
CALD1 promotes the expression of PD-L1 in bladder cancer via the JAK/STAT signaling pathway
Introduction
Bladder cancer (BC) is among the most commonly diagnosed cancers of the urinary system. As the ninth most common malignancy worldwide, BC is reported to cause approximately 20,000 deaths annually (1). Radical cystectomy and chemotherapy has long been applied as gold standard for muscle invasive BC, and has been stagnant for nearly three decades (2). Recent advances in immune checkpoint inhibitor therapy have seemingly provided a new approach for advanced BC treatment. However, mainstream initiation of immune checkpoint therapy is obstructed in BC due to the low overall response rate (13–24%) (3). Therefore, studies have focused on screening and identifying potential biomarkers to guide early detection, predict prognosis, and forecast treatment response, especially immune checkpoint blockade.
CALD1, which is located in 7q33, is a gene encoding caldesmon, and has two major isoforms. The high molecular weight caldesmon protein is restricted to visceral and vascular smooth muscle cells, while the low molecular weight isoforms are present in non-smooth muscle cells (4). Caldesmon can bind with calmodulin and actin and regulate smooth muscle contraction. It has also been found to regulate cell morphology, cell motility, and cytokinesis, and may therefore play a role in tumor metastasis and proliferation (5). The overexpression (OE) of CALD1 is associated with metastasis and worse prognosis in various tumor types, such as gastric, colorectal, and oral cancers (6-8). To our knowledge, the association between the CALD1 gene and BC has not been deeply investigated. PD-L1 is a famous immunomodulatory molecule that suppresses the immune response in both physiological and pathological pathways by interacting with its corresponding receptor (9). PD-1 is expressed on activated immune cells, while PD-L1 is expressed on tumor cells. PD-L1 expressed on cancer cells inactivate cytotoxic T cells and attenuate immunosurveillance in the tumor micro environment. PD-L1 blocking therapy provides a new direction for the new treatment of BC. However, there is currently no consistent method to evaluate the efficacy of PD-L1. Thus, it is important to further understand the mechanisms of resistance, identify biomarkers to guide treatment, and develop new combination therapies.
In the present study, we aim to demonstrate expression and prognostic contribution of the CALD1 gene in BC, and further investigate the mechanism of PD-L1 regulation by CALD1 in vitro and in vivo. With the development of multiple genetic and genomic datasets, we strive to provide pilot evidence for understanding the biological activity and therapeutic potential of CALD1 in BC.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4192).
Methods
Data processing
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The gene expression profiles dataset [433 cases, Workflow Type: HTSeqCounts, corrected by trusted platform module (TPM)] and corresponding clinical data (including survival status and survival time records of 385 BC patients) were downloaded from The Cancer Genome Atlas (TCGA) official website (https://portal.gdc.cancer.gov/) for Bladder Cancer projects (TCGA-BLCA). Gene expression data from expression profiles (GSE13507, GSE32894, GSE48075 and GSE31684) and corresponding clinical data were obtained from the Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/geo).
Bioinformatics analyses
The protein-coding gene sequences were filtered from the TCGA-BLCA dataset. By calculating the average expression value, expression values of all probes for the same gene were reduced to a single value for each sample. The Limma package from R Bioconductor35 (10) was used to identify differentially expressed genes (DEGs) between BC samples and adjacent non-tumor samples with the cut-off criteria of |log2 fold change (FC)| >1.5, P<0.05.
The weighed gene co-expression network analysis (WGCNA) R package in was used to construct a gene co-expression network based on the expression data profiles of the DEGs. A Pearson’s correlation coefficient matrix was primarily obtained for all pairwise genes. Subsequently, a weighted adjacency matrix was constructed using the power function: amn = |cmn|β(amn = adjacency between gene n and gene m; cmn = Pearson’ s correlation between gene n and gene m; and β is a soft-thresholding parameter that emphasizes high correlations at the expense of low correlations). The appropriate β value was determined when the degree of independence (R2) was 0.95. Following validation of β, the adjacency was transformed into a topological overlap matrix (TOM) and the corresponding dissimilarity (1 − TOM) was also calculated. Next, average linkage hierarchical clustering was performed according to the TOM-based dissimilarity measure. We used a minimum module size of 30 for the genes dendrogram and some highly similar modules with correlations higher than 0.8 were merged together. Finally, based on the TOM-based dissimilarity measure, genes with similar expression profiles were classified into gene modules through average linkage hierarchical clustering.
Modules related to the clinical characteristics of BC were selected using module eigengenes (MEs). All genes were represented by the expression of MEs in a given module, and modules with a high relevance were screened by evaluating the correlation between MEs and clinical characteristics. Core genes, which are highly associated with each clinical feature, were screened out from the module [cor.gene module membership (MM) >0.8 and cor.gene gene significance (GS) >0.15]. Overlapping core genes were then selected by Venn software online (http://bioinformatics.psb.ugent.be/webtools/Venn/).
The prognostic value and clinical correlation of core genes were investigated in TCGA database, and verified by survival analysis and meta-analysis in four GEO datasets (GSE13507, GSE32894, GSE48075, and GSE31684).
