Scientometric analysis of mTOR signaling pathway in liver disease
Introduction
The mammalian target of rapamycin (mTOR) signaling pathway senses and integrates a variety of environmental cues to regulate organismal growth and homeostasis. It has been recognized as playing a significant role in different metabolic diseases such as obesity(1), non-alcoholic fatty liver disease (NAFLD) (2-4), insulin resistance (5), neurological diseases (6-8), genetic disorders (9) and type 2 diabetes (10,11). Furthermore, it is an efficient regulator of cell growth and metabolism. In the liver, the mTOR pathway is vital for homeostasis, metabolism, transplantation, and regeneration (12-15). In addition, it is related to several pathological conditions such as cirrhosis (16), NAFLD (11), and hepatocellular carcinoma (HCC) (17).
Bibliometric is a new method for summarizing progress in a research field and further identifying hot spots or rising patterns in the research area by creating info graphics. Several biometric programs have been used to focus on the zone of scientometrics or social science (18-20), including CiteSpace (21), CitNetExplorer, VOSviewer (22), and HistCite (23). Many researchers have made similar endeavors within the field of biomedicine. Sugimoto et al. investigated factors affecting sex-related reporting in medical research (24); Shen et al. detected the interdisciplinary nature and topic hotspots of robotics in surgery (25). Liao et al. critically analyzed the most cited articles in coronary heart disease between 1970 and 2015 (26). This study aims to use bibliometric methods so as to analyze articles on the mTOR signaling pathway in liver diseases. CiteSpace, VOSviewer and some other tools were exploited to identify the contribution of countries, institutions and authors, milestone manuscripts, research sub-themes, and the evolution of scientific ideas in the research field.
Methods
Data collection
We searched the Web of Science Core Collection (WoSCC) for relevant literatures. Publications were searched by using key words “mTOR” and “liver” and extracted from WoSCC on May 21, 2019. The titles, keywords, author information, abstracts, and references were downloaded in TXT format.
Analysis methods
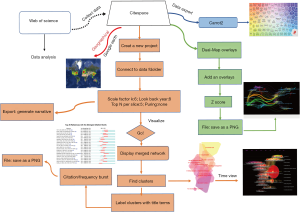
Distribution of countries/regions, institutions, journals, authors, and research areas of publications were analyzed based on the WOSCC database. CiteSpace software (Chaomei Chen, Drexel University, USA), carrort2 project (Dawid Weiss, Poznan University of Technology, Poland), and VOSviewer, a program operated by the Centre for Science and Technology Studies at Leiden University, were used to create data maps.
Co-citation analysis
A bibliometric program, CiteSpace (form 5.3.R4 64-bit), was initially utilized for recognizing co-cited authors/references, capturing keywords with strong citation bursts, and developing visualization maps of all the items mentioned above. The time trends of keywords were also analyzed by CiteSpace. A total of 2,232 articles were imported into CiteSpace. The individual network was derived from the 50 most highly cited papers in a period of 1 year (27). Scale factor k and the look back year were set as 6 to simplify the structure of the co-citation network. According to the definition in CiteSpace, each node is a cited paper, and the link between the two nodes represents the citation relationship. The size of each nod related to the citing count. After constructing the initial co-citation network, cluster analysis, citation burst detection, and the timeline or time zone view were applied. Then, dual-map overlay analysis was performed on publications. Association among journals, investigating collaboration networks involved with authors/institutes/countries, term clustering and quotation systems of profoundly cited authors, and journals were recognized by VOSviewer. Carrot2 was used for topical categories analysis.
Data analysis
The process of “clustering” of CiteSpace was used to recognize diverse research subtopics in all papers on “mTOR signaling pathway in liver diseases.” The name of each cluster was summarized from the titles of the references with the strategy of log-likelihood (LLR). Furthermore, changes and advances in scientific concepts over the years were evaluated. “Circles visualization” created by Carrot2 was utilized to extract keywords of significance and give relative impact to each keyword. Journal co-citation density analysis was performed by VOSviewer.
