
Expression of colorectal neoplasia differentially expressed in anaplastic thyroid carcinoma and its effect on cancer cell proliferation
Introduction
The incidence of anaplastic thyroid cancer (ATC) is high among human cancers, and also exhibits high invasiveness, metastasis, and uncontrolled tumor spread at the time of diagnosis (1). Due to the resistance of ATC to traditional thyroid cancer (THCA) treatment [including inhibition of thyroid stimulating hormone (TSH) and radioactive iodine], it has a poor prognosis, including a median survival of only 5 months and a 1-year survival rate of less than 20% (2). At present, there are not effective treatments for this problematic tumor. Exploration of the pathogenesis of ATC may provide new insights into developing an effective treatment.
Long-noncoding RNAs (lncRNAs) are endogenous RNA molecules with more than 200 nucleotides, which do not exert a protein encoding function, and play a critical role in multiple normal biological processes, such as cell development, differentiation, and motility (3). A previous study has shown that abnormal expression of lncRNAs may promote tumor development, acting as tumor suppressor genes and oncogenes to regulate tumor cell proliferation, invasion, metastasis, and other malignant progression. Targeted regulation of lncRNAs can significantly inhibit the malignant progression of tumors (4).
Through bioinformatics analysis of The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) data, we found that colorectal neoplasia differentially expressed (CRNDE) plays an important role in ATC progression (5-7). CRNDE is located on human chromosome 16, and can be used as a biomolecular marker (8). CRNDE is highly expressed in common tumors such as colorectal, liver, and lung cancers. It also promotes the proliferation, migration, invasion, and angiogenesis of tumor cells, and inhibits apoptosis, which is a potential molecular target for anti-tumor therapy (8-10). However, the expression of CRNDE in ATC and its mechanism of action remain unknown. In this study, TCGA was used to analyze the expression of CRNDE in ATC tissues as well as its relationship to patient prognosis, and explored whether CRNDE plays a role as a cancer-promoting factor in ATC, with the aim of obtaining an important basis of CRNDE as a potential molecular target for the treatment of ATC. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-945/rc).
Methods
Transcriptome data acquisition and pre-processing
We obtained ATC gene expression profiling datasets (11,12), GSE65144 GSE33630, GSE29265 (no citation), and GSE53157, from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database. The basic information of these four datasets is shown in Table 1. The THCA transcriptome data and corresponding clinical characteristics of patients were downloaded from the publicly available TCGA (http://cancergenome.nih.gov/) database (Table S1). A total of 510 THCA tissue samples and 58 adjacent normal tissues were extracted from TCGA. Patients in TCGA cohort who failed to meet the following criteria were eliminated: (I) THCA transcriptome data with basic survival information including, overall survival status and survival time; and (II) primary tumor samples (as opposed to metastatic tumor samples). Finally, 502 THCA samples and 58 adjacent normal tissues samples from TCGA were included. The clinical characteristics of THCA patients in the TCGA database are summarized in Table 2. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| GEO_ID | Samples | Case/control | Country | Platforms | Chip |
|---|---|---|---|---|---|
| GSE65144 | 34 | 16/18 | USA | GPL570 | HG-U133_Plus_2 |
| GSE33630 | 105 | 11/45 | Belgium | GPL570 | HG-U133_Plus_2 |
| GSE29265 | 49 | 9/20 | Belgium | GPL570 | HG-U133_Plus_2 |
ATC, anaplastic thyroid cancer; GEO, Gene Expression Omnibus.
Table 2
| Gene name | Ensembl.id | Chromosomal position | P | HR |
|---|---|---|---|---|
| LINC01116 | ENSG00000163364 | chr2: 176629572-176637931 (−) | 9.28E-01 | 0.96 |
| LINC00265 | ENSG00000188185 | chr7: 39733430-39793092 (+) | 7.72E-01 | 0.86 |
| APTR | ENSG00000214293 | chr7: 77657659-77696267 (−) | 8.37E-01 | 1.11 |
| LINC00271 | ENSG00000231028 | chr6: 135497422-135813990 (+) | 2.55E-01 | 0.56 |
| LINC00865 | ENSG00000232229 | chr10: 89829483-89840861 (+) | 9.50E-01 | 0.97 |
| LINC00886 | ENSG00000240875 | chr3: 156747343-156817062 (−) | 1.97E-01 | 1.95 |
| CRNDE | ENSG00000245694 | chr16: 54845189-54929189 (−) | 2.99E-02 | 3.51 |
| LINC01094 | ENSG00000251442 | chr4: 78638780-78683185 (+) | 6.72E-01 | 1.24 |
| RMST | ENSG00000255794 | chr12: 97430884-97598415 (+) | 1.52E-01 | 0.46 |
| LINC01003 | ENSG00000261455 | chr7: 152463786-152465549 (+) | 8.77E-01 | 1.08 |
| CHASERR | ENSG00000272888 | chr15: 92819540-92899701 (+) | 1.77E-01 | 0.50 |
+, positive strand; −, negative strand. LncRNA, long-noncoding RNA; THCA, thyroid cancer; HR, hazard rate.