Gene set enrichment analysis (GSEA)
GSEA was performed between high and low expression phenotypes of hub genes in TCGA database using c2.cp.kegg.v6.2.symbols.gmt. The GSEA software (version 6.3) is available on the GSEA website (http://software.broadinstitute.org/gsea/index.jsp). A false discovery rate (FDR) <0.25 and P value <0.05 were used as the cut-off criteria.
Cell culture
The human BC T24 cell line was obtained from the Chinese Academy of Sciences (Shanghai, China), and cultured in RPMI-1640 medium (Gibco), supplemented with 10% fetal calf serum (FCS; Gibco) and 1% penicillin/streptomycin (PS; Gibco), and cultured in a humidified incubator at 37 °C and 5% CO2.
Lentivirus packaging and cell infection
Small interfering RNA (siRNA) targeting the CALD1 (Gene ID: 800) sequence was transformed into short-hairpin RNA (shRNA) (stem-loop-stem structure) and cloned into a plenti-shRNA-GFP-Puro vector (Asia-Vector Biotechnology, Shanghai, China). The siRNA plasmids, together with four kinds of lentiviral packaging plasmids (Table 1), were transfected into 293T cells to generate the corresponding lentivirus. The lentivirus was collected and concentrated after 3 days of incubation. For cell infection, T24 cells were then incubated with CALD1 siRNA and non-silencing control (NC) lentiviruses. Quantitative real-time PCR was used on day 3 post transduction to validate the knockdown efficiency. The cell viability assay, transwell assay, apoptosis assays, and cell cycle analysis were subsequently conducted after confirming the knockdown efficiency.
Table 1
| Plasmid | Sense sequence |
|---|---|
| h-CALD1-sh1 | AGGACGAGCTCAGTTGTAG |
| h-CALD1-sh2 | AGAGGAATGACGATGATGA |
| h-CALD1-sh3 | AGAAAAGCAGTGGTGTCAA |
| h-CALD1-sh4 | CATGTATATTATCACTATA |
In addition, we also constructed a CALD1 OE plasmid. When grew to 80–90% on the six-well plate, the T24 cells were transfected with the CALD1 plasmid (CMV-MCS-3FLAG-SV40-Neomycin, purchased from Asia-Vector Biotechnology) using the transfection reagent Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. An empty pcDNA3.1 (−) plasmid was used as a vector control and was also added to normalize the amount of DNA in each transfection.
PD-L1 activation and JAK/STAT signaling pathway inhibition
IFN-γ is a secretory protein with immunomodulatory effect that can stimulate the expression of PD-L1 via the JAK/STAT3 signaling pathway (11,12). T24 cells were grown to approximately 80% confluence and stimulated with 20 ng/mL recombinant human IFN-γ (Novoprotein Scientific Inc., #C014) for 24 h, and then collected for detection. Ruxolitinib (INCB018424) was the first selective JAK1/2 inhibitor to be used clinically (13). Ruxolitinib (SELLECK, #S1378) was dissolved in dimethyl sulfoxide (DMSO) and added to the culture media at concentrations of 1 µM for 24 h.
Reverse transcription-quantitative PCR (RT-qPCR)
To test the knockdown efficiency of the lentiviruses, RT-qPCR was performed using SG Fast qPCR Master Mix (Sangon Biotech) and the Illumina eco Real-Time PCR System. Total RNA was isolated from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. Reverse transcription was performed using TransScript® II All-in-One First-Strand cDNA Synthesis SuperMix for PCR and TransScript® miRNA First-Strand cDNA Synthesis SuperMix (TransGen Biotech) according to the manufacturer’s instructions, and qPCR was performed in a thermocycling condition as follows: 95 °C for 3 min, followed by 45 cycles of 95 °C for 7 sec, at 57 °C for 10 sec, and at 72 °C for 15 sec. Relative mRNA levels were normalized against GAPDH. Data were analyzed using the 2−ΔΔCq method (14). Primers used in RT-qPCR were as follows: GAPDH forward, 5'-GGAGCGAGATCCCTCCAAAAT-3'; GAPDH reverse, 5'-GGCTGTTGTCATACTTCTCATGG-3'; CALD1 forward, 5'-TGGAGGTGAATGCCCAGAAC-3'; CALD1 reverse, 5'-GAAGGCGTTTTTGGCGTCTTT-3'.
Cell viability assays
Cell viability was measured using the Cell Counting Kit-8 (CCK-8; BBI Life Sciences Corporation). Briefly, phosphate buffered saline (PBS)-treated or transfected cells were seeded into 96-well plates at a density of 1×103 cells/well. When cultured to 0, 24, 48, or 72 h, 10 µL of the CCK-8 reagent was added into each well and further incubated at 37 °C for 2 h. Next, the absorbance rate was measured at 450 nm on a microplate spectrophotometer (BioTek Instruments Inc., Winooski, VT, USA).
