
Mitochondrial disorders
Introduction
Mitochondria are the quintessential innovator of survival in cellular evolution. Their survival as an organelle, utilizing an equally novel notion of endosymbiosis, speculates the timing of their origins around the same time as that of eukaryotes (1). It is believed that mitochondrial oxidative phosphorylation occurred due to mitochondrial genes with resultant huge increase in nuclear genome size, protein expression, rise to cellular complexity and function and evolutionary multicellular, multiorgan human being (2). Better understanding of mammalian mitochondrial genome and role of tRNA was followed by complete sequencing of the human mitochondrial genome (mtDNA) (3). Understanding its capabilities with gene coding, processing and expression and HeLA cells experiments further clarified dual control by mtDNA and nDNA in mitochondrial biogenesis (4). Subsequently, the discovery of a mtDNA mutation causing human disease gave insight into clinical variability and multi-systemic presentations (5). Typically organs with highest needs of energy such as brain, heart, eyes, muscles, GI are affected the most, leading to the initial clinical presentation and can present at any age. Adding to the complexity of these disorders is poor genotype-phenotype correlations: the same genotype can cause different phenotypes (i.e., m.3243A > G causes MELAS, MIDD, CPEO) and the same phenotype can be caused by many different genotypes (i.e., Leigh syndrome has >75 single gene etiologies). A new category of nuclear DNA disorders has emerged called the mitochondrial aminoacyl-tRNA synthetases (mtARSs); these enzymes are needed to attach amino acids with their corresponding tRNAs during mitochondrial translation. Pathogenic variants in these genes have led to severe phenotypes of primary mitochondrial disease including brain malformations, severe refractory fatal infantile lactic acidosis and other multi-systemic presentations. Collectively termed as “energy metabolism disorders”, the number of primary mitochondrial disorders grows with our improved understanding of the complex molecular role of two genomes, their communication and relationship with each other and in cell signaling, and ongoing discovery of new pathogenic variants. Inheritance of mitochondrial disorders can be nuclear: autosomal-dominant (AD), autosomal recessive (AR), X-linked or mtDNA (mitochondrial). Variable presentations within the same family can be due to heteroplasmy, best understood as percentage of mutation load of maternally inherited mtDNA. Each oocyte, and thereafter cellular lineages, produced by a carrier female will have a different mutation load after reduction or segregation by a phenomena called “bottleneck” (6). The number of mutant mtDNA to cause mitochondrial dysfunction in affected organ and its clinical presentation is known as the threshold effect. Clinical presentation can change within the same individual as they age from infancy to adulthood from a shift in the number of mutant mtDNA in daughter cells due to mitotic segregation (7). Currently our knowledge of mtDNA and nDNA mutations has widened our understanding of mitochondrial human diseases and current known variant information is compiled comprehensively in MITOMAP—a human mitochondrial genome database (8). This review will further elaborate phenotypes associated with mtDNA pathogenic variants and rearrangements and nDNA role in mitochondrial disorders, briefly touching on other secondary disorders that can occur in the mitochondria, making mitochondria a “Pandora’s box” of IEMs.
Primary energy metabolism defects
Energy production from glucose in the glycolytic pathway involves pyruvate metabolism and its role in continuing the biochemical reactions from cytosol into the mitochondria culminating in Krebs cycle. Defects in pyruvate metabolism predominantly present as primary lactic acidosis. Accumulating pyruvate usually converts into lactate and alanine and provide indirect evidence of such defects. Measurement of pyruvate levels is dependent on specimen integrity and timeliness of analysis and not a commonly available test. The ratio of lactate to pyruvate can help identify defects in pyruvate metabolism; which can be broadly categorized as pyruvate dehydrogenase complex (PDHC) defects and pyruvate carboxylase (PC) deficiency.
PDHC is a multienzyme unit in the mitochondrial matrix composed of 3 enzymes: E1 (pyruvate dehydrogenase-PDH), E2 (dihydrolipoamide acetyltransferase), E3 (dihydrolipoamide dehydrogenase). PDH E1 has four subunits, 2 alpha and 2 beta with central active E1 alpha, PDHA1 as its core with thiamine pyrophosphate (TPP) binding site essential for pyruvate decarboxylation.
PDHA1 deficiency has X-linked dominant inheritance and encoded by PDHA1 gene on chromosome Xp22.12, and PDHB deficiency encoded by PDHB gene on chromosome 3p14.3 can both present with fatal lactic acidemia to chronic central nervous system (CNS) presentations and structural anomalies often categorized as Leigh syndrome (9). High dose thiamine therapy showed improvement in neurological presentation and lactic acidemia in PDHA1 (10).
PDHE2 deficiency has AR inheritance and encoded by DLAT gene on chromosome 11q23.1, and presents with severe lactic acidemia and hyperammonemia. Lipoic acid, thiamine and ketogenic diet resulted in marked clinical improvement (11).
PDHE3 deficiency has AR inheritance and encoded by DLD gene on chromosome 7q31.1 and presents with deficiency branched chain alpha-keto acid dehydrogenase [similar to as seen in maple syrup urine disease (MSUD)], PDH complex and alpha-ketoglutarate dehydrogenase complex, with lactic acidosis, organic aciduria and neurologic deterioration (12).
