Abstract
The striking and complex phenotype of Cockayne syndrome (CS) patients combines progeria-like features with developmental deficits. Since the establishment of the in vitro culture of skin fibroblasts derived from patients with CS in the 1970s, significant progress has been made in the understanding of the genetic alterations associated with the disease and their impact on molecular, cellular, and organismal functions. In this review, we provide a historic perspective on the research into CS by revisiting seminal papers in this field. We highlighted the great contributions of several researchers in the last decades, ranging from the cloning and characterization of CS genes to the molecular dissection of their roles in DNA repair, transcription, redox processes and metabolism control. We also provide a detailed description of all pathological mutations in genes ERCC6 and ERCC8 reported to date and their impact on CS-related proteins. Finally, we review the contributions (and limitations) of many genetic animal models to the study of CS and how cutting-edge technologies, such as cell reprogramming and state-of-the-art genome editing, are helping us to address unanswered questions.
Keywords
Cockayne syndrome; transcription-coupled nucleotide excision repair; neurodegeneration; progeroid syndrome; DNA repair
The epidemiology of Cockayne Syndrome (CS) and the discovery of the CS genes
Edward Alfred Cockayne first described CS in 1936. He diagnosed it in two young siblings (born to healthy parents) that displayed a set of very similar characteristics that included skin photosensitivity, short stature, prominent superior maxillae, disproportionally large hands and feet, sunken eyes with retinal atrophy, hearing impairment, below-average intelligence, a severely limited speaking ability, and muscle contraction, conditions that would later become characteristic of CS patients (Cockayne, 1936Cockayne EA (1936) Dwarfism with retinal atrophy and deafness. Arch Dis Child 11:1–8., 1946Cockayne EA (1946) Dwarfism with retinal atrophy and deafness. Arch Dis Child 11:52–54.; Laugel, 2013Laugel V, Dalloz AC, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Gener B, Tobias ES et al. (2009) Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat 31:113–126.). Subsequent reports also confirmed that patients with CS feature complex and heterogeneous neuropathology that includes calcification of the basal ganglia, cerebellar atrophy, loss of Purkinje and granular cells, hyperchromatic macroglial cells, microcephaly, and patchy demyelination (Guardiola et al., 1999Guardiola A, Álvares-da-Silva CR, Grisolia J and Silbermann R (1999) Síndrome de Cockayne. Arq Neuro-Psiquiatr 57:106–110.; Karam et al., 2000Karam SM, Costa JC, Jardim L, Pires RF, Lehmann AR and Giugliani R (2000) Cockayne syndrome: Report of a Brazilian family with confirmation of impaired RNA synthesis after UV-irradiation. Genet Mol Biol 275:273–275.; Weidenheim et al., 2009Wilson BT, Stark Z, Sutton RE, Danda S, Ekbote AV, Elsayed SM, Gibson L, Goodship JA, Jackson AP, Keng WT et al. (2015) The Cockayne Syndrome Natural History (CoSyNH) study: Clinical findings in 102 individuals and recommendations for care. Genet Med 18:483–493.; Wilson et al., 2015Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA and Wilson DM (2007) Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res 35:4103–4113.; Karikkineth et al., 2017Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL and Bohr VA (2017) Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res Rev 33:3–17.).
CS is an autosomal recessive disorder with a prevalence of ~2.7 per million births in Western Europe and in Japan (Kleijer et al., 2008Kleijer WJ, Laugel V, Berneburg M, Nardo T, Fawcett H, Gratchev A, Jaspers NGJ, Sarasin A, Stefanini M and Lehmann AR (2008) Incidence of DNA repair deficiency disorders in western Europe: Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. DNA Repair (Amst) 7:744–750.; Kubota et al., 2015Kubota M, Ohta S, Ando A, Koyama A, Terashima H, Kashii H, Hoshino H, Sugita K and Hayashi M (2015) Nationwide survey of Cockayne syndrome in Japan: Incidence, clinical course and prognosis. Pedriatrics Int 57:339–347.). The phenotype of the patients can range from mild to very severe and is subdivided into three types. The classical type (type I) corresponds to the moderate phenotype in which life expectancy is 16 years. In type II (the most severe and with the earliest onset), life expectancy is 5 years, whereas in the third type (mild and atypical), the phenotype manifests itself later in life, with life expectancy above 30 years. In all cases, pneumonia/respiratory ailments are the most common causes of death (Natale, 2010Natale V (2010) A comprehensive description of the severity groups in Cockayne Syndrome. Am J Med Genet Part A 155:1081–1095.).
In vitro culture culture of skin fibroblasts derived from patients with CS in the 1970s was the first step toward the development of experimental models of the disease. CS fibroblasts are characterized by extreme sensitivity to ultraviolet light (UV) despite a normal ability to excise pyrimidine-dimers from the genome (Schmickel et al., 1977Schmickel RD, Chu EH, Trosko JE and Chang CC (1977) Cockayne syndrome: A cellular sensitivity to ultraviolet light. Pediatrics 60:135–139.; Andrews et al., 1978Andrews AD, Barrett SF, Yoder FW and Robbins JH (1978) Cockayne’s Syndrome fibroblasts have increased sensitivity to ultraviolet light but normal rates of unscheduled DNA synthesis. J Invest Dermatol 70:237–239.). In fact, CS cells display a marked defect in the recovery of RNA synthesis after UV irradiation (Mayne and Lehmann, 1982Mayne LV and Lehmann AR (1982) Failure of RNA synthesis to recover after UV irradiation: An early defect in cells from individuals with Cockayne’s syndrome and Xeroderma pigmentosum. Cancer Res 42:1473–1478.) owing to a failure in the repair of transcriptionally active genes (Venema et al., 1990Vermeij WP, Dollé MET, Reiling E, Jaarsma D, Payan-Gomez C, Bombardieri CR, Wu H, Roks AJM, Botter SM, van der Eerden BC et al. (2016) Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 537:427–431.; van Hoffen et al., 1993van Houten B, Hunter SE and Meyer JN (2016) Mitochondrial DNA damage induced autophagy, cell death, and disease. Front Biosci 21:42–54.). By evaluation of post-UV RNA synthesis recovery in multinucleated cells obtained by the fusion of cells from different CS donors, three complementation groups (A, B, and C) were initially identified (Tanaka et al., 1981Tantin D (1998) RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J Biol Chem 273:27794–27799.; Lehmann, 1982Lehmann AR (1982) Three complementation groups in Cockayne syndrome. Mutat Res 106:347–356.). Group C identified by Lehmann corresponded to a patient that had combined features of CS and xeroderma pigmentosum (XP). Patients that fall in this category (termed XP/CS) manifest, in addition to CS features, the classical XP characteristics (skin pigmentation and extremely high skin cancer predisposition) and harbor mutations in the genes XPB, XPD, XPG, or (more recently identified) XPF or ERCC1 (Weeda et al., 1990Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D and Hoeijmakers JHJ (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol 7:427–439.; Kashiyama et al., 2013Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L et al. (2013) Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, Xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet 92:807–819.; Lehmann et al., 2014Lehmann J, Schubert S, Schäfer A, Apel A, Laspe P, Schiller S, Ohlenbusch A, Gratchev A and Emmert S (2014) An unusual mutation in the XPG gene leads to an internal in-frame deletion and a XP/CS complex phenotype. Br J Dermatol 171:903–905.; Moriel-Carretero et al., 2015Moriel-Carretero M, Herrera-Moyano E and Aguilera A (2015) A unified model for the molecular basis of Xeroderma pigmentosum-Cockayne syndrome. Rare Dis 3:e1079362.).
In the 1990s, the genes corresponding to groups A and B were cloned, characterized, and termed CSA and CSB, respectively. CSB was originally termed ERCC6 (excision repair cross-complementation group 6) because it was found to complement the nucleotide excision repair (NER) defect of the Chinese Hamster Ovary mutant cell line UV61, a representative of complementation group 6 of rodent cell lines defective in excision repair (Troelstra et al., 1990Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D and Hoeijmakers JHJ (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71:939–953.). Two years later, Hoeijmaker’s group demonstrated that the expression of this gene could reverse UV sensitivity and rescue post-UV RNA synthesis in a cell line (CS1AN) assigned to CS group B but not in group A cells (Troelstra et al., 1992Troelstra C, Hesen W, Bootsma D and Hoeijmakers JHJ (1993) Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res 21:419–426.). Another two years later, using episomal vectors to drive the expression of a cDNA library, Friedberg’s group was able to identify the gene capable of reversing the UV sensitivity of CS cells from group A (but not B) and to reactivate the expression of a UV-inactivated reporter gene (Henning et al., 1995Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV et al. (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555–564.). They discovered that the CSA protein, encoded by ERCC8 (Itoh et al., 1996Itoh T, Shiomi T, Shiomi N, Harada Y, Wakasugi M, Matsunaga T, Nikaido O, Friedberg EC and Yamaizumi M (1996) Rodent complementation group 8 (ERCC8) corresponds to Cockayne syndrome complementation group A. Mutat Res 362:167–174.), can interact with the CSB protein. Both genes play a critical role in the transcription-coupled nucleotide excision repair (TC-NER) of damaged DNA, which is described below in more detail.
CSA and CSB in TC-NER
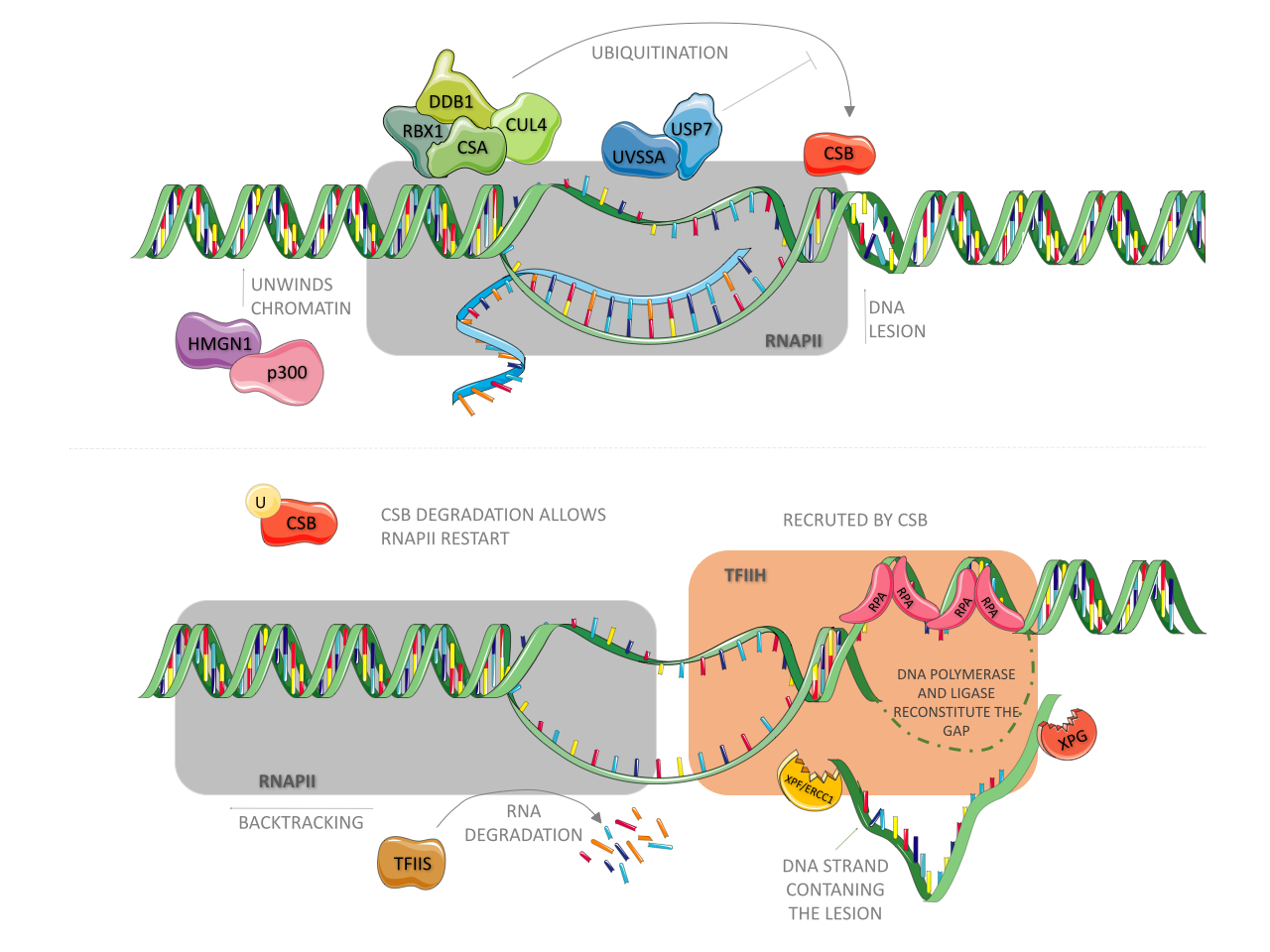
Cells evolved complex and refined mechanisms to prevent genome instability in response to the presence of exogenously and endogenously generated DNA lesions. One of these mechanisms, conserved from bacteria to humans, is the NER pathway (Schärer, 2013Schärer OD (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5:a012609.; Marteijn et al., 2014Marteijn JA, Lans H, Vermeulen W and Hoeijmakers JHJ (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15:465–481.; Gregersen and Svejstrup, 2018Gregersen LH and Svejstrup JQ (2018) The cellular response to transcription-blocking DNA damage. Trends Biochem Sci 43:327–341.). This system drives the repair of bulky distorting DNA lesions (such as those induced by UV and by some redox processes) in four sequential steps: i) detection of a lesion, ii) excision of a DNA single-strand fragment containing the lesion, iii) DNA synthesis by a polymerase to fill the gap, and iv) nick sealing by a ligase (Costa et al., 2003; Reardon and Sancar, 2005Reardon JT and Sancar A (2005) Nucleotide excision repair. Prog Nucleic Acid Res 79:183–235.; Menck and Munford, 2014Menck CFM and Munford V (2014) DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol 37:220–233.). In bacteria, three proteins (UvrA, UvrB, and UvrC) are critical for the detection and excision of the lesion (Seeberg and Strike, 1976Seeberg E and Strike P (1976) Excision repair of ultraviolet-irradiated deoxyribonucleic acid in plasmolyzed cells of Escherichia coli. J Bacteriol 125:787–795. ; Sancar and Rupp, 1983Sancar A and Rupp WD (1983) A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33:249–260.), whereas in humans, more than 30 proteins acting in an orchestrated manner are required for these steps, as reviewed elsewhere in detail (Menck and Munford, 2014Menck CFM and Munford V (2014) DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol 37:220–233.). NER is subdivided into two sub-pathways: global genome repair (GG-NER) and transcription-coupled repair (TC-NER). They differ in how DNA lesions are detected, although the excision and DNA re-synthesis steps are shared by the two pathways (Menck and Munford, 2014Menck CFM and Munford V (2014) DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol 37:220–233.). In humans, GG-NER is initiated by the XPC protein (Sugasawa et al., 1998Sunesen M, Selzer RR, Brosh Jr. RM, Balajee AS, Stevnsner T and Bohr VA (2000) Molecular characterization of an acidic region deletion mutant of Cockayne syndrome group B protein. Nucleic Acids Res 28:3151–3159.), which is constantly scanning the whole genome for the presence of helix-distorting lesions (Hoogstraten et al., 2008Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, Dinant C, Hoeijmakers JHJ, Vermeulen W et al. (2008) Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci 121:2850–2859.), and the detection of UV products is facilitated by XPE/DDB2 (Cleaver et al., 2009Cleaver JE, Lam ET and Revet I (2009) Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet 10:756–768.). In TC-NER, as shown in Figure 1, the triggering event is the arrest of RNA polymerase II (RNA pol II) owing to the presence of a lesion in the actively transcribed strand of a gene (Bohr et al., 1985Bohr VA, Smith CA, Okumoto DS and Hanawalt PC (1985) DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40:359–369.; Hanawalt and Spivak, 2008Hanawalt PC and Spivak G (2008) Transcription-coupled DNA repair: Two decades of progress and surprises. Nature 9:958–970.). It is in this pathway that the CSA and CSB proteins play a critical part. Upon RNA pol II blockade, the binding of CSB to RNA pol II is stabilized (Tantin et al., 1997Tirode F, Busso D, Coin F and Egly JM (1999) Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell 3:87–95.; van den Boom et al., 2004van der Horst GTJ, van Steeg H, Berg RJW, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR et al. (1997) Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer pre-disposition. Cell 89:425–435.; Fousteri et al., 2006Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.) and CSB then wraps DNA around itself, altering its conformation and recruiting histone acetyltransferase p300 and core NER factors (such as RPA, XPG, and TFIIH) to RNA pol II arrest sites (Fousteri et al., 2006Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.), as presented in Figure 1 (upper panel). CSB also recruits an E3-ubiquitin ligase complex, highlighted in green, composed of CSA (which contains WD motifs involved in protein-protein interactions), DDB1, Cullin 4A, and ROC1/Rbx1 proteins (Groisman et al., 2003Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K and Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357–367.; Fousteri et al., 2006Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.). Although CSA is not necessary for the recruitment of NER factors, it is required for recruiting HMGN1, XAB2, and TFIIS to RNA pol II arrest sites (Fousteri et al., 2006Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.). XAB2 is an XPA-interacting protein (Nakatsu et al., 2000Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo J, Khaw M, Saijo M, Kodo N et al. (2000) XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and rranscription. J Biol Chem 275:34931–34937.) and might act as a scaffolding factor for protein assembly during TC-NER (Fousteri and Mullenders, 2008Fousteri M and Mullenders LHF (2008) Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res 18:73–84.), while the nucleosome-binding protein, HMGN1, was suggested to promote chromatin changes that allow for the incision step (Fousteri and Mullenders, 2008Fousteri M and Mullenders LHF (2008) Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res 18:73–84.).