Apoptosis and cell cycle analysis
For apoptosis and cell cycle analysis, cells infected with lentivirus-mediated shCALD1 or NC siRNA and blank controls treated with PBS were inoculated into six-well plates, 1×106 cells in each well were harvested, and fixed in 70% ethanol for 1 h. After two washes, cell apoptosis was measured by flow cytometry using the Annexin V-FITC/PI apoptosis detection kit (40302ES20; YEASEN Biotechnology Co., Ltd., Shanghai, China), and cell cycle analysis was measured with the propidium iodide (PI; Sangon Biotech, E607306) in accordance with manufacturer’s guidelines.
Migration and invasion assay
For the assay, the upper chamber of the 24-well transwell chambers with an 8 µm pore size membrane (Corning, Lowell, MA, USA) was filled with 5×104 PBS treated cells or transfected cells and 200 µL culture medium without FCS, while the bottom chamber was filled with 800 µL culture medium containing 10% FCS. Cells were cultured for 24 h at 37 °C and 5% CO2. Cells remaining on the upper surface of the membrane were removed with a cotton swab, while migrating cells or invading cells were washed with ice-cold PBS, fixed with 4% methanol (A500684; BBI Life Sciences Corporation, Shanghai, China) for 30 min, stained with 0.1% crystal violet solution (E607309; BBI Life Sciences Corporation, Shanghai, China) for 30 min, and counted under a light microscope (Olympus Corporation, Tokyo, Japan) with 400 times visual field. The migration assay was carried out according to the above method, while the transwell filters were pre-coated with matrigel (BD Biosciences, Franklin Lakes, NJ, USA) for the cell invasion assay.
Nude mouse xenograft assay
Animal experiments performed at the Tongji University School of Medicine were carried out in accordance with the regulatory animal care guidelines of the United States National Institute of Health (Bethesda, MD, USA) and were approved by the Institutional Animal Care and Use Committee of Shanghai Tenth People’s Hospital (ID: SHSY-IEC-4.1/19-120/01). Briefly, 12 male nude BALB/c mice with an average age of 4–5 weeks were purchased from Sino-British SIPPR/BK Lab Animal Ltd. (Shanghai, China), group-housed in specific pathogen-free (SPF) barrier facilities at 24 °C on a fixed 12 h light and 12 h dark cycle, and fed with SPF chow with free access to sterile water. For the nude mouse xenograft assay, cells (1×106) were injected subcutaneously into the flanks of nude mice. Tumor growth was evaluated weekly post-injection with a vernier caliper and calculated as follows: length × width2/2. The injections were administered in a sterilized hood. After 40 days, mice were euthanized and tumor xenografts were collected for analysis.
Detection of PD-L1 surface-antigen
The cells were divided into six groups treated with IFN-γ (Table 2) and four groups treated with ruxolitinib (Table 3). Cells were isolated and resuspended in staining buffer [PBS containing 1% FCS and 2 mM ethylenediaminetetraacetic acid (EDTA)]. A specific cell surface-antigen antibody from Biolegend against CD117 and STRO-1 or an isotype control labeled by fluorescence were used to label the tested cells, and the percentage of PD-L1 surface-antigen positive cells was analyzed by flow cytometry, in order to compare the difference of PD-L1 cell surface-antigen expression among the experimental groups treated under different conditions.
Table 2
| Group | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| T24 | + | + | + | + | + | + |
| IFN-γ | − | + | − | + | − | + |
| NC | − | − | + | + | − | − |
| CALD1 shRNA | − | − | − | − | + | + |
NC, non-silencing control; shRNA, short-hairpin RNA.
Table 3
| Group | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Vector | + | + | − | − |
| Ruxolitinib | − | + | − | + |
| CALD1 OE | − | − | + | + |
OE, overexpression.
Western blot (WB) analysis
Total cellular proteins were isolated using RIPA buffer (Beyotime, Shanghai, China) and quantified with a BCA Protein Assay Kit (Beyotime). Proteins (40 µg per sample) were loaded in each lane and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) before being transferred to polyvinylidene fluoride (PVDF) membranes (Millipore) via the semi-dry transfer system (Bio-Rad). The membranes were washed with double distilled H2O (ddH2O) and blocked with 5% skim milk in PBS with 0.05% Tween-20 (PBST) for 30 min at room temperature, and then exposed to a primary antibody overnight at 4 °C. The primary antibodies were as follows: CALD1 (1:10,000, ab32330, Abcam, USA), PD-L1 (1:1,000, #13684, CST); p-JAK1 (1:1,000, ab13805, Abcam); JAK1 (1:1,000, ab133666, Abcam); p-JAK2 (1:1,000, #3771, CST); JAK2 (1:1,000, 3230, CST); p-STAT1 (1:1,000, ab109457, Abcam); and STAT1 (1:1,000, ab234400, Abcam). Endogenous GAPDH (1:20,000, #5174, CST) was performed as an internal standard for WB analysis. On the following day, after washing with tris-buffered saline with 0.1% Tween-20 (TBST) thrice for 10 min each time, the membranes were incubated with secondary antibodies for 1 h at room temperature and then visualized and quantified using the Odyssey two-color infrared laser imaging system (LI-COR Biosciences, Lincoln, NE, USA).