Experimental reagents and instruments
Human thyroid cancer cell line ARO, FRO, sw579 and 8505c were obtained from the Cell Bank of Shanghai Academy of Sciences (Shanghai, China). The culture medium, pancreatin, and fetal bovine serum (FBS) were purchased from Gibco (United States). The TRIzol reagent and RNA extraction kit were purchased from TaKaRa (Japan). CRNDE and internal reference glyceraldehyde-3phosphate dehydrogenase (GAPDH) primers were obtained from Jierui Bioengineering Co., Ltd. (Shanghai, China). The AMV (Avian Myeloblastosis Virus) One-step Reverse Transcription PCR (RT-PCR) kit was obtained from Shenggong Bioengineering Co., Ltd. (Shanghai, China). The CRNDE small interfering RNA (siRNA) was purchased from Ruibo Biological Co., Ltd. (Guangzhou, China). The Lip 2000 and Invitrogen reagents were obtained from Invitrogen (United States). The 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide MTT proliferation detection reagents were obtained from Airan Biotechnology Co., Ltd. (Beijing, China). The Giemsa staining solution, cell cycle detection kit, and bicinchoninic acid (BCA) protein concentration detection kit were purchased from Shanghai Biyuntian Biological Co., Ltd. Protein lysate was purchased from Solarbio (Beijing, China). The polyvinylidene difluoride (PVDF) membrane was purchased from Promega (United States). The efficient chemiluminescence (ECL) protein exposure kit was purchased from Huiying Biotechnology Co., Ltd. (Shanghai, China). Antibodies were purchased from Abcam (United Kingdom).
Cell culture
ARO, FRO, sw579, and 8505c of the ATC cell lines were removed from liquid nitrogen and quickly recovered in a 37 ℃ water bath. After centrifugation at 800 rpm for 5 minutes, the freezing solution was discarded, and the cells were resuspended in a medium containing 10% FBS and placed in a fully humidified incubator with 5% carbon dioxide (CO2) at 37 ℃. When a color change was observed in the cells, the medium was immediately replaced with fresh medium. The overgrown cells were washed with phosphate buffer saline (PBS) once, and trypsin was used for digestion according to the ratio of 1:2 for cells (13).
RNA extraction and qRT-PCR
Total RNA was extracted from the cell lines using TRIzol (Invitrogen, Carlsbad, CA, USA) as directed by the manufacturer’s instructions. The reverse transcription reaction was performed with the Moloney Leukemia Virus Reverse Transcriptase Kit using 1µg of total RNA as a template to create a high-quality cDNA. To evaluate the expression of lncRNA, the cDNA samples were amplified by qRT-PCR using the Green Mix SYBR (Promega), and GAPDH was employed as the internal control to standardize their expression. CRNDE, (forward, 5'-TGTCAAGGGTTTGAACTGCTAAT-3'; reverse, 5'-GATAAGCATGCTACAAGCTCTGACA-3') (14).
CRNDE siRNA transfection
The expression level of CRNDE in ATC cells was detected by qRT-PCR, and ATC cells with the highest and lowest CRNDE expression were selected for siRNA transfection. Cells in the logarithmic growth phase were collected by trypsinization after washing once with PBS. After counting the cells, they were inoculated into six-well plates at 2×105 cells/well, and divided into negative control (si-NC), si-CRNDE-1, si-CRNDE-2, and si-CRNDE-3 groups and placed in an incubator for culturing. After culturing for 12 h, Lip 2000 was mixed with siRNA in each group according to the manufacturer’s instructions, and transfected into each group of cells. After culturing for 8 h, the medium was replaced with fresh medium, and culturing was continued for 48 h. The expression of CRNDE in each group was detected by qRT-PCR to verify the effect of CRNDE siRNA transfection, and two siRNA sequences with obvious inhibitory effects were selected for a subsequent study (15).
MTT for cell proliferation ability
The ATC cells were collected by trypsin digestion, and 1,500 cells per well were seeded in a 96-well plate. Each group was set with six parallel multiple wells and placed in an incubator for culturing. After culturing for 12 h, transfection of siRNA was carried out according to the above transfection method. The 20 µL MTT reagent was added to each well at 0, 24, 48, and 72 h, respectively. After culturing in an incubator for 4 h, the culture medium was discarded, and 150 µL dimethyl sulfoxide (DMSO) was added for shock mixing. After the crystals are completely dissolved, a microplate reader (United States) was used to detect the cell optical density (OD) at 490 nm. All experiments were performed in triplicate (16).
Flow cytometry for cell cycle
ATC cells transfected for 48 h were collected by trypsinization, washed with PBS three times, and fixed with pre-cooled absolute ethanol (three in each group in parallel). After washing once with PBS, cell cycle staining buffer, propidium iodide (PI) staining solution, and RNase were added, mixed well, and incubated at 37 ℃ for 15 min. Flow cytometry was used to detect the cell cycle ratio.