PC deficiency has AR inheritance and encoded by PC gene on chromosome 11q13.2 and has three distinguished presentation: (I) simple group A or North American phenotype with lactic acidemia and intellectual disability; (II) severe group B or French form with neonatal congenital lactic acidosis, hyperammonemia, hyperketonemia and severe neurological symptoms with survival only to early infancy; (III) benign group C with mild episodic lactic acidosis with no neurodevelopmental or cognitive impairment (12).
Other energy metabolism disorders of Kreb’s cycle and mitochondrial transporter are well reviewed in various resources (13).
Respiratory chain or oxidative phosphorylation (OXPHOS) defects
The mitochondria plays a critical role in the generation of adenosine triphosphate (ATP) in a process known as oxidative phosphorylation (OXPHOS). Mitochondrial production of ATP utilizes energy released from the oxidation of various electron carriers (14). More specifically, OXPHOS relies on the transport of electrons generated from NADH that are sequentially transferred to molecular oxygen via enzymatic respiratory chain complexes known as (complexes I–V) embedded in the mitochondrial inner membrane. OXPHOS is initiated as complex I (NADH: coenzyme Q oxidoreductase) oxidizes NADH along with the reduction of coenzyme Q10 (CoQ) from its ubiquinone form to ubiquinol to thereby generate an electrochemical gradient across the inner mitochondrial membrane. Complex II (succinate-CoQ oxidoreductase) is then responsible for the oxidization of succinate and reduction of CoQ from its ubiquinone form to ubiquinol. Complex III (ubiquinol-cytochrome c oxidoreductase) catalyzes the reduction of cytochrome c by oxidation of ubiquinol and in the process, also generates an electrochemical gradient. Complex IV (cytochrome C oxidase) will then carry out the last enzymatic reaction of OXPHOS which involves the transfer of electrons (e−) to molecular oxygen which also results in the generation of an electrochemical gradient. Finally, complex V (ATP synthase) uses the transmembrane electrochemical proton gradient to catalyze the generation of ATP from ADP and phosphate. Shown in Figure 1 is the process of oxidative phosphorylation, whereby mitochondrial protein complexes I–V function to generate an electrochemical gradient via the transfer of electrons to ultimately generate ATP (15); with relevant genes being disguised and the distribution of mtDNA and nDNA encoded subunits of complex I–V (16). Table 1 provides an overview of complex I–V disorder associated genetic and clinical features. Mutations in both mtDNA and nDNA can have any pattern of inheritance ranging from de novo to maternal for mtDNA mutations to autosomal dominant, AR, X-linked for nDNA and with a wide neurocognitive and developmental presentations such as developmental regression, failure to thrive, seizures, spasticity, dystonia, ataxia, and nystagmus. In certain instances, genotype-phenotype prediction in many OXPHOS disorders has been linked to specific genes and mutations (17).
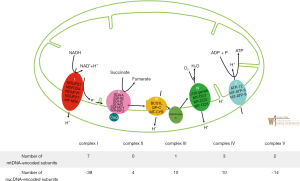
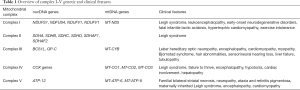
Full table
Complex I deficiency is most common with complex heterogeneity and prevalent clinical presentations such as fatal neonatal-onset lactic acidosis or encephalomyopathies, including Leigh syndrome, leukoencephalopathy, hypertrophic cardiomyopathy, and exercise intolerance.
Complex I is a multi-subunit structure consisting of 45 subunits. Among these subunits, 38 are encoded by nDNA and 7 are encoded by mtDNA. The mode of inheritance is dependent upon the causative gene mutation as mutations can occur in both mtDNA or in nDNA. Interestingly of significance, is that even though mtDNA is inherited maternally, pathogenic mtDNA mutations can often arise sporadically and can cause human disease (18). Non-invasive molecular analysis can help confirm diagnosis though invasive approaches include spectrophotometric assay of rotenone-sensitive complex I activity in biopsied skeletal muscle, cardiac or liver tissue.
No curative therapies exist but some patients, especially those with a myopathic presentation, can show a clinical response to supplementation with riboflavin (vitamin B2) (19).
Complex II deficiency typically presents with Leigh Syndrome, a progressive neurodegenerative disorder of infancy or childhood characterized by focal, necrotizing lesions of the basal ganglia, diencephalon, cerebellum or brainstem and clinical symptoms that include psychomotor delay or regression, muscular hypotonia, ataxia, respiratory insufficiency, and movement disorders (20).
Complex II is comprised of four succinate dehydrogenase (SDH) subunits, SDHA, SDHB, SDHC, and SDHD, which are all encoded by nuclear DNA with SDHB mutations being the most common. Complex II’s role is a critical component of both OXPHOS and the Krebs cycle.
Non-invasive molecular analysis can help confirm diagnosis though invasive approach includes histochemical analysis of muscle with positive staining of SDH. Measurement of complex II activity on fresh muscle or an immediately frozen sample remains the most reliable diagnostic confirmation (21).
No definitive treatment exists and the prognosis is poor. A clinical trial (NCT02352896), recently completed recruitment, to evaluate the safety and efficacy of a drug, EPI-743, on children ages 1 to 18 years old with Leigh syndrome (22).