Results
Annual publications and trend
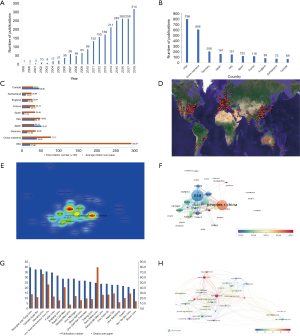
Data processing was displayed in a flowchart (Figure 1). According to the WOSCC database, the number of articles increased every year and annual publications grew from 17 in 2004 to 314 in 2018 (Figure 2). A total of 2,232 papers were published and these publications have been cited 51,795 times, and each paper was cited 23.21 times on average.
Contribution of countries and institutions
A total of 62 countries have participated in the publication of studies on mTOR in liver disease. Some articles were composed by researchers from different countries. The country location and density of the published articles were shown on a map created by CiteSpace and Google Earth (Figure 2D). The top 10 most productive countries produced 2,385 of the overall articles. The USA was the foremost productive country, with 796 articles published, followed by China (n=606), Germany (n=206), Japan (n=161), Italy (n=151), Spain (n=122), France (n=118), England (n=83), Switzerland (n=73), and Canada (n=69) (Figure 2B). The number of publications in the USA and China was much higher than in other countries (Figure 2B). A heat map created by VOSviewer showed that China and the USA were the countries with the highest article density (Figure 2E). The number of citations is the most commonly utilized tool to determine national efficiency in a specified investigation area. The top two countries with total citation numbers were USA (n=29,237) and China (n=7,777). The total citations of these two countries far exceed those of other countries (Figure 2C), while the average citation number per paper between Japan and Canada is higher than other nations (Figure 2C). The United States, Japan and some European countries, such as France, Germany, Italy and Spain, had the most publications concentrated in 2013 to 2014, while articles of from mainland China and India are mainly published after 2017 (Figure 2F).
A total of 2,239 institutions have published articles. VOSviewer software was used to analyze the citation network among these institutions. “Active institutions” were defined as one whose total citation number was higher than 100, and the total publication number was higher than 15. By this threshold, 36 out of the 2,239 institutions were selected. The average number of citations from of Harvard University, Pennsylvania State University, Penn University, California San Diego University, the University of Texas MD Anderson Cancer Center, and Baylor College of Medicine, was more than 50 times higher (Figure 2G). The top 10 organizations that published the most articles were shown in Table 1. Among these institutions, six belong to mainland China, three to the United States, and one in South Korea (Table 1). The number of Chinese organizations posting after 2016 has increased (Figure S1). Articles from the Penn State University were cited 80.20 times on average, which was much higher than others (Figure 2G). This was due to two profoundly cited researchers, Jefferson, Ls; Kimball, Sr; who recorded the College of Milan as their affiliation in numerous papers.

Full table
Article distribution among journals
The 2,232 papers were distributed in 730 SCI-E recorded journals. VOSviewer software was applied to analyze the citation network among these journals, and the total publication number was set higher than 5 to produce visible maps for 100 journals (Figure 3). The top 22 most productive journals distributed 674 articles, accounting for one-third of the total publications. PLoS One, Hepatology, and Journal of Hepatology were the top three journals with the most elevated numbers of distributed papers. Fifteen of these magazines were established in the United States (Table 2). The top 5 magazines with the highest average citation rate were Proceedings of the National Academy of Sciences of the United States of America (9.58), Human Molecular Genetics (4.544), Cell Metabolism (22.415), Nature (43.07), and Cancer Prevention Research (3.866). Advanced examination uncovered that articles published in these magazines have been cited over 500 times. Since New England Journal of Medicine has only published one related article: sirolimus for angiomyolipoma (28) in tuberous sclerosis complex or lymphangioleiomyomatosis, even though the number of citations reached 737, the magazine has not been selected by the program. Journals centering on the same investigate field tend to be within the same cluster. There were four clusters on the map. Distinctive color implied diverse clusters, and the line between each journal meant co-cited with each other (Figure 3A,B), suggesting that mTOR was included within the pathophysiological process in these fields.
