
CBX4 contributes to radioresistance by regulating autophagic activity in esophageal squamous cell carcinoma
Introduction
Esophageal carcinoma (EC) is the 8th most common malignancy, and represents the 6th deadliest cancer worldwide, with a 5-year overall survival of below 20% (1). The broad category of EC mainly comprises esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). As a major histopathologic subtype, ESCC accounts for approximately 80% of all EC cases, and is especially predominant among Asians (2). Treatment of ESCC commonly includes surgical, chemotherapeutic, and/or radiotherapeutic (RT) approaches; among these, RT is one of the most effective therapeutic options (3). Indeed, RT is very important in non-surgical treatment of ESCC; however, radioresistance contributes highly to recurrence and diminishes survival (4). Novel strategies aiming to improve ESCC radiosensitivity are essential (5).
Chromobox 4 [CBX4, or polycomb 2 (Pc2)] is a transcriptional repressor as well as a SUMO E3 ligase (6). The N-terminal chromodomain and 2 SUMO-interacting motifs (SIM) are responsible for the polycomb- and SUMO E3 ligase-dependent functions of CBX4, respectively (7). Previously, CBX4 overexpression has been detected in several types of malignant tumors, and it is therefore considered a therapeutic target. In hepatocellular carcinoma, CBX4 increases vascular endothelial growth factor (VEGF)-associated angiogenesis, with elevated CBX4 expression predicting poor survival (8). In breast cancer, CBX4 exerts oncogenic effects through the Notch1 pathway (9). In colorectal carcinoma, CBX4 blunts metastasis by recruiting HDAC3 to the Runx2 promoter and acts as a tumor suppressor (10). Genomic analysis has revealed several amplified genes in ESCC with potential oncologic roles, including CBX4 (11). However, the function and mechanism of CBX4 in radio-response remain largely undefined.
Autophagy, as a homeostatic mechanism, causes the bulk degradation of long-lived proteins and cellular organelles in eukaryotic cells (12). It has been reported that autophagy plays important anti-carcinogenic roles in early carcinogenesis through removal of dysfunctional mitochondria and abnormal protein aggregates responsible for reactive oxygen species (ROS) production (13). In addition, suppressing autophagy-associated genes damages the DNA and induces carcinogenesis (14). Besides, several studies have also suggested that autophagy plays important roles in tumor cell response to anti-cancer treatments, chemotherapy, and radiotherapy. It was reported that autophagy plays 2 overtly contrasting roles in cancer cell response to radiation-associated stress, exerting cyto-protective and cytotoxic effects (15). Cells under stress showing sustained DNA damage are normally cleared by autophagy. However, once this mechanism fails and cancer formation occurs, malignant cells utilize autophagy to endure stress associated with chemotherapy and/or radiotherapy.
In this manuscript, we investigated the influence of CBX4 on ESCC cells under radiation therapy both in vitro and in vivo, and showed that CBX4 increased ESCC radioresistance by regulating the autophagic activity. These findings suggest CBX4 as a potential therapeutic target for ESCC cells and provide new insights into the molecular mechanisms of radiosensitivity in ESCC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-3630).
Methods
ESCC cells and treatment
Human ESCC TE-13 and KYSE-150 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 µg/mL) (Gibco) in a humid environment with 5% CO2 at 37 ℃. The irradiation groups underwent 2, 4, 6, and 8 Gy X-ray irradiation using single energy 6 MV flattening filter (FF) beams, generated by a medical accelerator (Elekta Precise, Stockholm, Sweden). The autophagy inhibitor CQ was provided by Sigma Aldrich (St. Louis, MO, USA). Cell LC3-II protein levels were determined upon incubation with or without 10 µM chloroquine for 2 h (16).
Lentivirus-mediated CBX4 knockdown
Human CBX4-specific short hairpin RNA (shRNA) (GATGAAGATAGTCAAGAACAA) underwent cloning into the hU6-MCS-Ubiquitin-IRES-Puro vector to yield the hU6-MCS-Ubiquitin-IRES-Puro-CBX4-shRNA plasmid (GeneChem, Shanghai, China). Lentiviral transduction was performed based on a previous report (17). Transfection efficiency was determined by real-time reverse transcription polymerase chain reaction (RT-PCR) and immunoblot. We obtained ESCC cells with stable CBX4 knockdown by puromycin (0.5 mg/mL) selection for 10 days.