Complex III deficiency is one of the least common of OXPHOS defects, mimicking glycogen storage diseases with hypoglycemia, lactic acidosis, elevated creatine kinase, and urine organic acid abnormalities (23). Complex III forms the central part of the respiratory chain, oxidizing coenzyme Q and reducing cytochrome c while pumping protons from the matrix to the intermembrane space. There are three disease-causing genes identified: the mitochondrial gene encoding cytochrome B (MT-CYB) and the two nuclear genes BCS1L and QP-C.
Mutations in MT-CYB can present with Leber hereditary optic neuropathy (LHON), encephalopathy, cardiomyopathy and myopathy. Mutations in the BCS1L gene can present with Bjornstad syndrome, hair abnormalities, sensorineural hearing loss, encephalopathy, liver failure, and tubulopathy. There is no cure and treatment remains largely symptomatic and does not alter the disease course.
Complex IV deficiency has a wide range of clinical presentations from failure to thrive, encephalopathy, hypotonia, Leigh syndrome, cardiac involvement, and hepatopathy soon after birth or early infancy with generally poor prognosis. Some of the metabolic findings include aminoaciduria, anemia, decreased cytochrome C oxidase staining in muscle fibers, glycosuria, hyperphosphaturia, hypercalcuria, and lactic acidosis.
Complex IV, also known as cytochrome oxidase, consists of thirteen subunits that catalyze the transfer of electrons from ferro-cytochrome c to molecular oxygen which is coupled to the transfer of protons across the inner mitochondrial membrane. There have been a variety of genes implicated ranging from mitochondrial COX genes, such as MTCO1, MTCO2 and MTCO3, to nuclear COX genes such as COX6B1 to COX assembly genes (24).
Complex V deficiency clinical presentation seems to be determined by associated mtDNA or nDNA mutations. Mutations in ATP6 can cause familial bilateral striatal necrosis (FBSN), neuropathy, ataxia, and retinitis pigmentosa (NARP), maternally inherited Leigh syndrome (MILS), and severe encephalopathy. Mutations in ATP8 can cause cardiomyopathy and neuropathy. Mutations in ATP12 can cause encephalomyopathy. Plasma citrulline can be low. Though there is impaired respiration, the limiting step involves phosphorylation rather than the respiratory chain. Complex V is the final complex, and also commonly referred to as ATP synthase as it uses the proton gradient to generate ATP. It consists of two functional domains, F1, situated in the mitochondrial matrix, and Fo, located in the inner mitochondrial membrane (25). There is no cure, and treatment is largely symptomatic without change in disease progression.
Mitochondrial disorders due to mtDNA defects
mtDNA is a circular genome with multiple copies within each mitochondrion, containing 16,569 base pairs and 37 genes, with a great deal of information that can be linked to various human diseases and aging, mainly resulting from a maternal inheritance pattern. Heteroplasmy and homoplasmy play an important role in the development of mtDNA related disorders. Each mitochondrion has multiple copies of mtDNA. If a cell has a uniform collection of mitochondria, whether normal or mutant, then it displays homoplasmy. Heteroplasmy refers to the concept in which normal mitochondria and mutant mitochondria may be mixed at various proportions. The proportion or load of mutant mtDNA that leads to diseases presentation is known as threshold. A mother may have mild or no symptoms with her defective mitochondria, but her children may have mild to severe disease presentation due to heteroplasmy i.e., varying proportion of mutant mtDNA. Replicative segregation in which a small number of the mother’s mitochondria are randomly selected during oogenesis determines the varying number of mutant mtDNA in oocytes. At cell division, mtDNA replicates and sorts randomly among mitochondria such that mitochondria sort randomly among daughter cells. Each daughter cell may subsequently receive different proportions of mitochondria carrying normal and mutant mtDNA. Known pathogenic variants include point mutations, deletions, and mtDNA depletion (reduction in mtDNA copy number). Mitochondrial DNA is also prone to somatic mutations, which occur over the span of a person’s lifetime. Though these changes are not passed maternally to future generations, they have been linked to various cancers due to the increase in reactive oxygen species and limited capability to self-repair. Some of the known mtDNA defects and associated human disease are reviewed below.
Single, large scale mtDNA deletion syndromes (SLSMDS) include three clinical presentations, but may also include multi-systemic symptoms not fitting one of these classic syndromes: Pearson syndrome, Kearns-Sayre syndrome (KSS) and progressive external ophthalmoplegia (PEO).
Pearson syndrome, also commonly known as the Pearson marrow-pancreas syndrome, is a contiguous gene deletion/duplication syndrome with refractory sideroblastic anemia and exocrine pancreatic dysfunction in form of steatorrhea and can be fatal in infancy. It is believed that infants who survive later have childhood presentations of KSS.
KSS is a multi-systemic disorder which presents by age 20 with PEO, salt and pepper retinopathy, ataxia, cardiac conduction problems requiring prophylactic pacemaker, sensorineural hearing loss, myopathy, diabetes, hypoparathyroidism, renal tubular acidosis, growth problems and hearing loss.
PEO can present at any age without major KSS features, including ptosis (droopy eyelids) and generally considered benign.