Statistical analysis
SPSS software (version 22; SPSS, Inc., Chicago, IL, USA) and GraphPad (Version 5.0, GraphPad Prism Software Inc., San Diego, CA, USA) were used for all statistical analyses. The Wilcoxon rank-sum test was utilized to analyze the significance of difference between two groups, and one-way analysis of variance (ANOVA) was performed to test the significance of difference among three or multiple groups. The chi-square test was used to examine the relationship between the gene expression level and clinicopathologic characteristics. Survival analysis was performed according to Kaplan-Meier analysis and the log-rank test. Disease-free survival (DFS) was defined as the time from randomization to the first event of either disease recurrence or death due to any cause. Overall survival (OS) was defined as the time from randomization to death due to any cause. P values <0.05 were considered to indicate statistically significant differences.
Results
WGCNA and screening of CALD1
Protein-coding gene data (19,957 genes) was extracted from the gene expression data downloaded from TCGA-BLCA. In this study, 414 BC tissues and 19 adjacent non-tumor tissues were considered. By setting |log2 FC| ≥1.5 and adjusted P value <0.05 as the screening criteria, we found 3,083 significantly dysregulated genes, including 1,805 overexpressed and 1,278 downregulated genes.
The co-expression analysis included 358 BC patients with clinical information (Table 4) in the TCGA dataset. Following a quality check through the WGCNA package in R, 332 patients were selected for subsequent analysis. The power of β=5 (scale free R2=0.95) was selected as the soft-thresholding parameter (Figure 1A) to ensure a scale-free network. Average linkage hierarchical clustering identified 20 modules (Figure 1B,1C).
Table 4
| Clinical characteristics | Total (n=358) | % |
|---|---|---|
| Age at diagnosis (y) | ||
| Median [range] | 68.5 [34–90] | |
| <68.5 | 179 | 50 |
| >68.5 | 179 | 50 |
| Gender | ||
| Male | 264 | 73.74 |
| Female | 94 | 26.26 |
| Histological grade | ||
| Low | 19 | 5.31 |
| High | 339 | 94.69 |
| Stage | ||
| I | 1 | 0.28 |
| II | 100 | 27.93 |
| III | 129 | 36.03 |
| IV | 128 | 35.75 |
| T stage | ||
| T1 | 1 | 0.28 |
| T2 | 118 | 32.96 |
| T3 | 185 | 51.68 |
| T4 | 54 | 15.08 |
| N stage | ||
| N0 | 233 | 65.08 |
| N1 | 44 | 12.29 |
| N2 | 74 | 20.67 |
| N3 | 7 | 1.96 |
TCGA, The Cancer Genome Atlas.
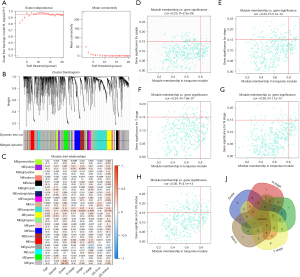
The turquoise module (containing 414 genes) was the only module that was positively associated with most of the crucial prognostic factors. Thus, the turquoise module was selected for subsequent analysis. Genes that were highly correlated with the turquoise module (cor.gene MM >0.8) and highly correlated with grade, stage, T stage, N stage, and survival status (cor.gene GS >0.15) (Figure 1D-1H) were regarded as core genes. In this way, CALD1 was the only overlapping gene (Figure 1I).
CALD1 was associated with the clinicopathologic features and prognosis of BC
We determined the association of CALD1 expression level with the clinicopathologic features and prognosis of BC in the TCGA dataset and four GEO databases. Next, we found that the increased expression of CALD1 was significantly correlated with histological grade, clinical stage, T stage, and lymphatic metastasis (Figure 2A-2D, all P<0.001). Kaplan-Meier survival curves showed that high CALD1 expression was associated with poor OS (Figure 2E, P=0.0024) and DFS (Figure 2F, P=0.013) in TCGA database, and with poor OS in the four GEO databases (Figure 2G, GSE13507, P=0.011; Figure 2H, GSE32894, P<0.001; Figure 2I, GSE48075, P=0.13; and Figure 2J, GSE31684, P=0.157).
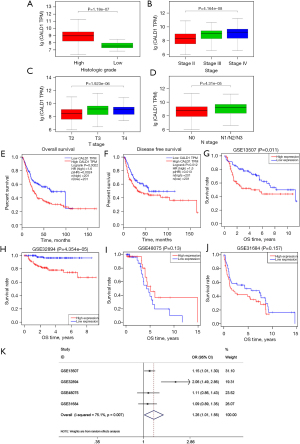
Furthermore, a total of 555 patients from these four BC cohorts in the GEO database were included to assess the correlation between CALD1 expression level and OS by meta-analysis. The random-effects model was expected to be adopted. Analysis showed the pooled odds ratio (OR) was 1.26 [95% confidence interval (CI): 1.01–1.56, P=0.007], which indicated that high expression of CALD1 was predictive of poor OS (Figure 2K).