A protocol was prepared before the study without registration.
Generation of a cell line stably overexpressing Beclin 1
For the generation of a cell line overexpressing Beclin 1, the pEZ-M02 vector was modified to precisely include the open reading frame of human Beclin 1 by GeneCopoeia (Rockville, MD, USA). Transfection was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as directed by the manufacturer.
Xenograft tumor model
Female 4–5-week old BALB/c nude mice (18–20 g) were assessed. Xenografts of TE-13 cells in mice were obtained by subcutaneously injecting 0.1 mL of normal TE-13 (n=12) or CBX4-knockdown TE-13 (n=12) cells (106 cells) into the right proximal hindlimb. The mice were randomly allocated to 4 groups (n=6), including the Control, CBX4-Knockdown, Control + IR, and CBX4-Knockdown + IR groups. Tumors were assessed at 3-day intervals with a Vernier caliper, and tumor volume (V) was determined as V=(a × b2) ×0.5 (a is the long diameter, and b is the short diameter). For irradiation groups, a RS-2000 biological irradiator (Rad Source Technologies, Suwanee, GA, USA) was employed for irradiation at 6 Gy by X-rays (2 Gy/min) when the tumor volume reached about 80 mm3. At 15 d following cancer cell injection, the mice were euthanized, followed by tumor extraction and weighing (18). Experiments were performed under a project license (No. 050432-4-1212B) granted by The Ethics Committee of Fudan University Shanghai Cancer Center, in compliance with Animal [Scientific Procedures] Act 1986, national or institutional guidelines for the care and use of animals.
Quantitative real-time polymerase chain reaction
Human CBX4 and Beclin 1 mRNA amounts were quantitated with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for normalization, by polymerase chain reaction (PCR) employing Assays-on-demand Gene Expression Products (PE Applied Biosystems, Darmstadt, Germany). Total RNA extraction from cells was carried out with TRIzol (Invitrogen) as directed by the manufacturer. The PrimeScript 1st Strand cDNA synthesis kit (Takara Bio, Kusatsu, Shiga, Japan) was employed for circular DNA (cDNA) production as instructed by the manufacturer. Amplification in 20-μL reactions was performed at 95 ℃ for 2 min, followed by 40 cycles of 94 ℃ (10 s), 59 ℃ (10 s), and 72 ℃ (40 s) on an ABI7900 real-time PCR System (Applied Biosystems, Waltham, MA, USA). The 2-ΔΔCT method was employed for data analysis. Primers for CBX4 and Beclin 1 amplification were purchased from All-in-One™ qPCR Primer (CBX4, HQP021189; Beclin 1, HQP114858; GeneCopoeia) and GAPDH’s primer sequences are listed below: GAPDH-Forward, GTCTTCACCACCATGGAGAAG; GAPDH-Reverse, GTCTTCACCAC CATGGAGAAG.
Immunoblot
Immunoblot was performed essentially as described in a previous report (19), with rabbit primary antibodies (1:1,000) targeting CBX4 (Catalog no. HQP021189, GeneCopoeia), Beclin 1 (Catalog no. HQP114858, GeneCopoeia), cleaved-PARP (Catalog no. 5625, Cell Signaling Technology, Danvers, MA, USA), Bcl-2 (Catalog no. 4223, Cell Signaling), Bax (Catalog no. 5023, Cell Signaling), Bad (Catalog no. 9239, Cell Signaling) and LC3B (Catalog no. 3868, Cell Signaling), as well as mouse anti-p62 antibodies (Catalog no. 88588, Cell Signaling). Mouse anti-GAPDH and rabbit anti-β-actin antibodies (1:500; Bioworld Technology, USA) were applied for normalization.
Immunohistochemistry
For animal tissue samples, immunohistochemistry (IHC) was carried out for detecting CBX4 using rabbit anti-CBX4 antibodies (Abcam, Cambridge, MA, 1:200), and Ki67 with rabbit anti-Ki67 antibodies (Abcam, 1:100). We performed IHC and scoring as described previously (20).
