
Integrated analysis of immunocyte infiltration and differential gene expression in tricuspid aortic valve-associated thoracic aortic aneurysms
Introduction
Thoracic aortic aneurysms (TAAs) are a class of vascular diseases with rapid progression and very high mortality and morbidity. Epidemiological studies have suggested that the overall incidence of TAAs is greater than 7.6/100,000 and that TAAs are identified in only 78% of patients before death (1,2). Even with surgical treatment, 16% of patients died within 30 days after surgery, and the 1-, 5-, and 10-year survival rates were 92%, 77%, and 57%, respectively, while the reoperation rate within 10 years was 7.8% (2,3). With the development of 3D printing technology and hybrid surgery, the treatment and prognosis of TAAs have greatly improved (4). However, the mechanisms of TAA occurrence and progression are still unclear.
In recent years, the vigorous development of high-throughput sequencing technology has provided a new opportunity for studying the mechanism of TAAs. Studies have shown that the development and progression of TAAs are closely related to immune regulatory pathways and genes. Kim et al. investigated gene expression profile differences between the thoracic aortas of TAA patients and normal thoracic aortas in organ transplant patients. They found that the differentially expressed genes (DEGs) associated with TAAs were mainly associated with ion transport, cell signal transduction, and immune inflammatory responses (5). Tang et al. analyzed the pathological process of vascular remodeling (changes in the vascular outer diameter) and intima dilation in ascending aorta specimens from TAAs. They found that the transmural inflammatory state of the aorta and the production of interferon-gamma (IFN-γ) in TAAs were closely related to increases in the outer diameter of the aneurysm, thickening of the intima, maintenance of the density of vascular smooth muscle cells, and decreases in matrix proteins (6). Similarly, Sprague et al. believed that the aneurysm dilation process was closely related to the inflammatory state of blood vessels. Vascular injury can stimulate the expression of endothelial cell adhesion molecules and promote the recruitment of inflammatory cells, growth factors, and cytokines, thus affecting the functions of vascular smooth muscle cells and endothelial cells. In addition, these cytokines can induce the production or activation of vasodilation mediators, such as nitric oxide, prostacyclin, endothelial-derived hyperpolarizing factors, and bradykinin, and vasoconstrictors, such as endothelin and angiotensin II, thereby regulating the pathological process of vascular dilation (7). However, aneurysm formation is not always associated with immune inflammation. By comparing the vascular tissue differential gene expression profiles and pathological mechanisms of bicuspid aortic valve (BAV)- and tricuspid aortic valve (TAV)-associated aortic aneurysm expansion, Folkersen et al. found that immune mediators were activated only in TAV tissues, whereas BAV tissues did not exhibit a significant immune process. However, the specific mechanism has not yet been elucidated (8). Based on this result, this study aimed to conduct an in-depth investigation of the pathological molecular mechanism of TAV-associated vascular dilation by analyzing DEGs from the whole gene expression profile, immune cell infiltration, and related enrichment pathways in the dilated and nondilated aortic intima-media (AMed) and outer aortic adventitia (AAdv) of TAV patients. This study will provide a new diagnostic or therapeutic target for this disease.
Methods
Data screening and acquisition
We downloaded the GSE26155 dataset from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database for subsequent analysis (9). These data were acquired from microarray expression profiling of AAdv and AMed tissues of TAAs and were published in the Advanced Study of Aortic Pathology (ASAP). A total of 83 vascular tissue specimens were selected, including 46 AMed tissue samples (17 dilated and 23 nondilated samples and 6 boundaries that needed to be excluded) and 37 AAdv tissue samples (12 dilated and 21 nondilated samples and 4 boundaries that needed to be excluded) (8). The corresponding microarray platform was the GPL570 (HG U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array platform (Affymetrix, Santa Clara, CA, USA). In addition, the clinical information corresponding to each sample was downloaded for further analysis.
Data analysis
The data processing flow was as follows: (I) detection of the CEL fluorescence intensity; (II) quality control; (III) background processing using the robust multiarray average (RMA) method; (IV) processing of missing probe values by log2 transformation and the k-nearest neighbor (kNN) algorithm; (V) gene annotation using probe names; (VI) differential expression analysis of expression profiles using linear models for microarray data (LIMMA) (10); and (VII) examination of the data structure by principal component analysis (PCA). Cross-checking was applied to identify DEGs, and then the Benjamini-Hochberg method was used to adjust the statistical P values of the false discovery rate (FDR) to calculate the expression fold change (FC) (a log2FC >1.0 and a corrected P<0.05 represented DEGs) (10). All of the data were obtained from the GEO database, and a research ethics application was not needed for this study.