Physical blockage of RNA polymerase II facilitates CSB binding to it. The following is responsible for recruiting p300, CSA and the other NER factors. CSA, together with CUL4, RBX1 and DDB1 are constantly ubiquitinating CSB, however UVSSA-USP7 complex are constantly removing ubiquitin tags from it. CSA also recruits HMGN1, which together with p300 unwinds chromatin upstream RNApol II, allowing it to backtrack and expose the lesion site to NER factors. TFIIS stimulates RNA cleavage by RNApol II during this process. NER factors unwinds DNA around the lesion. While RPA protects ssDNA from degradation, XPG and XPF-ERCC1 endonucleases cleave the strand containing the lesion. DNA polymerase and ligase then fill up the gap. CSB degradation is necessary to RNA synthesis recovery.
The fates of the nascent transcript and that of the stalled RNA pol II are still debated. One hypothesis is that transcript cleavage may occur in an elongation factor for RNA pol II (ELL)-dependent manner (Gregersen and Svejstrup, 2018Gregersen LH and Svejstrup JQ (2018) The cellular response to transcription-blocking DNA damage. Trends Biochem Sci 43:327–341.). As for the stalled RNA pol II — which occupies a space that ranges from 25 nucleotides downstream of the lesion to 10 nucleotides upstream (Spivak and Ganesan, 2014Spivak G and Ganesan AK (2014) The complex choreography of transcription-coupled repair. DNA Repair (Amst) 19:64–70. Stadler J and Richly H (2017) Regulation of DNA repair mechanisms: How the chromatin environment regulates the DNA damage response. Int J Mol Sci 18:1–15.), thus impairing the assembly of NER factors — it may undergo reverse translocation/backtracking (Donahue et al., 1994Donahue BA, Yint S, Taylort JS, Reines D and Hanawalt PC (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A 91:8502–8506.; Fousteri et al., 2006Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.) or be targeted for proteasomal degradation by ubiquitination (Harreman et al., 2009Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CAS, Conaway RC, Conaway JW et al. (2009) Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A 106:20705–20710.), as shown in Figure 1 (lower panel).
Upon lesion resolution, the CSA-E3 ubiquitin ligase complex performs a critical function in the recovery of transcription by targeting CSB for proteasomal degradation (Groisman et al., 2006Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A and Nakatani Y (2006) CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20:1429–1434.). To avoid premature degradation of CSB, UVSSA, which binds firmly to stalled RNA pol II, recruits USP7, an enzyme that promotes deubiquitination of CSB (Schwertman et al., 2012Schwertman P, Lagarou A, Dekkers DHW, Raams A, van der Hoek A, Laffeber C, Hoeijmakers JH, Demmers JAA, Fousteri M, Vermeulen W et al. (2012) UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 44:598-602.), highlighted in blue in Figure 1. Therefore, once the DNA lesion is removed, USP7-mediated deubiquitination of CSB ceases, and CSB is finally targeted for degradation.
In the absence of the CSA or CSB protein, the arrest of RNA pol II persists, an event that leads to p53 activation and cell death, thus explaining the extreme sensitivity of CS cells to UV damage (Ljungman and Zhang, 1996Ljungman M and Zhang F (1996) Blockage of RNA polymerase as a possible trigger for UV light-induced apoptosis. Oncogene 13:823–831.). Besides its participation in TC-NER, proteins CSA and CSB play several other important roles. In the following sections, we provide detailed descriptions of the structures of these proteins, their functions outside of TC-NER, and a comprehensive review of pathological mutations, their consequences for protein function, and their association with patients’ clinical characteristics.
The structure and functions of proteins CSA and CSB, or how pathological mutations are (not) associated with clinical phenotypes
CSB is a 168 kDa protein composed of 1493 amino acid residues and is encoded by the ERCC6 gene located in chromosomal region 10q11 (Troelstra et al., 1992Troelstra C, Hesen W, Bootsma D and Hoeijmakers JHJ (1993) Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res 21:419–426., 1993Tuo J, Müftüoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Rodriguez H, Dizdaroglu M and Bohr VA (2001) The Cockayne syndrome group b gene product is involved in general genome base excision repair of 8-Hydroxyguanine in DNA. J Biol Chem 276:45772–45779.). It belongs to the SWI2/SNF2 family of helicases, and just as all the proteins in this family, it does not have the capacity to open the DNA double helix (Selby and Sancar, 1997aSelby CP and Sancar A (1997a) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A 94:11205–11209.). In contrast, the SWI2/SNF2 proteins temporarily modify DNA conformation via ATP hydrolysis, thereby altering the DNA contact with histones and nucleosome positioning. Therefore, SWI2/SNF2 proteins are considered chromatin remodelers (Lusser and Kadonaga, 2003Lusser A and Kadonaga JT (2003) Chromatin remodeling by ATP-dependent molecular machines. BioEssays 25:1192–1200.; Beerens et al., 2005Beerens N, Hoeijmakers JHJ, Kanaar R, Vermeulen W and Wyman C (2005) The CSB protein actively wraps DNA. J Biol Chem 280:4722–4729.). In addition to its known classic function in TC-NER, CSB takes part in the regulation of transcription and assists with nuclear and mitochondrial base excision repair (BER). It has been reported that CSB interacts with proteins XPB, XPD, XPG, TFIIH, RNA pol I and II, and glycosylases (Tantin, 1998Tantin D, Kansal A and Carey M (1997) Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol 17:6803–6814.; Bradsher et al., 2002Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I and Egly JM (2002) CSB Is a Component of RNA Pol I Transcription. Mol Cell 10:819–829.; Tuo et al., 2002; Sarker et al., 2005Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E and Cooper PK (2005) Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: Insights for transcription-coupled repair and Cockayne syndrome. Mol Cell 20:187–198.; Stevnsner et al., 2008Stichel CC, Zhu XR, Bader V, Linnartz B, Schmidt S and Lübbert H (2007) Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum Mol Genet 16:2377–2393.; Kamenisch et al., 2010Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dollé ME, Kuiper R, Majora M et al. (2010) Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207:379–90.), although some of these interactions were described only once and need additional confirmation of their biological relevance.
The CSB structure mainly includes the following domains: an acidic domain, SNF2/ATPase region, ubiquitin-binding domain, and a nuclear localization signal (Liu et al., 2015Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y and Zhao Y (2015) Sequence analysis IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361.). The acidic domain comprising amino acid residues 356 to 394 is located in the N-terminal portion, which is mostly negatively charged (Troelstra et al., 1992Troelstra C, Hesen W, Bootsma D and Hoeijmakers JHJ (1993) Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res 21:419–426.). In other proteins, this domain facilitates protein-protein interactions, especially those of nuclear and DNA-binding proteins, such as transcriptional activators and chromatin remodelers (Melcher, 2000Melcher K (2000) The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional protiens. J Mol Biol 301:1097–1112.; Carpenter et al., 2005Carpenter AE, Memedula S, Plutz MJ and Belmont AS (2005) Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol 25:958–968.; Wu et al., 2017Xu C and Min J (2011) Structure and function of WD40 domain proteins. Protein Cell 2:202–214.). Given that CSB is classified as a chromatin remodeler, it has been hypothesized that its acidic domain facilitate this activity, but the possible underlying mechanisms have not yet been fully elucidated (Brosh et al., 1999Brosh RM Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L and Bohr VA (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell 10:3583–3594.).
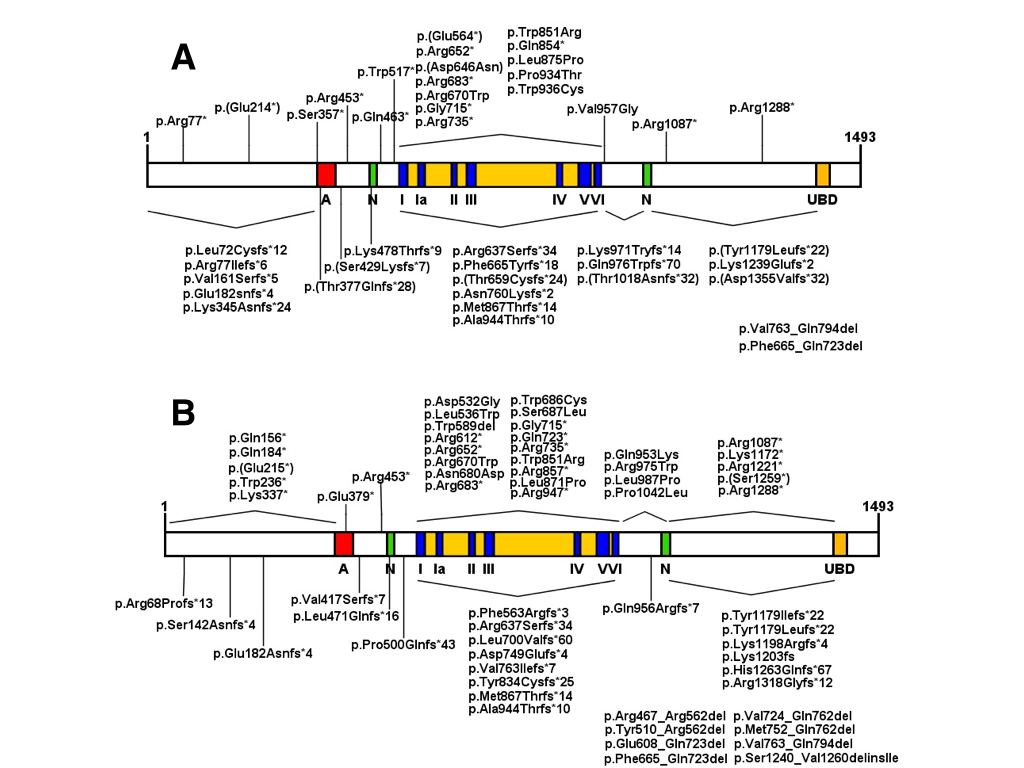
Mutations in the acidic domain of CSB in UV61 cells do not compromise the ability to repair lesions caused by UV, 4-QNO (4-Nitroquinoline 1-oxide), and NA-AAF (N-acetoxy-2-acetylaminofluorene), or cell viability after exposure to these agents (Brosh et al., 1999Brosh RM Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L and Bohr VA (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell 10:3583–3594.; Sunesen et al., 2000Taghdiri M, Dastsooz H, Fardaei M, Mohammadi S, Fard MAF and Faghihi MA (2017) A novel mutation in ERCC8 gene causing Cockayne syndrome. Front Pediatr 5:169.). These data indicate that the integrity of this domain is not essential for this protein’s function in TC-NER. Similar results were obtained by Lake et al. (2010)Lake RJ, Geyko A, Hemashettar G, Zhao Y and Fan HY (2010) UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell 37:1–20. with UV irradiation of CS1AN-SV cells expressing CSB protein lacking the first 454 amino acid residues in the N-terminal portion, demonstrating that the absence of this region does not compromise the ability of the protein to associate with chromatin but instead makes such associations much more frequent even without UV exposure. It was also observed that this deletion increases the ATPase activity of CSB, indicating that the N-terminal portion acts as a negative regulator of its association with chromatin via ATP hydrolysis (Lake et al. (2010)Lake RJ, Geyko A, Hemashettar G, Zhao Y and Fan HY (2010) UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell 37:1–20.). CSB protein structure and homozygous and heterozygous pathological alterations are illustrated in Figure 2 A and B, whereas Table S1 Table S1 - Homozygous and heterozygous ERCC6 (CSB) mutations and their effects on patients phenotype. lists all ERCC6 mutations reported in the literature.
Representation of CSB protein and domains. Acidic region (A), nuclear localization signal (N), helicase motifs (I, Ia -VI) and ubiquitin binding domain (UBD). (A) Homozygous mutations are indicated: frameshifts and nonsense mutations are indicated above the protein, while deletions and missense mutations are indicated below the protein. (B) Represents heterozygous mutations.
The nuclear localization signal is found within regions 466–481 and 1038–1055 (amino acid positions) (Lange et al., 2007Lange A, Mills RE, Lange CJ, Stewart M, Devine SE and Corbett AH (2007) Classical nuclear localization signals: Definition, function, and interaction with importin α. J Biol Chem 282:5101–5105.). In a recent work, Iyama et al. (2018)Iyama T, Okur MN, Golato T, McNeill DR, Lu H, Hamilton R, Raja A, Bohr VA and Wilson III DM (2018) Regulation of the intranuclear distribution of the Cockayne syndrome proteins. Sci Rep 8:1–12. identified through computational analysis the existence of a third region of nuclear localization signal, in addition to the three nucleolar localization signals that cooperate for the distribution of the protein between the nucleus and nucleolus.
Among these regions there is also the SNF2/ATPase domain, which is highly conserved in the SWI2/SNF2 family (Pazin and Kadonaga, 1997Pazin MJ and Kadonaga JT (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88:737–740.). This domain extends from amino acid residue 510 to residue 960 and contains seven ATPase motifs: I, Ia, II, II, IV, V, and VI (Troelstra et al., 1992Troelstra C, Hesen W, Bootsma D and Hoeijmakers JHJ (1993) Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res 21:419–426.), essential for the functioning of the protein (Brosh et al., 1999Brosh RM Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L and Bohr VA (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell 10:3583–3594.; Selzer et al., 2002Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM Jr and Bohr VA (2002) Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res 30:782–793.). The function of the ATPase region is the most relevant for the activity of CSB, because this function provides energy for its association with (and remodeling of) chromatin by altering the positioning of nucleosomes (Citterio et al., 2000Citterio E, van den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JHJ and Vermeulen W (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol 20:7643–7653.). Through this activity, CSB enables the repair of DNA lesions by promoting the access of other proteins, such as CSA and NER factors, to the site of stalled RNA pol II (Stadler and Richly, 2017Stevnsner T, Muftuoglu M, Aamann MD and Bohr VA (2008) The role of Cockayne syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev 129:441–448.). During the transcription process, CSB alters in situ chromatin conformation, favoring the binding of transcription factors (Lake et al., 2014Lake RJ, Boetefuer EL, Tsai PF, Jeong J, Choi I, Won KJ and Fan HY (2014) The sequence-specific transcription factor c-jun targets Cockayne syndrome protein B to regulate transcription and chromatin structure. PLoS Genet 10:e1004284.).