CALD1 promoted BC growth and inhibited tumor cell apoptosis in vitro and in vivo
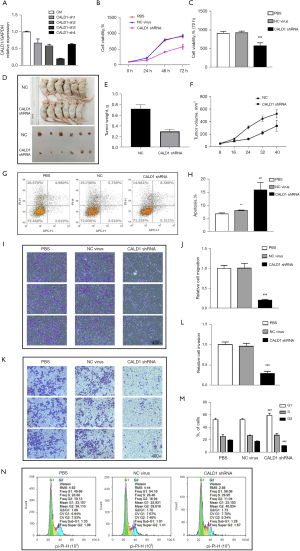
We assessed the effects of CALD1 expression on BC cell proliferation. Transfection of shCALD1 mimics into T24 cells significantly decreased CALD1 expression compared to the PBS blank control cells and siRNA-NC control cells (Figure 3A), and thus, CALD1-sh3, which had the best silencing effect, was selected for further experimentation. We measured the effect of CALD1 silencing on tumor cell growth and found that it could significantly reduce cell viability (Figure 3B,3C) in T24 cells. The nude mouse xenograft assay revealed that the growth of tumor cell xenografts after CALD1 silencing was also suppressed compared to the NC group (Figure 3D-3F). Furthermore, the apoptosis rate of CALD1 silencing tumor cells was upregulated compared to the negative controls (Figure 3G,3H).
CALD1 promoted BC cell migration, invasion, and cell cycle
We evaluated the effect of CALD1 on BC cell migration and invasion capacity. Our data showed that CALD1 silencing significantly reduced T24 cell migration (Figure 3I,3J) and invasion (Figure 3K,3L) compared to the negative controls. Furthermore, we detected the cell cycle using flow cytometry. Compared with NC group cells, in, the number of T24 cells with CALD1 silencing in the G1 phase significantly increased, those in the G2 phase significantly decreased, and the cell cycle was blocked in G1 phase (Figure 3M,3N).
CALD1 expression was correlated with immune infiltration levels and immune checkpoint blockade marker
We assessed the correlation of CALD1 expression with diverse immune cells infiltration levels in BC from the Tumor Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/). The results (Figure 4A) showed that CALD1 expression was negatively correlated with tumor purity (r=−0.565, P=1.53e−32) and positively correlated with the levels of infiltrating immune cells, including B cells (r=−0.184, P=4.25e−04), CD8+T cells (r=0.238, P=4.24e−06), CD4+ T cells (r=0.271, P=1.47e−07), macrophages (r=0.446, P=3.12e−19), neutrophils (r=0.305, P=2.99e−09), and dendritic cells (r=0.284, P=3.19e−08).
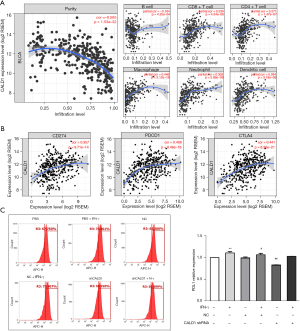
We then investigated the correlation between CADL1 expression and CD274, PDCD1, and CTLA4, which are the most commonly targeted genes by immune checkpoint inhibitors. Interestingly, we found that CADL1 expression was positively correlated with increased CD274 (r=0.357, P=9.71e−14), PDCD1 (r=0.408, P=8.46e−18), and CTLA4 (r=0.441, P=8.36e−21) levels (Figure 4B). To further confirm the correlation between CALD1 and PD-L1 in BC cells, we employed IFN-γ to activate the expression of PD-L1, and detected the PD-L1 cell surface-antigen expression by flow cytometry. We found that the expression of the PD-L1 surface-antigen increased in the IFN-γ treatment group compared to the blank control group (P<0.01) and the shRNA negative control group (P<0.05), and decreased in the CALD1 shRNA transfection group compared to the shRNA negative control group (P<0.05) (Figure 4C).
CALD1 promoted the expression of PD-L1 via the JAK/STAT signaling pathway
So far, we have revealed the pro-tumor activity of CALD1 in BC using bioinformatics methods and cell experiments in vitro, as well as its correlation with immune checkpoint blockade markers. To investigate the potential biological signaling pathways of CALD1 in BC, we performed GSEA between high and low expression subtypes in TCGA database. In this study, we uncovered that CALD1 was also correlated with signaling pathways of BC malignant progression. Our results showed that the CALD1 high expression phenotype was associated with KEGG_BLADDER CANCER, KEGG_PATHWAYS_IN_CANCER, and KEGG_JAK_STAT_SIGNALING_PATHWAY by GSEA (Figure 5A).
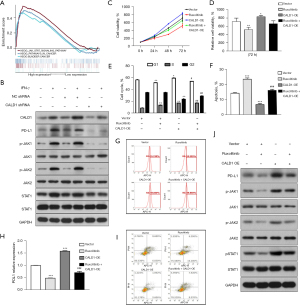
Furthermore, we performed WB analysis to detect the proteins of CALD1, PD-L1, and JAK/STAT signaling pathway-related proteins. We found that the expression of CALD1, PD-L1, p-JAK1/JAK1, p-JAK2/JAK2, and p-STAT1/STAT1 increased in the IFN-γ treatment group compared to the blank control group, and decreased in the CALD1 shRNA transfection group compared to the shRNA negative control group (Figure 5B).