Gene Ontology (GO) and pathway enrichment analysis of gene sets
The GO of the AMed- and AAdv-associated DEGs was obtained based on analysis using the clusterProfiler algorithm (11). The results for co-DEG-related GO and Kyoto Encyclopedia of Genes and Genomes (KEGG)-enriched pathways were obtained based on an analysis of the MetaScape gene annotation and retrieval platform (http://metascape.org/gp/index.html) (12).
Analysis of immune infiltration
CIBERSORT (https://cibersort.stanford.edu/) is an immune cell subtype infiltration calculation algorithm that was developed based on linear support vector regression (13). Users can comprehensively estimate the infiltration level of each cell subtype from chip expression profile and RNA-seq expression data. The parameters applied in this study were as follows: (I) gene expression values corrected by the RMA algorithm; (II) 1,000 deconvolutions (Perm); and (III) P<0.05 for differential subtypes.
Association analysis between core genes and differentially infiltrating immune cell subtypes
In addition, by constructing the co-DEG-related KEGG pathway network, pathway-rich genes of interest were selected for protein-protein interaction (PPI) network analysis. The node correlation degree in the network was calculated using the STRING database (https://string-db.org/) (14) and CytoScape software (15) to identify candidate regulatory factors. To further clarify the associations between core genes and immune genes, we performed a Pearson correlation clustering analysis of candidate genes and differential cell subtype infiltration values.
Results
Data acquisition and pretreatment
The results of the difference analysis suggested that (I) in the AAdv tissues, compared with the nondilated group, a total of 1,190 differential mRNAs (3 downregulated and 1,187 upregulated) were present in the dilated group; and (II) in the AMed tissues, 173 DEGs were present between the dilated group and nondilated group (7 downregulated and 166 upregulated). The distributions of DEGs in AMed and AAdv tissue samples are shown in Figure 1A and http://cdn.amegroups.cn/static/application/d1026d3979cb63f6749fde5efbe1d50d/atm.2020.03.05-1.pdf.
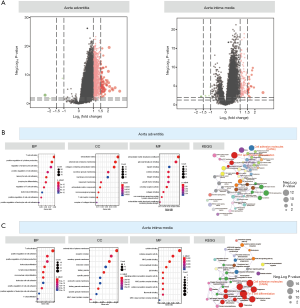
Functional enrichment analysis of DEGs
In the enrichment analysis, we found that GO:0042110~T cell activation (P=1.24E-19, n=75), GO:0002694~regulation of leukocyte activation (P=1.25E-18, n=78), and GO:0001819~positive regulation of cytokine production (P=1.38E-16, n=70) were closely related to the biological process (BP) of DEGs associated with AAdv dilation, whereas GO:0031012~extracellular matrix (P=9.49E-11, n=60), GO:0009897~external side of the plasma membrane (P=2.05E-10, n=35), and GO:0062023~collagen-containing extracellular matrix (P=3.32E-10, n=53) were closely related to the cellular component (CC) of DEGs associated with AAdv dilation. GO:0005201~extracellular matrix structural constituent (P=1.60E-10, n=32), GO:0005539~glycosaminoglycan binding (P=4.68E-05, n=26), and GO:0008201~heparin binding (P=5.88E-04, n=19) were closely related to the molecular function (MF) of DEGs associated with AAdv dilation (Figure 1B and http://cdn.amegroups.cn/static/application/5a2217b4614bc4137ef3fa5e5d5d32d7/atm.2020.03.05-2.pdf). Similarly, GO:0042110~T cell activation (P=6.61E-17, n=27), GO:0001819~positive regulation of cytokine production (P=7.35E-15, n=25), and GO:0045785~positive regulation of cell adhesion (P=3.03E-14, n=23) were closely related to the BP of DEGs associated with AMed dilation, while GO:0009897~external side of the plasma membrane (P=4.76E-11, n=15), GO:0030667~secretory granule membrane (P=1.01E-08, n=15), and GO:0043235~receptor complex (P=7.26E-08, n=14) were closely related to the CC of DEGs associated with AMed dilation. GO:0042287~major histocompatibility (MHC) protein binding (P=5.78E-07, n=6), GO:0019955~cytokine binding (P=1.55E-06, n=8), and GO:0004896~cytokine receptor activity (P=4.12E-05, n=6) were closely related to the MF of DEGs associated with AMed dilation (Figure 1C and http://cdn.amegroups.cn/static/application/5a2217b4614bc4137ef3fa5e5d5d32d7/atm.2020.03.05-2.pdf).