Cell proliferation
Cell Counting Kit-8 (CCK-8) was performed for evaluating ESCC cell proliferation. We used 96-well plates for the assay, with cell density at seeding of 4×103 cells/well. A CCK-8 cell proliferation and cytotoxicity assay kit (Obio, Toronto, Ontario, Canada) was employed to determine cell viability upon treatment, and optical density was read at 450 nm.
Colony formation assessments
Colony formation assessment was based on a previous report (21). Briefly, cells (0 Gy, 300 cells; 2 Gy, 600 cells; 4 Gy, 1,200 cells; 6 Gy, 3,000 cells; 8 Gy, 6,000 cells) were seeded in 6-well plates and incubated at 37 ℃ for 14 d with X-ray treatment at 2, 4, 6, and 8 Gy, respectively, for clonogenic survival assay after radiation. Colony staining was carried out with 1% crystal violet staining for 30 s following 10% formalin fixation (5 min). Colonies with ≥50 cells were numbered under an IX51 light microscope (Olympus, Shinjuku, Tokyo, Japan). Plating efficiency (PE) was calculated as the number of colonies divided by that of seeded cells. The surviving fraction (SF) of each radiation group was corrected by the PE of the non-radiated control group. Dose-response clonogenic survival curves were plotted on a log-linear scale. Cell survival curves based on the mean survival rates of the cell line were fitted to a multi-target single-hit model: S =1− (1– e − D/D0)N, where S is the fraction of cells surviving a dose; D0 is the mean lethal dose. The Dq was defined as the intercept of the extrapolated high dose; N was the extrapolation number; and SER (sensitivity enhancement ratio) was derived as follows: SER = (D0 of control-shRNA treated group)/(D0 of CBX4-knockdown group).
Transmission electron microscopy (TEM)
Fresh cells were washed with chilled phosphate-buffered saline (PBS), fixation with pre-cooled 2% glutaraldehyde (2 hours), and post-fixation with 1% osmium tetroxide. Graded ethanol was employed for sample dehydration, followed by Epon 812 embedding. We used Leica Ultracut R (Leica, Wetzlar, Germany) for sectioning, and ultrathin sections underwent staining with uranyl acetate and lead citrate, followed by observation under a Philips-CM120 (Philips, Amsterdam, Netherlands) transmission electron microscope. The TEM was operated according to a previous report (22).
Immunofluorescence
Cell seeding in 6-well plates was performed at 2×105 cells per well on sterilized coverslips. Upon irradiation, cells underwent PBS washes, 4% formalin fixation, and permeabilization (0.2% Triton X-100 in PBS, 15 min). Then, 2% bovine serum albumin (BSA) in PBS was employed to block non-specific binding (1 h at an ambient temperature), followed by successive incubations with primary antibodies targeting γ-H2AX (Cell Signaling; overnight at 4 ℃) and Alexa Fluor 568-linked donkey anti-rabbit IgG (1:500, Invitrogen; 1 h, ambient) (23). Counterstaining was carried out with Hoechst 33342 for 10 min. A Zeiss LSM 700 Meta confocal microscope (Zeiss, Oberkochen, Germany) was used for sample analysis at 100×.
Autophagy detection
A GFP-mRFP-LC3 lentiviral system (GeneChem, China) was used for autophagosome detection. We conducted ESCC cell transfection with the GFP-mRFP-LC3 lentivirus following the manufacturer’s instructions. Cell culture was performed on glass-bottom dishes, followed by irradiation with a single 8 Gy dose. Autophagosomes were located and quantitated under an LSM5 confocal microscope (Zeiss; 63x oil-immersion objective).
Cell cycle assay
Cell seeding in 6-well plates was performed at 2×105 cells per well. Following overnight culture, cells were irradiated, collected by trypsin treatment, and fixed with chilled 70% ethanol at −20 ℃ overnight. Next, the cells were resuspended in propidium iodide (PI) staining solution (eBioscience, San Diego, CA, USA) and submitted to >2 h incubation at 4 ℃ away from light. A total of 104 events were assessed per sample, and cell cycle distribution was evaluated on a BD FACSVerse with the ModFit LT software (Becton Dickinson and Co., Franklin Lakes, NJ, USA). Triplicate assays were carried out.