Scratch test for cell migration ability
The transfected ATC cells of each group were collected by trypsinization and seeded into a six-well plate at 4×105 cells/well, with three parallel multiple wells in each group, and placed in an incubator for culturing. After culturing for 12 h, a scratcher was used to make a scratch in a six-well plate. After using PBS to wash off the exfoliated cells, serum-free medium was added, and the width of the cell scratch was recorded by taking pictures, which was recorded as 0 h. After culturing in an incubator for 48 h and washing off the exfoliated cells with PBS, a photo was taken to record the width of the cell scratch, which was recorded as 48 h. The cell migration ability was expressed by cell migration rate (17).
Western blotting
The ATC cells in each group, which were transfected for 48 h, were collected by trypsinization, washed with PBS three times, and lysed with protein lysate. After ultrasonic lysis for 20 min, high-speed centrifugation was performed at 4 ℃ for 30 min. The supernatant was then collected and the protein concentration was detected using the BCA detection kit. Then, the loading buffer was added and boiled for 3 min, and the denatured protein was separated by gel electrophoresis. The protein was transferred to the PVDF membrane via wet transfer, and the PVDF membrane was sealed with skimmed milk. After washing once with TBST, the PVDF membrane was incubated with the target primary antibody at 4 ℃ overnight and the secondary antibody at room temperature for 1 h. After washing with TBST three times, the ECL kit was used to expose the protein bands (17).
Econazole regulates CRNDE to inhibit ATC cell proliferation
ATC cells were collected by trypsin digestion, and 1,500 cells per well were seeded in a 96-well plate, divided into NC, econazole, overexpression (oe)-CRNDE, and econazole + oe-CRNDE groups (with six parallel multiple wells in each group), and placed in the incubator for cultivation. After culturing for 12 h, the NC group was transfected with unrelated sequence therapy. The econazole group was transfected with the concentration of half maximal inhibitory concentration (IC50) econazole, the oe-CRNDE group was transfected with CRNDE overexpression plasmid, and the econazole + oe-CRNDE group was transfected with IC50 econazole at the same time. After incubation for 48 h, 20 µL of MTT reagent was added to each well. After culturing in an incubator for 4 h, the culture medium was discarded, and a further 150 µL of DMSO was added for shock mixing. When the crystals were completely dissolved, the absorbance (OD) at 490 nm was detected by a microplate reader. All experiments were performed in triplicate (17).
The Connectivity Map
The Connectivity Map (CMap, https://clue.io/) has collected 1,309 compounds and contains more than 7,000 gene expression profiles, and reveals the connection between diseases, genes, and drugs. It is used to discover potential treatments for diseases drug. The Score value in CMap was between −1 and 1; a positive value indicated that the drug has a disease-promoting effect, while a negative value suggested that the drug has an inhibitory effect on the disease. A larger absolute value signified a stronger correlation.
Statistical analysis
SPSS17.0 statistical software (https://www.ibm.com/products/spss-statistics) was used for analysis. All P value calculations were performed using an unpaired t-test or non-parametric one-way analysis of variance (ANOVA) and other special analyses. P<0.05 was considered to be indicative of statistical significance.
Results
Screening of different expression of LncRNA in ATC
For ATC data, we obtained the GSE65144, GSE33630, and GSE29265 datasets from the GEO database (Table 1). According to the threshold of |Log2(FC)|>1, P<0.0001, a total of 33 significant probes in the GSE65144 were determined, involving 30 lncRNAs with different expressions (Figure 1A). Also, the GSE33630 involved 74 differentially-expressed lncRNAs, and the GSE29265 involved 47 differentially expressed lncRNAs. The Venn diagram results showed that a total of 11 lncRNAs were differentially expressed in THCA and control tissues (Figure 1B,1C).
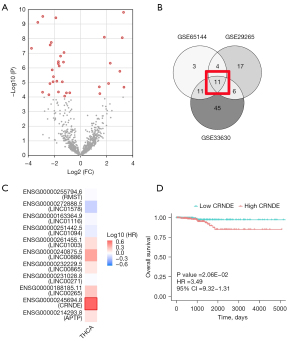
Using TCGA data, a univariate analysis of 11 genes was performed (as shown in Table 2), which showed that CRNDE expression was the most significant, and thus, a survival curve was drawn for it (Figure 1D). At the same time, multivariate Cox regression analysis showed that high CRNDE expression was an important factor for the poor prognosis of THCA patients, as shown in Table 3. Figure 2A showed that compared with 58 normal thyroid tissues in TCGA dataset, CRNDE was highly expressed in 502 THCA tissues,
Table 3
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| CRNDE expression | 2.12 | 1.33–3.40 | 1.70E-03 | 2.08 | 1.17–3.72 | 1.31E-02 | |
| Tumor stage (stage III_IV vs. I_II) | 7.14 | 2.3–22.19 | 6.82E-04 | 0.52 | 0.09–3.03 | 4.68E-01 | |
| T stage (T3_T4 vs. T1_T2) | 3.11 | 1.08–8.96 | 3.56E-02 | 0.54 | 0.10–2.86 | 4.71E-01 | |
| N stage (N1 vs. N0) | 1.47 | 0.48–4.51 | 4.97E-01 | – | – | – | |
| M stage (M1 vs. M0) | 4.74 | 1.01–22.22 | 4.84E-02 | – | – | – | |
| Residual tumor (R1_R2 vs. R0) | 3.69 | 1.11–12.25 | 3.32E-02 | 11.55 | 2.14–62.44 | 4.50E-03 | |
| Gender (male vs. female) | 1.93 | 0.7–5.34 | 2.05E-01 | – | – | – | |
| Race (white vs. others) | 1.41 | 0.32–6.22 | 6.48E-01 | – | – | – | |
| Age | 1.16 | 1.10–1.22 | 2.08E-08 | 1.19 | 1.10–1.28 | 3.52E-06 | |
| Disease history (yes vs. no) | 0.16 | 0.02–1.23 | 7.89E-02 | – | – | – | |
| Location (isthmus vs. bilateral) | 1.11 | 0.1–12.47 | 9.31E-01 | – | – | – | |
| Location (left lobe vs. bilateral) | 0.95 | 0.19–4.81 | 9.52E-01 | – | – | – | |
| Location (right lobe vs. bilateral) | 0.90 | 0.18–4.53 | 9.02E-01 | – | – | – | |
CRNDE, colorectal neoplasia differentially expressed; THCA, thyroid cancer; HR, hazard rate; CI, confidence interval.