Analysis of the MetaScape database showed that AAdv-associated DEGs were mainly associated with type 1 T helper (Th1) and Th2 cell differentiation (enrichment score =5.31, P=2.51E-11, n=25), the PI3K-Akt signaling pathway (enrichment score =2.77, P=4.00E-10, n=46), the tumor necrosis factor (TNF) signaling pathway (enrichment score =4.0, P=4.62E-08, n=21), and other pathways (Figure 1B and http://cdn.amegroups.cn/static/application/f7d732aa65558464c10f0cf7a8a8a686/atm.2020.03.05-3.pdf). AMed-associated DEGs were mainly associated with Th17 cell differentiation (enrichment score =22.50, P=1.80E-16, n=15), cell adhesion molecules (CAMs) (enrichment score =14.14, P=7.23E-15, n=17), leishmaniasis (enrichment score =23.92, P=9.49E-14, n=12), and other pathways (Figure 1C and http://cdn.amegroups.cn/static/application/f7d732aa65558464c10f0cf7a8a8a686/atm.2020.03.05-3.pdf).
Identification and functional enrichment of co-expressed DEGs in the dilation process of AMed and AAdv tissues
By evaluating the intersection set (Figure 2), 107 DEGs shared by both AMed and AAdv tissues were obtained (Figure 2A). PCA showed that co-DEGs could significantly distinguish whether AAdv and AMed tissues contained dilated blood vessels (Figure 2C). Based on cluster analysis, the 107 DEGs were divided into five categories. Functional enrichment analysis showed that GO:0030334~regulation of cell migration, GO:0045785~positive regulation of cell adhesion, GO:0002322~B cell proliferation involved in the immune response, GO:0045321~leukocyte activation, and GO:0002253~activation of the immune response were closely related to the functional enrichment of each gene set (Figure 2B and http://cdn.amegroups.cn/static/application/c039b80ae29ac404d82338a29dc47044/atm.2020.03.05-4.pdf).
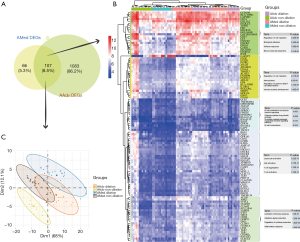
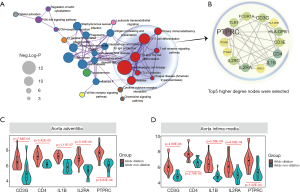
Core gene identification
Based on the KEGG pathway enrichment network, we found that the co-DEGs of AAdv and AMed tissues were closely associated with T cell-associated immune pathways (Figure 3A). We further constructed a PPI network for genes enriched in T cell-associated immune pathways and found that protein tyrosine phosphatase receptor type C (PTPRC) (degree =11), interleukin-1B (IL1B) (degree =7), CD4 (degree =7), CD3G (degree =7), and IL-2 receptor alpha chain (IL2RA) (degree =11) were closely related to the progression of aortic dilation (Figure 3B). In addition, compared with the nondilated group, these 5 core genes were highly expressed in both the AAdv tissues and the AMed tissues in the dilated group (all P<0.05; Figure 3C,D).