To understand and characterize the functional importance of each ATPase motif, several cell lines carrying mutations in different and highly conserved regions in these motifs have been created. In general, amino acid substitutions in these regions decrease cell survival, RNA synthesis recovery, and DNA repair after exposure to UV, as well as increase the sensitivity to 4-NQO (Brosh et al., 1999Brosh RM Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L and Bohr VA (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell 10:3583–3594.; Muftuoglu et al., 2002Muftuoglu M, Selzer R, Tuo J, Brosh RM and Bohr VA (2002) Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene 283:27–40.; Selzer et al., 2002Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM Jr and Bohr VA (2002) Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res 30:782–793.). Notably, mutations in domains I and II, named “Walker A” and “Walker B,” respectively, can completely inactivate the ATPase activity (Citterio et al., 1998Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH and Vermeulen W (1998) Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem 273:11844–11851.; Christiansen et al., 2003Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM and Bohr VA (2003) Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res 31:963–973.). Mutations in motifs V and VI also compromise the ATPase activity, although to a lesser extent (Christiansen et al., 2003Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM and Bohr VA (2003) Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res 31:963–973.). Different motifs can also contribute in different ways to other activities carried out by CSB. Tuo et al. (2001)Tuo J, Chen C, Zeng X, Christiansen M and Bohr VA (2002a) Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 1:913–927. demonstrated that cells mutated in motifs V and VI are more sensitive to γ-radiation than wildtype cells, and DNA lesions such as 7,8-dyhydro-2’-deo-xyguanosine (8-oxoGua) accumulate in CSB-null and VI mutant-CSB cells after exposure to γ-radiation, indicating a possible relation between CSB and the BER pathway (Tuo et al., 2002aTuo J, Jaruga P, Rodriguez H, Dizdaroglu M and Bohr VA (2002b) The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem 277:30832–30837.).
The ubiquitin-binding domain (UBD) is located in the C-terminal region of the CSB protein (amino acid residues 1400–1428). UBD-CSB-deficient cells have a phenotype similar to that of cells that do not express the CSB protein at all. Although the TC-NER complex is fully assembled around the lesion and RNA pol II in these cells, the repair does not proceed because of the inability of CSB to leave the lesion site (Anindya et al., 2010Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM and Svejstrup JQ (2010) A ubiquitin-binding domain in cockayne syndrome b required for transcription-coupled nucleotide excision repair. Mol Cell 38:637–648.). The replacement of the CSB UBD by another UBD, such as UBA2 of Rad23, an otherwise unrelated Saccharomyces cerevisiae DNA repair gene, also enables CSB dissociation from the lesion region and progression of the repair process, thereby demonstrating the need for CSB ubiquitination for the correct functioning of the protein in this TC-NER (Anindya et al., 2010Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM and Svejstrup JQ (2010) A ubiquitin-binding domain in cockayne syndrome b required for transcription-coupled nucleotide excision repair. Mol Cell 38:637–648.). Cells lacking UBD in the CSB protein are sensitive to oxidatively induced DNA damage (Ranes et al., 2016Ranes M, Boeing S, Wang Y, Wienholz F, Menoni H, Walker J, Encheva V, Chakravarty P, Mari PO, Stewart A et al. (2016) A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic Acids Res 44:5246–5255.), suggesting that this domain is important for the repair of this kind of lesion. The conserved amino acid lysine at position 911 was recently found to be a ubiquitination site that is also required for this function, but is dispensable for TC-NER (Ranes et al., 2016Ranes M, Boeing S, Wang Y, Wienholz F, Menoni H, Walker J, Encheva V, Chakravarty P, Mari PO, Stewart A et al. (2016) A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic Acids Res 44:5246–5255.).
By constructing several CSB mutants with different deletions in the C-terminal region, Sin et al. (2016)Sin Y, Tanaka K and Saijo M (2016) The C-terminal region and SUMOylation of Cockayne syndrome group B protein play critical roles in transcription-coupled nucleotide excision repair. J Biol Chem 291:1387–1397. found that the integrity of the amino acid sequence in this region is important for this sumoylation of this protein and association with chromatin. Aside from this region, a functional UBD domain is necessary for RNA Pol II interaction and CSA recruitment to the nucleus (Sin et al., (2016)Sin Y, Tanaka K and Saijo M (2016) The C-terminal region and SUMOylation of Cockayne syndrome group B protein play critical roles in transcription-coupled nucleotide excision repair. J Biol Chem 291:1387–1397.). In addition, Groisman (2006) demonstrated that the degradation of CSB depends on the action of an E3-ubiquitin ligase complex that contains CSA.
CSA is a 44 kDa protein 396 amino acid residues long and is encoded by the ERCC8 gene located in chromosomal region 5q12.1 (Henning et al., 1995Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV et al. (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555–564.). It belongs to the WD-repeat family because it contains 7 WD40 domains that are repeated in its structure. These domains consist of approximately 40 amino acid residues that start with a conserved glycine and histidine sequence and terminate in tryptophan and aspartic acid (WD), a seven-bladed propeller structure with its N terminus attached to DDB1 via a helix-loop-helix motif (Fischeret al., 2011Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F et al. (2011) The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147:1024–1039.). Proteins with the WD40 domain characteristically interact with other proteins and are typically known for their ability to form protein complexes (Xu and Min, 2011Yu S, Chen L, Ye L, Fei L, Tang W and Tian Y (2014) Identification of two missense mutations of ERCC6 in three chinese sisters with Cockayne syndrome by Whole Exome Sequencing. PloS One 9:1–11.). Although they do not have a catalytic activity, they are involved in a variety of cellular functions, such as the regulation of transcription and chromatin conformation, apoptosis, signal transduction, and cell cycle control, among others (Xu and Min, 2011Yu S, Chen L, Ye L, Fei L, Tang W and Tian Y (2014) Identification of two missense mutations of ERCC6 in three chinese sisters with Cockayne syndrome by Whole Exome Sequencing. PloS One 9:1–11.). CSA protein structure and homozygous/heterozygous pathological alterations are illustrated in Figure 3A and B. Table S2 Table S2 - Homozygous and heterozygous ERCC8 (CSA) mutations and their effects on patients phenotype. shows all the ERCC8 mutations reported in the literature.
Representation of CSA protein and its seven WD-repeat domains. (A) Homozygous mutations are indicated: frameshifts and nonsense mutations are indicated above the protein, while deletions and missense mutations are indicated below the protein. (B) Represents heterozygous mutations.
It is known that CSA is part of the E3 ubiquitin ligase complex, along with DDB1, RBX1, and CUL4A (Groisman et al., 2003Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K and Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357–367.; Fischer et al., 2011Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F et al. (2011) The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147:1024–1039.), responsible for the ubiquitination and degradation of TC-NER proteins when the repair is finalized (Groisman et al., 2006Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A and Nakatani Y (2006) CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20:1429–1434.). In addition, CSA interacts and complexes with several proteins involved in transcription, ribosomal biogenesis, and TC-NER (reviewed by Aamann et al., 2014Aamann MD, Hvitby C, Popuri V, Muftuoglu M, Lemminger L, Skeby CK, Keijzers G, Ahn B, Bjoras M, Bohr VA et al. (2014a) Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity. Mech Ageing Dev 135:1–14.; Koch et al., 2014Koch S, Gonzalez OG, Assfalg R, Schelling A, Schäfer P, Scharffetter-Kochanek K and Iben S (2014) Cockayne syndrome protein A is a transcription factor of RNA polymerase I and stimulates ribosomal biogenesis and growth. Cell Cycle 13:2029–2037.). Nonetheless, the mechanisms via which these interactions occur, and which CSA regions are involved, are not yet completely understood.
The three-dimensional structure and conserved amino acid residues in specific regions of the protein are of great importance for the regulatory activity and the interactions. Mutations in these regions that lead to the alteration of one of these factors may inevitably impair these functions (Muftuoglu et al., 2002Muftuoglu M, Selzer R, Tuo J, Brosh RM and Bohr VA (2002) Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene 283:27–40.:; Christiansen et al., 2003Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM and Bohr VA (2003) Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res 31:963–973.).
To identify a possible relation between the mutations and the phenotype of patients with CS, we mapped all the mutations in the ERCC8 and ERCC6 genes and their impact on proteins CSA and CSB, respectively, that is already reported in the literature. In total, we found 102 mutations in ERCC6 (50 homozygous and 52 heterozygous) and 37 mutations in ERCC8 (23 homozygous and 14 heterozygous). These numbers indicate that 70% of all the cases of the disease are caused by CSB mutations and 30% by CSA mutations. Analyzing only homozygous mutations, we observed the lack of an obvious correlation between the type of mutation or the affected region with the severity of CS (Tables S1 Table S1 - Homozygous and heterozygous ERCC6 (CSB) mutations and their effects on patients phenotype. and S2 Table S2 - Homozygous and heterozygous ERCC8 (CSA) mutations and their effects on patients phenotype. ).
Nevertheless, as discussed by Laugel (2013)Laugel V (2013) Cockayne syndrome: The expanding clinical and mutational spectrum. Mech Ageing Dev 134:161–170., the large prevalence of type I CS clinical classification (moderate phenotype) is noted in CSA patients, with most of the mutations located in the WD domains. Regarding CSB, mutations are predominant among the domains (such as domains III and IV, for example) and are mostly nonsense mutations and frameshifts, with lower prevalence of missense mutations and deletions. Unlike CSA cases, type II CS clinical manifestation is predominant among CSB patients (Laugel, 2013Laugel V (2013) Cockayne syndrome: The expanding clinical and mutational spectrum. Mech Ageing Dev 134:161–170.; Calmelset al., 2018Calmels N, Botta E, Jia N, Fawcett H, Nardo T, Nakazawa Y, Lanzafame M, Moriwaki S, Sugita K, Kubota M et al. (2018) Functional and clinical relevance of novel mutations in a large cohort of patients with Cockayne syndrome. J Med Genet 55:329-343.).
The mutations that give rise to truncated CS proteins and the phenotype of the patients are also non-correlating variables. Proteins with mutations in the amino acid end sequences, such as R1087X, all have the integral ATPase domains but still lead to the manifestation of type II CS (more severe phenotype). On the other hand, the most striking example is the total absence of functional CSB resulting from the R77X mutation, as reported by Horibata et al. (2004)Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M and Tanaka K (2004) Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A 101:15410–15415., not leading to CS manifestation but instead causing UV-sensitive syndrome (UVSS) (Spivak, 2005Spivak G (2005) UV-sensitive syndrome. Mutat Res 577:162–169.). Another interesting observation is that the same mutation may cause different forms of CS in different patients. Other examples of one specific mutation resulting in different phenotypes exist in the literature. For example, Colella et al. (2000)Colella S, Nardo T, Botta E, Lehmann AR and Stefanini M (2000) Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-Cacchione variant of Xeroderma pigmentosum. Hum Mol Genet 9:1171–1175. described two siblings with the R735X mutation in CSB that do not cause CS but instead induce De Sanctis-Cacchione syndrome (a variant form of XP), whereas Mallery et al. (1998)Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M and Lehmann AR (1998) Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet 62:77–85. described an association of the same mutation with type I of CS. In addition, Jaakkola et al. (2010)Jaakkola E, Mustonen A, Olsen P, Miettinen S, Savuoja T, Raams A, Jaspers NG, Shao H, Wu BL and Ignatius J (2010) ERCC6 founder mutation identified in Finnish patients with COFS syndrome. Clin Genet 78:541–547. discovered the R1288X mutation (in CSB), which causes a severe neurological disorder known as cerebro-ocular-facial-skeletal syndrome (COFS) in several members of the same family, while the same mutation was also associated with type II CS manifestation (Laugel et al., 2009Laugel V (2013) Cockayne syndrome: The expanding clinical and mutational spectrum. Mech Ageing Dev 134:161–170.). This unexpected difference in phenotypes suggests that the genetic background may be a key factor that also needs to be taken into account in studies on the development, diversification of forms, and manifestations of the disease.
CS as a transcription syndrome
A deficiency in TC-NER and the consequent inability to recover RNA synthesis upon DNA damage in actively transcribed strands of genes could contribute to the neurological CS phenotype. Nonetheless, patients with UVSS (which can be caused by mutations in genes CSA, CSB, or UVSSA) lack TC-NER and are sun sensitive but do not manifest any of the neurological symptoms of CS (Itoh et al., 1996; Horibata et al., 2004Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M and Tanaka K (2004) Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A 101:15410–15415.; Spivac, 2005Spivak G (2005) UV-sensitive syndrome. Mutat Res 577:162–169.; Nardo et al., 2009Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC et al. (2009) A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A 106:6209–6214.; Zhang et al., 2012; Brooks, 2013Brooks PJ (2013) Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 12:656–671.).
In 1997, both the Hoeijmakers and Sancar groups discovered that CSB associates with RNA pol II, suggesting that CSB is implicated in transcription (Selby and Sancar, 1997aSelby CP and Sancar A (1997a) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A 94:11205–11209.,bSelby CP and Sancar A (1997b) Human transcription-repair coupling factor CSB / ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem 272:1885–1890.; van Gool et al., 1997van Hoffen A, Natarajan AT, Mayne LV, Zeeland AA Van, Mullendersl LHF and Venemal J (1993) Deficient repair of the transcribed strand of active in Cockayne’s syndrome cells. Nucleic Acids Res 21:5890–5895.). In fact, CSB was shown to increase the rate of transcription elongation up to threefold in an in vitro assay that reconstituted the transcription system (Selby and Sancar, 1997bSelby CP and Sancar A (1997b) Human transcription-repair coupling factor CSB / ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem 272:1885–1890.). Similar observations were made by Balajee et al. (1997)Balajee AS, May A, Dianov GL, Friedberg EC and Bohr VA (1997) Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci U S A 94:4306–4311.. These results prompted the authors to suggest that CS may be mainly a transcription, rather than a DNA repair related syndrome.
Five years later, Bradsher et al. (2002)Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I and Egly JM (2002) CSB Is a Component of RNA Pol I Transcription. Mol Cell 10:819–829. stated that CSB was found in the nucleolus in a complex containing RNA pol I, which regulates ribosomal RNA (rRNA) transcription. The authors noticed that a CSB mutant cell line showed 8- to 10-fold lower rates of rRNA synthesis than did wild-type cells, confirming a critical role of CSB not only in mRNA synthesis (by RNA pol II) but also in rRNA synthesis (by RNA pol I). Of note, the CSA protein turned out to be associated with RNA pol I in the nucleolus, and a knockdown of CSA reduced rRNA synthesis (Koch et al., 2014Koch S, Gonzalez OG, Assfalg R, Schelling A, Schäfer P, Scharffetter-Kochanek K and Iben S (2014) Cockayne syndrome protein A is a transcription factor of RNA polymerase I and stimulates ribosomal biogenesis and growth. Cell Cycle 13:2029–2037.). This malfunction in RNA pol I transcription has also been linked to endoplasmic-reticulum stress, leading to an unfolded protein response and the loss of proteostasis, which may be linked to the CS phenotype (Alupei et al., 2018Alupei MC, Maity P, Esser P, Krikki I, Tuorto F, Parlato R, Penzo M, Schelling A, Laugel V, Montanaro L et al. (2018) Loss of proteostasis is a pathomechanism in Cockayne syndrome. Cell Rep 23:1612–1619.).