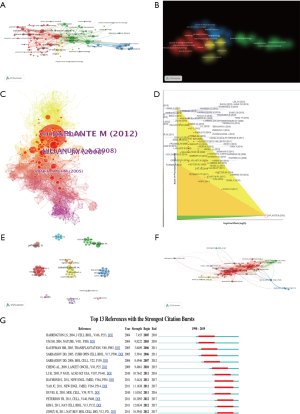

Full table
Overview of landmark manuscripts and authors
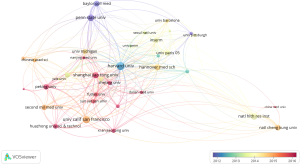
There were up to 13,701 authors among the 2,232 publications, and the average number of researchers per paper was 5.75. The most highly cited authors and collaboration between authors were revealed in our research. Co-authorship and citation network among authors were also analyzed by VOSviewer and CiteSpace. “Key researcher” is defined as one whose papers have received over 400 citations (global citation score >400). Some major research teams were disclosed. Each node on the graph represents an author, the size of the circle means the number of articles issued by the researcher, the lines between the circles shows the cooperation between the authors (Figure 3E,F). Each author’s entire citations were also revealed in our study. Point size indicates the number of citation (Figure 3E). Calvisi DF, Jefferson LS, Kimball SR, Wang C, Wu G, made extraordinary contributions to this research area.
The average number of articles per author is 13.4. Calvisi DF published 20 articles in this field. Jefferson LS, Sabatini DM, Llovet JM, Kimball SR, Bruix J each reached 1,000 citations. Sabatini DM and Laplante M collaborated on numerous papers, whereas their collaboration with Llovet JM, Jefferson LS was not as frequent. The 2,232 compositions about mTOR in liver disease also cited each other or a few other studies in their references. Combined with their 71,544 references, a streamlined co-citation network of the compositions was made by setting g-index to 6.5 articles according to the esteem of add up to citations were marked (Figure 3C). The article with the highest citation rate was also proceeded with in-depth analysis (Figure 3D). Additionally, CiteSpace software was used to analyze the co-citation network and received the top 13 strongest citation bursts in Figure 3G. The number of citations of these articles suddenly increased in a certain period after publication, indicating the rapid acceptance and dissemination in the field of the presented research results, making a significant contribution to the knowledge structure (29).
Key concepts and topics in the research area
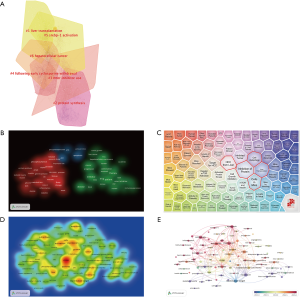
The articles indicated that within the co-citation network there were into 11 diverse clusters in Figure 4. The clusters are labeled by extricating terms from the titles of the cited publications. Clustering analysis of mTOR co-citation networks was based on VOSviewer (Figure 4B). To recognize key topics about mTOR in liver disease, the Carrot2 program was utilized to display key concepts. The top 6 keywords with the highest rate of occurrence are in the red circle, “inhibition of protein” “HCC from liver” “liver transplantation” “cell autophagy” “inhibition tumor”, and “cells in mice” (Figure 4C). Also, network visualization of keywords was based on VOSviewer (Figure 4E).
Timeline view and time zone view of mTOR in liver diseases co-citation network
Changes in research trends over a period of time can be shown with a timeline view: the publication date is shown at the top of the view; the most recent articles are placed closer to the right; nodes of a cluster share a horizontal line. The link between two nodes represents citing, while the number of nodes in a cluster reflects the significance of research area. More nodes equal more importance. The occurrence, popularity, and decline of research topics with the most representative articles were also shown in the view (Figure 5A). Furthermore, this picture shows the temporal characteristics of the research fields that clusters reflect. The development of cluster 2 (protein synthesis) occurred earliest, suggesting that early considerations of mTOR in liver research focused on protein synthesis. Cluster 1 (liver transplantation), cluster5 (srebp-1 activation), and cluster 8 (new treatment option) are current research hotspots (Figure 5A). The keywords timeline visualization also shows decreasing mtorc1 stability, hepatic stellate cell proliferation, liver transplantation, liver development; hepatocarcinoma cell line and non-alcoholic steatohepatitis related-cirrhosis (Figure 5B). Current research focuses on treatment and new development in the field of liver disease. The time zone cluster comprised of nodes that represent when articles were cited by the co-citation network (Figure S2). The advancement of the research area over time can be clearly illustrated through this visualization. It may be a critical period if there are more articles in a time zone. Fewer articles demonstrate periods that affect the result less. The central connection between time zones shows the extent to which information is left behind or information is acknowledged, thereby stimulating investigations and driving other revelations (Figure S2).
