
Bromodomain containing 4 transcriptionally activated Deltex E3 ubiquitin ligase 2 contributes to glioma progression and predicts an unfavorable prognosis
Introduction
Gliomas are the most common brain tumors. Under the World Health Organization (WHO) 2016 classification system, there are 5 subtypes of glioma, which are categorized based on tumor morphology and molecular genetics (1). Glioblastoma multiforme (GBM) is the most common type of glioma, and the most aggressive brain malignancy in adults (2,3). Diagnostic (4,5) and therapeutic (6-10) advances have been made, but patients with GBM still have a poor prognosis (2,11). Molecular alterations [e.g., isocitrate dehydrogenase 1 (IDH1) mutations, telomerase reverse transcriptase promoter mutations, and 1p/19q co-deletions] have been observed in glioma (12,13), and glioma patients with distinct molecular subtypes show different outcomes and therapeutic responses (14). The identification of novel survival status-related markers and the validation of their functions in glioma could extend current understandings of glioma and shed light on drug development.
It is well established that disturbed signaling transduction mediated by oncogenes is a contributor to cancer cell malignant behaviors and the pathogenesis of glioma (15). Additionally, oncogenes have prognostic significance as biomarkers (16,17). The human Deltex protein family, which functions as ubiquitin E3 ligases, comprises 5 members [i.e., Deltex E3 ubiquitin ligase (DTX) 1, DTX2, DTX3, DTX3L, and DTX4] (18). The DTX family members act as Notch signaling modifiers that control cell fate determination (19), and the pathogenesis and progression of various tumors (18,20,21). The effects of DTX2 on cell differentiation were demonstrated, DTX2 changes the methylation status of H3K9 in the distal regulatory region of the MyoD promoter and directly inhibits demethylase activity of Jumonji domain-containing 1C (JMJD1C) by monoubiquitination to reduce MyoD expression (22). However, studies of DTX2 in cancer are limited, and very few studies of the role of DTX2 in glioma have been conducted. In colon cancer patients, DTX2 has been shown to serve as a predictor of relapse in high-risk stage II and III cancer samples (23). A recent study found that DTX2 recognizes and recruits ADP-ribosylated proteins (24). The role of DTX2 in glioma is yet unknown.
In the present study, we first conducted a survival-status analysis to identify survival status-related genes in The Cancer Genome Atlas (TCGA) GBM and low-grade glioma (LGG) data sets. Subsequently, we identified 3 novel prognostic makers, among which DTX2 had the best performance in the validated Chinese Glioma Genome Atlas (CGGA) data sets. To examine the function of DTX2 in glioma, short-hairpin ribonucleic acids (shRNAs) were designed to silence DTX2 expression, and the growth and migration capabilities of 2 cell lines were examined. Finally, the molecular basis for increased DTX2 in glioma was investigated, and DTX2 was found to be transcriptionally activated by bromodomain containing 4 (BRD4). This study first uncovered the clinical significance, in-vitro functions and mechanism of DTX2 in glioma, which could provide new options for glioma prognosis prediction and treatment. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-555/rc).
Methods
TCGA and CGGA data analysis
The survival-status analysis was conducted with the online tool, LinkedOmics (25). We used the LinkFinder in the LinkedOmics (http://www.linkedomics.org/) module to screen RNA expressions significantly correlated with the clinical parameter of survival status included in TCGA GBM and LGG data sets (http://www.linkedomics.org/login.php#dataSource). The gene expression, survival heatmaps, and Kaplan-Meier survival plots of the genes were analyzed with Gene Expression Profiling Interactive Analysis 2 (GEPIA 2.0) (26). For the CGGA data sets, 3 array data sets (i.e., Array 325, Array 301, and Array693) were used for the analysis (27). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Single-cell data and tumor-immune infiltration analysis
The single-cell data analysis was conducted with the online tool, Single-cell portal (https://singlecell.broadinstitute.org/single_cell). For the tumor-immune infiltration analysis, the Tumor Immune Estimation Resource (TIMER; cistrome.shinyapps.io/timer) was used (28).
Cell lines and cell cultures
DBTRG and U251 cell lines were preserved at our laboratory, and cultured in a humidified atmosphere of 5% carbon dioxide at 37 ℃. DBTRG and U251 were cultured in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum (FBS). Thawed cells from liquid nitrogen were used for the first 3 passages.