Statistical analysis
The software SPSS 22.0 (SPSS Inc., IBM Corp., Chicago, IL, USA) was used for data analysis, with 2-sided P<0.05 indicating statistical significance. Normally distributed data were expressed as mean ± standard deviation (SD), and comparisons were carried out by one-way analysis of variance (ANOVA) with the Tukey post-hoc test. Data with skewed distribution were expressed as median ± interquartile range (IQR), and comparisons were performed by the Wilcoxon rank test for paired samples.
Results
CBX4 contributes to ESCC proliferation
Previous data had identified CBX4 amplification in ESCC by whole-genome and whole-exome sequencing, with functional studies revealing its contribution to cell proliferation, colony formation, and cell invasion in selected cell lines (11). We successfully generated stable CBX4-knockdown cell lines of both TE-13 and KYSE-150 cells by utilizing a specific shRNA against CBX4 (Figure 1A,1B). In comparison with cells administered control shRNA, the CBX4 knockdown groups had significantly decreased proliferation rates for both TE-13 and KYSE-150 cells detected by CCK-8 (Figure 1C).
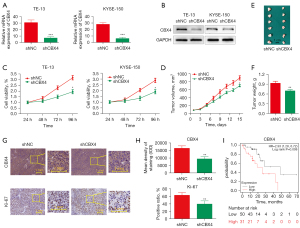
To further demonstrate that CBX4 contributes to tumor growth in vivo, TE-13 cells with stable CBX4 knockdown were subcutaneously administered to BALB/c nude mice. The results showed that tumors grown from CBX4-knockdown cells were much smaller than those generated from control cells. After euthanasia, tumor sizes/volumes in the CBX4-knockdown group were reduced in comparison with control values (Figure 1D,1E). Similar results were obtained for xenograft weights (Figure 1F). Tumor tissues collected from the CBX4-knockdown group had fewer Ki67-positive cells compared with control values (Figure 1G,1H). Moreover, high CBX4 mRNA expression was significantly associated with poor prognosis in ESCC data retrieved from the Kaplan Meier (KM)-plot survival prediction database (http://kmplot.com/analysis/index.php?P=service&start=1; Figure 1I). Collectively, the above data demonstrated that CBX4 was highly correlated with proliferation in ESCC.
CBX4 affects radiosensitivity in ESCC
To investigate the potential regulation of CBX4 involved in radiation response, CBX4 expression levels in ESCC cells were assessed upon treatment with different irradiation doses (0, 2, 4, 6, and 8 Gy). The level of CBX4 in TE-13 and KYSE-150 cells was significantly and dose-dependently upregulated after cell exposure to irradiation (Figure 2A,2B). After CBX4 knockdown, ESCC cell susceptibility to irradiation was dramatically increased. The survival clones were assessed after crystal violet staining, and colony formation data were further submitted to a multi-target single-hit model (Figure 2C,2D). After CBX4 knockdown, the SER values for TE-13 and KYSE-150 cells were 1.13 and 1.22, respectively. These data suggested that CBX4 silencing enhanced ESCC radiosensitivity.
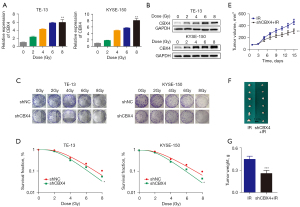
To further demonstrate that knockdown of CBX4 could contribute to enhancing the radiosensitivity of ESCC in vivo, TE-13 cells with stable CBX4 knockdown were subcutaneously administered to BALB/c nude mice, followed by 6 Gy X-ray irradiation. Tumors grown from CBX4-knockdown cells were smaller than those generated by control-shRNA treated cells with radiotherapy. After euthanasia, tumor sizes/volumes in the CBX4-knockdown group were reduced in comparison with those of the control group upon irradiation (Figure 2E,2F). Similar results were acquired for xenograft weights (Figure 2G). The above findings indicated that genetically altering CBX4 markedly influenced radioresistance in ESCC cells both in vitro and in vivo.