Elevated lactate in body fluids can be seen at rest. Diagnostic confirmation includes mtDNA molecular analysis with deletion up to 10 kb, typically around 5 kb. Symptomatic treatment addressing various presentations, cardiac and endocrine surveillance, and use of antioxidants can help. Avoidance of mitochondrial toxic medications like valproic acid, aminoglycosides, chloramphenicol, linezolid, nucleoside reverse transcriptase inhibitors is helpful (26,27).
mtDNA point mutations associated with human disease are usually familial and due to pathogenic missense mutations in the mtDNA polypeptide genes as well as tRNA or rRNA genes. Many of them present with neuromuscular degenerative characteristics, known for their clinical presentations and constellation of symptoms resulted in their nomenclature and associated abbreviations as their diagnostic identity e.g., LHON, mitochondrial encephalomyopathy, lactic acidosis, and stroke like episodes (MELAS), myoclonic epilepsy with ragged red fibers (MERRF), NARP and Leigh syndrome (subacute necrotizing encephalomyopathy).
MITOMAP as noted above has the current comprehensive information on all associated variants.
LHON is associated with mutations in the mitochondrial genes MT-ND1, MT-ND4, MT-ND4L, and MT-ND6. Specifically, there are point mutations at nucleotide positions m.3460, m.11778, and m.14484, which lead to mutant proteins impairing oxidative phosphorylation in complexes I, III, and IV and therefore the generation of ATP and increased ROS.
Symptoms primarily affect males and include early childhood blurring & central vision loss affecting one eye at a time in the acute phase followed by optic nerve atrophy leading to permanent visual loss. Neurological symptoms (LHON-plus) include myopathy, movement disorders, peripheral neuropathy and tremor. A multiple sclerosis-like illness is often seen in women (28). These mutations may also cause deafness and dystonia and/or Leigh syndrome.
MELAS is associated with mutations in MT-ND1, MD-ND5, MT-TH, MT-TV, and MT-TL1. Though majority (>80%) of all MELAS cases are due to MT-TL1 point mutations m.3243A > G, 7–15% are due to m.3271T > C (29).
Common childhood initial presentations include growth delay, short stature, generalized clonic tonic seizure, migraine headaches, recurrent vomiting. Subsequent learning problems, transient hemiparesis or cortical blindness result from stroke like episodes associated with seizures, with gradual impairment in physical and cognitive abilities, hearing loss; progressive neurologic dysfunction and dementia are commonly seen by age 40. Elevated lactate is a biochemical hallmark and common illnesses can be trigger. Symptomatic treatment, with multidisciplinary surveillance with audiology, cardiology, neurology, endocrinology, integrated behavioral health, physical medicine and rehabilitation and nutrition with use of coenzyme Q10, arginine and avoidance of mito-toxic medications can help improve quality of life (30).
MERRF is associated with at least four mitochondrial gene mutations, including MT-TK, MT-TL1, MT-TH, and MT-TS1. The MT-TK m.8344A>G mutation is responsible for 80% of all cases, alters the tRNA molecule for lysine.
MERRF is characterized by myoclonus (muscle twitches) alongside generalized epilepsy, ataxia, and myopathy and cardiomyopathy with Wolff-Parkinson-White (WPW) syndrome, with hearing loss, short stature, optic atrophy, and pigmented retinopathy also seen. The common m.8344A>G pathogenic variant may also cause Leigh syndrome. Treatment and surveillance are similar to those in MELAS (31).
Leigh syndrome (subacute necrotizing encephalomyopathy) includes bilateral, symmetric necrotizing lesions with spongey changes and microcysts in the thalamus, brainstem, spinal cord, and basal ganglia, easily identified in brain imaging studies. The predominant associated mutations are noted in MT-ATP6 gene, which encodes the proteins necessary for complex V of oxidative phosphorylation and ATP production, but can include multiple mtDNA and nDNA genes. Symptoms often manifest in infancy and early childhood with failure to thrive, feeding problems as presentation, followed by developmental delay, myopathy and weakness, cardiomyopathy, hypotonia, movement disorder, and ultimately fatality from respiratory or cardiac failure. Atypical and non-neurologic features can include diabetes, anemia, and diarrhea (32).
NARP is characterized by a mutation in the MT-ATP6 gene mutations affecting ATP production, with m.8993T>G being most common, which is also seen with the Leigh syndrome phenotype, depending on threshold of heteroplasmy levels. Characteristic features include proximal neurogenic myopathy with sensory neuropathy, ataxia and pigmented retinopathy. Ataxia and learning difficulties may be initial symptoms with common illness associated deterioration and characteristic manifestations (32).