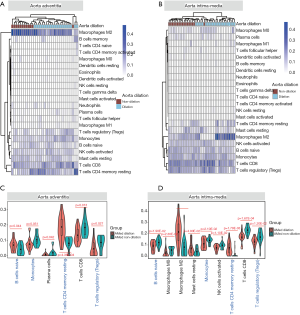
Analysis of immune infiltration
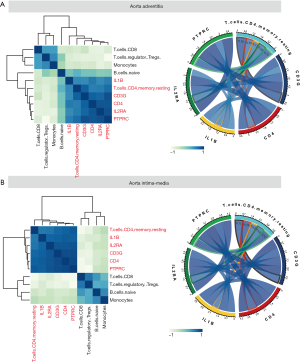
The overall immune infiltration profiles of AAdv and AMed vascular tissues are shown in Figure 4A,B. Resting memory CD4 T cells (AAdv P=4.36E-04, AMed P=1.79E-03), regulatory T cells (Tregs) (AAdv P=0.027, AMed P=1.20E-02), naïve B cells (AAdv P=0.044, AMed P=1.90E-02), and monocytes (AAdv P=0.051, AMed P=2.10E-02) demonstrated significant differential infiltration in both dilated and nondilated AAdv and AMed tissues (Figure 4C,D, http://cdn.amegroups.cn/static/application/1466096de4b886deaaabe22d4e91095d/atm.2020.03.05-5.pdf).
Through correlation analysis, we also found that the core regulatory genes (including PTFRC, IL1B, CD4, CD3G, and IL2RA) had strong correlations with the degree of infiltration of resting memory CD4 T cells (AAdv: PTPRC coefficient =0.71, IL1B coefficient =0.52, CD4 coefficient =0.59, CD3G coefficient =0.78, IL2RA coefficient =0.60; AMed: PTPRC coefficient =0.82, IL1B coefficient =0.62, CD4 coefficient =0.78, CD3G coefficient =0.73, IL2RA coefficient =0.64) in AAdv and AMed tissues (Figure 5A,B, http://cdn.amegroups.cn/static/application/559e390a40e3d65f1f41288170134c84/atm.2020.03.05-6.pdf).
Discussion
The association between the pathogenesis of TAAs and the immune inflammatory response has always been a popular research topic. Our study found that both AAdv and AMed tissues were closely associated with T cell activation during the process of vascular dilation. By further building a network of disease mechanisms, our results also suggested that PPTRC, IL1B, CD4, CD3G, and IL2RA may be the core regulatory genes of vascular dilation; these genes were closely related to the degree of infiltration of resting memory CD4 T cells in AAdv and AMed vascular tissues, indicating that these genes may be important regulatory mediators in TAA pathogenesis.
An increasing number of researchers believe that TAAs are immune inflammatory diseases, and the risk of disease increases with increasing age (16). Compared with other aneurysms, the pathological changes in the outer adventitia and media of the aortic wall are more closely associated with the immune inflammatory response, especially those in TAV-associated TAA vascular tissues (16,17). Among these changes, T cell activation is the most important molecular mechanism. Itani et al. found that angiotensin II can promote the infiltration of leukocytes [CD45 (+)], memory T cells [CD3 (+)/CD45 Ro (+)) and T lymphocytes (CD3 (+) and CD4 (+)] in thoracic aortic tissues, increase activated CD4+ and CD8+ T cells in the circulation, and increase the production of IL17a and IFN-γ, suggesting that functional activation of T cells and their subpopulations is associated with hypertension-induced vascular remodeling and dilation (18). Similarly, Ju et al. found that the cytokine IL6 can induce Th17 lymphocytes to aggregate in dilated vascular tissues through the transcription-3 signaling pathway in an angiotensin II perfusion-induced vascular dissection model. At the same time, these lymphocytes promoted macrophage recruitment and mediated the development of vascular dissection through the transcription-3 signaling pathway (19). Ye et al. found that CD4 T cell infiltration was closely related to aortic root inflammation and the degree of root dilation in TAA patients (20).
The comparison between dilated and nondilated vascular walls revealed that resting memory CD4+ T cells were significantly infiltrated in the dilated AAdv and AMed tissues, especially the AAdv tissues. Crotty believes that memory CD4 T cells have a degree of plasticity and can differentiate into other subtypes of T cells; however, no experimental data have confirmed the plasticity or differentiation ability of resting memory CD4 T cells (21). McKinstry et al. suggested that memory CD4 T cells can not only differentiate into subcells such as secretory T cells (Th) but also secrete a large amount of cytokines to recruit immune cells and enhance the immune response, facilitating the immune response of CD8− T cells and B cells (22). Sbrana et al. found that the degree of CD4+ T lymphocyte infiltration was increased and the production of IFN-γ, IL-17a, and IL-21 was increased in the vascular tissues of patients with ascending aortic dilation (23). Jones et al. found that compared with other immune cells, effector memory and central memory CD4+ T cells showed higher levels of glycolysis and oxidative phosphorylation and a higher metabolic capacity, and regulation of this metabolic capacity and cell recruitment were closely related to early activation of naïve CD4+ T cells (24).