In addition, we employed the JAK inhibitor, ruxolitinib, to treat the Vector group and CALD1 OE group cells. The results of the CCK-8 assay (Figure 5C,5D) showed that the relative cell activity of the CALD1 OE group increased compared to the Vector group (P<0.05), and the relative cell activity decreased significantly after ruxolitinib inhibitor treatment (P<0.01). The results of cell cycle analysis (Figure 5E) showed that the number of CALD1 OE group cells in the G1 phase (P<0.05) and the S phase (P<0.01) increased, while the number of cells in G2 phase (P<0.01) decreased. After ruxolitinib inhibitor treatment, the number of cells in the G1 phase decreased. The apoptosis assay (Figure 5F,5G) showed that cell apoptosis in the CALD1 OE group decreased (P<0.001), and significantly increased after ruxolitinib inhibitor treatment (P<0.001). Using flow cytometry to detect PD-L1 surface antigen (Figure 5H,5I), we found that the PD-L1 surface antigen of CALD1 OE group increased compared to the Vector group (P<0.001), and decreased significantly after ruxolitinib inhibitor treatment (P<0.001). The expression of PD-L1, pJAK1, JAK1, pJAK2, JAK2, pSTAT1, and STAT1 was detected by WB. The results of (Figure 5J) showed that the expression of PD-L1, pJAK1, pJAK2, and pSTAT1 increased after CALD1 OE, and decreased significantly after ruxolitinib inhibitor treatment.
Discussion
The role of CALD1 in the progression and prognosis of BC has been reported in several previous studies (15,16); however, the underlying mechanism was not evaluated. In this study, we found that CALD1 and PD-L1 were overexpressed in BC. Both CALD1 and PD-L1 may serve as blocking targets for the treatment of cancer. Furthermore, CALD1 may promote the expression of PD-L1 by activating the JAK/STAT pathway, thus affecting the progression and prognosis of BC.
In the present study, we first employed the gene expression data of 433 BC samples downloaded from TCGA database. By differential analysis and WGCNA, CALD1 was screened as the only overlapping hub gene associated with BC prognosis. The increased expression of CALD1 in BC was associated with advanced clinicopathologic characteristics (higher grade, clinical stage, T stage, and positive lymphatic metastasis). Meanwhile, we investigated and validated the prognostic value of CALD1 gene in TCGA dataset and four GEO datasets (GSE13507, GSE32894, GSE48075, and GSE31684). Furthermore, the meta-analysis results also demonstrated that BC patients with high CALD1 expression were more susceptible to a shorter OS time.
CALD1 is a novel target of TEA domain family member 4 (17). The TEAD family of transcription factors is necessary for developmental processes (18). CALD1 encodes the caldesmon1 protein, which is an actin- and Ca2+-calmodulin binding protein that was first discovered from chicken gizzard (19), and is involved in actomyosin-based actin filament contraction and stability (20). Previous studies have demonstrated that caldesmon isoforms are differentially expressed in colorectal and gastric cancers (6,21), and are closely related to the migration and invasion of cancer. A subset of the tumor-specific splicing alterations of CALD1 was also found in colon, bladder, and prostate cancers, which may represent a series of general cancer-related splicing events (22). Zheng et al. (23) reported that splicing variants of CALD1 are differentially expressed in glioma neovascularization versus normal brain microvasculature. The activated isoforms of CALD1 can induce protein OE and have a synergetic effect on the cell contractility of vascular components, enhancing the permeability of microvessels, and thereby promoting the extravasation and migration of tumor cells (24). The above findings point to a potentially important role of the CALD1 gene in carcinogenesis.
Although combination chemotherapy is widely applied in BC, the recurrence rate of is exceptionally high and the prognosis after tumor metastasis is extremely poor (25). Immune checkpoint inhibitor therapy has continued to demonstrate clinical response and survival benefit in patients with variety of tumors (26). Immunotherapy blocking of PD-L1 is a promising approach. The PD-1/PD-L1 pathway has been shown to be a crucial immune checkpoint, and anti-PD-L1 therapy has been used in a variety of cancers, including non-small cell lung carcinoma, Hodgkin’s lymphoma, renal cell carcinoma, and BC (27). Several anti-PD-L1 antibodies, such as atezolimumab, pembrolizumab, durvalumab, nivolumab and avelumab, have been approved for the treatment of metastatic BC (28). The binding of PD-L1 to PD-1 alters the immune activity to block the immune response of the patients and continue growing (29). Multiple solid tumor types generate an immunosuppressive tumor microenvironment and avoid T cell cytolysis by expressing PD-L1 (30). PD-L1 OE in cancers is also associated with poor clinical outcomes and predicts a poorer clinical prognosis across tumor histologies (31). Studies have shown that PD-L1 was significantly associated with clinicopathological parameters that indicate a more aggressive phenotype of BC, such as more advanced and higher tumor grades. In addition, the high level of PD-L1 expression was also related to the decreased survival rate of BC patients (32). However, PD-L1 expression was found in about 20% to 30% of bladder urothelial carcinomas (33). For PD-L1 positive patients, the effectiveness of anti-PD-L1 therapy is of significance. When evaluating the role of PD-1/PD-L1 as a predictor of ICI treatment, a composite assessment of patient selection should be made. In addition, the combination of anti-PD-1/PD-L1 therapies and more classic drugs, including trials of intravesical BCG or chemotherapy, is also opening up new methods for the treatment of aggressive BC (34). Therefore, therapeutic intervention targeting PD-L1 expression has becoming very promising.