Mitochondrial disorders due to nDNA defects
It is important to distinguish between two genomes, namely, the nuclear genome (nDNA) and the circular mitochondrial genome (mtDNA). These two genetic components of the organelle work in concert to control the process of oxidative phosphorylation and the crucial generation of ATP—necessities for cell survival and human life. The nDNA genes specifically contribute to the expression of proteins including major electron transport chain subunits, oxidative phosphorylation assembly proteins and ancillary proteins, proteins governing mitochondrial DNA expression, and proteins involved in fusion, fission, and synthesis related to mitochondrial replication and maintenance (33). Figure 2 shows the defects in many steps of these processes representing significant group of severe disorders also known mitochondrial DNA depletion syndromes (MDS) and are reviewed below. The usual pathogenesis involves mutations in nDNA. Affected genes responsible for mtDNA maintenance result in depletion of the mtDNA, disrupting normal mtDNA function, resulting impairment in the electron transport chain and energy production (34). While the nucleoside salvage pathway, governed by nDNA genes, is especially important for the proper replication and synthesis of mtDNA (35,36). Rather than one particular manifestation, the mitochondrial DNA depletion syndromes represent a group of disorders caused by defects in the nDNA-based maintenance, replication, and synthesis of mitochondrial DNA with a plethora of manifestations in various organs and can present with devastating effects quite early on in life. Some are linked closely with certain phenotypes, while others are more general in the energy metabolism issues that they cause. Tables 2 and 3 show associated genetic, diagnostic and clinical features and are typically grouped four clinical categories: myopathic, encephalomyopathic, hepatocerebral, and neurogastrointestinal. The neurogastrointestinal form of MDS is generally referred to as MNGIE, discussed below.
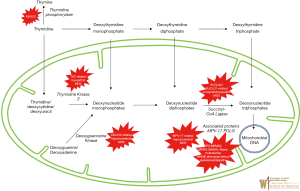
Full table
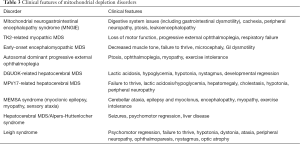
Full table
Mitochondrial neurogastrointestinal encephalomyopathy syndrome (MNGIE) is an AR disorder involving mtDNA deoxynucleotide/dNTP pools due to mutations in TYMP gene, located on chromosome 22q13.33, affecting the normal catabolism of thymidine to thymine and 2-deoxy d-ribose 1-phosphate (by the enzyme thymidine phosphorylase) and altering proper communication between the dual genomes for mtDNA synthesis. It typically presents with severe gastrointestinal dysmotility, dysphagia, early satiety, and abdominal pain, cachexia, and can include ptosis, ophthalmoplegia, peripheral neuropathy, and leukoencephalopathy; often prior to 20 years age and progresses to death by early/middle adulthood. Diagnostic confirmation can include thymidine phosphorylase enzyme activity or plasma levels of thymidine and deoxyuridine with molecular analysis of mitochondrial nuclear genome. Management is mainly supportive for gastrointestinal symptoms and nutrition with antibiotic use for intestinal bacterial overgrowth (37); a clinical trial using hematopoietic bone marrow transplant is available (see clinicaltrials.gov).
Myopathic MDS results from AR mutations in the thymidine kinase 2 (TK2) gene, located on chromosome 16q21 which plays a crucial part in pyrimidine nucleoside salvage. As a result, impaired mitochondrial DNA synthesis presents as myopathic features including hypotonia, muscle weakness, bulbar weakness, and an increase in serum creatine kinase. Early childhood presentation tends not to affect cognitive function. However, the myopathy progressively leads to respiratory failure and most often, death within a few years (38). A clinical trial using nucleoside replacement therapy is available (see clinicaltrials.gov).
Encephalomyopathic MDS results from mutations in several genes, including SUCLA1, SUCLG1, and RRM2B—genes associated with succinyl CoA ligase and subunits of ribonucleotide reductase that ultimately causes issues with mtDNA synthesis, with AR and variable inheritance. These diseases, specifically those of SUCLA2 and SUCLG1 origin, show myopathic involvement as well as issues with psychomotor delay, movement, sensorineural hearing complications, seizures, and liver dysfunction, among other manifestations. They tend to present in infancy with quite poor prognosis and with significant methylmalonic aciduria (39).
Hepatocerebral MDS is associated with mutations in DGUOK, MPV17, POLG, and C10orf2 (encoding the twinkle protein). These genes together are involved in various processes involved in maintenance of mtDNA synthesis and replication, although the exact function of MPV17 is still largely unknown. Clinical symptoms include hepatic dysfunction, along with other ocular, neurological, and atrophic manifestations depending on the particular gene mutation. It is important to mention that mutations in each of genes on their own have roles in many mitochondrial diseases, thus it is difficult to assign a particular disorder to each; it may be more suitable to describe the hepatocerebral category as a continuum of phenotypes. A reason for this may be due to the fact that genes such as POLG and C10orf2 are so crucial, that mutations can cause so many variable presentations. POLG in particular, is involved in a subunit of DNA polymerase gamma, known to be the only DNA polymerase involved in replication and repair of mtDNA. In regards to its role in hepatocerebral MDS, the POLG-associated manifestation may also be referred to as Alpers-Huttenlocher syndrome. This syndrome is known to be very severe, causing seizures, encephalopathy, visual changes, and hypotonia, in addition to hepatopathy. Valproic acid may provoke life-threatening fulminant hepatic failure. Individuals may present at age 2–4 years old with another peak in young adulthood, with variable life expectancy and symptoms (40).