Real-time PCR
Total RNA was extracted from DBTRG and U251 cells using TRIzol® RNA Isolation Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Reverse transcription was performed using the PrimeScriptTM RT reagent kit (Takara, Dalian, China). All messenger RNA levels were normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The following DTX2 primers were used in this study: 5'-GTCCACAGCGTCTGGATACAG-3' (F) and 5'-CCTTATTGCCGTTGCAGTACA-3' (R). The following GAPDH primers were used in this study: 5'-TGACTTCAACAGCGACACCCA-3' (F) and 5'-CACCCTGTTGCTGTAGCCAAA-3' (R). All samples were treated under the same conditions and analyzed by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) using Synergy Brands (SYBR) Premix ExTaqTM (Takara, Dalian, China) in accordance with the manufacturer’s protocol.
DTX2 knockdown with shRNAs
The 3 shRNAs targeting DTX2 were designed and synthesized by Shanghai Sangon Biotech (Shanghai, China). DBTRG and U251 cells were transfected with DTX2 shRNA lentiviral vectors and their control vectors. The DTX2 shRNA target sequences were as follows: shRNA1, 5'-GCTTCATCGAGCAGCAGTTTG-3'; shRNA2, 5'-GCAGCAGTTTGTCCAGCAGAA-3'; and shRNA3, 5'-GGTACAACTACACTGTCAACT-3'.
CCK8 assays
Cell Counting Kit 8 (CCK8) (Sigma, product number: 96992) was used to measure cell viability. Briefly, 7500 DTBRG and U251 cells/well were plated in a 96-well plate and incubated overnight. The cells were then washed with phosphate buffered solution, and then underwent shRNA transfection as indicated. Following transfection, all the media were removed and replaced with fresh medium containing CCK8 reagent. The plate was then incubated for 1 h, and absorbance was measured at 450 nm using FLUOstar Omega (BMG Labtech, Offenburg, Germany).
Wound-healing assays
The DBTRG and U251 cells were seeded into 12-well plates with 1% FBS. The cells achieved 100% confluence. The yellow pipette tips were applied for the straight scratch. The wound distance was measured at the indicated time points, and normalized with the starting time point.
Luciferase assays
Luciferase reporter assays were performed according to a standard protocol as previously described (29). Briefly, U251 cells (3×104 cells/well) were seeded into 24-well plates in triplicate, and allowed to attach for 24 h. The DTX2 promoter wild-type (WT) and mutant (MUT) sequences were as follows: WT, 5'-TGGGCCCTACCGGGACTACATTTCCC-3'; and MUT, 5'-CCCAAGTACAACTTACAAGACGTAAGAAA-3'. The indicated plasmids (DTX2 promoter WT and MUT) and 1.5 ng of pRL-TK Renilla plasmid were transfected using Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The cells were treated with dimethyl sulfoxide (DMSO) and JQ1 [1 µM (29)] compounds for 24 h. After 48 h of treatment, the luciferase and Renilla signals were assessed using a Dual-Luciferase Reporter Assay Kit (Promega, cat. No. E1980) in accordance with the manufacturer’s instructions.
Statistical analysis
GraphPad 8.0 software was employed to analyze the data. The values are shown as the mean ± standard deviation. A one-way analysis of variance and Student’s t-test were used for comparisons between 2 groups. All assays were replicated 3 times. A P value <0.05 was considered statistically significant.
Results
The survival-status analysis identified 3 novel unfavorable prognostic markers in glioma
To acquire the gene expression signatures associated with glioma prognosis, we applied the online tool, LinkedOmics (25), to generate survival status-related positive and negative genes in TCGA GBM multiforme and LGG data sets, respectively. As a result, 1,748 and 10,754 genes showed significant (P<0.05) survival status-related performance in GBM and LGG, respectively. The top 50 positive and negative genes in GBM and LGG are shown in Figure 1. By using a relative value of 0.7 and –0.7, 116 and 3,014 differential genes were acquired for GBM and LGG, respectively. Next, we compared the 116 and 3,014 differential genes, and 24 common genes were found to be related in LGG and GBM (see Table 1 and Figure 2A). Next, we constructed overall survival (OS) map showing the prognostic performance of the 24 common genes in the GBM and LGG data sets (see Figure 2B). To further explore the clinical significance of these 24 common genes, the expression of these genes was investigated. GEPIA database was analyzed the expression and OS of 24 common genes in GBM and LGG. The expression of Williams-Beuren syndrome chromosomal region 16 protein (WBSCR16), paxillin (PXN), cytochrome P450 oxidoreductase (POR), membrane-associated RING CH 5 (MARCH5), family with sequence similarity 33, member A (FAM35A), Deltex E3 ubiquitin ligase 2 (DTX2), cleft lip and palate transmembrane 1-like (CLPTM1L), and a previously uncharacterized Rho GTPase-activating protein (ARHGAP12) were upregulated in GBM and LGG tumor tissues compared with normal tissues (see Figure 2C). Finally, Kaplan-Meier survival plots results revealed that the high expression of AGK, CLPTMM1L, DTX2, POR, PXN, and WBSCR16 were also correlated with an unfavorable prognosis in GBM and LGG patients; the low expression of ARHGAP12, FAM35A, MARCH5 and PPAGB were also correlated with good prognosis in GBM and LGG patients (see Figure 2D) (30).