In agreement with the idea that CS is a transcription syndrome, Newman et al. (2006)Newman JC, Bailey AD and Weiner AM (2006) Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci U S A 103:9613–9618. found that CSB-null fibroblasts feature a gene dysregulation pattern similar to that induced by HDAC inhibitors. Wang et al. (2014)Weeda G, van Ham RCA, Vermeulen W, Bootsma D, van der Eb AJ and Hoeijmakers JHJ (1990) A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders Xeroderma pigmentosum and Cockayne’s syndrome. Cell 62:777–791. reported dysregulation of several genes (linked to neurons) in CSB-mutant and CSA-mutant fibroblasts and in post-mortem brain tissue of patients. The authors also noticed that in vitro transdifferentiation of fibroblasts into neurons and neuroblast differentiation are impaired in CSB-deficient cells. These results strongly indicate that CSB is critical for neuronal differentiation and maintenance, and that gene expression defects might underlie the neurodegenerative and the neurodevelopmental defects observed in patients.
By taking advantage of cell reprogramming, Andrade et al. (2012)Andrade LN, Nathanson JL, Yeo GW, Menck CFM and Muotri AR (2012) Evidence for premature aging due to oxidative stress in iPSCs from Cockayne syndrome. Hum Mol Genet 21:3825–3834. for the first time reprogrammed CSB-mutated primary fibroblasts into induced-pluripotent stem cells. Aside from making it possible to obtain a cell type that can be used to model CS development in vitro, the authors noticed dysregulation of hundreds of targets (including p53 and TXNIP) in these cells (Andrade et al., 2012Andrade LN, Nathanson JL, Yeo GW, Menck CFM and Muotri AR (2012) Evidence for premature aging due to oxidative stress in iPSCs from Cockayne syndrome. Hum Mol Genet 21:3825–3834.). More recently, by combining cell reprogramming with neuron differentiation protocols, Vessoni et al. (2016)Wallace DC (2012) Mitochondria and cancer. Nat Rev Cancer 12:685–698. were, for the first time, able to obtain live neurons from patients’ skin fibroblasts. By RNA sequencing, the authors were able to find that pathological mutations in the ERCC6 gene changed the expression of almost 5000 transcripts in neurons of CSB-deficient patients compared to unaffected controls. Pathways related to axonogenesis, the action potential of neurons, neurotransmission, as well as transcripts related to the growth hormone–IGF-1 pathway were found to be dysregulated in the CSB-deficient neurons. Collectively, these results confirm that CSB deficiency heavily impacts the transcriptional process of the cell types relevant for the disease, even in the absence of exogenous DNA-damaging agents. Such extensive transcriptional dysregulation may underlie the complex and heterogeneous CS phenotype.
Nonetheless, as mentioned before, some mutations in genes XPB, XPD, XPF, XPG, or ERCC1, which participate in NER (Menck and Munford, 2014Menck CFM and Munford V (2014) DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol 37:220–233.), may result in a combined phenotype of CS and XP (Lehmann 1982Lehmann AR (1982) Three complementation groups in Cockayne syndrome. Mutat Res 106:347–356., 2014Lehmann J, Schubert S, Schäfer A, Apel A, Laspe P, Schiller S, Ohlenbusch A, Gratchev A and Emmert S (2014) An unusual mutation in the XPG gene leads to an internal in-frame deletion and a XP/CS complex phenotype. Br J Dermatol 171:903–905.; Weeda et al., 1990Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D and Hoeijmakers JHJ (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol 7:427–439.; Moriel-Carretero et al., 2015Moriel-Carretero M, Herrera-Moyano E and Aguilera A (2015) A unified model for the molecular basis of Xeroderma pigmentosum-Cockayne syndrome. Rare Dis 3:e1079362.). How can we explain the CS phenotype in all these cases? A likely answer to this question may depend on a multiprotein complex, TFIIH, which is indispensable for NER and for transcription. TFIIH consists of two functional subcomplexes (Core and CAK) that participate in initiation, promoter escape, and early elongation (Compe and Egly, 2012Compe E and Egly JM (2012) TFIIH: When transcription meets DNA repair. Nat Rev Mol Cell Biol 13:343–354.). The core subcomplex consists of seven proteins, including 3’ to 5’ ATP-dependent helicase XPB, and XPD, a 5’ to 3’ ATP-dependent helicase that binds to the core and CAK complexes together and facilitates optimal transcription (Tirode et al., 1999Titus TA, Yan YL, Wilson C, Starks AM, Frohnmayer JD, Canestro C, Rodriguez-Mari A, He X and Postlethwait JH (2009) The Fanconi anemia/BRCA gene network in zebrafish: Embryonic expression and comparative genomics. Mutat Res 668:117–132.; Egly and Coin, 2011Egly JM and Coin F (2011) A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 10:714–721.). In this sense, two mutations in the XPB gene (associated with the XP/CS phenotype) reduce transcriptional activity in a reconstituted transcription assay in vitro (Coin et al., 1999Coin F, Bergmann E, Tremeau-Bravard A and Egly J (1999) Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J 18:1357–1366.). Moreover, XPD or XPB mutations associated with CS were found to disrupt the interactions among CSB, TFIIH, and RNA pol I (Bradsher et al., 2002Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I and Egly JM (2002) CSB Is a Component of RNA Pol I Transcription. Mol Cell 10:819–829.). The XPG nuclease was also found to associate with and stabilize TFIIH, and mutations in the XPG gene related to an XP-G/CS phenotype abrogate the XPG–TFIIH interaction (Ito et al., 2007Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM and Tanaka K (2007) XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/Patients with CS. Mol Cell 26:231–243.; Lehmann et al., 2014Lehmann J, Schubert S, Schäfer A, Apel A, Laspe P, Schiller S, Ohlenbusch A, Gratchev A and Emmert S (2014) An unusual mutation in the XPG gene leads to an internal in-frame deletion and a XP/CS complex phenotype. Br J Dermatol 171:903–905.; Narita et al., 2015Narita T, Narita K, Takedachi A, Saijo M and Tanaka K (2015) Regulation of transcription elongation by the XPG-TFIIH complex is implicated in Cockayne syndrome. Mol Cell 35:3178–3188.). The recently described XP/CS patients with mutations in XPF or ERCC1 (Kashiyama et al., 2013Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L et al. (2013) Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, Xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet 92:807–819.) pose a challenge to the “CS as a transcription syndrome” point of view, because XPF/ERCC1 is implicated in NER and not in basal transcription. Although XPF is recruited to the promoter of inducible genes (to facilitate chromatin modification for transcription) in the absence of exogenous DNA damage, other NER factors are recruited as well, including XPC, in which mutations do not result in neurological abnormalities (Le May et al., 2010Le May N, Mota-Fernandes D, Vélez-Cruz R, Iltis I, Biard D and Egly JM (2010) NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell 38:54–66.). XPF mutations that cause the XP/CS phenotype were recently found to cause persistent recruitment of NER proteins to DNA damage sites, which may induce the stalling of RNA and DNA polymerases, thereby interfering with the transcription and replication processes (Sabatella et al., 2018Sabatella M, Theil AF, Ribeiro-Silva C, Slyskova J, Thijssen K, Voskamp C, Lans H and Vermeulen W (2018) Repair protein persistence at DNA lesions characterizes XPF defect with Cockayne syndrome features. Nucleic Acids Res 46:9563–9577.). Nevertheless, endogenous levels of DNA lesions that are substrates for NER have been reported to accumulate in mammalian cells and tissues to the levels that would not be consistent with the notion of defective TC-NER as a cause of the neurological symptoms of CS (Brooks, 2013Brooks PJ (2013) Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 12:656–671.). Therefore, more studies are needed to clarify the mechanism behind XPF/ERCC1 mutations and the development of the CS phenotype in these patients (Kashiyama et al., 2013Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L et al. (2013) Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, Xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet 92:807–819.).
CS and redox processes
The manifestation of CS occurs only when some NER proteins are mutated, while the complete deactivation of this pathway, via mutations in XPA, leads to the development of XP. Consequently, the roles of proteins CSA and CSB in addition to those known classic functions come into question. In addition, the main symptoms of patients with CS, e.g., neurological aberrations, cannot be explained only by the inefficient repair of UV damage because neurons are not exposed to this type of radiation. Due to the systemic presence of reactive species and their ability to chemically and structurally modify biomolecules, especially DNA, these compounds have been investigated as possible contributing factors of CS.
Experiments with keratinocytes derived from patients with CS suggest that these cells contain high concentrations of reactive oxygen species (ROS) with redox balance alterations under baseline conditions, characteristics that are related to the senescence phenotype of these cells (Cordisco et al., 2018Cordisco S, Tinaburri L, Teson M, Orioli D, Cardin R, Degan P, Stefanini M, Zambruno G, Guerra L and Dellambra E (2018) Cockayne syndrome type A protein protects primary human keratinocytes from senescence. J Invest Dermatol 139:1–13. Costa RM, Chiganças V, Galhardo RS, Carvalho H and Menck CFM (2003) The eukaryotic nucleotide excision repair pathway. Biochimie 85:1083–1099.). The induction of redox processes by the exposure of mice and CSB-/- mouse embryonic fibroblasts to ionizing radiation or paraquat revealed high sensitivity to the toxic effects of these agents, while such high sensitivity is not observed in CSA-mutant mice (de Waard et al., 2003de Waard H, de Wit J, Gorgels TGMF, van den Aardweg G, Andressoo JO, Vermeij M, Van Steeg H, Hoeijmakers JHJ and van der Horst GTJ (2003) Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2:13–25., 2004de Waard H, de Wit J, Andressoo JO, van Oostrom CTM, Riis B, Weimann A, Poulsen HE, van Steeg H, Hoeijmakers JHJ and van der Horst GTJ (2004) Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol Cell Biol 24:7941–7948.). The sensitivity to the redox processes was also observed in CSA-/- keratinocytes and fibroblasts treated with potassium bromide (D’Errico et al., 2007D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM et al. (2007) The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 26:4336–4343.) and in CSB-/- cells exposed to MMS (methyl methansulfonate) and 5-hydroxymethyl-2’deoxyuridine (Wong et al., 2007Wu M, Wei W, Chen J, Cong R, Shi T, Li J, Wong J and Du JX (2017) Acidic domains differentially read histone H3 lysine 4 methylation status and are widely present in chromatin-associated proteins. Sci China Life Sci 60:138–151.).
Moreover, lipid peroxidation products such as HNE (4-hydroxynonenal) at high cellular concentrations perform direct modifications on the CSB protein, compromising its ATPase activity required for the DNA repair by TC-NER (Maddukuri et al., 2009Maddukuri L, Speina E, Christiansen M, Dudzinska D, Zaim J, Obtulowicz T, Kabaczyk S, Komisarki M, Bukowy Z, Szczegielniak J et al. (2009) Cockayne syndrome group B protein is engaged in processing of DNA adducts of lipid peroxidation product trans-4-hydroxy-2-nonenal. Mutat Res 666:23–31.). On the other hand, Boetefuer et al. (2018) demonstrated that this activity is not needed for CSB–chromatin association when CS1AN-sv cells are exposed to menadione. In that condition, these associations are loci-specific and are stimulated by the PARP1 protein, thereby indicating possible participation of CSB in the transcriptional regulation in response to oxidative stress (Boetefuer et al., 2018aBoetefuer EL, Lake RJ, Dreval K and Fan HY (2018a) Poly(ADP-ribose) polymerase 1 (PARP1) promotes oxidative stress–induced association of Cockayne syndrome group B protein with chromatin. J Biol Chem 293:17863-17874.). These findings are in agreement with previous results that point to the involvement of a CSB function in this process, thereby showing that CSB localization and interaction with transcriptional repressor CTCF in promoter regions are greater in cells under oxidative stress (Lake et al., 2016Lake RJ, Boetefuer EL, Won KJ and Fan HY (2016) The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res 44:2125–2135.; Boetefuer et al., 2018bBoetefuer EL, Lake RJ and Fan HY (2018b) Mechanistic insights into the regulation of transcription and transcription-coupled DNA repair by Cockayne syndrome protein B. Nucleic Acids Res 46:7471–7479.).
Although DNA oxidation products are typically repaired by the BER pathway, it has been demonstrated that BER and NER proteins not only show crosstalk, but also that some oxidatively generated lesions are substrates for the NER pathway (D’Errico et al., 2007D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM et al. (2007) The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 26:4336–4343.; Berra et al., 2013Berra CM, de Oliveira CS, Garcia CCM, Rocha CRR, Lerner LK, Lima LC, Baptista M da S and Menck CFM (2013) Nucleotide excision repair activity on DNA damage induced by photoactivated methylene blue. Free Radic Biol Med 61:343–356.). For example, 8-oxoGua, thymine glycol, malondialdehyde, and etheno adducts induce distorting modifications in the double helix and have the potential to block transcription (reviewed by Tornaletti, 2005Trapp C, Reite K, Klungland A and Epe B (2007) Deficiency of the Cockayne syndrome B (CSB) gene aGG-NERavates the genomic instability caused by endogenous oxidative DNA base damage in mice. Oncogene 26:4044–4048.; Chaim et al., 2017Chaim IA, Gardner A, Wu J, Iyama T, Wilson DM and Samson LD (2017) A novel role for transcription-coupled nucleotide excision repair for the in vivo repair of 3,N4-ethenocytosine. Nucleic Acids Res 45:3242–3252.), with cyclopurines being DNA oxidation products that are repaired only by NER (Brooks et al., 2000Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K et al. (2000) The oxidative DNA lesion 8,5’-(S)-Cyclo- 2’-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem 275:22355–22362.). Numerous experiments have confirmed the importance of the CSA and CSB proteins for the repair of oxidized bases, by demonstrating that in the absence of CSA or CSB there is accumulation of 8-oxoGua in DNA (Dianov et al., 1999Dianov G, Bishoff C, Sunesen M and Bohr VA (1999) Repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Nucleic Acids Res 27:1365–1368.; Tuo et al., 2001Tuo J, Chen C, Zeng X, Christiansen M and Bohr VA (2002a) Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 1:913–927.; D’Errico et al., 2007D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM et al. (2007) The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 26:4336–4343.; Aamann et al., 2014aAamann MD, Hvitby C, Popuri V, Muftuoglu M, Lemminger L, Skeby CK, Keijzers G, Ahn B, Bjoras M, Bohr VA et al. (2014a) Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity. Mech Ageing Dev 135:1–14.; Cordisco et al., 2018Cordisco S, Tinaburri L, Teson M, Orioli D, Cardin R, Degan P, Stefanini M, Zambruno G, Guerra L and Dellambra E (2018) Cockayne syndrome type A protein protects primary human keratinocytes from senescence. J Invest Dermatol 139:1–13. Costa RM, Chiganças V, Galhardo RS, Carvalho H and Menck CFM (2003) The eukaryotic nucleotide excision repair pathway. Biochimie 85:1083–1099.). 8-OH-Ade, 5-hydroxycytosine, and cyclopurines are also lesions that are inefficiently repaired in cells harboring mutations in CS genes (Tuo et al., 2002bTuo J, Jaruga P, Rodriguez H, Bohr VA and Dizdaroglu M (2003) Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J 17:668–674., 2003van den Boom V, Citterio E, Hoogstraten D, Zotter A, Egly JM, van Cappellen WA, Hoeijmakers JHJ, Houtsmuller AB and Vermeulen W (2004) DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J Cell Biol 166:27–36.; D’Errico et al., 2013D’Errico M, Pascucci B, Iorio E, van Houten B and Dogliotti E (2013) The role of CSA and CSB protein in the oxidative stress response. Mech Ageing Dev 134:261–269.). Because of the inefficient repair in the cells of important tissues such as the brain, the accumulation of these lesions in DNA may be one of the factors that cause and aggravate the neurological symptoms of the disease, as seen in patients with neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, Huntington’s disease, and amyotrophic lateral sclerosis (Ayala-Peña, 2013Ayala-Peña S (2013) Role of oxidative DNA damage in mitochondrial dysfunction and Huntington’s disease pathogenesis. Free Radic Biol Med 62:102–110.; Coppedè et al., 2016Coppedè F, Tannorella P, Stoccoro A, Chico L, Siciliano G, Bonuccelli U and Migliore L (2016) Methylation analysis of DNA repair genes in Alzheimer’s disease. Mech Ageing Dev 161:105–111.; Abolhassani et al., 2017Abolhassani N, Leon J, Sheng Z, Oka S, Hamasaki H, Iwaki T and Nakabeppu Y (2017) Molecular pathophysiology of impaired glucose metabolism, mitochondrial dysfunction, and oxidative DNA damage in Alzheimer’s disease brain. Mech Ageing Dev 161:95–104.).