The activation of different signaling pathways has been demonstrated to induce the expression of PD-L1 in cancer cells (35). Tumor-activated neutrophils in gastric cancer induce PD-L1 expression via the GM-CSF-PD-L1 pathway (36). The Hippo Pathway component TAZ can promote immune evasion through PD-L1 (37). Also, FGFR2 promotes expression of PD-L1 in colorectal cancer via the JAK/STAT3 signaling pathway (38). The expression of PD-L1 is related to immune surveillance. In the process of cell transformation and tumorigenesis, the up-regulation of PD-L1 by abnormal signaling pathways or gene mutations weakens the activity of immune cells, makes tumor cells evade immune surveillance and enhances their survival and metastatic potential.
Activation of the JAK/STAT pathway is common in BC and promotes cellular proliferation and motility (39). In this study, the results of GSEA showed that the CALD1 high expression phenotype was associated with the JAK/STAT signaling pathway. Based these studies and our bioinformatics results, we hypothesized that CALD1 is involved in regulating PD-L1 expression in BC via the JAK/STAT signaling pathway. Activation of the JAK/STAT pathways can drive PD-L1 expression (36). CALD1 signals effectively activated the JAK/STAT signaling pathway accompanied by increased PD-L1 expression in vitro. In addition, we also observed that knocking down of CALD1 in human BC cells increased the rate of apoptosis and decreased the migration and invasion capability of T24 cells. IFN-γ plays a crucial role in anti-tumor responses and also induces expression of PD-L1 (40). In the process of stimulating PD-L1 expression with INF-γ, we found that PD-L1 was up-regulated and knocking down CALD1 could reduce the upregulation of PD-L1. In addition, after suppressing the JAK/STAT signaling pathway with the JAK-inhibitor, ruxolitinib, OE of CALD1 did not promote the expression of PD-L1. These results further confirmed that CALD1 upregulated PD-L1 expression by activating the JAK/STAT3 signaling pathway. However, the mechanism of the CALD1-JAK/STAT axis requires further investigation. For immunotherapy, combining CALD1 inhibitors with PD-L1 inhibitors may lead to increased sensitivity and a better therapeutic effect.
Acknowledgments
Funding: This study was funded by the Shanghai Youth Science and Technology Talents Sailing Program (20YF1437200), National Natural Science Foundation of China (81802554), Shanghai Municipal Health Commission Fund (202040179), Shanghai Science Committee Foundation (19411967700), National Natural Science Foundation of China (81602469), Shanghai Science Committee Foundation (20PJ1412400) and Shanghai Natural Science Foundation (20ZR1443000).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4192
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4192
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4192). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The animal trials in this study had previously received approval from the Shanghai Tenth People’s Hospital Ethics Committee (ID: SHSY-IEC-4.1/19-120/01). Animal experiments performed at the Tongji University School of Medicine were carried out in accordance with the regulatory animal care guidelines of the United States National Institute of Health (Bethesda, MD, USA).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Richters A, Aben KKH, Kiemeney LALM. The global burden of urinary bladder cancer: an update. World J Urol 2020;38:1895-904. [Crossref] [PubMed]
- Massari F, Di Nunno V, Cubelli M, et al. Immune checkpoint inhibitors for metastatic bladder cancer. Cancer Treat Rev 2018;64:11-20. [Crossref] [PubMed]
- Hayashi K, Yano H, Hashida T, et al. Genomic structure of the human caldesmon gene. Proc Natl Acad Sci U S A 1992;89:12122-6. [Crossref] [PubMed]
- Mayanagi T, Sobue K. Diversification of caldesmon-linked actin cytoskeleton in cell motility. Cell Adh Migr 2011;5:150-9. [Crossref] [PubMed]
- Hou Q, Tan HT, Lim KH, et al. Identification and functional validation of caldesmon as a potential gastric cancer metastasis-associated protein. J Proteome Res 2013;12:980-90. [Crossref] [PubMed]
- Chang KP, Wang CL, Kao HK, et al. Overexpression of caldesmon is associated with lymph node metastasis and poorer prognosis in patients with oral cavity squamous cell carcinoma. Cancer 2013;119:4003-11. [Crossref] [PubMed]
- Kim KH, Yeo SG, Kim WK, et al. Up-regulated expression of l-caldesmon associated with malignancy of colorectal cancer. BMC Cancer 2012;12:601. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Polpitiya AD, Qian WJ, Jaitly N, et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 2008;24:1556-8. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Zhang X, Zeng Y, Qu Q, et al. PD-L1 induced by IFN-γ from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol 2017;22:1026-33. [Crossref] [PubMed]