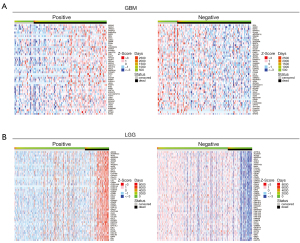
Table 1
| Gene | LGG | GBM | |||
|---|---|---|---|---|---|
| Statistic | P value | Statistic | P value | ||
| ABCB7 | 0.794051 | 0.001293 | −0.72152 | 0.016578 | |
| AGK | 0.893597 | 0.000152 | 0.804206 | 0.00504 | |
| ARHGAP12 | −0.98946 | 3.07E−11 | −0.73121 | 0.000422 | |
| CLPTM1L | 0.892507 | 0.000843 | 0.873131 | 0.001093 | |
| CUL1 | 1.085598 | 4.24E−05 | 1.127012 | 0.000152 | |
| DEFA5 | −1.19452 | 0.000762 | −1.96127 | 0.033578 | |
| DTX2 | 0.914772 | 3.05E−10 | 0.768018 | 0.00157 | |
| DUSP21 | 2.393109 | 0.000269 | 1.549411 | 0.035974 | |
| EXOC3 | −1.10342 | 4.59E−05 | 0.731373 | 0.034279 | |
| FAM35A | −0.90858 | 8.87E−07 | −0.82252 | 0.000819 | |
| IL23R | 1.102708 | 0.002592 | 1.144196 | 0.002301 | |
| MARCH5 | −2.30162 | 1E−16 | −0.82393 | 0.003978 | |
| POR | 0.950188 | 2.89E−05 | 0.946453 | 0.000196 | |
| PXN | 0.91187 | 2.61E−05 | 0.704881 | 0.001214 | |
| RNF10 | 0.795993 | 0.022542 | 0.770314 | 0.049849 | |
| RRAGB | −1.30144 | 1.01E−14 | −0.73706 | 0.000422 | |
| RXRB | −1.86159 | 8.75E−14 | −0.75101 | 0.006494 | |
| SAMM50 | −1.61489 | 9.24E−08 | −0.76129 | 0.002265 | |
| SCO1 | 1.4827 | 4.25E−05 | 0.723089 | 0.024169 | |
| TFAM | −0.83161 | 0.000483 | −0.70665 | 0.004929 | |
| UBE2Z | 0.832866 | 0.014292 | 0.865427 | 0.018523 | |
| VPS52 | −2.2241 | 5.75E−12 | −0.87765 | 0.006009 | |
| WBSCR16 | 1.150865 | 3.37E−07 | 0.971678 | 6.72E−05 | |
| ZNF705D | 1.128865 | 0.009759 | 0.773717 | 0.015574 | |
LGG, low-grade glioma; GBM, glioma multiforme.
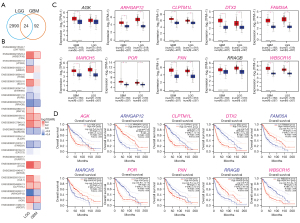
Of the above identified 5 markers, CLPTM1L has been reported to predict poor survival outcomes and resistance to temozolomide therapy in glioma (31). Similarly, PXN has also been proven to serve as an oncogene in glioma progression, and may be a new potential target for therapy (32). As studies on DTX2, POR, and RCC1L (WBSCR16) are limited in glioma, we conducted a further analysis of these 3 genes.