CBX4 regulates radiation-associated DNA damage, cell cycle distribution and apoptosis
To explore the mechanisms by which CBX4 affects radiosensitivity in ESCC cell lines, different cellular responses induced by irradiation, including DNA damage, cell cycle arrest, and apoptosis, were measured. First, to determine whether CBX4 is involved in double-strand DNA break repair, the expression pattern of phospho-H2AX (γH2AX), a well-recognized sensitive molecular marker of DNA damage and repair, was detected in CBX4-knockdown and shRNA-control treated cells before and after irradiation. Representative images of γH2AX immunostaining (Figure 3A) and related quantitative analysis (Figure 3B) indicated that γH2AX signals were significantly increased in a short period of time after irradiation, and then slowly dropped in both TE-13 and KYSE-150 cell lines. Meanwhile, the expression levels of γH2AX (Figure 3B) were higher in CBX4-knockdown groups compared with control-shRNA treated counterparts at different time-points after irradiation. In agreement, the expression levels of c-PARP (another indicator of double-strand DNA break), detected by western blot, were higher in CBX4-knockdown cells than control-shRNA treated cells (Figure 3C).
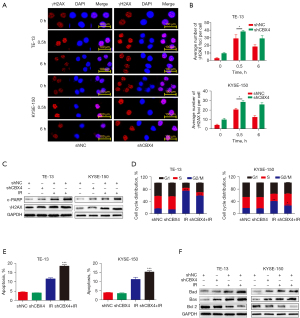
Besides double-strand DNA breaks, cell cycle arrest is another significant parameter for assessing the cell response to irradiation. Next, cell cycle distribution was examined in both CBX4-knockdown and control-shRNA groups before and after irradiation. As shown in Figure 3D and Figure S1A, the percentages of G2/M phase cells were remarkably increased after irradiation in both cell lines; meanwhile, the percentages of G2/M phase cells were markedly reduced in CBX4-knockdown groups and those of S phase cells were increased, in comparison with shRNA-control counterparts. This indicated that CBX4 silencing might help cells escape irradiation-induced G2/M arrest.
Apoptosis is another important cellular response to irradiation. We found that knockdown of CBX4 did not affect cellular apoptosis under normal conditions but increased apoptotic levels after irradiation in both TE-13 and KYSE-150 cell lines (Figure 3E and Figure S1B). In agreement, CBX4 silencing increased BAD and BAX expression levels and decreased BCL2 expression after irradiation (Figure 3F). In general, knockdown of CBX4 increased double-strand DNA damage response, reduced cell cycle arrest, and enhanced cellular apoptosis upon irradiation.
CBX4-knockdown promotes radiation-associated cell death by suppressing autophagosome formation
To further explore the mechanism by which CBX4 regulates radiosensitivity in ESCC, Gene Set Enrichment Analysis (GSEA) was performed to examine the potential pathways or biological processes involved. Interestingly, low CBX4 amounts were correlated with reduced expression levels of certain components of the autophagy pathways [with false discovery rate (FDR)-q value of 0.195], suggesting that low CBX4 expression might negatively regulate autophagic activity in ESCC (Figure 4A).