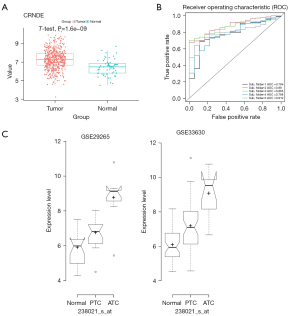
To assess the efficacy of CRNDE in diagnosing THCA, we performed five-fold cross-validation using a logistic regression model in TCGA database. The 502 tumor samples and 58 normal samples were separately randomly selected to form training and validation sets (7:3), which avoided the size imbalance of the two groups. The area under the receiver operating characteristic (ROC) curve (AUC) was used to evaluate the sensitivity (true positive rate) and specificity (true negative rate) of CRNDE for the diagnosis of THCA in TCGA database using the ROCR package (https://cran.r-project.org/web/packages/ROCR/index.html). As a result, the AUC values were >0.75 in each fold (Figure 2B). The average values of accuracy, precision, recall, and F1-score were 0.89, 0.89, 1.00, and 0.94, respectively (Table 4). In summary, CRNDE showed the greatest influence, and the hazard rate (HR) of CRNDE was the largest (Table 2), so this gene was selected for further study.
Table 4
| Group | Accuracy | Precision | Recall | F1-score | AUC |
|---|---|---|---|---|---|
| Sub-folder 1 | 0.89 | 0.89 | 0.99 | 0.94 | 0.78 |
| Sub-folder 2 | 0.89 | 0.89 | 1.00 | 0.94 | 0.88 |
| Sub-folder 3 | 0.89 | 0.89 | 0.99 | 0.94 | 0.86 |
| Sub-folder 4 | 0.89 | 0.89 | 0.99 | 0.94 | 0.79 |
| Sub-folder 5 | 0.89 | 0.89 | 1.00 | 0.94 | 0.81 |
| Average | 0.89 | 0.89 | 1.00 | 0.94 | 0.83 |
CRNDE, colorectal neoplasia differentially expressed; AUC, area under the receiver operating characteristic curve.
Moreover, the expression level of CRNDE was analyzed in ATC’s GEO-meta database. The results of both datasets (GSE33630 and GSE29265) showed that CRNDE was specifically and highly expressed in ATC tissues (Figure 2C). These results indicated that CRNDE may play an important role in ATC. Therefore, CRNDE was selected for follow-up research.
CRNDE promotes the proliferation and migration of ATC cells
In order to verify the above results, qRT-PCR was performed, which showed that CRNDE expression was relatively high in ARO, and was relatively low in sw579 (Figure 3A). To explore the molecular mechanism of CRNDE and its effect on tumor proliferation, we selected for CRNDE siRNA transfection for subsequent experimental studies. Following transfection of CRNDE siRNA with ARO and sw579 cells for 48 h, we found that the relative expression of CRNDE in the si-CRNDE-1, si-CRNDE-2, and si-CRNDE-3 groups of ARO and sw579 cells were significantly decreased compared with the si-NC group (P<0.05). Also, the inhibitory effect of CRNDE in the ARO and sw579 cells si-CRNDE-1 and si-CRNDE-2 groups was more significant. Thus, both were selected for subsequent functional experiments (Figure 3B).
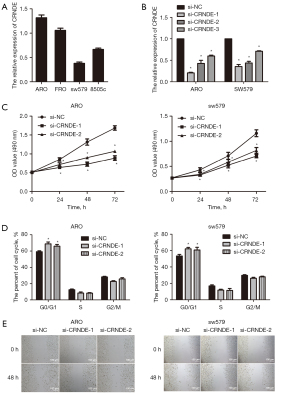
MTT was used to determine the effect of CRNDE siRNA on the proliferation ability of ATC cells. The results showed that compared with the si-NC group, the proliferation ability of ARO and sw579 cells in the si-CRNDE-1 and si-CRNDE-3 groups were markedly reduced (P<0.05, Figure 3C). Flow cytometry was used to detect the effect of CRNDE siRNA on the ATC cell cycle. The results showed that compared with the si-NC group, the ratio of G0/G1 cells in the si-CRNDE-1 and siCRNDE-2 groups of ARO and sw579 cells was also significantly increased (P<0.05, Figure 3D). The scratch test was used to evaluate the effect of CRNDE siRNA on the migration ability of ATC cells. The results showed that the migration ability of ARO and sw579 cells in the si-CRNDE-1 and the si-CRNDE-3 groups was notably reduced compared with the si-NC group (P<0.05, Figure 3E).