DTX2 is a novel prognostic marker and is positively correlated with immune cells in LGG
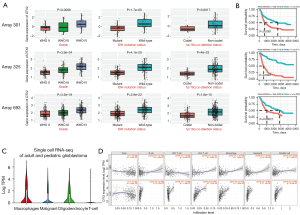
Next, we validated the expression of these 3 genes in the CGGA database (27). Of the 3 data sets included in the CGGA database, DTX expression level was correlated with WHO grade, IDH mutation status and 1p/19q co-deletion status (see Figure 3A). Additionally, glioma patients with higher DTX2 expression also had worse survival outcomes, which suggests that DTX2 is an unfavorable prognostic marker (see Figure 3B). An examination of the expression of DTX2 in the single-cell data of the glioma samples revealed that DTX2 was highly expressed in both malignant and immune cells (see Figure 3C). As the tumor immune microenvironment plays an important role in glioma progression and therapeutic responses, we analyzed the expression significance of tumor immune infiltrating cells using TIMER. No strong correlation was observed in GBM, but the correlation coefficients in LGG were higher in B cells (Corr =0.429), CD4+ T cells (Corr =0.647), macrophages (Corr =0.573), neutrophils (Corr =0.495), and dendritic cells (Corr =0.563) (see Figure 3D).
Silencing DTX2 significantly repressed glioma cell growth and migration in vitro
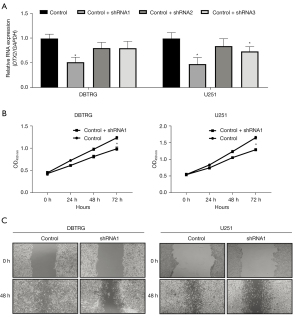
To study the function of DTX2 in glioma cell malignant behavior, the expression of DTX2 was silenced with 3 shRNAs (shRNA 1–3) in the 2 glioma cell lines of DBTRG and U251 cell lines. The knockdown efficiency was examined with qRT-PCR, and shRNA1 had the highest knockdown efficiency (see Figure 4A), and thus was used in subsequent studies. Next, CCK8 assays were conducted to study the growth difference between the control and DTX2-silenced glioma cells. As Figure 4B shows, the cell growth capability of DTX2-silenced glioma cells was significantly lower at 72 h in both cell lines compared with control, which suggests that repressing DTX2 expression may provide a glioma growth repressing strategy. Finally, the migration capabilities of the cells were examined using wound-healing assays, DTX2 silencing was found to significantly decrease the migration distance compared to that of the control cells (see Figure 4C). These in-vitro data suggest that DTX2 contributes to cancer cell growth and migration, and DTX2 could serve as a therapeutic target in glioma.
DTX2 is transcriptionally activated by BRD4 in glioma
Finally, to investigate the transcriptional reasons for the significantly higher expression of DTX2 in glioma, we first analyzed transcriptional factors with the chromatin immunoprecipitation (ChIP)-sequencing data provided in ChIP-atlas. The transcription activation signals (DNase-Seq, RNA polymerase II, histoneH3lysine 4 trimethylation (H3K4me3), andhistoneH3lysine 27 acetylation (H3K27ac) in the DTX2 promoter region suggested a transcriptional activation status in glioma tissues and cell lines (see Figure 5A). Of all the transcription factors included, 3 transcriptional regulons (i.e., MAX, BRD4, and BRD2) had binding signals at the DTX2 promoter. Next, the expression correlations of MAX, BRD4, and BRD2 with DTX2 were validated with TCGA and the CGGA data sets. As Figure 5B,5C show, the expression level BRD4 was positively correlated with DTX2 expression in both data sets (Corr =0.42 for TCGA and Corr =0.514 for the CGGA). Furthermore, thus, we used BRD4 in the subsequent assays.
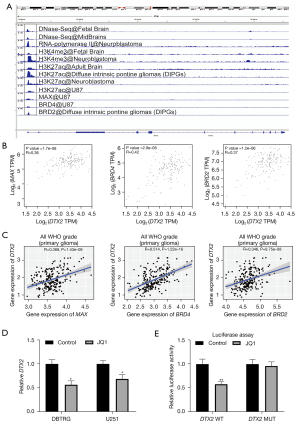
Multiple studies have proven that BRD4 serves as an oncogene and a potential therapeutic target in glioma (33,34). JQ1 is one of the most widely used BRD4 inhibitors (34). Thus, the expression of DTX2 was examined with qRT-PCR after BRD4 treatment, and JQ1 treatment was found to decrease the abundance of DTX2 in both cell lines (see Figure 5D). Finally, a luciferase assay was conducted to validate the transcriptional regulation of BRD4 in DTX2, and while BRD4 treatment significantly reduced luciferase intensity in the DTX2 promoter WT group, no significance was observed in the MUT group (see Figure 5E). In conclusion, these results suggest that DTX2 contributes to cell growth and migration, serves as an unfavorable marker, and is transacted by BRD4 in glioma.