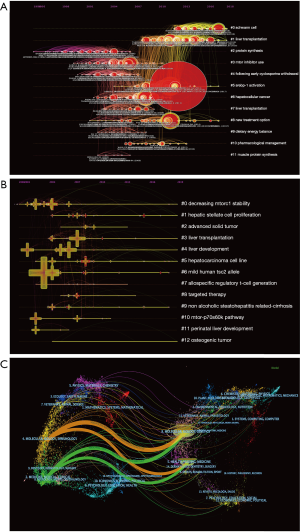
Dual-mapping analysis (Dual-Map Overlay) was designed to reveal overall scientific contributions. The interactions of more than 10,000 journals further divided into different areas were shown in Dual-map overlays. Publications and citations in this field can be described at the disciplinary level. CiteSpace’s dual-map overlay function was used to construct a citation dual-map. The left half side is the citing outline, the right half side is the cited outline, and the curve is the quotation association line from the outside to the proper side. This connection illustrates the flow of knowledge and connections of different research areas (Figure 5C).
Discussion
Different from the traditional reviews, systemic analysis of all literature, and related papers in this field using bibliometric analysis can be more visual. This method can show visual results, which is beneficial to researchers who are new to the field. It can help them grasp the integral tendency of the field investigated. It also can reveal current research hotspots and developing future trends, as well as milestone manuscripts. Moreover, the current bibliometric analysis software is simple and user-friendly.
According to the results, the number of publications on mTOR signaling in liver diseases is increasing annually. A total of 2,232 manuscripts with 51,795 references were searched using WoSCC. Among 62 countries, studies executed in the USA (n=796) and China (n=606) accounted for 58.8% of the publications. Among the top 10 most productive institutions, 5 institutions are in China (Table 1). China is the only developing country among the top 10 most productive countries/regions, with the total number of publications second only to the USA (Figure 2B). Though China’s total citation rate is much lower than that of the United States, the average citation rate is higher than the USA, which may be due to the excess self-citations in China (Figure 2C). Among the top 20 institutions, the citation rate of each paper in Chinese institutions is lower than that of all other institutions (Figure 2G,H). These results indicate that current research about the mTOR signaling pathway in liver diseases are mainly concentrated on European and American countries. The field of liver disease research in China is developing rapidly, and the gap between the United States and Japan is gradually narrowing. But the quality of articles still needs to be improved. The publication year of the articles in China, India, and Thailand is concentrated after 2016 (Figure 2F), showing that even more developing countries will be involved in further research in this area.