In addition, our study suggested that IL1B, CD3G, CD4, IL2RA, and PTPRC may be the core regulatory genes of disease progression in TAV-associated TAAs and that these genes are positively correlated with the degree of infiltration of resting memory CD4 T cells. Cochain et al. performed single-cell sequencing on mouse aortic arch CD45+ macrophages in a low-fat diet group and a high-fat diet group and found that IL1B was closely associated with the inflammatory status of aortic arch endothelial cells (25). Yang et al. found that miR-30c could participate in the process of vascular dilation of abdominal aortic aneurysms (AAAs) by targeting IL1B, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta (PIK3CD), and Ras-related C3 botulinum toxin substrate 2 (RAC2) (26). CD3G is an important regulatory factor during the process of T cell development and differentiation (27) and is highly expressed in the torn vascular tissues of patients with acute aortic dissection (D13). Goudy et al. showed that IL2RA mutations can significantly affect the function of regulatory and effector T cells, which are associated with lymphocyte proliferation and T cell activation and are the key mediators of immune function and homeostasis in the body (28). Similarly, a study by Belot et al. also found that methylation of the IL2RA promoter region can affect IL2RA expression and T cell activation (29). PTPRC (CD45) is an important marker of macrophage and leukocyte activation. Gallo et al. found that high CD45 expression in TAA patients was positively correlated with increased monocyte infiltration in the vascular walls and increased IFN-γ, IFN-inducible protein 10, and IFN-induced T cell α chemokine levels in the circulation (30).
Conclusions
In summary, our study found that during the process of AAdν and AMed vascular dilation in TAV-associated TAAs, PPTRC, IL1B, CD4, CD3G, and IL2RA were involved in the regulation of resting memory CD4 T cell tissue infiltration, which was closely related to the process of vascular dilation. Most of those candidate regulators were verified in previous studies. However, several limitations remain. First, although the correlations among these candidate markers and immunocyte infiltration in TAV-associated TAAs were identified, further experimental evidence concerning the mechanism is still needed. Second, the mechanism of TAV-associated vascular dilation is complicated; thus, immunocyte infiltration may be important but not essential.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Guangdong Province, China (grant number 2016A030313792), the Medical Science Research Foundation of Guangdong Province, China (grant number 2016115114137325), the Chinese Medicine Research Foundation of Guangdong Province, China (grant number 20161003), and the National Natural Science Foundation of China (NSFC) (grant numbers 81372114 and 81900285).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All of the microarray data were obtained from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database, and a research ethics application was not needed for this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McClure RS, Brogly SB, Lajkosz K, et al. Epidemiology and management of thoracic aortic dissections and thoracic aortic aneurysms in Ontario, Canada: A population-based study. J Thorac Cardiovasc Surg 2018;155:2254-64.e4. [Crossref] [PubMed]
- Olsson C, Thelin S, Stahle E, et al. Thoracic aortic aneurysm and dissection: increasing prevalence and improved outcomes reported in a nationwide population-based study of more than 14,000 cases from 1987 to 2002. Circulation 2006;114:2611-8. [Crossref] [PubMed]
- Olsson C, Eriksson N, Stahle E, et al. Surgical and long-term mortality in 2634 consecutive patients operated on the proximal thoracic aorta. Eur J Cardiothorac Surg 2007;31:963-9; discussion 969. [Crossref] [PubMed]
- Sun X, Zhang H, Zhu K, et al. Patient-specific three-dimensional printing for Kommerell's diverticulum. Int J Cardiol 2018;255:184-7. [Crossref] [PubMed]
- Kim JH, Na CY, Choi SY, et al. Integration of gene-expression profiles and pathway analysis in ascending thoracic aortic aneurysms. Ann Vasc Surg 2010;24:538-49. [Crossref] [PubMed]
- Tang PC, Yakimov AO, Teesdale MA, et al. Transmural inflammation by interferon-gamma-producing T cells correlates with outward vascular remodeling and intimal expansion of ascending thoracic aortic aneurysms. Faseb j 2005;19:1528-30. [Crossref] [PubMed]
- Sprague AH, Khalil RA. Inflammatory cytokines in vascular dysfunction and vascular disease. Biochem Pharmacol 2009;78:539-52. [Crossref] [PubMed]
- Folkersen L, Wagsater D, Paloschi V, et al. Unraveling divergent gene expression profiles in bicuspid and tricuspid aortic valve patients with thoracic aortic dilatation: the ASAP study. Mol Med 2011;17:1365-73. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59.