Secondary disorders involving mitochondria
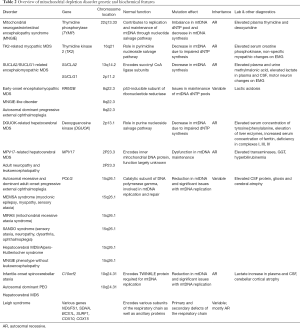
The remainder of the discussion will include disorders that are characterized by a deficiency in a mitochondrial-resident enzyme. While these enzymes are not encoded by mitochondrial DNA and are not directly involved in oxidative phosphorylation, their functions are nevertheless carried out within the mitochondria. As such, metabolic derangements caused by deficiencies in these enzymes have the ability to disrupt mitochondrial function in a number of ways. Given the various metabolic pathways that take their course through the mitochondria, these disorders vary greatly in both quality and severity. Deficiencies involve enzyme defect of the tricarboxylic acid (TCA) cycle, branched-chain amino acid (BCAA) breakdown, the urea cycle, ketone body (KB) metabolism, heme synthesis, and fatty acid (FA) metabolism are shown in Figure 3 and their genetic and clinical features noted in Table 4. Most TCA cycle defects are reviewed above. BCAA and FA metabolism disorders are discussed as separate articles in this issue. Rest of the secondary disorders involving mitochondria in the TCA cycle, urea cycle, KB metabolism, heme synthesis, is briefly reviewed below.

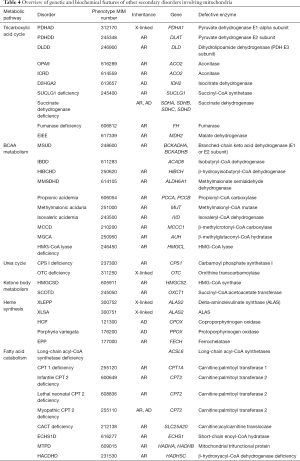
Full table
Optic atrophy-9 (OPA9) results from aconitase deficiency and is characterized by optic neuropathy which presents in early childhood and progresses well into adulthood, resulting in marked vision loss.
Infantile cerebellar-retinal degeneration (ICRD) also results from aconitase deficiency but is more severe than OPA9 with onset of symptoms at or before 6 months of age and characterized by hypotonia, writhing muscle movements, ophthalmologic degeneration, and delayed development of the CNS. Diagnostic studies may be normal, or may show metabolic acidosis and hyperglycemia.
D-2-hydroyglutaric aciduria 2 (D2HGA2) results from isocitrate dehydrogenase deficiency enzyme and manifests as D-2-hydroxyglutaric aciduria alone, or with any combination of the following symptoms: hypotonia, cardiomyopathy, epilepsy, and/or mental retardation. The main diagnostic feature of this disorder is an elevated urinary D-2-hydroxyglutaric acid level.
SDH deficiency does not lead to one disease in particular, a deficiency in any of the four subunits of mitochondrial enzyme complex II, SDH can lead to a cancer predisposition. Associated malignancies include gastrointestinal stromal tumor, paraganglioma/gastric stromal sarcoma, pheochromocytoma, Carney’s triad, Merkel cell carcinoma, and Cowden syndrome.
Fumarase deficiency (FMRD) is characterized by fumaric aciduria, intellectual disability, hypotonia, cerebral atrophy, and delayed childhood development, and in the most severe cases, leads to infant fatality. In less severe cases, the disease will lead to disability into the patient’s adolescent/adult years. In addition to fumaric aciduria, elevated lactate and pyruvate may be present. This disorder has an increased incidence in members of the Fundamentalist Church of Jesus Christ of Latter Day Saints (a group among which consanguinity is common), located in the southwestern United States.
Early infantile epileptic encephalopathy 51 (EIEE 51) (OMIM 154100) results from the malate dehydrogenase enzyme deficiency and can lead to intellectual disability, hypotonia, and epilepsy. The disease may or may not be fatal in infancy or early childhood but lactate elevations can be present in these patients.
Mitochondrial deficiencies of the urea cycle
Carbamoyl phosphate synthetase I (CPS I) is a mitochondrial enzyme responsible for catalyzing the committed step of the urea cycle. CPS I deficiency is characterized primarily by hyperammonemia. Other symptoms may include encephalopathy and respiratory alkalosis (as in all urea cycle disorders). CPS I deficiency can cause either a lethal neonatal disease, or a milder disease characterized by onset later in life. Treatment management includes use of with ammonia scavengers, arginine supplementation as well as hemodialysis as a last-resort to address hyperammonemia.
Ornithine transcarbamoylase (OTC) is the second mitochondrial enzyme involved in the urea cycle. OTC deficiency is an X-linked disorder characterized by hyperammonemia, encephalopathy, and respiratory alkalosis (similar to other urea cycle deficiencies). OTC deficiency can be differentiated from CPS I deficiency by the presence of orotic aciduria. Treatment is similar as noted in CPSI deficiency and includes dietary protein restriction.
Mitochondrial deficiencies of KB metabolism
HMG-CoA synthase mediates the first step in KB synthesis; deficiency is characterized by hypoketotic hypoglycemia, the onset of which is generally triggered by prolonged fasting or by infection. During the fed state, there is no evidence of metabolic dysfunction in these patients, so with strict dietary habits, quality of life is minimally impacted.
Succinyl-CoA:acetoacetate transferase (SCOT) is an enzyme involved in the catabolism of ketone bodies. SCOT deficiency results in the inability to catabolize ketone bodies for energy, and results in hypoglycemic ketoacidosis. Depending on the level of SCOT activity, patients may experience ketoacidotic episodes of varying frequency and severity. Premature death may occur in severe cases. Increased levels of β-hydroxybutyrate and acetoacetate may be found in the urine at the time of diagnosis. Treatment includes dietary protein restriction and avoidance of the ketotic state.