- Heine A, Held SA, Daecke SN, et al. The JAK-inhibitor ruxolitinib impairs dendritic cell function in vitro and in vivo. Blood 2013;122:1192-202. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Liu Y, Wu X, Wang G, et al. CALD1, CNN1, and TAGLN identified as potential prognostic molecular markers of bladder cancer by bioinformatics analysis. Medicine (Baltimore) 2019;98:e13847 [Crossref] [PubMed]
- Lee MS, Lee J, Kim JH, et al. Overexpression of caldesmon is associated with tumor progression in patients with primary non-muscle-invasive bladder cancer. Oncotarget 2015;6:40370-84. [Crossref] [PubMed]
- Lim B, Park JL, Kim HJ, et al. Integrative genomics analysis reveals the multilevel dysregulation and oncogenic characteristics of TEAD4 in gastric cancer. Carcinogenesis 2014;35:1020-7. [Crossref] [PubMed]
- Zhou Y, Huang T, Cheng AS, et al. The TEAD Family and Its Oncogenic Role in Promoting Tumorigenesis. Int J Mol Sci 2016;17:138. [Crossref] [PubMed]
- Mayanagi T, Morita T, Hayashi K, et al. Glucocorticoid receptor-mediated expression of caldesmon regulates cell migration via the reorganization of the actin cytoskeleton. J Biol Chem 2008;283:31183-96. [Crossref] [PubMed]
- Sobue K, Muramoto Y, Fujita M, et al. Purification of a calmodulin-binding protein from chicken gizzard that interacts with F-actin. Proc Natl Acad Sci U S A 1981;78:5652-5. [Crossref] [PubMed]
- Bisognin A, Pizzini S, Perilli L, et al. An integrative framework identifies alternative splicing events in colorectal cancer development. Mol Oncol 2014;8:129-41. [Crossref] [PubMed]
- Thorsen K, Sørensen KD, Brems-Eskildsen AS, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics 2008;7:1214-24. [Crossref] [PubMed]
- Zheng PP, Sieuwerts AM, Luider TM, et al. Differential expression of splicing variants of the human caldesmon gene (CALD1) in glioma neovascularization versus normal brain microvasculature. Am J Pathol 2004;164:2217-28. [Crossref] [PubMed]
- Helfman DM, Levy ET, Berthier C, et al. Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol Biol Cell 1999;10:3097-112. [Crossref] [PubMed]
- Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet 2009;374:239-49. [Crossref] [PubMed]
- U Gandhy S. Madan RA, Aragon-Ching JB. The immunotherapy revolution in genitourinary malignancies. Immunotherapy 2020;12:819-31. [Crossref] [PubMed]
- Voena C, Chiarle R. Advances in cancer immunology and cancer immunotherapy. Discov Med 2016;21:125-33. [PubMed]
- Lopez-Beltran A, Cimadamore A, Blanca A, et al. Immune Checkpoint Inhibitors for the Treatment of Bladder Cancer. Cancers (Basel) 2021;13:131. [Crossref] [PubMed]
- Barclay J, Creswell J, León J. Cancer immunotherapy and the PD-1/PD-L1 checkpoint pathway. Arch Esp Urol 2018;71:393-9. [PubMed]
- Blank C, Mackensen A. Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: an update on implications for chronic infections and tumor evasion. Cancer Immunol Immunother 2007;56:739-45. [Crossref] [PubMed]
- Wang X, Teng F, Kong L, et al. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther 2016;9:5023-39. [Crossref] [PubMed]
- Huang Y, Zhang SD, McCrudden C, et al. The prognostic significance of PD-L1 in bladder cancer. Oncol Rep 2015;33:3075-84. [Crossref] [PubMed]
- Faraj SF, Munari E, Guner G, et al. Assessment of tumoral PD-L1 expression and intratumoral CD8+ T cells in urothelial carcinoma. Urology 2015;85:703.e1-6. [Crossref] [PubMed]
- Rebola J, Aguiar P, Blanca A, et al. Predicting outcomes in non-muscle invasive (Ta/T1) bladder cancer: the role of molecular grade based on luminal/basal phenotype. Virchows Arch 2019;475:445-55. [Crossref] [PubMed]
- Xue W, Li W, Zhang T, et al. Anti-PD1 up-regulates PD-L1 expression and inhibits T-cell lymphoma progression: possible involvement of an IFN-γ-associated JAK-STAT pathway. Onco Targets Ther 2019;12:2079-88. [Crossref] [PubMed]
- Wang TT, Zhao YL, Peng LS, et al. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut 2017;66:1900-11. [Crossref] [PubMed]
- Janse van Rensburg HJ, Azad T, Ling M, et al. The Hippo Pathway Component TAZ Promotes Immune Evasion in Human Cancer through PD-L1. Cancer Res 2018;78:1457-70. [Crossref] [PubMed]
- Li P, Huang T, Zou Q, et al. FGFR2 Promotes Expression of PD-L1 in Colorectal Cancer via the JAK/STAT3 Signaling Pathway. J Immunol 2019;202:3065-75. [Crossref] [PubMed]
- Li Y, Shan Z, Liu C, et al. MicroRNA-294 Promotes Cellular Proliferation and Motility through the PI3K/AKT and JAK/STAT Pathways by Upregulation of NRAS in Bladder Cancer. Biochemistry (Mosc) 2017;82:474-82. [Crossref] [PubMed]
- Gao Y, Yang J, Cai Y, et al. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer 2018;143:931-43. [Crossref] [PubMed]