CRNDE promoted ATC proliferation via the Wnt/β-catenin signaling pathway
To explore the molecular mechanism of CRNDE, RNA-seq data analysis of 502 THCA tissues in the TCGA was conducted to identify the possible biological functions of CRNDE in THCA. The Gene Ontology (GO) enrichment analysis of 230 related genes (|R|>0.4) showed that CRNDE affected cell growth and proliferation (P<0.005, Table 5), and that the signaling pathways enriched by the high-expressing CRNDE group include the WNT (wnt signaling pathway), transforming growth factor-beta signaling pathway (TGF-beta), and cancer-related pathways (Figure 4A,4B).
Table 5
| Gene set | Size | Overlap | Expect | Enrichment ratio | P value | Description |
|---|---|---|---|---|---|---|
| GO:0040008 | 670 | 12 | 5.869434777 | 2.044489879 | 0.014836175 | Regulation of growth |
| GO:0061061 | 610 | 11 | 5.343813753 | 2.058454974 | 0.018479803 | Muscle structure development |
| GO:0040007 | 961 | 15 | 8.418696748 | 1.781748464 | 0.021382547 | Growth |
| GO:1901137 | 712 | 12 | 6.237369495 | 1.923887948 | 0.022694452 | Carbohydrate derivative biosynthetic process |
| GO:0008285 | 667 | 11 | 5.843153726 | 1.882545029 | 0.032733124 | Negative regulation of cell proliferation |
| GO:0007423 | 531 | 9 | 4.65174607 | 1.934757372 | 0.044170929 | Sensory organ development |
| GO:0048589 | 623 | 10 | 5.457698308 | 1.832274236 | 0.047470184 | Developmental growth |
TCGA, The Cancer Genome Atlas; CRNDE, colorectal neoplasia differentially expressed.
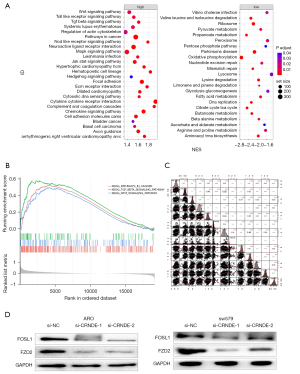
Cross 150 signal genes from the KEGG database (c2.cp.kegg.v7.4.symbols.gmt) and positively related genes were intersected to obtain 47 genes, as shown in Figure 4C and Table S2. After CRNDE siRNA transfected ARO and sw579 cells for 48 h, compared with the si-NC group, the relative expressions of Wnt3a, β-catenin and CyclinD1 protein in the si-CRNDE-1 group and si-CRNDE-2 group of ARO and sw579 cells were significantly reduced (P<0.05, Figure 4D).
Screening of small molecule drugs with potential efficacy using the Connectivity Map (CMap)
The differentially expressed genes (differential genes between CRNDE expression groups, P<0.05, |log2FC| >0.5) were uploaded to the CMap database, which screened out the small molecule compounds that were negatively correlated with the correlation coefficient (Table 6). Previous studies have shown that econazole and terazosin exert anti-tumor effects (18,19). The IC50 value of econazole on ARO and sw579 cells was the lowest detected by MTT (P<0.05, Figure 5A). Lastly, the MTT test showed that econazole inhibited the proliferation of ARO and sw579 cells in a dose-dependent manner (P<0.05, Figure 5B).
Table 6
| Drug name | Molecular formula | Mean connective score | n | Enrichment | P value | Specificity |
|---|---|---|---|---|---|---|
| STOCK1N-35696 | C17H18N4O3 | −0.791 | 2 | −0.98 | 8.70E-04 | 0.01 |
| Ambroxol | C13H18Br2N2O | −0.353 | 4 | −0.82 | 1.89E-03 | 0.01 |
| Econazole | C18H15Cl3N2O | −0.578 | 4 | −0.72 | 1.22E-02 | 0.10 |
| Etoposide | C29H32O13 | −0.543 | 4 | −0.72 | 1.30E-02 | 0.09 |
| Antazoline | C17H19N3 | −0.601 | 4 | −0.71 | 1.34E-02 | 0.01 |
| Terazosin | C19H25N5O4 | −0.323 | 4 | −0.70 | 1.82E-02 | 0.09 |
| Benzbromarone | C17H12Br2O3 | −0.499 | 3 | −0.79 | 1.97E-02 | 0.04 |
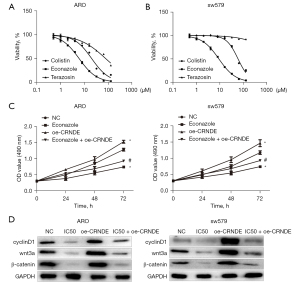
Econazole regulated the expression of CRNDE and inhibited the proliferation of ATC cells
In order to explore the targeting effect of drugs on CRNDE, we used MTT to determine the effect of econazole on the reversal the effect of CRNDE in ATC cell proliferation. The results showed that compared with the NC group, the proliferation of ARO and sw579 cells in the econazole group and econazole + oe-CRNDE group were reduced, and the proliferation ability of ARO and sw579 cells in the oe-CRNDE group was increased. Compared with the econazole group, the proliferation ability of ARO and sw579 cells in the econazole + oe-CRNDE group was increased (Figure 5C). Therefore, we believe that econazole exerts an anti-tumor effect by reversing CRNDE.