Mitochondrial deficiencies of Heme synthesis
Delta-aminolevulinate synthase (ALAS) catalyzes the first, and committed, step in heme synthesis. A deficiency in this enzyme X-linked erythropoietic protoporphyria (XLEPP) was characterized by childhood onset of photosensitivity, decreased iron reserves, and possible hepatic disease. Iron supplementation is thought to be a suitable treatment, and one clinical trial is currently underway with the goal of shedding light on this possible treatment.
Similar to XLEPP, X-linked sideroblastic anemia (XLSA) is caused by a deficiency in the ALAS enzyme. This is a hypochromic, microcytic anemia characterized by ringed sideroblasts on a blood marrow smear. This disease can present in utero, or can present at older than ninety years of age. Some patients may respond to treatment with vitamin B6. Phlebotomy and iron-chelation therapy are also indicated to prevent iron overload.
Hereditary coproporphyria (HCP) is caused by coproporphyrinogen oxidase (CPOX) deficiency and characterized by intermittent episodes of neurologic dysfunction. These exacerbations often occur following the medication use, fasting, menstrual cycle, or infection. As with many porphyrias, skin photosensitivity can occur. May see abnormally high amounts of coproporphyrin III in the urine and feces. In fact, an increased fecal coproporphyrin III:I ratio is diagnostic for the disease.
Porphyria variegata results from protoporphyrinogen oxidase (PPOX) deficiency, in one of the final steps of heme synthesis and manifests as mostly cutaneous findings, though patients may have frequent episodes of abdominal pain, dark urine, neurologic, and psychologic findings during acute attacks. Common symptoms include marked skin photosensitivity, skin fragility, skin hyperpigmentation, as well as increased urinary porphobilinogen and delta-aminolevulinic acid during acute attacks.
Erythropoietic protoporphyria (EPP) is caused by ferrochelatase deficiency in the final step of heme synthesis, where it catalyzes the addition of iron to protoporphyrin. EPP patients experience skin photosensitivity starting in early childhood. These patients experience episodes of acute itching/burning following exposure to bright light. Treatment includes avoidance of sunlight.
Discussion
Primary mitochondrial diseases continue to grow in terms of newly described disorders based on new gene discovery, and expansion of the known classic phenotypes. The group of disorders which encompass secondary mitochondrial dysfunction also continues to grow, as the mitochondria have several metabolic responsibilities. Although there is no current FDA-approved treatment or cure, clinical trials for several different genotypes and phenotypes are currently open, and treatment may need to be personalized for best efficacy (clinicaltrials.gov). To optimize care and management of these patients, organizations such as the Mitochondrial Medicine Society (www.mitosoc.org) are working with Patient Advocacy Groups, to advance education, global collaboration, clinical care, standardize diagnosis and treatment guidelines, and form clinical networks such as the Mitochondrial Care Network in the US, in addition to other world-wide efforts (i.e., MetabERN in Europe) (41-43). As with other inborn errors of metabolism (IEMs), international consortia, registries, and well-designed multicenter trials are imperative to discover efficacious treatments for these complicated and rare disorders.
Acknowledgements
None.
Footnote
Conflicts of Interest: A Goldstein is an active past president of Mitochondrial Medicine Society (mitosoc.org), Scientific and Medical Advisory Board member of UMDF (umdf.org). The other authors have no conflicts of interest to declare.
References
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature 1998;392:37-41. [Crossref] [PubMed]
- Lane N, Martin W. The energetics of genome complexity. Nature 2010;467:929-34. [Crossref] [PubMed]
- Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature 1981;290:457-65. [Crossref] [PubMed]
- Attardi G, Schatz G. Biogenesis of Mitochondria. Annu Rev Cell Biol 1988;4:289-333. [Crossref] [PubMed]
- Wallace DC. Diseases of the Mitochondrial DNA. Annu Rev Biochem 1992;61:1175-212. [Crossref] [PubMed]
- Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol 2013;5:a021220. [Crossref] [PubMed]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 2014;15:634-46. [Crossref] [PubMed]
- MITOMAPMITOMAP. Available online: https://www.mitomap.org/
- Brown GK, Otero LJ, LeGris M, et al. Pyruvate dehydrogenase deficiency. J Med Genet 1994;31:875-9. [Crossref] [PubMed]
- Naito E, Ito M, Yokota I, et al. Thiamine-responsive pyruvate dehydrogenase deficiency in two patients caused by a point mutation (F205L and L216F) within the thiamine pyrophosphate binding region. Biochim Biophys Acta - Mol Basis Dis 2002;1588:79-84.