Discussion
Glioblastoma patients have a poor prognosis, and the identification of novel makers may benefit patients by predicting their survival status and time. By conducting a survival-status and time analysis, we identified 10 genes related to prognosis. A subsequent expression analysis revealed 3 novel genes; that is, DTX2, POR, and RCC1L. POR, also known as CYPOR, encodes an endoplasmic reticulum membrane, oxidoreductase, which is essential in multiple metabolic processes (35). In breast cancer, POR has been revealed to be a novel and independent prognostic biomarker (36). WBSCR16 is located in a large deletion region of Williams-Beuren syndrome (WBS), and encodes a protein containing regulator of chromosome condensation 1-like repeats. Studies have shown that WBSCR16 is essential for mitochondrial function (37-39), and is involved in the regulation of oxidative phosphorylation (40,41). In cancer, WBSCR16 has been found to be significantly correlated with the survival of adrenal cortical carcinoma (42). The role of DTX3L in glioma remains largely unexplored (18). DTX3L has been shown to be upregulated in glioma, and is associated with glioma progression (43). This study investigated the role of DTX2 in glioma, and our team intends to examine the role of the other 2 genes in the future.
We first characterized DTX2 as a novel prognostic marker in the GBM and LGG samples. Notably, DTX2 showed higher expression with advanced WHO grades III and IV compared with WHO grades II, which also suggests that it has progression-related clinical significance. The results of the subsequent cellular assays also supported this finding, as DTX2-silenced cell lines exhibited repressed growth and migration capabilities. Additionally, the expression of DTX2 was also increased in IDH WT and 1p/19q co-deletion glioma samples, which implies that its role in glioma may be maintained irrespective of these molecular changes. The initial functional evaluation of DTX2 in the 2 glioma cell lines suggests that DTX2 could be a therapeutic target for glioma, but further in-vivo studies and more gain-of-function studies need to be conducted to validate this in-vitro phenotype. Besides, it is better to validate the expression of DTX2 by real world data, and explore the effects of DTX2 on apoptosis. We will carry out these experiments in the future. Recently, Ahmed et al. showed that the C-terminal domain harbors an adenosine diphosphate (ADP)-ribose-binding pocket and recruits poly-ADP-ribose (PAR)-modified proteins (e.g., PARP1) for ubiquitination (24). It has been revealed that ubiquitination of NOTCH2 by DTX3 suppresses the proliferation and migration of human esophageal carcinoma (44). Given that the substrates of E3 ligases differ with cancer types and pathological conditions, the specific substrate of DTX2 in glioma still needs to be explored.
Finally, BRD4 was proposed to be a transcriptional regulator of DTX2 in glioma. JQ1, which is one of the most widely used BRD4 inhibitors (34), reduced the expression of DTX2. Notably, BRD4 inhibitors may overcome resistance to PARP inhibitors in multiple cancer types (45-47). Given that DTX2 is transcriptionally activated by BRD4, and may mediate the ubiquitination of PARP1, we propose the regulation axis of BRD4-DTX2-PARP1 in glioma, which may provide an alternative option for BRD4 and PARP inhibitors in glioma.