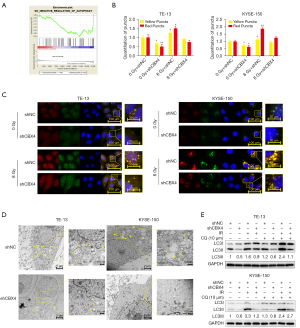
To determine whether CBX4 knockdown would affect the autophagic flux, GFP-mRFP-LC3 was transfected into TE-13 and KYSE-150 cell lines. After puromycin selection, these cells were exposed to 8 Gy irradiation. It was found that autophagosome numbers were significantly decreased both in TE-13 and KYSE-150 CBX4-knockdown cell lines without treatment, in comparison with control-shRNA cells. Similarly, fewer autophagosomes were found both in TE-13 and KYSE-150 CBX4-knockdown cell lines after irradiation in comparison with the control-shRNA groups (Figure 4B,4C). Corroborating data were obtained by TEM for detecting autophagosome formation (Figure 4D). Microtubule-associated protein 1 light chain 3 (MAP1LC3/LC3) represents an autophagy marker. During autophagy, the fat-insoluble form of LC3 (LC3-I) interacts with phosphatidylethanolamine (PE), undergoing transformation to yield the fat-soluble LC3-II, which contributes to the generation of autophagosomes (24). The LC3-II is rapidly degraded by lysosomes; thus, autophagy flux can be easily measured by alterations in the amount of LC3-II (25). To further investigate the role of CBX4 in regulating cellular autophagic activity, transition from LC3 type I to type II was assessed by immunoblot. The protein levels of LC3-II were significantly increased in control-shRNA groups compared with CBX4 knockdown groups with or without irradiation, or after chloroquine (CQ) administration. In comparison with irradiation or CQ administration as monotherapy, the combined administration of irradiation and CQ resulted in more pronounced transition of LC3-II at the protein level in both control-shRNA and CBX4-knockdown groups (Figure 4E).
CBX4 regulates autophagic activity after irradiation by targeting Beclin 1
Beclin 1 controls autophagosome formation and maturation (26-28). The expression of Beclin 1 was examined in CBX4-knockdown and control-shRNA ESCC cell lines after irradiation or not. The data revealed that Beclin 1 levels were dramatically decreased after CBX4 knockdown. Meanwhile, p62, which is considered a specific autophagosome cargo protein, was remarkably upregulated after CBX4 knockdown (Figure 5A).
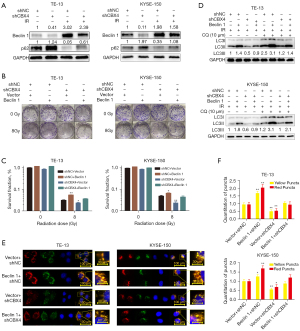
To further determine the role of Beclin 1 in regulating CBX4 for radioresistance and autophagy inhibition in ESCC, Beclin 1 was overexpressed in ESCC cell lines. The results clearly demonstrated that Beclin 1 overexpression increased colony formation after irradiation (Figure 5B,5C) and reversed autophagy inhibition induced by CBX4 silencing according to LC3-II levels (Figure 5D). In agreement, autophagosome formation and maturation were significantly enhanced after Beclin 1 overexpression in ESCC cell lines (Figure 5E,5F). Collectively, the above findings indicated that radioresistance could be partly attributed to irradiation-associated autophagy activation.
Discussion
With recent advances in physics and biology, radiation therapy has moved into the era of precision medicine. Precision radiation radiotherapy at the biological level is also very promising. However, in multiple malignancies, recurrence is linked to acquired radioresistance (29). Developing novel approaches or identifying new biomarkers to overcome resistance is the key to improve the response rate in tumor radiotherapy, fulfilling the requirements of precision radiotherapy at the biological level. Radiotherapy is critical to non-operative treatment of ESCC (30). However, radioresistance prolifically causes recurrence and reduces survival in the ESCC treatment process (31). Moreover, a great deal of ESCC cases show local recurrence upon radiation therapy, with relapsed ESCC being generally more aggressive. Multiple reports have assessed potential target molecules for overcoming ESCC resistance to radiotherapy, few of which have shown beneficial clinical effects (32). As shown above, CBX4 played an oncogenic role by contributing to ESCC growth, and was upregulated by radiation in a dose-dependent manner. Meanwhile, we also investigated the role of CBX4 in radiosensitivity both in cultured cells and nude mice. We firstly demonstrated that CBX4 silencing in ESCC cells could improve radio-response. These findings imply that targeting CBX4 could be a promising approach for overcoming radioresistance in this challenging malignancy.