The mutational impact of oxidatively induced DNA damage in CS was investigated by Lodato et al. (2018)Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Lovelace LJ, Yandava CN et al. (2018) Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359:555–559.. By means of single-cell whole-genome sequencing followed by genome-wide somatic single-nucleotide variant identification, they detected an increased number of mutations in the neurons of patients with CS as compared to the control. In addition, C > A variants, a signature for mutations induced by oxidatively induced DNA damage, are found in a higher frequency among patients with CS (Lodato et al., 2018Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Lovelace LJ, Yandava CN et al. (2018) Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359:555–559.).
It is noteworthy that in response to irradiation or hydrogen peroxide, CSA is translocated to the nuclear matrix by a CSB-dependent mechanism (Kamiuchi et al., 2002Kamiuchi S, Saijo M, Citterio E, de Jager M, Hoeijmakers JH and Tanaka K (2002) Translocation of Cockayne syndrome group A protein to the nuclear matrix: Possible relevance to transcription-coupled DNA repair. Proc Natl Acad Sci U S A 99:201–206.), but as discussed above, these two proteins perform different functions in chromatin remodeling and in recruitment of the factors associated with the repair and blocking of RNA pol II in vivo (Fousteri et al., 2006Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.). This cooperation between CSA and CSB was not observed in response to treatment with alkylating agents and is independent of XPA and XPC, thus suggesting that TC-NER plays a key part in this cellular response mechanism (Kamiuchi et al., 2002Kamiuchi S, Saijo M, Citterio E, de Jager M, Hoeijmakers JH and Tanaka K (2002) Translocation of Cockayne syndrome group A protein to the nuclear matrix: Possible relevance to transcription-coupled DNA repair. Proc Natl Acad Sci U S A 99:201–206.).
Brain biopsies of CS and XPA patients yielded distinct results in response to the accumulation of oxidized bases in DNA and SOD expression, with only XPA-mutant patients showing upregulation of 8-oxoGua in the nucleus and alteration in SOD expression (Hayashi et al., 2005Hayashi M, Araki S, Kohyama J, Shioda K and Fukatsu R (2005) Oxidative nucleotide damage and superoxide dismutase expression in the brains of Xeroderma pigmentosum group A and Cockayne syndrome. Brain Dev 27:34–38.).
In addition to the participation of these proteins in the repair of oxidation-induced lesions through the NER pathway, there is also a contribution to the removal of DNA damage via direct and indirect activity in the BER pathway (Khobta and Epe, 2013Khobta A and Epe B (2013) Repair of oxidatively generated DNA damage in Cockayne syndrome. Mech Ageing Dev 134:253–260.). This contribution is mediated by the direct modulation through interactions of CSB with BER protein glycosylases, APE1, NEIL1 and NEIL2, and association with the OGG1 complex, which stimulates the incision activity of these proteins and drives the repair (Wong et al., 2007Wu M, Wei W, Chen J, Cong R, Shi T, Li J, Wong J and Du JX (2017) Acidic domains differentially read histone H3 lysine 4 methylation status and are widely present in chromatin-associated proteins. Sci China Life Sci 60:138–151.; Muftuoglu et al., 2009Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M and Bohr VA (2009) Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem 284:9270–9279.; Aamann et al., 2014bAamann MD, Muftuoglu M, Bohr VA and Stevnsner T (2014b) Multiple interaction partners for Cockayne syndrome proteins. Mech Ageing Dev 134:212–224.). Csbm/m/Ogg1-/- mice show high concentrations of 8-oxoGua in comparison with Ogg1-/- animals, thereby confirming the importance of the cooperation between these two proteins in the removal of these lesions (Osterod et al., 2002Osterod M, Larsen E, Le Page F, Hengstler JG, van der Horst GTJ, Boiteux S, Klungland A and Epe B (2002) A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene 21:8232–8239.; Trapp et al., 2007Troelstra C, Odijk LH, de Wit J, Westerveld A, Thompson LH, Bootsma D and Hoeijmakers JHJ (1990) Molecular cloning of the human DNA excision repair. Mol Cell Biol 10:5806–5813.). Pastoriza-Gallego et al. (2007)Pastoriza-Gallego M, Armier J and Sarasin A (2007) Transcription through 8-oxoguanine in DNA repair-proficient and Csb-/Ogg1- DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis 22:343–351. also demonstrated that the 8-oxoGua lesions exert different effects on gene expression depending upon the promoter and sequence context, and that both proteins, Csb and Ogg1, are required for full repair.
Recent evidence also indicates that CSB recruits the XRCC1 protein to single-strand break regions after oxidatively induced DNA damage for subsequent processing by BER (Menoni et al., 2018Menoni H, Wienholz F, Theil AF, Janssens RC, Lans H, Campalans A, Radicella JP, Marteijn JA and Vermeulen W (2018) The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res 46:7747–7756.). In the indirect modulation of the BER pathway, CSB acts as an important factor in the expression of hOGG1, inducing its transcription. It has also been observed that in CSB-deficient cells, the amounts of OGG1 mRNA and protein are low (Dianov et al., 1999Dianov G, Bishoff C, Sunesen M and Bohr VA (1999) Repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Nucleic Acids Res 27:1365–1368.; Tuo et al., 2002aTuo J, Jaruga P, Rodriguez H, Dizdaroglu M and Bohr VA (2002b) The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem 277:30832–30837.; Aamann et al., 2014bAamann MD, Muftuoglu M, Bohr VA and Stevnsner T (2014b) Multiple interaction partners for Cockayne syndrome proteins. Mech Ageing Dev 134:212–224.).
Some studies point to a possible implication of CSB in the repair of oxidized bases in genomic and mitochondrial DNA, to CSB migration to mitochondria, and modulation of p53 activity in response to a redox process (Stevnsner et al., 2008Stichel CC, Zhu XR, Bader V, Linnartz B, Schmidt S and Lübbert H (2007) Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum Mol Genet 16:2377–2393.; Frontini and Proietti-De-Santis, 2009Frontini M and Proietti-De-Santis L (2009) Cockayne syndrome B protein (CSB): Linking p53, HIF-1 and p300 to robustness, lifespan, cancer and cell fate decisions. Cell Cycle 8:693–696.; Aamann et al., 2010Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, Stevnsner T et al. (2010) Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J 24:2334–2346.).
The repair of oxidized bases also has a fundamental role for the mitochondria. These organelles produce ATP through the electron transport chain, a process that not only provides cells with energy, but also produces ROS as a by-product of respiration (reviewed by Kowaltowski et al., 2009Kowaltowski AJ, de Souza-Pinto NC, Castilho RF and Vercesi AE (2009) Mitochondria and reactive oxygen species. Free Radic Biol Med 47:333–343.). For this reason, mitochondria are also considered the primary source of ROS in cells, having a much more oxidizing environment within the mitochondrial matrix as compared with the cellular cytosol (Hu et al., 2008Hu J, Dong L and Outten CE (2008) The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem 283:29126–29134.).
Because of proximity to the initial site of formation and constant exposure to ROS, mitochondrial DNA (mtDNA) is the main target of these molecules, and just as nuclear DNA, it is susceptible to oxidation in the DNA bases and undergoes other modifications in its structure (reviewed by Muftuoglu et al., 2014Muftuoglu M, Mori MP and de Souza-Pinto NC (2014) Formation and repair of oxidative damage in the mitochondrial DNA. Mitochondrion 17:164–181.). Besides, the accumulation of lesions in mtDNA causes instability and compromises its functioning, which can lead to mutations and affect genes that are important for mitochondrial metabolism. In addition, various aging-related diseases, such as cardiovascular diseases, Parkinson’s disease, Alzheimer’s disease, and cancer are associated with significant amounts of lesions in mitochondria, thus being considered one of the causes of the pathologies related to mitochondrial dysfunction (Mecocci et al., 1994Mecocci P, MacGarvey U and Beal MF (1994) Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann Neurol 36:747–751.; Ide et al., 2001Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura KI, Utsumi H, Hamasaki N and Takeshita A (2001) Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res 88:529–535.; Stichel et al., 2007Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC and Bohr VA (2005) Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res 33:3722–3732.; Wallace, 2012Wang W, Xu J, Chong J and Wang D (2018) Structural basis of DNA lesion recognition for eukaryotic transcription-coupled nucleotide excision repair. DNA Repair (Amst) 71:43–55.).
Mitochondria have some specific repair pathways, such as BER and mismatch repair (MMR). As in the repair of nuclear DNA, the CSA and CSB proteins are present and participate in this process, even though there is no mitochondrial NER (Kamenisch et al., 2010Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dollé ME, Kuiper R, Majora M et al. (2010) Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207:379–90.). It has been demonstrated that CSA and CSB, after induction of redox processes by the exposure to H2O2, UV, or menadione, are directed toward mitochondria and interact with OGG1 (Aamann et al., 2010Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, Stevnsner T et al. (2010) Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J 24:2334–2346.; Kamenisch et al., 2010Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dollé ME, Kuiper R, Majora M et al. (2010) Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207:379–90.; reviewed by Prates Mori and de Souza-Pinto, 2018Prates Mori M and de Souza-Pinto NC (2018) Role of mitochondrial dysfunction in the pathophysiology of DNA repair disorders. Cell Biol Int 42:643–650.). In mitochondria, CSB may act as a modulator of the BER pathway, in association with the inner mitochondrial membrane for mtDNA lesion removal (Stuart et al., 2005Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker APM, Hanaoka F, Bootsma D and Hoeijmakers JHJ (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 2:223–232.; Aamann et al., 2010Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, Stevnsner T et al. (2010) Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J 24:2334–2346.). In fact, CSBm/m mice accumulate 2.5-fold more 8-oxoGua in mtDNA than wild-type animals (Osenbroch et al., 2009Osenbroch PO, Auk-Emblem P, Halsne R, Strand J, Forstrøm RJ, van der Pluijm I and Eide L (2009) Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J 276:2811–2821.). The mutation in CSBm/m mice is the same as that in the ERCC6 gene of the CS1AN patient, K337 → stop, which generates a truncated nonfunctional protein (Troelstra et al., 1992Troelstra C, Hesen W, Bootsma D and Hoeijmakers JHJ (1993) Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res 21:419–426.; van der Horst et al., 1997van der Horst GTJ, Meira L, Gorgels TG, de Wit J, Velasco-Miguel S, Richardson JA, Kamp Y, Vreeswijk MPG, Smit B, Bootsma D et al. (2002) UVB radiation-induced cancer pre-disposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair (Amst) 1:143–157.).
In addition to the influence on DNA repair, CSB is involved in gene expression by engaging in the elongation by RNA polymerase II (Selby and Sancar, 1997aSelby CP and Sancar A (1997a) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A 94:11205–11209.). In in vitro transcription, through interactions with proteins of the mitochondrial nucleoid complex, CSB promotes POLRMT transcriptional elongation and allows it to access mtDNA by TFAM removal from the region to be transcribed (Berquist et al., 2012Berquist BR, Canugovi C, Sykora P, Wilson DM and Bohr VA (2012) Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res 40:8392–8405.).
Regarding CS mitochondrial metabolism, Scheibye-Knudsen et al. (2012)Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, Cooper M, Spencer R, de Cabo R et al. (2012) Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med 209:855–869. reported a significant increase of this metabolism in both CSBm/m mice and CSB-deficient cells, and this phenomenon is also manifested in CSA- or CSB- deficient fibroblasts and CSA-/-/XPA-/- mice (Pascucci et al., 2012Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F et al. (2012) An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell 11:520–529.; Brace et al., 2016Brace LE, Vose SC, Stanya K, Gathungu RM, Marur VR, Longchamp A, Treviño-Villarreal H, Mejia P, Vargas D, Inouye K et al. (2016) Increased oxidative phosphorylation in response to acute and chronic DNA damage. Aging Mech Dis 2:16022.). This increase in energy metabolism is accompanied by a large production of ROS through electron leakage (Pascucci et al., 2012Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F et al. (2012) An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell 11:520–529.; Cleaver et al., 2014Cleaver JE, Brennan-Minnella AM, Swanson RA, Fong K, Chen J, Chou KM, Chen YW, Revet I and Bezrookove V (2014) Mitochondrial reactive oxygen species are scavenged by Cockayne syndrome B protein in human fibroblasts without nuclear DNA damage. Proc Natl Acad Sci U S A 111:13487-13492.), and is the main source of DNA damage induction (Scheibye-Knudsen et al., 2012Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, Cooper M, Spencer R, de Cabo R et al. (2012) Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med 209:855–869.) and of changes in the membrane potential, excessive fragmentation, and mitochondrial fission (Pascucci et al., 2012Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F et al. (2012) An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell 11:520–529., 2016Pascucci B, D’Errico M, Romagnoli A, De Nuccio C, Savino M, Pietraforte D, Lanzafame M, Calcagnile AS, Fortini P, Baccarini S et al. (2016) Overexpression of Parkin rescues the defective mitochondrial phenotype and the increased apoptosis of Cockayne Syndrome A cells. Oncotarget 8:102852–102867.).
Under normal cell conditions, organelles and defective proteins are eliminated by autophagy. On the other hand, dysfunctional or damaged mitochondria are subjected to a special form of autophagy, called mitophagy (Fivenson et al., 2017Fivenson EM, Lautrup S, Sun N, Sheibye-Knudsen M, Stevnsner T, Nilsen H, Bohr VA and Fang EF (2017) Mitophagy in neurodegeneration and aging. Neurochem Int 109:202–209.). This mechanism contributes to cell homeostasis, and its malfunction is present in several aging-related diseases (reviewed by Redmann et al., 2014). Human and murine CSB-mutant fibroblasts feature a dysfunction in this pathway owing to defects in the recruitment of P62, an essential player in mitophagy (Scheibye-Knudsen et al., 2012Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, Cooper M, Spencer R, de Cabo R et al. (2012) Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med 209:855–869.).
Thus, dysfunctional mitochondria, with large amounts of mtDNA lesions, can produce more ROS via their metabolism. Consequently, ROS accumulate in the cells, promoting apoptosis (Pinto and Moraes, 2015Pinto M and Moraes CT (2015) Mechanisms linking mtDNA damage and aging. Free Radic Biol Med 85:250–258.; van Houten et al., 2016Venema J, Mullenders LH, Natarajan AT, van Zeeland AA and Mayne LV (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci U S A 87:4707–4711.), which might be associated with reduced amounts of subcutaneous fat in CSA-/- and CSBm/m mice (Kamenisch et al., 2010Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dollé ME, Kuiper R, Majora M et al. (2010) Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207:379–90.). Treatment with pharmacological activators of autophagy, such as rapamycin (Scheibye-Knudsen et al., 2012Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, Cooper M, Spencer R, de Cabo R et al. (2012) Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med 209:855–869.) and an HDAC inhibitor (Majora et al., 2018Majora M, Sondenheimer K, Knechten M, Uthe I, Esser C, Schiavi A, Ventura N and Krutmann J (2018) HDAC inhibition improves autophagic and lysosomal function to prevent loss of subcutaneous fat in a mouse model of Cockayne syndrome. Sci Transl Med 10:1–13.), attenuates the accumulation of damaged mitochondria in CSB-deficient cells and the loss of subcutaneous fat in CSBm/m mice.