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Pisano C, Balistreri CR, Ricasoli A, et al. Cardiovascular Disease in Ageing: An Overview on Thoracic Aortic Aneurysm as an Emerging Inflammatory Disease. Mediators Inflamm 2017;2017:1274034.
- Balistreri CR, Buffa S, Allegra A, et al. A Typical Immune T/B Subset Profile Characterizes Bicuspid Aortic Valve: In an Old Status? Oxid Med Cell Longev 2018;2018:5879281.
- Itani HA, McMaster WG Jr, Saleh MA, et al. Activation of Human T Cells in Hypertension: Studies of Humanized Mice and Hypertensive Humans. Hypertension 2016;68:123-32. [Crossref] [PubMed]
- Ju X, Ijaz T, Sun H, et al. Interleukin-6-signal transducer and activator of transcription-3 signaling mediates aortic dissections induced by angiotensin II via the T-helper lymphocyte 17-interleukin 17 axis in C57BL/6 mice. Arterioscler Thromb Vasc Biol 2013;33:1612-21. [Crossref] [PubMed]
- Ye P, Chen W, Wu J, et al. GM-CSF contributes to aortic aneurysms resulting from SMAD3 deficiency. J Clin Invest 2013;123:2317-31. [Crossref] [PubMed]
- Crotty S. Do Memory CD4 T Cells Keep Their Cell-Type Programming: Plasticity versus Fate Commitment? Complexities of Interpretation due to the Heterogeneity of Memory CD4 T Cells, Including T Follicular Helper Cells. Cold Spring Harb Perspect Biol 2018;10. [PubMed]
- McKinstry KK, Strutt TM, Swain SL. The potential of CD4 T-cell memory. Immunology 2010;130:1-9. [Crossref] [PubMed]
- Sbrana S, Tiwari KK, Bevilacqua S, et al. Relationships Between Phenotype and Function of Blood CD4+ T-Cells and Ascending Thoracic Aortic Aneurysm: an Experimental Study. Braz J Cardiovasc Surg 2019;34:8-16. [Crossref] [PubMed]
- Jones N, Vincent EE, Cronin JG, et al. Akt and STAT5 mediate naive human CD4+ T-cell early metabolic response to TCR stimulation. Nat Commun 2019;10:2042. [Crossref] [PubMed]
- Cochain C, Vafadarnejad E, Arampatzi P, et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res 2018;122:1661-74. [Crossref] [PubMed]
- Yang P, Cai Z, Wu K, et al. Identification of key microRNAs and genes associated with abdominal aortic aneurysm based on the gene expression profile. Exp Physiol 2020;105:160-73. [Crossref] [PubMed]
- Flanagan BF, Wotton D, Tuck-Wah S, et al. DNase hypersensitivity and methylation of the human CD3G and D genes during T-cell development. Immunogenetics 1990;31:13-20. [Crossref] [PubMed]
- Goudy K, Aydin D, Barzaghi F, et al. Human IL2RA null mutation mediates immunodeficiency with lymphoproliferation and autoimmunity. Clin Immunol 2013;146:248-61. [Crossref] [PubMed]
- Belot MP, Castell AL, Le Fur S, et al. Dynamic demethylation of the IL2RA promoter during in vitro CD4+ T cell activation in association with IL2RA expression. Epigenetics 2018;13:459-72. [Crossref] [PubMed]
- Gallo A, Saad A, Ali R, et al. Circulating interferon-gamma-inducible Cys-X-Cys chemokine receptor 3 ligands are elevated in humans with aortic aneurysms and Cys-X-Cys chemokine receptor 3 is necessary for aneurysm formation in mice. J Thorac Cardiovasc Surg 2012;143:704-10. [Crossref] [PubMed]