Acknowledgments
Funding: This study was supported by a grant from the Natural Science Foundation of Fujian Province (No. 2019J01500).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-555/rc).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-555/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Molinaro AM, Taylor JW, Wiencke JK, et al. Genetic and molecular epidemiology of adult diffuse glioma. Nat Rev Neurol 2019;15:405-17. [Crossref] [PubMed]
- Tan AC, Ashley DM, López GY, et al. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin 2020;70:299-312. [Crossref] [PubMed]
- Lah TT, Novak M, Breznik B. Brain malignancies: Glioblastoma and brain metastases. Semin Cancer Biol 2020;60:262-73. [Crossref] [PubMed]
- Butler M, Pongor L, Su YT, et al. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020;6:380-91. [Crossref] [PubMed]
- Gatto L, Franceschi E, Di Nunno V, et al. Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives. Oncologist 2021;26:865-78. [Crossref] [PubMed]
- Yan G, Wang Y, Chen J, et al. Advances in drug development for targeted therapies for glioblastoma. Med Res Rev 2020;40:1950-72. [Crossref] [PubMed]
- Wang H, Xu T, Huang Q, et al. Immunotherapy for Malignant Glioma: Current Status and Future Directions. Trends Pharmacol Sci 2020;41:123-38. [Crossref] [PubMed]
- Wen PY, Stein A, van den Bent M, et al. Dabrafenib plus trametinib in patients with BRAFV600E-mutant low-grade and high-grade glioma (ROAR): a multicentre, open-label, single-arm, phase 2, basket trial. Lancet Oncol 2022;23:53-64. [Crossref] [PubMed]
- Dunn GP, Cloughesy TF, Maus MV, et al. Emerging immunotherapies for malignant glioma: from immunogenomics to cell therapy. Neuro Oncol 2020;22:1425-38. [Crossref] [PubMed]
- Sulman EP, Ismaila N, Armstrong TS, et al. Radiation Therapy for Glioblastoma: American Society of Clinical Oncology Clinical Practice Guideline Endorsement of the American Society for Radiation Oncology Guideline. J Clin Oncol 2017;35:361-9. [Crossref] [PubMed]
- Reardon DA, Brandes AA, Omuro A, et al. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol 2020;6:1003-10. [Crossref] [PubMed]
- Tejada Neyra MA, Neuberger U, Reinhardt A, et al. Voxel-wise radiogenomic mapping of tumor location with key molecular alterations in patients with glioma. Neuro Oncol 2018;20:1517-24. [Crossref] [PubMed]
- Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019;565:654-8. [Crossref] [PubMed]
- Molinaro AM, Hervey-Jumper S, Morshed RA, et al. Association of Maximal Extent of Resection of Contrast-Enhanced and Non-Contrast-Enhanced Tumor With Survival Within Molecular Subgroups of Patients With Newly Diagnosed Glioblastoma. JAMA Oncol 2020;6:495-503. [Crossref] [PubMed]
- Tomiyama A, Ichimura K. Signal transduction pathways and resistance to targeted therapies in glioma. Semin Cancer Biol 2019;58:118-29. [Crossref] [PubMed]
- Berzero G, Bellu L, Baldini C, et al. Sustained Tumor Control With MAPK Inhibition in BRAF V600-Mutant Adult Glial and Glioneuronal Tumors. Neurology 2021;97:e673-83. [Crossref] [PubMed]
- Xie Z, Janczyk PŁ, Zhang Y, et al. A cytoskeleton regulator AVIL drives tumorigenesis in glioblastoma. Nat Commun 2020;11:3457. [Crossref] [PubMed]
- Wang L, Sun X, He J, et al. Functions and Molecular Mechanisms of Deltex Family Ubiquitin E3 Ligases in Development and Disease. Front Cell Dev Biol 2021;9:706997. [Crossref] [PubMed]
- Miyamoto K, Fujiwara Y, Saito K. Zinc finger domain of the human DTX protein adopts a unique RING fold. Protein Sci 2019;28:1151-6. [Crossref] [PubMed]
- Wang L, Huang Y, Liu C, et al. Deltex3 inhibits Epithelial Mesenchymal Transition in Papillary Thyroid Carcinoma via promoting ubiquitination of XRCC5 to regulate the AKT signal pathway. J Cancer 2021;12:860-73. [Crossref] [PubMed]
- Bachmann SB, Frommel SC, Camicia R, et al. DTX3L and ARTD9 inhibit IRF1 expression and mediate in cooperation with ARTD8 survival and proliferation of metastatic prostate cancer cells. Mol Cancer 2014;13:125. [Crossref] [PubMed]
- Luo D, de Morree A, Boutet S, et al. Deltex2 represses MyoD expression and inhibits myogenic differentiation by acting as a negative regulator of Jmjd1c. Proc Natl Acad Sci U S A 2017;114:E3071-80. [Crossref] [PubMed]