Models for CS study
To better understand the mechanisms of CS progression, several animal models have been developed for research on the disease, e.g., mice, Caenorhabditis elegans, and zebrafish. In general, such models carry knockout mutations or mutations identical to those found in humans, enabling a more accurate analysis in different cells and tissues of an organism.
The first models generated to study CS were created on the basis of mice with the C57BL/6 background - CSBm/m mice (van der Horst et al., 1997van der Horst GTJ, Meira L, Gorgels TG, de Wit J, Velasco-Miguel S, Richardson JA, Kamp Y, Vreeswijk MPG, Smit B, Bootsma D et al. (2002) UVB radiation-induced cancer pre-disposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair (Amst) 1:143–157.), and CSA-/- mice (van der Horst et al., 2002van der Pluijm I, Garinis GA, Brandt RMC, Gorgels TG, Wijnhoven SW, Diderich KEM, De Wit J, Mitchell JR, van Oostrom C, Beems R et al. (2007) Impaired genome maintenance suppresses the growth hormone-insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5:0023–0038.). In CSA-/- cells, the interruption of the gene sequence in exon 2 of the Ercc8 gene results in protein absence (van der Horst et al., 2002van der Pluijm I, Garinis GA, Brandt RMC, Gorgels TG, Wijnhoven SW, Diderich KEM, De Wit J, Mitchell JR, van Oostrom C, Beems R et al. (2007) Impaired genome maintenance suppresses the growth hormone-insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5:0023–0038.). When exposed to UV, the fibroblasts of these animals manifest higher UV sensitivity, and they have an inability to resume RNA synthesis and defective TC-NER, these being the classic characteristic of CS (Lehmann, 1982Lehmann AR (1982) Three complementation groups in Cockayne syndrome. Mutat Res 106:347–356.; van der Horst et al., 1997van der Horst GTJ, Meira L, Gorgels TG, de Wit J, Velasco-Miguel S, Richardson JA, Kamp Y, Vreeswijk MPG, Smit B, Bootsma D et al. (2002) UVB radiation-induced cancer pre-disposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair (Amst) 1:143–157., 2002van der Pluijm I, Garinis GA, Brandt RMC, Gorgels TG, Wijnhoven SW, Diderich KEM, De Wit J, Mitchell JR, van Oostrom C, Beems R et al. (2007) Impaired genome maintenance suppresses the growth hormone-insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5:0023–0038.).
The CSBm/m and CSA-/- models have similar phenotypic characteristics, although neither has the severe neurodegenerative phenotype and lifespan reduction seen in patients with CS. Despite differences between them, these mouse models have a few characteristics that resemble their human counterpart, such as the tendency toward reduced body weight via a decrease in the amounts of subcutaneous and visceral fat (Gorgels et al., 2007Gorgels TG, van der Pluijm I, Brandt RM, Garinis GA, van Steeg H, van den Aardweg G, Jansen GH, Ruijter JM, Bergen AA, van Norren D et al. (2007) Retinal degeneration and ionizing radiation hypersensitivity in a mouse model for Cockayne syndrome. Mol Cell Biol 27:1433–1441.; Scheibye-Knudsen et al., 2012Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, Cooper M, Spencer R, de Cabo R et al. (2012) Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med 209:855–869.), mild neurological dysfunction with changes in myelin (Jaarsma et al., 2011Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, Rijksen Y, Maas A, van Steeg H, Hoeijmakers JHJ et al. (2011) Age-related neuronal degeneration: Complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet 7:e1002405.), activation of glial cells in white-matter regions (Jaarsma et al., 2011Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, Rijksen Y, Maas A, van Steeg H, Hoeijmakers JHJ et al. (2011) Age-related neuronal degeneration: Complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet 7:e1002405.), progressive deafness (Nagtegaal et al., 2015Nagtegaal AP, Rainey RN, van der Pluijm I, Brandt RM, van der Horst GT, Borst JG and Segil N (2015) Cockayne syndrome group B (Csb) and group A (Csa) deficiencies predispose to hearing loss and cochlear hair cell degeneration in mice. J Neurosci 35:4280–4286.), photophobia and sensitivity to UV exposure (van der Horst et al., 1997, 2002). In contrast to what is manifested in humans, the mouse models are prone to skin and eye cancers when exposed to UV, a feature that can be explained by the inefficiency of murine GG-NER in dealing with UV-induced cyclobutane pyrimidine dimer-like lesions, which are preferentially repaired by TC-NER (van der Horst et al., 1997van der Horst GTJ, Meira L, Gorgels TG, de Wit J, Velasco-Miguel S, Richardson JA, Kamp Y, Vreeswijk MPG, Smit B, Bootsma D et al. (2002) UVB radiation-induced cancer pre-disposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair (Amst) 1:143–157., 2002van der Pluijm I, Garinis GA, Brandt RMC, Gorgels TG, Wijnhoven SW, Diderich KEM, De Wit J, Mitchell JR, van Oostrom C, Beems R et al. (2007) Impaired genome maintenance suppresses the growth hormone-insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5:0023–0038.; de Boer and Hoeijmakers, 1999de Boer J and Hoeijmakers JH (1999) Cancer from the outside, aging from the inside: Mouse models to study the consequences of defective nucleotide excision repair. Biochimie 81:127–137.).
CSBm/m cells (from several tissues, mainly the brain and kidneys) also contain high levels of formamidopyrimidines and 8-oxoGua in DNA when compared to wild-type mice (Muftuoglu et al., 2009Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M and Bohr VA (2009) Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem 284:9270–9279.). In addition to nuclear DNA damage, mtDNA damage and mutational load in the mtDNA of CSA-/- and CSBm/m mice increased during aging and is related to subcutaneous fat loss, one of the main characteristics of CS, probably mediated by the apoptosis caused by mitochondrial dysfunction (Kamenisch et al., 2010Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dollé ME, Kuiper R, Majora M et al. (2010) Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207:379–90.) and by cell senescence, a feature commonly present in progeroid models (Carrero et al., 2016Carrero D, Soria-Valles C and López-Otín C (2016) Hallmarks of progeroid syndromes: Lessons from mice and reprogrammed cells. Dis Model Mech 9:719–735.).
The combined deletion of other NER proteins such as XPA in these CS mice leads to a more severe phenotype. These animals manifest severe neurological symptoms, ataxia, compromised growth, low weight, a lack of motor coordination, kyphosis, and abnormal behavior and development with premature death within 20 days (Murai et al., 2001Murai M, Enokido Y, Inamura N, Yoshino M, Nakatsu Y, van der Horst GT, Hoeijmakers JH, Tanaka K and Hatanaka H (2001) Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc Natl Acad Sci U S A 98:13379–13384.; van der Pluijm et al., 2007van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D and Hoeijmakers JH (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J 16:5955–5965.). Inactivation of GG-NER by an XPC knockout in CSBm/m mice leads to a similar phenotype, with dysmyelination, slow development, and low body weight: characteristics that resemble those of patients with CS (Laposa et al., 2007Laposa RR, Huang EJ and Cleaver JE (2007) Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc Natl Acad Sci U S A 104:1389–1394.; van der Pluijm et al., 2007van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D and Hoeijmakers JH (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J 16:5955–5965.; Revet et al., 2012Revet I, Feeney L, Tang A, Huang EJ and Cleaver JE (2012) Dysmyelination not demyelination causes neurological symptoms in preweaned mice in a murine model of Cockayne syndrome. Proc Natl Acad Sci U S A 109:4627–32.). CSA-/-/XPA-/- mice also show evidence of neurological problems, life expectancy reduced to ~1 month, an increase in cellular oxidative phosphorylation, and lipodystrophy (Brace et al., 2013Brace LE, Vose SC, Vargas DF, Zhao S, Wang XP and Mitchell JR (2013) Lifespan extension by dietary intervention in a mouse model of Cockayne syndrome uncouples early postnatal development from segmental progeria. Aging Cell 12:1144–1147., 2016Brace LE, Vose SC, Stanya K, Gathungu RM, Marur VR, Longchamp A, Treviño-Villarreal H, Mejia P, Vargas D, Inouye K et al. (2016) Increased oxidative phosphorylation in response to acute and chronic DNA damage. Aging Mech Dis 2:16022.).
The mechanism that underlies this aggravation of the CS phenotype by the double inactivation of CS and XP genes in mice has yet to be uncovered. Since CS and XP proteins act in the same DNA repair pathway, it is assumed that this severe CS phenotype in mice is due to the disruption of both NER-related and other functions of the CS proteins, such as transcription regulation (Brooks, 2013Brooks PJ (2013) Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 12:656–671.) and/or interactions with other DNA repair pathways (Murai et al., 2001Murai M, Enokido Y, Inamura N, Yoshino M, Nakatsu Y, van der Horst GT, Hoeijmakers JH, Tanaka K and Hatanaka H (2001) Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc Natl Acad Sci U S A 98:13379–13384.). Inactivation of NER by an XPC or XPA gene knockout can result in the accumulation of transcription-blocking DNA lesions, and the loss of either CS protein may further interfere with transcription and further increase the DNA damage load, which can in turn have detrimental effects, such as cell death or early cell senescence, both of which have been observed in progeroid mice (Weeda et al., 1997Wei L, Nakajima S, Böhm S, Bernstein KA, Shen Z and Tsang M (2015) DNA damage during the G0 / G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc Natl Acad Sci U S A 112:E3495–E3504.; van der Pluijm et al., 2007van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D and Hoeijmakers JH (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J 16:5955–5965.; Brooks, 2013Brooks PJ (2013) Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 12:656–671.).
Combined inactivation of TC-NER and NER in neurons has also been reported to generate an age-related progressive neurodegenerative phenotype. CSBm/m mice featuring a neuron-specific conditional XPA knockout have a shorter lifespan, behavioral abnormalities, and brain atrophy. These characteristics are possibly related to a synergistic effect of the TC-NER and NER pathways or to other functions of the inactivated proteins, such as chromatin remodeling or other functions in transcription (Newman et al., 2006Newman JC, Bailey AD and Weiner AM (2006) Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci U S A 103:9613–9618.; Jaarsma et al., 2011Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, Rijksen Y, Maas A, van Steeg H, Hoeijmakers JHJ et al. (2011) Age-related neuronal degeneration: Complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet 7:e1002405.; Wang et al., 2018Wang Y, Chakravarty P, Ranes M, Kelly G, Brooks PJ, Neilan E, Stewart A, Schiavo G and Svejstrup JQ (2014) Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease. Proc Natl Acad SciUSA 111:14454–14459.). Cell type–specific conditional knockouts may also help to better understand the participation of other processes in the progression of the disease, along with their molecular mechanisms, e.g., the role of oligodendrocytes (Howng et al., 2010Howng SY, Avila RL, Emery B, Traka M, Lin W, Watkins T, Cook S, Bronson R, Davisson M, Barres BA et al. (2010) ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev 24:301–311.) or other glial cells (Raj et al., 2014Raj DD, Jaarsma D, Holtman IR, Olah M, Ferreira FM, Schaafsma W, Brouwer N, Meijer MM, de Waard MC, van der Pluijm I et al. (2014) Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol Aging 35:2147–2160.) in CS dysmyelination and in the neurodegenerative phenotype.
Mutations in XPG or ERCC1 can yield mice with the characteristics similar to those of CS and other diseases on the CS spectrum, such as XP/CS and COFS (JaarsmaJaarsma D, van der Pluijm I, van der Horst GTJ and Hoeijmakers JH (2013) Cockayne syndrome pathogenesis: Lessons from mouse models. Mech Ageing Dev 134:180–195.et al., 2013). These NER nuclease–deficient models have several neurodegenerative features, altered metabolism, and a reduced lifespan (Weeda et al., 1997Wei L, Nakajima S, Böhm S, Bernstein KA, Shen Z and Tsang M (2015) DNA damage during the G0 / G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc Natl Acad Sci U S A 112:E3495–E3504.; Barnhoorn et al., 2014Barnhoorn S, Uittenboogaard LM, Jaarsma D, Vermeij WP, Tresini M, Weymaere M, Menoni H, Brandt RM, de Waard MC, Botter SM et al. (2014) Cell-autonomous progeroid changes in conditional mouse models for repair endonuclease XPG Deficiency. PLoS Genet 10:e1004686.). Notably, despite the lipodystrophy observed in all these animal models, dietary restriction and methionine restriction (which are metabolic interventions known to increase the lifespan of several species) reversed some of the neurodegeneration indicators of the CSA-/-/XPA-/- and ERCC1Δ/- models, with dietary restriction nearly doubling the lifespan of ERCC1Δ/- mice. This finding indicates that metabolic interventions are a possible therapy for NER-related progeroid diseases (Brace et al., 2016Brace LE, Vose SC, Stanya K, Gathungu RM, Marur VR, Longchamp A, Treviño-Villarreal H, Mejia P, Vargas D, Inouye K et al. (2016) Increased oxidative phosphorylation in response to acute and chronic DNA damage. Aging Mech Dis 2:16022.; Vermeij et al., 2016Vessoni AT, Herai RH, Karpiak JV, Leal AMS, Trujillo CA, Quinet A, Lima LFA, Menck CFM and Muotri AR (2016) Cockayne Syndrome-derived neurons display reduced synapse density and altered neural network synchrony. Hum Mol Genet 25:1271–1280.).
The current murine models for CS research do not fully reproduce the phenotype seen in humans because these animals manifest milder symptoms of the disease when only a single TC-NER protein is mutated. One of the possible explanations is the adaptation of NER to deal with DNA damage that would be repaired initially by CSA and CSB. This explains the severe symptoms when both TC-NER and NER are inactivated (van der Pluijm et al., 2007van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D and Hoeijmakers JH (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J 16:5955–5965.). In this case, it is important to consider the evolutionary distance that separates mice from humans. This distance entails differences in gene expression and metabolic and physiological profiles, among others (Seok et al., 2013Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L et al. (2013) Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110:3507–3512.; Lin et al., 2014Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC et al. (2014) Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 111:17224–17229.). Such differences contribute to the following phenomenon: the phenotype resulting from the same mutation and the activity of the repair pathway are not identical between the two species. Table 1 summarizes the mouse models carrying mutations in CS genes and other single-NER-mutation models that recapitulate CS-like phenotypes.
Reported neurological and metabolic characteristics of Cockayne Syndrome and related mouse models.
Different animal models of lower complexity have also been developed to study the role of CSA and CSB proteins, and are an interesting alternative for the research into protein functions. Among these models are Caenorhabditis elegans and zebrafish.