Journals focusing on the same investigative area tend to be in the same cluster. The top 100 journals were detected and divided into four clusters based on VOSviewer (Figure 3A,B). Using the clustering function of CiteSpace, the entire science field was divided into twelve subtopics. The largest six clusters are shown in Figure 4A, cluster 1 “liver transplantation”, cluster2 “protein synthesis”, cluster 3 “mTOR inhibitor”, cluster 4 “following early cyclosporine withdrawal”, cluster 5 “srebp-1 activation”, and cluster 6 “hepatocellular cancer”. This means that mTOR research in the field of liver disease is mainly concentrated in these six directions. Cluster 3 “mTOR inhibitor” and cluster 6 “hepatocellular cancer” suggest that mTOR plays a pivotal role in HCC and inhibition of mTOR may suppress malignancies. HCC is a leading cause of cancer-related death worldwide and it is expected to increase further in the future (30). The prognosis of this tumor has not improved significantly in the past few years, despite advances in classic therapies such as surgery, transplantation, radiofrequency use and transarterial embolization. Even though the molecular mechanisms that lead to the occurrence and development of HCC are largely unclear, mTOR inhibitors for treating HCC have recently become the focus of attention (31,32). In 40–50% of HCCs (33,34), the mTORC1 and mTORC2 pathways are up-regulated. Furthermore, several landmark papers were determined during co-citation analysis by CiteSpace (Figure 3C). These publications are included in the top 5 in the center, as well as the top 13 in burstiness. For example, Llovet JM discovered the pivotal role of mTOR signaling in HCC pathogenesis (35). Yao et al. also evaluated the everolimus an oral inhibitor of mTOR in a prospective randomized phase 3 study (36). The work by Zoncu et al. has the biggest citation burst: mTOR: from growth signal integration to cancer, diabetes and aging (37). Zhu et al. assessed the efficacy of everolimus in patients with advanced HCC and for whom sorafenib treatment failed (38). The work of Kauffman et al. was included among the top 5 in centrality and the top 13 in burstiness (Figure 3G). This paper elaborated mTOR inhibitors and can reduce incidence of de novo malignancies (39). These publications were included in cluster 1 “liver transplantation” and cluster 4 “following early cyclosporine withdrawal”. Included in cluster 2 “protein synthesis” and cluster 5 “srebp-1 activation” was Kim et al. depicting autophagy inhibited by mTOR (40). A paper published in 2016: mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2 was also included (3).
These landmark articles allow readers to have an authoritative understanding of the mechanism of mTOR in liver cancer and fatty liver lesions and their application in the treatment of liver diseases. The development process of the mTOR signaling pathway in liver disease over a certain period of time is clearly illustrated in Figure 5A,B. Current research hotspots are mainly focused on liver transplantation (41-43) and new treatment options. Srebp-1 activation is a research focus and research has revealed the mechanism that mTORC1 to lipogenesis via the SREBPs (44-46). The study of the mTOR pathway in liver disease might start with protein synthesis related to mTOR. Time trends of keywords were also analyzed (Figure 4E). Carrot2 and VOSviewer were used to identify keywords of significance (Figure 4B,C,D). Liver transplantation, hepatic stellate cell proliferation, NAFLD, therapy of HCC, cell growth, and autophagy were research hotspots and may be promising in the next few years.
In the dual-map overlay analysis (Figure 5C), we concluded that the mTOR pathway in liver disease is limited to the areas of medicine and biology, but has little relation with other disciplines. With the in-depth study of the clinical application of mTOR in liver disease, other disciplines will be involved. For example, Chiew et al. have described a 3D co-culture model by using fluorescent proteins and fluorescent resonance energy transfer (FRET) to detect the protein Akt/mTOR (47). Hence, the potential for cross-cooperation exists in many fields in the future. However, this study has two limitations. First, the co-citation analysis can only analyze papers searched from WOSCC. Not all related papers were included. Second, some clusters that were not directly related to the liver were detected, such as “Schwann cell” “muscle protein synthesis”. The reason might be liver kinase B1 and mTORC1 from two signaling systems that coordinate metabolic and cellular activity with changes in the environment (48).