- Giampieri R, Scartozzi M, Loretelli C, et al. Cancer stem cell gene profile as predictor of relapse in high risk stage II and stage III, radically resected colon cancer patients. PLoS One 2013;8:e72843. [Crossref] [PubMed]
- Ahmed SF, Buetow L, Gabrielsen M, et al. DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination. Sci Adv 2020;6:eabc0629. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Zhao Z, Zhang KN, Wang Q, et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics Proteomics Bioinformatics 2021;19:1-12. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Gong L, Song J, Lin X, et al. Serine-arginine protein kinase 1 promotes a cancer stem cell-like phenotype through activation of Wnt/β-catenin signalling in NSCLC. J Pathol 2016;240:184-96. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Bhawe K, Felty Q, Yoo C, et al. Nuclear Respiratory Factor 1 (NRF1) Transcriptional Activity-Driven Gene Signature Association with Severity of Astrocytoma and Poor Prognosis of Glioblastoma. Mol Neurobiol 2020;57:3827-45. [Crossref] [PubMed]
- Sun LH, Yang FQ, Zhang CB, et al. Overexpression of Paxillin Correlates with Tumor Progression and Predicts Poor Survival in Glioblastoma. CNS Neurosci Ther 2017;23:69-75. [Crossref] [PubMed]
- Nagaraja S, Vitanza NA, Woo PJ, et al. Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017;31:635-652.e6. [Crossref] [PubMed]
- Gusyatiner O, Bady P, Pham MDT, et al. BET inhibitors repress expression of interferon-stimulated genes and synergize with HDAC inhibitors in glioblastoma. Neuro Oncol 2021;23:1680-92. [Crossref] [PubMed]
- Vonada A, Tiyaboonchai A, Nygaard S, et al. Therapeutic liver repopulation by transient acetaminophen selection of gene-modified hepatocytes. Sci Transl Med 2021;13:eabg3047. [Crossref] [PubMed]
- Pedersen MH, Hood BL, Ehmsen S, et al. CYPOR is a novel and independent prognostic biomarker of recurrence-free survival in triple-negative breast cancer patients. Int J Cancer 2019;144:631-40. [Crossref] [PubMed]
- Reyes A, Favia P, Vidoni S, et al. RCC1L (WBSCR16) isoforms coordinate mitochondrial ribosome assembly through their interaction with GTPases. PLoS Genet 2020;16:e1008923. [Crossref] [PubMed]
- Huang G, Massoudi D, Muir AM, et al. WBSCR16 Is a Guanine Nucleotide Exchange Factor Important for Mitochondrial Fusion. Cell Rep 2017;20:923-34. [Crossref] [PubMed]
- Koyama M, Sasaki T, Sasaki N, et al. Crystal structure of human WBSCR16, an RCC1-like protein in mitochondria. Protein Sci 2017;26:1870-7. [Crossref] [PubMed]
- Proust B, Radić M, Vidaček NŠ, et al. NME6 is a phosphotransfer-inactive, monomeric NME/NDPK family member and functions in complexes at the interface of mitochondrial inner membrane and matrix. Cell Biosci 2021;11:195. [Crossref] [PubMed]
- Arroyo JD, Jourdain AA, Calvo SE, et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab 2016;24:875-85. [Crossref] [PubMed]
- Jang HN, Moon SJ, Jung KC, et al. Mass Spectrometry-Based Proteomic Discovery of Prognostic Biomarkers in Adrenal Cortical Carcinoma. Cancers (Basel) 2021;13:3890. [Crossref] [PubMed]
- Xu P, Tao X, Zhao C, et al. DTX3L is upregulated in glioma and is associated with glioma progression. Int J Mol Med 2017;40:491-8. [Crossref] [PubMed]
- Ding XY, Hu HY, Huang KN, et al. Ubiquitination of NOTCH2 by DTX3 suppresses the proliferation and migration of human esophageal carcinoma. Cancer Sci 2020;111:489-501. [Crossref] [PubMed]
- Sun C, Yin J, Fang Y, et al. BRD4 Inhibition Is Synthetic Lethal with PARP Inhibitors through the Induction of Homologous Recombination Deficiency. Cancer Cell 2018;33:401-416.e8. [Crossref] [PubMed]
- Zhang P, Li R, Xiao H, et al. BRD4 Inhibitor AZD5153 Suppresses the Proliferation of Colorectal Cancer Cells and Sensitizes the Anticancer Effect of PARP Inhibitor. Int J Biol Sci 2019;15:1942-54. [Crossref] [PubMed]
- Wang SP, Li Y, Huang SH, et al. Discovery of Potent and Novel Dual PARP/BRD4 Inhibitors for Efficient Treatment of Pancreatic Cancer. J Med Chem 2021;64:17413-35. [Crossref] [PubMed]