C. elegans has advantages, such as the ease of laboratory maintenance, of genetic manipulation, of tissue differentiation, rapid reproduction with several offspring, a generally fixed and genetically determined number of cells, and a short life cycle, allowing for the study of its development within short periods. It has been demonstrated that the NER pathway is well conserved in C. elegans, resembling the repair mechanisms of mammals (Meyer et al., 2007Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH and van Houten B (2007) Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol 8:R70.; reviewed by Lans and Vermeulen, 2011Lans H and Vermeulen W (2011) Nucleotide excision repair in Caenorhabditis elegans. Mol Biol Int 2011:1–12.), and because they do not repair DNA by photoreactivation (Hartman et al., 1989Hartman PS, Hevelone J, Dwarakanath V and Mitchell DL (1989) Excision repair of UV radiation-induced DNA damage in Caenorhabditis elegans. Genetics 122:379–385.). Rather, the lesions caused by UV are repaired exclusively by NER. CSA and CSB analogs, csa-1 (Babu et al., 2014Babu V, Hofmann K and Schumacher B (2014) A C. elegans homolog of the Cockayne syndrome complementation group A gene. DNA Repair (Amst) 24:57–62.) and csb-1 (CeCSB) (Lee et al., 2002Lee MH, Ahn B, Choi IS and Koo HS (2002) The gene expression and deficiency phenotypes of Cockayne syndrome B protein in Caenorhabditis elegans. FEBS Lett 522:47–51.), respectively, were found to be a part of this pathway. Animals mutated in csa-1 are hypersensitive to UV-B light exposure (Babu et al., 2014Babu V, Hofmann K and Schumacher B (2014) A C. elegans homolog of the Cockayne syndrome complementation group A gene. DNA Repair (Amst) 24:57–62.), and csb-1–deficient animal germ cells show apoptosis induction and morphological abnormalities after exposure to this agent (Lee et al., 2002Lee MH, Ahn B, Choi IS and Koo HS (2002) The gene expression and deficiency phenotypes of Cockayne syndrome B protein in Caenorhabditis elegans. FEBS Lett 522:47–51.). Moreover, a knockout of either csa-1 or csb-1 in C. elegans resulted in increased oxygen consumption and in changes in the transcription of genes related to mitochondrial ATP production, ubiquitin pathways, and transcriptional regulation (Scheibye-Knudsen et al., 2016Scheibye-Knudsen M, Tseng A, Jensen MB, Scheibye-Alsing K, Fang EF, Iyama T, Bharti SK, Marosi K, Froetscher L, Kassahun H et al. (2016) Cockayne syndrome group A and B proteins converge on transcription-linked resolution of non-B DNA. Proc Natl Acad Sci U S A 113:12502–12507.).
In general, C. elegans is a good model for the investigation of the DNA damage response to UV via the NER pathway, because this species makes it possible to reproduce and examine the effects of irradiation or other DNA-damaging agents on an entire organism and throughout its developmental stages within a short period of time.
The zebrafish is a vertebrate model that is widely used in research on the effects of exposure to genotoxic agents, carcinogenesis processes, and mainly embryonic development (Spitsbergen and Kent, 2003Spitsbergen JM and Kent ML (2003) The state of the art of the zebrafish model for toxicology and toxicologic pathology research - advantages and current limitations. Toxicol Pathol 31:62–87.; Titus et al., 2009Tornaletti S (2005) Transcription arrest at DNA damage sites. Mutat Res 577:131–145.). This animal has orthologous DNA repair genes in the pathways present in higher eukaryotes, e.g., BER, NER, MMR, Non-homologous end joining and Homologous recombination (Pei and Strauss, 2013Pei DS and Strauss PR (2013) Zebrafish as a model system to study DNA damage and repair. Mutat Res 743–744:151–159.). In NER, 44 genes are responsible for the functioning mechanisms of damage removal. Despite this observation, there are still few zebrafish studies where these characteristics are exploited from the perspective of DNA repair alone.
Zebrafish with the CSB depleted by morpholino oligonucleotides at the larval stage show an increased frequency of morphological abnormalities, which may recapitulate some of the congenital and developmental manifestations seen in patients with CS. Ionizing radiation can further increase morphological aberrations in CSB-depleted zebrafish, thereby pointing to an important role of CSB in the defense against oxidative DNA damage (Wei et al., 2015Weidenheim KM, Dickson DW and Rapin I (2009) Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech Ageing Dev 130:619–636.).
Due to the complexity of CS and the unique characteristics of patients with CS, which cannot be fully recapitulated in any animal model, a combination of human cell models with various animal models offers complimentary approaches to elucidate the various characteristics of this syndrome. In this sense, the use of somatic cell reprogramming coupled with genome editing allows investigators to obtain relevant and functional cell types (such as neurons) carrying patient-specific mutations. Moreover, this method offers an opportunity to investigate how the genetic background of different patients interacts with the one given pathological mutation. Such an approach may clarify why certain patients that share the same mutation in a CS gene have different phenotypes (Colella et al., 2000Colella S, Nardo T, Botta E, Lehmann AR and Stefanini M (2000) Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-Cacchione variant of Xeroderma pigmentosum. Hum Mol Genet 9:1171–1175.).
Conclusion
Almost 60 years of accumulated research in the NER field, especially regarding progeroid CS, provides extensive knowledge about the structure of proteins CSA and CSB and their participation in TC-NER and other mechanisms, such as transcription, repair, and mitochondrial functioning. For the advances made so far, human cellular models and animal models have been invaluable. They have revealed that the CS phenotype is likely a combination of altered gene transcription, metabolic adjustment, redox imbalance, and DNA repair defects, although the relative importance of each of these mechanisms for the disease is still largely unknown. Moreover, the full spectrum of the syndrome is not completely mirrored in animals, and this situation undoubtedly hinders further research. This difficulty is also associated with the impossibility of correlation between the mutations in ERCC6/ERCC8 genes and the phenotype of the patients, suggesting that the genetic background can heavily influence the manifestation of the symptoms.
Recently, the ability to generate pluripotent stem cells from patients with CS enabled for the first time the recapitulation of the full genetic signature of a patient in a cell type relevant for the disease, e.g., neurons. This observation expands and complements the previous CS models because it enables the investigation of metabolism, of the DNA damage response, and of gene expression in a patient’s cells. These data were previously impossible or very difficult to obtain. Still, one needs to keep in mind that CS is a systemic disease, and for this reason, investigating how dysfunction in one tissue/organ impacts others is critical. All these difficulties reveal how complex and diverse CS is and imply that a multimodel approach will therefore help to better recapitulate certain characteristics of CS. Moreover, although not entirely equivalent to normal aging, CS has clinical and cellular similarities to some aging-related diseases. Thus, improvements of (and new approaches to) CS models may have a broad impact on the study of these diseases as well.
Acknowledgments
We acknowledge the scholarship to CCCG from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES Brasil – Finance code 001), the scholarships to LLSN and GSK from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and CAPES. This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, grant APQ00828-14) and the Pró-Reitoria de Pesquisa da Universidade Federal de Ouro Preto. Last but not least, we thank Prof. Dr. Carlos Menck for critical reading and encouragement.
References
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, Stevnsner T et al. (2010) Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J 24:2334–2346.
- Aamann MD, Hvitby C, Popuri V, Muftuoglu M, Lemminger L, Skeby CK, Keijzers G, Ahn B, Bjoras M, Bohr VA et al. (2014a) Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity. Mech Ageing Dev 135:1–14.
- Aamann MD, Muftuoglu M, Bohr VA and Stevnsner T (2014b) Multiple interaction partners for Cockayne syndrome proteins. Mech Ageing Dev 134:212–224.
- Abolhassani N, Leon J, Sheng Z, Oka S, Hamasaki H, Iwaki T and Nakabeppu Y (2017) Molecular pathophysiology of impaired glucose metabolism, mitochondrial dysfunction, and oxidative DNA damage in Alzheimer’s disease brain. Mech Ageing Dev 161:95–104.
- Alupei MC, Maity P, Esser P, Krikki I, Tuorto F, Parlato R, Penzo M, Schelling A, Laugel V, Montanaro L et al. (2018) Loss of proteostasis is a pathomechanism in Cockayne syndrome. Cell Rep 23:1612–1619.
- Andrade LN, Nathanson JL, Yeo GW, Menck CFM and Muotri AR (2012) Evidence for premature aging due to oxidative stress in iPSCs from Cockayne syndrome. Hum Mol Genet 21:3825–3834.
- Andrews AD, Barrett SF, Yoder FW and Robbins JH (1978) Cockayne’s Syndrome fibroblasts have increased sensitivity to ultraviolet light but normal rates of unscheduled DNA synthesis. J Invest Dermatol 70:237–239.
- Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM and Svejstrup JQ (2010) A ubiquitin-binding domain in cockayne syndrome b required for transcription-coupled nucleotide excision repair. Mol Cell 38:637–648.
- Ayala-Peña S (2013) Role of oxidative DNA damage in mitochondrial dysfunction and Huntington’s disease pathogenesis. Free Radic Biol Med 62:102–110.
- Babu V, Hofmann K and Schumacher B (2014) A C. elegans homolog of the Cockayne syndrome complementation group A gene. DNA Repair (Amst) 24:57–62.
- Balajee AS, May A, Dianov GL, Friedberg EC and Bohr VA (1997) Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci U S A 94:4306–4311.
- Barnhoorn S, Uittenboogaard LM, Jaarsma D, Vermeij WP, Tresini M, Weymaere M, Menoni H, Brandt RM, de Waard MC, Botter SM et al. (2014) Cell-autonomous progeroid changes in conditional mouse models for repair endonuclease XPG Deficiency. PLoS Genet 10:e1004686.
- Beerens N, Hoeijmakers JHJ, Kanaar R, Vermeulen W and Wyman C (2005) The CSB protein actively wraps DNA. J Biol Chem 280:4722–4729.
- Berquist BR, Canugovi C, Sykora P, Wilson DM and Bohr VA (2012) Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res 40:8392–8405.
- Berra CM, de Oliveira CS, Garcia CCM, Rocha CRR, Lerner LK, Lima LC, Baptista M da S and Menck CFM (2013) Nucleotide excision repair activity on DNA damage induced by photoactivated methylene blue. Free Radic Biol Med 61:343–356.
- Boetefuer EL, Lake RJ, Dreval K and Fan HY (2018a) Poly(ADP-ribose) polymerase 1 (PARP1) promotes oxidative stress–induced association of Cockayne syndrome group B protein with chromatin. J Biol Chem 293:17863-17874.
- Boetefuer EL, Lake RJ and Fan HY (2018b) Mechanistic insights into the regulation of transcription and transcription-coupled DNA repair by Cockayne syndrome protein B. Nucleic Acids Res 46:7471–7479.
- Bohr VA, Smith CA, Okumoto DS and Hanawalt PC (1985) DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40:359–369.
- Brace LE, Vose SC, Vargas DF, Zhao S, Wang XP and Mitchell JR (2013) Lifespan extension by dietary intervention in a mouse model of Cockayne syndrome uncouples early postnatal development from segmental progeria. Aging Cell 12:1144–1147.
- Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I and Egly JM (2002) CSB Is a Component of RNA Pol I Transcription. Mol Cell 10:819–829.
- Brace LE, Vose SC, Stanya K, Gathungu RM, Marur VR, Longchamp A, Treviño-Villarreal H, Mejia P, Vargas D, Inouye K et al. (2016) Increased oxidative phosphorylation in response to acute and chronic DNA damage. Aging Mech Dis 2:16022.
- Brooks PJ (2013) Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 12:656–671.
- Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K et al. (2000) The oxidative DNA lesion 8,5’-(S)-Cyclo- 2’-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem 275:22355–22362.
- Brosh RM Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L and Bohr VA (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell 10:3583–3594.
- Calmels N, Botta E, Jia N, Fawcett H, Nardo T, Nakazawa Y, Lanzafame M, Moriwaki S, Sugita K, Kubota M et al. (2018) Functional and clinical relevance of novel mutations in a large cohort of patients with Cockayne syndrome. J Med Genet 55:329-343.
- Carpenter AE, Memedula S, Plutz MJ and Belmont AS (2005) Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol 25:958–968.
- Carrero D, Soria-Valles C and López-Otín C (2016) Hallmarks of progeroid syndromes: Lessons from mice and reprogrammed cells. Dis Model Mech 9:719–735.
- Chaim IA, Gardner A, Wu J, Iyama T, Wilson DM and Samson LD (2017) A novel role for transcription-coupled nucleotide excision repair for the in vivo repair of 3,N4-ethenocytosine. Nucleic Acids Res 45:3242–3252.
- Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM and Bohr VA (2003) Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res 31:963–973.
- Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH and Vermeulen W (1998) Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem 273:11844–11851.
- Citterio E, van den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JHJ and Vermeulen W (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol 20:7643–7653.
- Cleaver JE, Lam ET and Revet I (2009) Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet 10:756–768.
- Cleaver JE, Brennan-Minnella AM, Swanson RA, Fong K, Chen J, Chou KM, Chen YW, Revet I and Bezrookove V (2014) Mitochondrial reactive oxygen species are scavenged by Cockayne syndrome B protein in human fibroblasts without nuclear DNA damage. Proc Natl Acad Sci U S A 111:13487-13492.
- Cockayne EA (1936) Dwarfism with retinal atrophy and deafness. Arch Dis Child 11:1–8.
- Cockayne EA (1946) Dwarfism with retinal atrophy and deafness. Arch Dis Child 11:52–54.
- Coin F, Bergmann E, Tremeau-Bravard A and Egly J (1999) Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J 18:1357–1366.
- Colella S, Nardo T, Botta E, Lehmann AR and Stefanini M (2000) Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-Cacchione variant of Xeroderma pigmentosum. Hum Mol Genet 9:1171–1175.
- Compe E and Egly JM (2012) TFIIH: When transcription meets DNA repair. Nat Rev Mol Cell Biol 13:343–354.
- Coppedè F, Tannorella P, Stoccoro A, Chico L, Siciliano G, Bonuccelli U and Migliore L (2016) Methylation analysis of DNA repair genes in Alzheimer’s disease. Mech Ageing Dev 161:105–111.
- Cordisco S, Tinaburri L, Teson M, Orioli D, Cardin R, Degan P, Stefanini M, Zambruno G, Guerra L and Dellambra E (2018) Cockayne syndrome type A protein protects primary human keratinocytes from senescence. J Invest Dermatol 139:1–13. Costa RM, Chiganças V, Galhardo RS, Carvalho H and Menck CFM (2003) The eukaryotic nucleotide excision repair pathway. Biochimie 85:1083–1099.
- D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM et al. (2007) The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 26:4336–4343.
- D’Errico M, Pascucci B, Iorio E, van Houten B and Dogliotti E (2013) The role of CSA and CSB protein in the oxidative stress response. Mech Ageing Dev 134:261–269.
- de Boer J and Hoeijmakers JH (1999) Cancer from the outside, aging from the inside: Mouse models to study the consequences of defective nucleotide excision repair. Biochimie 81:127–137.
- de Waard H, de Wit J, Gorgels TGMF, van den Aardweg G, Andressoo JO, Vermeij M, Van Steeg H, Hoeijmakers JHJ and van der Horst GTJ (2003) Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2:13–25.
- de Waard H, de Wit J, Andressoo JO, van Oostrom CTM, Riis B, Weimann A, Poulsen HE, van Steeg H, Hoeijmakers JHJ and van der Horst GTJ (2004) Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol Cell Biol 24:7941–7948.
- Dianov G, Bishoff C, Sunesen M and Bohr VA (1999) Repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Nucleic Acids Res 27:1365–1368.
- Donahue BA, Yint S, Taylort JS, Reines D and Hanawalt PC (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A 91:8502–8506.
- Durik M, Kavousi M, van der Pluijm I, Isaacs A, Cheng C, Verdonk K, Loot AE, Oeseburg H, Bhaggoe UM, Leijten F et al. (2013) Nucleotide Excision DNA Repair is Associated with Age-Related Vascular Dysfunction. Circulation 126:468–478.
- Egly JM and Coin F (2011) A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 10:714–721.
- Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F et al. (2011) The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147:1024–1039.
- Fivenson EM, Lautrup S, Sun N, Sheibye-Knudsen M, Stevnsner T, Nilsen H, Bohr VA and Fang EF (2017) Mitophagy in neurodegeneration and aging. Neurochem Int 109:202–209.
- Fousteri M and Mullenders LHF (2008) Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res 18:73–84.