Western blotting was used to detect the effect of econazole on regulating the expression of CRNDE and inhibiting the proliferation of ATC cells. The results showed that compared with the NC group, the expression of wnt3a, β-catenin, and cyclinD1 in ARO and sw579 cells in the econazole and the econazole + oe-CRNDE groups were significantly reduced. Importantly, the expression of wnt3a, β-catenin, and cyclinD1 proteins increased in ARO and sw579 cells in the CRNDE group. Lastly, compared with the econazole group, the expression of wnt3a, β-catenin, and cyclinD1 proteins in the econazole + oe-CRNDE group were markedly increased (Figure 5D).
Discussion
THCA is the most common endocrine cancer, and approximately 67,400 cases of THCA were diagnosed in the United States in 2019 (11). THCA is divided into papillary thyroid carcinoma (PTC) and follicular thyroid cancer (FTC), poorly differentiated (PDC), and undifferentiated THCA (ATC) according to the histological characteristics. The transmissibility of THCA increases with age, and ATC is the most transmissible thyroid cancer. Although it accounts for only 2% of all THCA, ATC has a high mortality rate and was the main cause of THCA death (2). Moreover, epidemiological surveys showed that the incidence of ATC increased from 1973 to 2014 (12). Therefore, it is clinically significant to explore effective treatment methods to improve the prognosis of ATC patients. ATC treatments include chemotherapy, radiotherapy, immunotherapy, etc.; however, the existing treatments cannot improve the survival of ATC patients (2). As a new treatment, targeted molecular therapy has become a potent method to reduce the mortality of malignant tumor patients (20). Therefore, exploring the pathogenesis of ATC and finding new molecular therapeutic targets may reveal new strategies for ATC therapy.
The molecular mechanisms of tumor pathogenesis are complex. Oncogene activation, tumor suppressor gene inactivation, and epigenetic changes play key roles in the development of tumors. However, the underlying epigenomic and transcriptome changes that lead to ATC have been relatively poorly studied (21). Epigenetic modification refers to the regulation of gene function through histone modification, DNA methylation, chromatin structure remodeling, and non-coding RNA regulation, which affects tumor progression (22). With the rapid development of second-generation sequencing technology, increasingly abnormally expression of lncRNAs has gradually been discovered in tumors, including ATC (5). The GEO and TCGA contain public databases, such as differentially-expressed genes, MicroRNA (miRNA), and lncRNA in tumor diseases, which are important bioinformatics analysis data for obtaining tumor-related molecules (23).
The GEO database contains four ATC datasets. The sample size of the GSE53157 dataset was relatively small and was eliminated. There are 11 differentially expressed lncRNAs in the other three THCA datasets. By analyzing the effects of differentially-expressed lncRNAs in ATC on the prognosis of patients, the results showed that among 11 lncRNAs, only CRNDE was highly expressed in THCA, leading to a decline in patient survival. This suggests that CRNDE may play a key role in promoting THCA. Further analysis of the expression level of CRNDE in the GEO database showed the specific high expression of CRNDE. Also, the expression level of CRNDE in normal tissue (NT), papillary thyroid carcinoma (PTC), and ATC tissues increased in turn, suggesting that CRNDE may exert important biological functions in ATC.
CRNDE is a lncRNA that is closely related to tumors, and was originally discovered in colorectal cancer. Under normal conditions, CRNDE is highly expressed in normal tissues such as skin, breast, and testis, but is lowly or not expressed in colorectal mucosa (8). Under pathological conditions, high expression of CRNDE leads to the progression of a variety of tumors, and promotes the proliferation, migration, and invasion of tumor cells, while inhibiting cell apoptosis (9,10).
We combined the reported results and used the RNA-seq data from the TCGA database to verify the possible biological functions of CRNDE in THCA. The results showed that inhibition of CRNDE reduced the proliferative ability of ATC cells. Uncontrolled cell growth leading to malignant proliferation is an important feature of tumors, and abnormal cell cycle regulation is closely related to cell malignant proliferation (24). After inhibiting the expression of CRNDE, the two ATC cell cycles were blocked in the G0/G1 phase, which is consistent with previous studies on CRNDE in non-small cell lung cancer, acute lymphoblastic leukemia, and tongue squamous cell carcinoma (10,13,25). Under normal circumstances, the cell cycle is strictly regulated by cyclins. CyclinD1 is a key protein that promotes cells from the G0/G1 phase to S phase (14). The western blotting results showed that after inhibiting CRNDE expression, the expression of the cyclinD1 protein in the two ATC cells was significantly reduced. Wnt/β-catenin is one of the classic signaling pathways regulating tumor cell proliferation (15), and the cyclinD1 protein is its downstream target gene (26). The wnt3a gene leads to the accumulation of β-catenin and its ectopic localization to the nucleus. The combination of β-catenin and T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors drives the expression of downstream target genes such as cyclinD1 (26). The results showed that the expression of wnt3a and β-catenin protein in the two ATC cells was markedly reduced. This indicates that CRNDE may regulate cell cycle proteins by inhibiting the Wnt/β-catenin signaling pathway, inducing cell cycle arrest, and subsequently inhibiting cell proliferation (27,28).
