed that TIMP3 could act as an important anticancer molecule in many kinds of tumors. Our previous studies showed that TIMP3 expression was significantly correlated with Ki-67 expression and the invasiveness of pituitary adenomas. The expression of TIMP3 mRNA and protein were reduced in all types of pituitary adenomas compared to adjacent non-tumorous pituitary tissues (20). In this study, we investigated its clinical significance in pituitary adenomas and confirmed that TIMP3 could inhibit the proliferation and migration of the pituitary adenoma cell line AtT-20. This indicated that TIMP3 might play an important role in the occurrence and development of pituitary adenomas as a tumor suppressor. We also confirmed that miR-30d targeted TIMP3 to regulate the proliferation and migration of pituitary adenoma cells. Therefore, miR-30d and TIMP3 have potential as important molecular markers to evaluate the invasiveness of pituitary adenomas.
Generally, serum miRNA detection has received increasing attention since its discovery because of its advantages of stability, specificity, minimal trauma, repeatability, and simplicity. Serum miRNA detection has been used not only in research on highly malignant cancers but also in research on benign tumors such as pituitary adenomas. In this study, we found that the miR-30d/TIMP3 signaling pathway plays an important regulatory role in pituitary adenomas. The findings also provide a theoretical basis for future clinical applications.
Acknowledgments
Funding: This work was financially supported by the Beijing Science and Nature Fund (No. 7172057) and Scientific research and cultivation plan of Beijing Municipal Hospital (No. PX2018006).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-720
Data Sharing Statement: Available at https://dx.doi.org/10.21037/gs-21-720
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-720). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Informed consent for the present study was provided by all participants, and the present study was approved by the Research Ethics Committee of Beijing Tongren Hospital (No. TRECKY2018-012). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ezzat S, Asa SL, Couldwell WT, et al. The prevalence of pituitary adenomas: a systematic review. Cancer 2004;101:613-9. [Crossref] [PubMed]
- Fernandez A, Karavitaki N, Wass JA. Prevalence of pituitary adenomas: a community-based, cross-sectional study in Banbury (Oxfordshire, UK). Clin Endocrinol (Oxf) 2010;72:377-82. [Crossref] [PubMed]
- Kasuki L, Raverot G. Definition and diagnosis of aggressive pituitary tumors. Rev Endocr Metab Disord 2020;21:203-8. [Crossref] [PubMed]
- Selbach M, Schwanhäusser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature 2008;455:58-63. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Sharma PC, Gupta A. MicroRNAs: potential biomarkers for diagnosis and prognosis of different cancers. Transl Cancer Res 2020;9:5798-818. [Crossref]
- Bottoni A, Piccin D, Tagliati F, et al. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol 2005;204:280-5. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Bottoni A, Zatelli MC, Ferracin M, et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol 2007;210:370-7. [Crossref] [PubMed]
- Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 2005;102:13944-9. [Crossref] [PubMed]
- Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 2006;24:4677-84. [Crossref] [PubMed]
- Amaral FC, Torres N, Saggioro F, et al. MicroRNAs differentially expressed in ACTH-secreting pituitary tumors. J Clin Endocrinol Metab 2009;94:320-3. [Crossref] [PubMed]
- He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A 2005;102:19075-80. [Crossref] [PubMed]
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70. [Crossref] [PubMed]
- Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007;67:8699-707. [Crossref] [PubMed]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001;294:862-4. [Crossref] [PubMed]
- Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83-6. [Crossref] [PubMed]
- O'Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005;435:839-43. [Crossref] [PubMed]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [Crossref] [PubMed]
- Sun B, Liu X, Yang Y, et al. The Clinical Utility of TIMP3 Expression in ACTH-Secreting Pituitary Tumor. J Mol Neurosci 2016;58:137-44. [Crossref] [PubMed]
- He Y, Chen X, Yu Y, et al. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Mol Carcinog 2018;57:772-83. [Crossref] [PubMed]
- Lin ZY, Chen G, Zhang YQ, et al. MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol Cancer 2017;16:48. [Crossref] [PubMed]
- Zeng Q, Dai Y, Duan C, et al. Long noncoding RNA POU3F3 enhances cancer cell proliferation, migration and invasion in non-small cell lung cancer (adenocarcinoma) by downregulating microRNA-30d-5p. BMC Pulm Med 2020;20:185. [Crossref] [PubMed]
- Yan L, Qiu J, Yao J. Downregulation of microRNA-30d promotes cell proliferation and invasion by targeting LRH-1 in colorectal carcinoma. Int J Mol Med 2017;39:1371-80. [Crossref] [PubMed]
- Nassar FJ, Chamandi G, Tfaily MA, et al. Peripheral Blood-Based Biopsy for Breast Cancer Risk Prediction and Early Detection. Front Med (Lausanne) 2020;7:28. [Crossref] [PubMed]
- Yin S, Yang M, Li X, et al. Peripheral blood circulating microRNA-4636/-143 for the prognosis of cervical cancer. J Cell Biochem 2020;121:596-608. [Crossref] [PubMed]
- Necula L, Matei L, Dragu D, et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol 2019;25:2029-44. [Crossref] [PubMed]
- Jackson HW, Hojilla CV, Weiss A, et al. Timp3 deficient mice show resistance to developing breast cancer. PLoS One 2015;10:e0120107. [Crossref] [PubMed]
- Das AM, Seynhaeve AL, Rens JA, et al. Differential TIMP3 expression affects tumor progression and angiogenesis in melanomas through regulation of directionally persistent endothelial cell migration. Angiogenesis 2014;17:163-77. [Crossref] [PubMed]
- Adissu HA, McKerlie C, Di Grappa M, et al. Timp3 loss accelerates tumour invasion and increases prostate inflammation in a mouse model of prostate cancer. Prostate 2015;75:1831-43. [Crossref] [PubMed]
- Qi JH, Ebrahem Q, Moore N, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 2003;9:407-15. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