It was suggested that autophagy is important in regulating carcinogenesis and tumor progression, as well as in determining tumor cell response to antitumor treatment. However, autophagy plays sophisticated roles in these cancer-associated processes, with conflicted consequences in cancer cells (33). To determine the cellular mechanism by which CBX4 modulates radioresistance, we further examined whether this molecule is important in inducing autophagy and participates in resistance to apoptosis associated with stress. As shown above, CBX4 regulated Beclin 1 protein expression. Improved radiosensitivity was observed after Beclin 1 overexpression, indicating that CBX4 contributes to cellular autophagy and radioresistance by regulating Beclin 1. A multi-domain protein, Beclin 1 controls autophagy both positively and negatively. As a gene required for embryo survival and development, Beclin 1 also has a tumor-suppression function with one allele suppressed in multiple malignancies (34,35). Several Beclin 1 domains modulate communication among many ligands that could change its conformation and binding features, thereby making Beclin 1 a critical molecular platform for regulating autophagy (36). Meanwhile, Beclin 1’s BH3 domain mediates its binding to the anti-apoptotic molecule Bcl2. This interaction sterically inhibits PI3K complex synthesis (37), which further explains why CBX4 decreases radiation-induced apoptosis. Modifications could alter Beclin 1/Bcl2 interaction, allowing cells to regulate autophagy in response to several internal and/or external stressors such as irradiation (38). Consequently, CBX4 triggers cyto-protective autophagy after irradiation by targeting Beclin 1 and causes radioresistance in ESCC cells.
As shown above, CBX4 was correlated with radioresistance in ESCC, and regulated radio-response by interacting with autophagic activity. However, the limitations of this study should be addressed. Although we demonstrated that CBX4 might regulate autophagic activity after irradiation by targeting Beclin 1, the underlying molecular mechanism was not thoroughly investigated in this research. It remains unknown whether CBX4 directly interacts with Beclin 1 or binds to other components for indirect regulation to subsequently control autophagic activity. Moreover, for potential clinical application, combining radiotherapy and autophagic activators such as rapamycin should be assessed in further research.
Conclusions
In ESCC radioresistance is promoted by CBX4, and its targeting may be a promising approach to improve tumor response to radiation therapy. The mechanisms underpinning CBX4’s effects deserve further attention, and such effects should be verified in experimental studies and clinical trials.
Acknowledgments
This research was partly presented at ESTRO 38, Milan, Italy.
Funding: This work was supported by National Natural Science Foundation of China (82102827, 82102831,81872454), Shanghai Sailing Program (19YF1409200), Chinese Society of Clinical Oncology/Beijing Xisike Clinical Oncology Research Foundation (Y-Young2020-0003), Beijing Bethune Charitable Foundation (flzh202119), and grants from FUSCC and Shanghai Key Laboratory of Radiation Oncology.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-3630/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-3630/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-3630/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. 050432-4-1212B) granted by The Ethics Committee of Fudan University Shanghai Cancer Center, in compliance with Animal [Scientific Procedures] Act 1986, national or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen W, Xia C, Zheng R, et al. Disparities by province, age, and sex in site-specific cancer burden attributable to 23 potentially modifiable risk factors in China: a comparative risk assessment. Lancet Glob Health 2019;7:e257-69. [Crossref] [PubMed]
- Lagergren J, Smyth E, Cunningham D, et al. Oesophageal cancer. Lancet 2017;390:2383-96. [Crossref] [PubMed]
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400-12. [Crossref] [PubMed]
- Cohen DJ, Leichman L. Controversies in the treatment of local and locally advanced gastric and esophageal cancers. J Clin Oncol 2015;33:1754-9. [Crossref] [PubMed]
- Di Costanzo A, Del Gaudio N, Conte L, et al. The HDAC inhibitor SAHA regulates CBX2 stability via a SUMO-triggered ubiquitin-mediated pathway in leukemia. Oncogene 2018;37:2559-72. [Crossref] [PubMed]