The CMap database has collected 1,309 compounds and contains more than 7,000 gene expression profiles, revealing the connection between diseases, genes, and drugs, which can be used to discover potential drugs for the treatment of diseases. The CMap database is used to discover new molecular targeted drugs for the treatment of THCA (16,17). Previous results have shown that there are nine small molecule drugs that can be used as targeted therapeutics for CRNDE in THCA (29-31), however the anti-tumor effect in THCA remains unclear. Tests have shown that the IC50 value of econazole on THCA cells is lower than other compounds. Therefore, econazole may have good therapeutic potential. Econazole is an imidazole compound, and as an effective broad-spectrum antifungal agent, it has shown anti-inflammatory effects. In recent decades, the anti-tumor effect of econazole has gradually been discovered, including in leukemia and other blood system tumors (32) as well as gastric cancer (18), and inhibits tumor proliferation, invasion, drug resistance, and other malignant biological behaviors (33,34). The MTT results confirmed that econazole significantly inhibited the proliferation of ATC cells, indicating an anti-tumor effect in THCA. Lastly, restoring the expression of CRNDE markedly weakened the inhibitory effect of econazole on the proliferation of ATC cells, and the inhibitory effect of econazole on the expression of wnt3a, β-catenin, and cyclinD1 proteins in ATC cells. It is suggested that econazole exerted its anti-cancer effect by regulating CRNDE.
In conclusion, this study systematically assessed the specific expression of CRNDE in ATC tissues and its potential prognostic value in ATC. It was experimentally verified that CRNDE plays a key role in promoting the development of ATC, providing a potential prognostic gene for ATC. Meanwhile, econazole inhibited the expression of CRNDE, which may be a potential therapeutic strategy for the treatment of ATC.
Acknowledgments
Funding: This study was funded by the National Natural Science Foundation of China (No. 82073491) and the National Natural Science Foundation of China (No. 81872560).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-945/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-945/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-945/coif). All authors report that this study was funded by the National Natural Science Foundation of China (No. 82073491) and the National Natural Science Foundation of China (No. 81872560). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Molinaro E, Romei C, Biagini A, et al. Anaplastic thyroid carcinoma: from clinicopathology to genetics and advanced therapies. Nat Rev Endocrinol 2017;13:644-60. [Crossref] [PubMed]
- Zhang R, Zhang Y, Tan J, et al. Antitumor Effect of 131I-Labeled Anti-VEGFR2 Targeted Mesoporous Silica Nanoparticles in Anaplastic Thyroid Cancer. Nanoscale Res Lett 2019;14:96. [Crossref] [PubMed]
- Charles Richard JL, Eichhorn PJA. Platforms for Investigating LncRNA Functions. SLAS Technol 2018;23:493-506. [Crossref] [PubMed]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci 2018;109:2093-100. [Crossref] [PubMed]
- Tan X, Wang P, Lou J, et al. Knockdown of lncRNA NEAT1 suppresses hypoxia-induced migration, invasion and glycolysis in anaplastic thyroid carcinoma cells through regulation of miR-206 and miR-599. Cancer Cell Int 2020;20:132. [Crossref] [PubMed]
- Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1. Cancer Biol Ther 2019;20:1355-65. [Crossref] [PubMed]
- Wang Y, Hou Z, Li D. Long noncoding RNA UCA1 promotes anaplastic thyroid cancer cell proliferation via miR-135a-mediated c-myc activation. Mol Med Rep 2018;18:3068-76. [Crossref] [PubMed]
- Lu Y, Sha H, Sun X, et al. CRNDE: an oncogenic long non-coding RNA in cancers. Cancer Cell Int 2020;20:162. [Crossref] [PubMed]
- Li Z, Wu G, Li J, et al. lncRNA CRNDE promotes the proliferation and metastasis by acting as sponge miR-539-5p to regulate POU2F1 expression in HCC. BMC Cancer 2020;20:282. [Crossref] [PubMed]
- Jing H, Xia H, Qian M, et al. Long noncoding RNA CRNDE promotes non-small cell lung cancer progression via sponging microRNA-338-3p. Biomed Pharmacother 2019;110:825-33. [Crossref] [PubMed]
- Pan Z, Li L, Fang Q, et al. Integrated Bioinformatics Analysis of Master Regulators in Anaplastic Thyroid Carcinoma. Biomed Res Int 2019;2019:9734576. [Crossref] [PubMed]