Conclusions
Various scientific methods were applied to reveal scientific productivity, collaboration, and research hotspots for the mTOR signaling pathway in liver disease. Liver transplantation, hepatic stellate cell proliferation, NAFLD, therapy of HCC, cell growth and autophagy were research hotspots and may prove promising in the next few years and further studies are needed. Future research directions could be provided for new researcher who have just entered this research field.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Alemán JO, Eusebi LH, Ricciardiello L, et al. Mechanisms of obesity-induced gastrointestinal neoplasia. Gastroenterology 2014;146:357-73. [Crossref] [PubMed]
- Uno K, Yamada T, Ishigaki Y, et al. A hepatic amino acid/mTOR/S6K-dependent signalling pathway modulates systemic lipid metabolism via neuronal signals. Nat Commun 2015;6:7940. [Crossref] [PubMed]
- Kim K, Qiang L, Hayden MS, et al. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun 2016;7:10255. [Crossref] [PubMed]
- Han J, Li E, Chen L, et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 2015;524:243-6. [Crossref] [PubMed]
- Ardestani A, Lupse B, Kido Y, et al. mTORC1 Signaling: A Double-Edged Sword in Diabetic β Cells. Cell Metab 2018;27:314-31. [Crossref] [PubMed]
- Crino PB. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat Rev Neurol 2016;12:379-92. [Crossref] [PubMed]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron 2014;84:275-91. [Crossref] [PubMed]
- Bockaert J, Marin P. mTOR in Brain Physiology and Pathologies. Physiol Rev 2015;95:1157-87. [Crossref] [PubMed]
- Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab 2014;19:373-9. [Crossref] [PubMed]
- Blandino-Rosano M, Barbaresso R, Jimenez-Palomares M, et al. Loss of mTORC1 signalling impairs β-cell homeostasis and insulin processing. Nat Commun 2017;8:16014. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- He J, Chen J, Wei X, et al. Mammalian Target of Rapamycin Complex 1 Signaling Is Required for the Dedifferentiation From Biliary Cell to Bipotential Progenitor Cell in Zebrafish Liver Regeneration. Hepatology 2019;70:2092-106. [Crossref] [PubMed]
- Matter MS, Decaens T, Andersen JB, et al. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol 2014;60:855-65. [Crossref] [PubMed]
- Guri Y, Colombi M, Dazert E, et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell 2017;32:807-823.e12. [Crossref] [PubMed]
- Waidmann O, Hofmann WP, Zeuzem S, et al. mTOR inhibitors and sorafenib for recurrent heptocellular carcinoma after orthotopic liver transplantation. J Hepatol 2011;54:396-8. [Crossref] [PubMed]
- Neef M, Ledermann M, Saegesser H, et al. Low-dose oral rapamycin treatment reduces fibrogenesis, improves liver function, and prolongs survival in rats with established liver cirrhosis. J Hepatol 2006;45:786-96. [Crossref] [PubMed]
- Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010;29:4989-5005. [Crossref] [PubMed]
- Molléri JS, Petersen K, Mendes E. Towards understanding the relation between citations and research quality in software engineering studies. Scientometrics 2018;117:1453-78. [Crossref] [PubMed]
- Palacios-Callender M, Roberts SA. Scientific collaboration of Cuban researchers working in Europe: understanding relations between origin and destination countries. Scientometrics 2018;117:745-69. [Crossref] [PubMed]
- van den Besselaar P, Sandström U, Schiffbaenker H. Studying grant decision-making: a linguistic analysis of review reports. Scientometrics 2018;117:313-29. [Crossref] [PubMed]
- Synnestvedt MB, Chen C, Holmes JH. CiteSpace II: visualization and knowledge discovery in bibliographic databases. AMIA Annu Symp Proc 2005.724-728. [PubMed]
- van Eck NJ, Waltman L. Citation-based clustering of publications using CitNetExplorer and VOSviewer. Scientometrics 2017;111:1053-70. [Crossref] [PubMed]
- Gao Y, Ge L, Shi S, et al. Global trends and future prospects of e-waste research: a bibliometric analysis. Environ Sci Pollut Res Int 2019;26:17809-20. [Crossref] [PubMed]
- Sugimoto CR, Ahn YY, Smith E, et al. Factors affecting sex-related reporting in medical research: a cross-disciplinary bibliometric analysis. Lancet 2019;393:550-9. [Crossref] [PubMed]