- Ning B, Zhao W, Qian C, et al. USP26 functions as a negative regulator of cellular reprogramming by stabilising PRC1 complex components. Nat Commun 2017;8:349. [Crossref] [PubMed]
- Li J, Xu Y, Long XD, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 2014;25:118-31. [Crossref] [PubMed]
- Zeng JS, Zhang ZD, Pei L, et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol 2018;95:1-8. [Crossref] [PubMed]
- Wang X, Li L, Wu Y, et al. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer Res 2016;76:7277-89. [Crossref] [PubMed]
- Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015;96:597-611. [Crossref] [PubMed]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007;8:931-7. [Crossref] [PubMed]
- Chen X, Wang P, Guo F, et al. Autophagy enhanced the radioresistance of non-small cell lung cancer by regulating ROS level under hypoxia condition. Int J Radiat Biol 2017;93:764-70. [Crossref] [PubMed]
- Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019;9:1167-81. [Crossref] [PubMed]
- Peng WX, Wan YY, Gong AH, et al. Egr-1 regulates irradiation-induced autophagy through Atg4B to promote radioresistance in hepatocellular carcinoma cells. Oncogenesis 2017;6:e292. [Crossref] [PubMed]
- Mauthe M, Orhon I, Rocchi C, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018;14:1435-55. [Crossref] [PubMed]
- He G, Di X, Yan J, et al. Silencing human epidermal growth factor receptor-3 radiosensitizes human luminal A breast cancer cells. Cancer Sci 2018;109:3774-82. [Crossref] [PubMed]
- Zhu H, Yang X, Ding Y, et al. Recombinant human endostatin enhances the radioresponse in esophageal squamous cell carcinoma by normalizing tumor vasculature and reducing hypoxia. Sci Rep 2015;5:14503. [Crossref] [PubMed]
- Yang X, Yang B, Cai J, et al. Berberine enhances radiosensitivity of esophageal squamous cancer by targeting HIF-1α in vitro and in vivo. Cancer Biol Ther 2013;14:1068-73. [Crossref] [PubMed]
- Li J, Zhang N, Song LB, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res 2008;14:3319-26. [Crossref] [PubMed]
- Ding YQ, Zhu HC, Chen XC, et al. Sunitinib modulates the radiosensitivity of esophageal squamous cell carcinoma cells in vitro. Dis Esophagus 2016;29:1144-51. [Crossref] [PubMed]
- Huang Y, Hou JK, Chen TT, et al. PML-RARα enhances constitutive autophagic activity through inhibiting the Akt/mTOR pathway. Autophagy 2011;7:1132-44. [Crossref] [PubMed]
- Qin Q, Cheng H, Lu J, et al. Small-molecule survivin inhibitor YM155 enhances radiosensitization in esophageal squamous cell carcinoma by the abrogation of G2 checkpoint and suppression of homologous recombination repair. J Hematol Oncol 2014;7:62. [Crossref] [PubMed]
- Moretti J, Roy S, Bozec D, et al. STING Senses Microbial Viability to Orchestrate Stress-Mediated Autophagy of the Endoplasmic Reticulum. Cell 2017;171:809-823.e13. [Crossref] [PubMed]
- Di Malta C, Siciliano D, Calcagni A, et al. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science 2017;356:1188-92. [Crossref] [PubMed]
- Liang C, Lee JS, Inn KS, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008;10:776-87. [Crossref] [PubMed]
- Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009;11:385-96. [Crossref] [PubMed]
- Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009;11:468-76. [Crossref] [PubMed]
- Bristow RG, Alexander B, Baumann M, et al. Combining precision radiotherapy with molecular targeting and immunomodulatory agents: a guideline by the American Society for Radiation Oncology. Lancet Oncol 2018;19:e240-51. [Crossref] [PubMed]
- Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol 2017;27:215-21. [Crossref] [PubMed]
- Schaaf MB, Jutten B, Keulers TG, et al. Canonical autophagy does not contribute to cellular radioresistance. Radiother Oncol 2015;114:406-12. [Crossref] [PubMed]
- Chen GZ, Zhu HC, Dai WS, et al. The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J Thorac Dis 2017;9:849-59. [Crossref] [PubMed]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer 2007;7:961-7. [Crossref] [PubMed]
- Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998;72:8586-96. [Crossref] [PubMed]
- Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003;100:15077-82. [Crossref] [PubMed]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010;22:140-9. [Crossref] [PubMed]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005;122:927-39. [Crossref] [PubMed]
- Fernández ÁF, Sebti S, Wei Y, et al. Author Correction: Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018;561:E30. [Crossref] [PubMed]