- Wang S, Wu J, Guo C, et al. Identification and Validation of Novel Genes in Anaplastic Thyroid Carcinoma via Bioinformatics Analysis. Cancer Manag Res 2020;12:9787-99. [Crossref] [PubMed]
- Pérez-Benavente B, Nasresfahani AF, Farràs R. Ubiquitin-Regulated Cell Proliferation and Cancer. Adv Exp Med Biol 2020;1233:3-28. [Crossref] [PubMed]
- Ren L, Yang S, Cao Q, et al. CRNDE Contributes Cervical Cancer Progression by Regulating miR-4262/ZEB1 Axis. Onco Targets Ther 2021;14:355-66. [Crossref] [PubMed]
- Ren Y, He W, Chen W, et al. CRNDE promotes cell tongue squamous cell carcinoma cell growth and invasion through suppressing miR-384. J Cell Biochem 2019;120:155-63. [Crossref] [PubMed]
- Ding Q, Mo F, Cai X, et al. LncRNA CRNDE is activated by SP1 and promotes osteosarcoma proliferation, invasion, and epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway. J Cell Biochem 2020;121:3358-71. [Crossref] [PubMed]
- Huan J, Xing L, Lin Q, et al. Long noncoding RNA CRNDE activates Wnt/β-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am J Transl Res 2017;9:1977-89. [PubMed]
- Choi EK, Park EJ, Phan TT, et al. Econazole Induces p53-Dependent Apoptosis and Decreases Metastasis Ability in Gastric Cancer Cells. Biomol Ther (Seoul) 2020;28:370-9. [Crossref] [PubMed]
- Alberti C. Apoptosis induction by quinazoline-derived alpha1-blockers in prostate cancer cells: biomolecular implications and clinical relevance. Eur Rev Med Pharmacol Sci 2007;11:59-64. [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Janz TA, Neskey DM, Nguyen SA, et al. Is the incidence of anaplastic thyroid cancer increasing: A population based epidemiology study. World J Otorhinolaryngol Head Neck Surg 2019;5:34-40. [Crossref] [PubMed]
- Mohan DR, Lerario AM, Finco I, et al. New strategies for applying targeted therapies to adrenocortical carcinoma. Curr Opin Endocr Metab Res 2019;8:72-9. [Crossref] [PubMed]
- Hu S, Liao Y, Chen L. Identification of Key Pathways and Genes in Anaplastic Thyroid Carcinoma via Integrated Bioinformatics Analysis. Med Sci Monit 2018;24:6438-48. [Crossref] [PubMed]
- Zhu LY, Zhu YR, Dai DJ, et al. Epigenetic regulation of alternative splicing. Am J Cancer Res 2018;8:2346-58. [PubMed]
- Wan Y, Zhang X, Leng H, et al. Identifying hub genes of papillary thyroid carcinoma in the TCGA and GEO database using bioinformatics analysis. PeerJ 2020;8:e9120. [Crossref] [PubMed]
- Ding C, Wei R, Rodríguez RA, et al. LncRNA PCAT-1 plays an oncogenic role in epithelial ovarian cancer by modulating cyclinD1/CDK4 expression. Int J Clin Exp Pathol 2019;12:2148-56. [PubMed]
- Tan SH, Barker N. Wnt Signaling in Adult Epithelial Stem Cells and Cancer. Prog Mol Biol Transl Sci 2018;153:21-79. [Crossref] [PubMed]
- Benard O, Qian X, Liang H, et al. p21CIP1 Promotes Mammary Cancer-Initiating Cells via Activation of Wnt/TCF1/CyclinD1 Signaling. Mol Cancer Res 2019;17:1571-81. [Crossref] [PubMed]
- Kai J, Wang S. Recent progress on elucidating the molecular mechanism of plasmid-mediated colistin resistance and drug design. Int Microbiol 2020;23:355-66. [Crossref] [PubMed]
- Sun GC, Liang WZ, Jan CR. Mechanisms underlying the effect of an oral antihyperglycaemic agent glyburide on calcium ion (Ca2+) movement and its related cytotoxicity in prostate cancer cells. Clin Exp Pharmacol Physiol 2020;47:111-8. [Crossref] [PubMed]
- Gao X, Abdelkarim H, Albee LJ, et al. Partial agonist activity of α1-adrenergic receptor antagonists for chemokine (C-X-C motif) receptor 4 and atypical chemokine receptor 3. PLoS One 2018;13:e0204041. [Crossref] [PubMed]
- Zhang Y, Berger SA. Increased calcium influx and ribosomal content correlate with resistance to endoplasmic reticulum stress-induced cell death in mutant leukemia cell lines. J Biol Chem 2004;279:6507-16. [Crossref] [PubMed]
- Sebastian J, Rathinasamy K. Sertaconazole induced toxicity in HeLa cells through mitotic arrest and inhibition of microtubule assembly. Naunyn Schmiedebergs Arch Pharmacol 2021;394:1231-49. [Crossref] [PubMed]
- Dong C, Chen Y, Ma J, et al. Econazole nitrate reversed the resistance of breast cancer cells to Adriamycin through inhibiting the PI3K/AKT signaling pathway. Am J Cancer Res 2020;10:263-74. [PubMed]
(English Language Editor: A. Kassem)