- Head RA, Brown RM, Clayton PT, et al. Clinical and genetic spectrum of pyruvate dehydrogenase deficiency: Dihydrolipoamide acetyltransferase (E2) deficiency. Ann Neurol 2005;58:234-41. [Crossref] [PubMed]
- Robinson B. Lactic Acidemia: Disorders of Pyruvate Carboxylase and Pyruvate Dehydrogenase | The Online Metabolic and Molecular Bases of Inherited Disease | OMMBID | McGraw-Hill Medical [cited 2018 Nov 13]. Available online: https://ommbid-mhmedical-com.ezproxy.med.wmich.edu/content.aspx?bookid=971§ionid=62633368
- Pascual JM, DiMauro S. Disorders of Energy Metabolism | Rudolph’s Pediatrics, 23e | AccessPediatrics | McGraw-Hill Medical [cited 2018 Nov 13]. Available online: https://accesspediatrics.mhmedical.com/content.aspx?bookid=2126§ionid=171566998#1154120224
- Smeitink JA, Zeviani M, Turnbull DM, et al. Mitochondrial medicine: A metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab 2006;3:9-13. [Crossref] [PubMed]
- Gorman GS, Chinnery PF, DiMauro S, et al. Mitochondrial diseases. Nat Rev Dis Prim 2016;2:16080. [Crossref] [PubMed]
- DiMauro S, Schon EA. Mitochondrial Respiratory-Chain Diseases. N Engl J Med 2003;348:2656-68. [Crossref] [PubMed]
- Kirby DM, Thorburn DR. Approaches to Finding the Molecular Basis of Mitochondrial Oxidative Phosphorylation Disorders. Twin Res Hum Genet 2008;11:395-411. [Crossref] [PubMed]
- Tucker EJ, Hershman SG, Köhrer C, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab 2011;14:428-34. [Crossref] [PubMed]
- Fassone E, Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J Med Genet 2012;49:578-90. [Crossref] [PubMed]
- Hoekstra AS, Bayley JP. The role of complex II in disease. Biochim Biophys Acta - Bioenerg 2013;1827:543-51.
- Jain-Ghai S, Cameron JM, Robinson B, et al. Complex II deficiency-A case report and review of the literature. Am J Med Genet Part A 2013;161A:285-94. [Crossref] [PubMed]
- Long-Term Safety and Efficacy Evaluation of EPI-743 in Children With Leigh Syndrome - Full Text View - ClinicalTrials.gov [cited 2018 Nov 13]. Available online: https://clinicaltrials.gov/ct2/show/NCT02352896?term=NCT02352896&rank=1
- Mori M, Shoffner J, Koeberl DD, et al. Complex III deficiency due to an in-frame MT-CYB deletion presenting as ketotic hypoglycemia and lactic acidosis. Mol Genet Metab Rep 2015;4:39-41. [Crossref] [PubMed]
- Rak M, Bénit P, Chrétien D, et al. Mitochondrial cytochrome c oxidase deficiency. Clin Sci (Lond) 2016;130:393-407. [Crossref] [PubMed]
- Jonckheere AI, Smeitink JAM, Rodenburg RJT. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis 2012;35:211-25. [Crossref] [PubMed]
- DiMauro S, Hirano M. Mitochondrial DNA Deletion Syndromes GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 17]. Available from online: http://www.ncbi.nlm.nih.gov/pubmed/20301382
- Home - UMDF [cited 2018 Nov 17]. Available online: https://www.umdf.org/
- Yu-Wai-Man P, Chinnery PF. Leber Hereditary Optic Neuropathy GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301353
- Stenqvist L, Paetau A, Valanne L, et al. A Juvenile Case of MELAS with T3271C Mitochondrial DNA Mutation. Pediatr Res 2005;58:258-62. [Crossref] [PubMed]
- DiMauro S, Hirano M. MELAS GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301411
- DiMauro S, Hirano M. MERRF GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301693
- Thorburn DR, Rahman J, Rahman S. Mitochondrial DNA-Associated Leigh Syndrome and NARP GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301352
- Angelini C, Bello L, Spinazzi M, et al. Mitochondrial disorders of the nuclear genome. Acta Myol 2009;28:16-23. [PubMed]
- Poole O V, Hanna MG, Pitceathly RDS. Mitochondrial disorders: disease mechanisms and therapeutic approaches. Discov Med 2015;20:325-31. [PubMed]
- El-Hattab AW, Craigen WJ, Wong LJC, et al. Mitochondrial DNA Maintenance Defects Overview GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/29517884
- El-Hattab AW, Scaglia F. Mitochondrial DNA depletion syndromes: review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics 2013;10:186-98. [Crossref] [PubMed]
- Hirano M. Mitochondrial Neurogastrointestinal Encephalopathy Disease GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301358
- Wang J, El-Hattab AW, Wong LJC. TK2-Related Mitochondrial DNA Maintenance Defect, Myopathic Form GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23230576
- Spinazzola A, Invernizzi F, Carrara F, et al. Clinical and molecular features of mitochondrial DNA depletion syndromes. J Inherit Metab Dis 2009;32:143-58. [Crossref] [PubMed]
- Cohen BH, Chinnery PF, Copeland WC. POLG-Related Disorders GeneReviews®. University of Washington, Seattle; 1993 [cited 2018 Nov 18]. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301791
- Parikh S, Goldstein A, DiMauro S, et al. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med 2015;17:689-701. [Crossref] [PubMed]
- Parikh S, Goldstein A, Chinnery PF, et al. Patient care standards for primary mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med 2017;19. [Crossref] [PubMed]
- Muraresku CC, McCormick EM, Falk MJ. Mitochondrial Disease: Advances in clinical diagnosis, management, therapeutic development, and preventative strategies. Curr Genet Med Rep 2018;6:62-72. [Crossref] [PubMed]