- Shen L, Wang S, Dai W, et al. Detecting the Interdisciplinary Nature and Topic Hotspots of Robotics in Surgery: Social Network Analysis and Bibliometric Study. J Med Internet Res 2019;21:e12625. [Crossref] [PubMed]
- Liao J, Wang J, Liu Y, et al. The most cited articles in coronary heart disease: A bibliometric analysis between 1970 and 2015. Int J Cardiol 2016;222:1049-52. [Crossref] [PubMed]
- Chen C. Searching for intellectual turning points: progressive knowledge domain visualization. Proc Natl Acad Sci U S A 2004;101:5303-10. [Crossref] [PubMed]
- Bissler JJ, Mccormack FX, Young LR, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med 2008;358:140-51. [Crossref] [PubMed]
- Lu K, Yu S, Yu M, et al. Bibliometric Analysis of Tumor Immunotherapy Studies. Med Sci Monit 2018;24:3405-14. [Crossref] [PubMed]
- Giordano S, Columbano A. Met as a therapeutic target in HCC: facts and hopes. J Hepatol 2014;60:442-52. [Crossref] [PubMed]
- Jondal DE, Thompson SM, Butters KA, et al. Single-Dose Neoadjuvant AKT Pathway Inhibitor Reduces Growth of Hepatocellular Carcinoma after Laser Thermal Ablation in Small-Animal Model. Radiology 2019;292:752-9. [Crossref] [PubMed]
- Liu X, Hu J, Song X, et al. Combined Treatment with MEK and mTOR Inhibitors is Effective in In Vitro and In Vivo Models of Hepatocellular Carcinoma. Cancers (Basel) 2019. [Crossref] [PubMed]
- Sahin F, Kannangai R, Adegbola O, et al. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res 2004;10:8421-5. [Crossref] [PubMed]
- Sieghart W, Fuereder T, Schmid K, et al. Mammalian target of rapamycin pathway activity in hepatocellular carcinomas of patients undergoing liver transplantation. Transplantation 2007;83:425-32. [Crossref] [PubMed]
- Villanueva A, Chiang DY, Newell P, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 2008;135:1972-83, 1983.e1-11.
- Yao JC, Shah MH, Ito T, Bohas CL, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med 2011;364:514-23. [Crossref] [PubMed]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011;12:21-35. [Crossref] [PubMed]
- Zhu AX, Kudo M, Assenat E, et al. Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib: the EVOLVE-1 randomized clinical trial. JAMA 2014;312:57-67. [Crossref] [PubMed]
- Kauffman HM, Cherikh WS, Cheng Y, et al. Maintenance immunosuppression with target-of-rapamycin inhibitors is associated with a reduced incidence of de novo malignancies. Transplantation 2005;80:883. [Crossref] [PubMed]
- Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132-41. [Crossref] [PubMed]
- Adams DH, Sanchez-Fueyo A, Samuel D. From immunosuppression to tolerance. J Hepatol 2015;62:S170-85. [Crossref] [PubMed]
- Guerrero M, Ferrín G, Rodríguez-Perálvarez M, et al. mTOR Expression in Liver Transplant Candidates with Hepatocellular Carcinoma: Impact on Histological Features and Tumour Recurrence. Int J Mol Sci 2019. [Crossref] [PubMed]
- Dumortier J, Couchonnal E, Lacaille F, et al. mTOR inhibitors in pediatric liver transplant recipients. Clin Res Hepatol Gastroenterol 2019;43:403-9. [Crossref] [PubMed]
- Chen H, Shen F, Sherban A, et al. DEP domain-containing mTOR-interacting protein suppresses lipogenesis and ameliorates hepatic steatosis and acute-on-chronic liver injury in alcoholic liver disease. Hepatology 2018;68:496-514. [Crossref] [PubMed]
- Bakan I, Laplante M. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol 2012;23:226-34. [Crossref] [PubMed]
- Lewis CA, Griffiths B, Santos CR, et al. Regulation of the SREBP transcription factors by mTORC1. Biochem Soc Trans 2011;39:495-9. [Crossref] [PubMed]
- Chiew GGY, Wei N, Sultania S, et al. Bioengineered three-dimensional co-culture of cancer cells and endothelial cells: A model system for dual analysis of tumor growth and angiogenesis. Biotechnol Bioeng 2017;114:1865-77. [Crossref] [PubMed]
- Beirowski B. The LKB1-AMPK and mTORC1 Metabolic Signaling Networks in Schwann Cells Control Axon Integrity and Myelination: Assembling and upholding nerves by metabolic signaling in Schwann cells. Bioessays 2019;41:e1800075. [PubMed]