- Fousteri M, Vermeulen W, van Zeeland AA and Mullenders LHF (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23:471–482.
- Frontini M and Proietti-De-Santis L (2009) Cockayne syndrome B protein (CSB): Linking p53, HIF-1 and p300 to robustness, lifespan, cancer and cell fate decisions. Cell Cycle 8:693–696.
- Fusser M, Nesse GJ, Khobta A, Xia N, Li H, Klungland A and Epe B (2011) Spontaneous mutagenesis in csbm/mogg1-/- mice is attenuated by dietary resveratrol. Carcinogenesis 32:80–85.
- Gorgels TG, van der Pluijm I, Brandt RM, Garinis GA, van Steeg H, van den Aardweg G, Jansen GH, Ruijter JM, Bergen AA, van Norren D et al. (2007) Retinal degeneration and ionizing radiation hypersensitivity in a mouse model for Cockayne syndrome. Mol Cell Biol 27:1433–1441.
- Gregersen LH and Svejstrup JQ (2018) The cellular response to transcription-blocking DNA damage. Trends Biochem Sci 43:327–341.
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K and Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357–367.
- Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A and Nakatani Y (2006) CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20:1429–1434.
- Guardiola A, Álvares-da-Silva CR, Grisolia J and Silbermann R (1999) Síndrome de Cockayne. Arq Neuro-Psiquiatr 57:106–110.
- Hanawalt PC and Spivak G (2008) Transcription-coupled DNA repair: Two decades of progress and surprises. Nature 9:958–970.
- Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CAS, Conaway RC, Conaway JW et al. (2009) Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A 106:20705–20710.
- Hartman PS, Hevelone J, Dwarakanath V and Mitchell DL (1989) Excision repair of UV radiation-induced DNA damage in Caenorhabditis elegans. Genetics 122:379–385.
- Hayashi M, Araki S, Kohyama J, Shioda K and Fukatsu R (2005) Oxidative nucleotide damage and superoxide dismutase expression in the brains of Xeroderma pigmentosum group A and Cockayne syndrome. Brain Dev 27:34–38.
- Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV et al. (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555–564.
- Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, Dinant C, Hoeijmakers JHJ, Vermeulen W et al. (2008) Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci 121:2850–2859.
- Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M and Tanaka K (2004) Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A 101:15410–15415.
- Howng SY, Avila RL, Emery B, Traka M, Lin W, Watkins T, Cook S, Bronson R, Davisson M, Barres BA et al. (2010) ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev 24:301–311.
- Hu J, Dong L and Outten CE (2008) The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem 283:29126–29134.
- Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura KI, Utsumi H, Hamasaki N and Takeshita A (2001) Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res 88:529–535.
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM and Tanaka K (2007) XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/Patients with CS. Mol Cell 26:231–243.
- Itoh T, Shiomi T, Shiomi N, Harada Y, Wakasugi M, Matsunaga T, Nikaido O, Friedberg EC and Yamaizumi M (1996) Rodent complementation group 8 (ERCC8) corresponds to Cockayne syndrome complementation group A. Mutat Res 362:167–174.
- Iyama T, Okur MN, Golato T, McNeill DR, Lu H, Hamilton R, Raja A, Bohr VA and Wilson III DM (2018) Regulation of the intranuclear distribution of the Cockayne syndrome proteins. Sci Rep 8:1–12.
- Jaakkola E, Mustonen A, Olsen P, Miettinen S, Savuoja T, Raams A, Jaspers NG, Shao H, Wu BL and Ignatius J (2010) ERCC6 founder mutation identified in Finnish patients with COFS syndrome. Clin Genet 78:541–547.
- Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, Rijksen Y, Maas A, van Steeg H, Hoeijmakers JHJ et al. (2011) Age-related neuronal degeneration: Complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet 7:e1002405.
- Jaarsma D, van der Pluijm I, van der Horst GTJ and Hoeijmakers JH (2013) Cockayne syndrome pathogenesis: Lessons from mouse models. Mech Ageing Dev 134:180–195.
- Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dollé ME, Kuiper R, Majora M et al. (2010) Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207:379–90.
- Kamiuchi S, Saijo M, Citterio E, de Jager M, Hoeijmakers JH and Tanaka K (2002) Translocation of Cockayne syndrome group A protein to the nuclear matrix: Possible relevance to transcription-coupled DNA repair. Proc Natl Acad Sci U S A 99:201–206.
- Karam SM, Costa JC, Jardim L, Pires RF, Lehmann AR and Giugliani R (2000) Cockayne syndrome: Report of a Brazilian family with confirmation of impaired RNA synthesis after UV-irradiation. Genet Mol Biol 275:273–275.
- Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL and Bohr VA (2017) Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res Rev 33:3–17.
- Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L et al. (2013) Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, Xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet 92:807–819.
- Khobta A and Epe B (2013) Repair of oxidatively generated DNA damage in Cockayne syndrome. Mech Ageing Dev 134:253–260.
- Kleijer WJ, Laugel V, Berneburg M, Nardo T, Fawcett H, Gratchev A, Jaspers NGJ, Sarasin A, Stefanini M and Lehmann AR (2008) Incidence of DNA repair deficiency disorders in western Europe: Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. DNA Repair (Amst) 7:744–750.
- Koch S, Gonzalez OG, Assfalg R, Schelling A, Schäfer P, Scharffetter-Kochanek K and Iben S (2014) Cockayne syndrome protein A is a transcription factor of RNA polymerase I and stimulates ribosomal biogenesis and growth. Cell Cycle 13:2029–2037.
- Kowaltowski AJ, de Souza-Pinto NC, Castilho RF and Vercesi AE (2009) Mitochondria and reactive oxygen species. Free Radic Biol Med 47:333–343.
- Kubota M, Ohta S, Ando A, Koyama A, Terashima H, Kashii H, Hoshino H, Sugita K and Hayashi M (2015) Nationwide survey of Cockayne syndrome in Japan: Incidence, clinical course and prognosis. Pedriatrics Int 57:339–347.
- Lake RJ, Geyko A, Hemashettar G, Zhao Y and Fan HY (2010) UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell 37:1–20.
- Lake RJ, Boetefuer EL, Tsai PF, Jeong J, Choi I, Won KJ and Fan HY (2014) The sequence-specific transcription factor c-jun targets Cockayne syndrome protein B to regulate transcription and chromatin structure. PLoS Genet 10:e1004284.
- Lake RJ, Boetefuer EL, Won KJ and Fan HY (2016) The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res 44:2125–2135.
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE and Corbett AH (2007) Classical nuclear localization signals: Definition, function, and interaction with importin α. J Biol Chem 282:5101–5105.
- Lans H and Vermeulen W (2011) Nucleotide excision repair in Caenorhabditis elegans. Mol Biol Int 2011:1–12.
- Laposa RR, Huang EJ and Cleaver JE (2007) Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc Natl Acad Sci U S A 104:1389–1394.
- Laugel V (2013) Cockayne syndrome: The expanding clinical and mutational spectrum. Mech Ageing Dev 134:161–170.
- Laugel V, Dalloz AC, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Gener B, Tobias ES et al. (2009) Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat 31:113–126.
- Le May N, Mota-Fernandes D, Vélez-Cruz R, Iltis I, Biard D and Egly JM (2010) NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell 38:54–66.
- Lee MH, Ahn B, Choi IS and Koo HS (2002) The gene expression and deficiency phenotypes of Cockayne syndrome B protein in Caenorhabditis elegans. FEBS Lett 522:47–51.
- Lehmann AR (1982) Three complementation groups in Cockayne syndrome. Mutat Res 106:347–356.
- Lehmann J, Schubert S, Schäfer A, Apel A, Laspe P, Schiller S, Ohlenbusch A, Gratchev A and Emmert S (2014) An unusual mutation in the XPG gene leads to an internal in-frame deletion and a XP/CS complex phenotype. Br J Dermatol 171:903–905.
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC et al. (2014) Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 111:17224–17229.
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y and Zhao Y (2015) Sequence analysis IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361.
- Ljungman M and Zhang F (1996) Blockage of RNA polymerase as a possible trigger for UV light-induced apoptosis. Oncogene 13:823–831.
- Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Lovelace LJ, Yandava CN et al. (2018) Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359:555–559.
- Lusser A and Kadonaga JT (2003) Chromatin remodeling by ATP-dependent molecular machines. BioEssays 25:1192–1200.
- Maddukuri L, Speina E, Christiansen M, Dudzinska D, Zaim J, Obtulowicz T, Kabaczyk S, Komisarki M, Bukowy Z, Szczegielniak J et al. (2009) Cockayne syndrome group B protein is engaged in processing of DNA adducts of lipid peroxidation product trans-4-hydroxy-2-nonenal. Mutat Res 666:23–31.
- Majora M, Sondenheimer K, Knechten M, Uthe I, Esser C, Schiavi A, Ventura N and Krutmann J (2018) HDAC inhibition improves autophagic and lysosomal function to prevent loss of subcutaneous fat in a mouse model of Cockayne syndrome. Sci Transl Med 10:1–13.
- Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M and Lehmann AR (1998) Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet 62:77–85.
- Marteijn JA, Lans H, Vermeulen W and Hoeijmakers JHJ (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15:465–481.
- Mayne LV and Lehmann AR (1982) Failure of RNA synthesis to recover after UV irradiation: An early defect in cells from individuals with Cockayne’s syndrome and Xeroderma pigmentosum. Cancer Res 42:1473–1478.
- McWhir J, Selfridge J, Harrison DJ, Squires S and Melton DW (1993) Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and dia before weaning. Nat Genet 5:217–224.
- Mecocci P, MacGarvey U and Beal MF (1994) Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann Neurol 36:747–751.
- Melcher K (2000) The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional protiens. J Mol Biol 301:1097–1112.
- Menck CFM and Munford V (2014) DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol 37:220–233.
- Menoni H, Wienholz F, Theil AF, Janssens RC, Lans H, Campalans A, Radicella JP, Marteijn JA and Vermeulen W (2018) The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res 46:7747–7756.
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH and van Houten B (2007) Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol 8:R70.
- Moriel-Carretero M, Herrera-Moyano E and Aguilera A (2015) A unified model for the molecular basis of Xeroderma pigmentosum-Cockayne syndrome. Rare Dis 3:e1079362.
- Muftuoglu M, Selzer R, Tuo J, Brosh RM and Bohr VA (2002) Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene 283:27–40.
- Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M and Bohr VA (2009) Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem 284:9270–9279.
- Muftuoglu M, Mori MP and de Souza-Pinto NC (2014) Formation and repair of oxidative damage in the mitochondrial DNA. Mitochondrion 17:164–181.
- Murai M, Enokido Y, Inamura N, Yoshino M, Nakatsu Y, van der Horst GT, Hoeijmakers JH, Tanaka K and Hatanaka H (2001) Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc Natl Acad Sci U S A 98:13379–13384.
- Nagtegaal AP, Rainey RN, van der Pluijm I, Brandt RM, van der Horst GT, Borst JG and Segil N (2015) Cockayne syndrome group B (Csb) and group A (Csa) deficiencies predispose to hearing loss and cochlear hair cell degeneration in mice. J Neurosci 35:4280–4286.
- Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo J, Khaw M, Saijo M, Kodo N et al. (2000) XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and rranscription. J Biol Chem 275:34931–34937.
- Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC et al. (2009) A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A 106:6209–6214.
- Narita T, Narita K, Takedachi A, Saijo M and Tanaka K (2015) Regulation of transcription elongation by the XPG-TFIIH complex is implicated in Cockayne syndrome. Mol Cell 35:3178–3188.
- Natale V (2010) A comprehensive description of the severity groups in Cockayne Syndrome. Am J Med Genet Part A 155:1081–1095.
- Nevedomskaya E, Meissner A, Goraler S, De Waard M, Ridwan Y, Zondag G, Van Der Pluijm I, Deelder AM and Mayboroda OA (2010) Metabolic profiling of accelerated aging ERCC1d/- mice. J Proteome Res 9:3680–3687.
- Newman JC, Bailey AD and Weiner AM (2006) Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci U S A 103:9613–9618.
- Osenbroch PO, Auk-Emblem P, Halsne R, Strand J, Forstrøm RJ, van der Pluijm I and Eide L (2009) Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J 276:2811–2821.
- Osterod M, Larsen E, Le Page F, Hengstler JG, van der Horst GTJ, Boiteux S, Klungland A and Epe B (2002) A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene 21:8232–8239.
- Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F et al. (2012) An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell 11:520–529.
- Pascucci B, D’Errico M, Romagnoli A, De Nuccio C, Savino M, Pietraforte D, Lanzafame M, Calcagnile AS, Fortini P, Baccarini S et al. (2016) Overexpression of Parkin rescues the defective mitochondrial phenotype and the increased apoptosis of Cockayne Syndrome A cells. Oncotarget 8:102852–102867.
- Pastoriza-Gallego M, Armier J and Sarasin A (2007) Transcription through 8-oxoguanine in DNA repair-proficient and Csb-/Ogg1- DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis 22:343–351.
- Pazin MJ and Kadonaga JT (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88:737–740.
- Pei DS and Strauss PR (2013) Zebrafish as a model system to study DNA damage and repair. Mutat Res 743–744:151–159.
- Pinto M and Moraes CT (2015) Mechanisms linking mtDNA damage and aging. Free Radic Biol Med 85:250–258.
- Prates Mori M and de Souza-Pinto NC (2018) Role of mitochondrial dysfunction in the pathophysiology of DNA repair disorders. Cell Biol Int 42:643–650.
- Raj DD, Jaarsma D, Holtman IR, Olah M, Ferreira FM, Schaafsma W, Brouwer N, Meijer MM, de Waard MC, van der Pluijm I et al. (2014) Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol Aging 35:2147–2160.
- Ranes M, Boeing S, Wang Y, Wienholz F, Menoni H, Walker J, Encheva V, Chakravarty P, Mari PO, Stewart A et al. (2016) A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic Acids Res 44:5246–5255.
- Reardon JT and Sancar A (2005) Nucleotide excision repair. Prog Nucleic Acid Res 79:183–235.
- Revet I, Feeney L, Tang A, Huang EJ and Cleaver JE (2012) Dysmyelination not demyelination causes neurological symptoms in preweaned mice in a murine model of Cockayne syndrome. Proc Natl Acad Sci U S A 109:4627–32.
- Sabatella M, Theil AF, Ribeiro-Silva C, Slyskova J, Thijssen K, Voskamp C, Lans H and Vermeulen W (2018) Repair protein persistence at DNA lesions characterizes XPF defect with Cockayne syndrome features. Nucleic Acids Res 46:9563–9577.
- Sancar A and Rupp WD (1983) A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33:249–260.
- Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E and Cooper PK (2005) Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: Insights for transcription-coupled repair and Cockayne syndrome. Mol Cell 20:187–198.
- Schärer OD (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5:a012609.
- Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, Cooper M, Spencer R, de Cabo R et al. (2012) Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med 209:855–869.
- Scheibye-Knudsen M, Tseng A, Jensen MB, Scheibye-Alsing K, Fang EF, Iyama T, Bharti SK, Marosi K, Froetscher L, Kassahun H et al. (2016) Cockayne syndrome group A and B proteins converge on transcription-linked resolution of non-B DNA. Proc Natl Acad Sci U S A 113:12502–12507.
- Schmickel RD, Chu EH, Trosko JE and Chang CC (1977) Cockayne syndrome: A cellular sensitivity to ultraviolet light. Pediatrics 60:135–139.
- Schwertman P, Lagarou A, Dekkers DHW, Raams A, van der Hoek A, Laffeber C, Hoeijmakers JH, Demmers JAA, Fousteri M, Vermeulen W et al. (2012) UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 44:598-602.
- Seeberg E and Strike P (1976) Excision repair of ultraviolet-irradiated deoxyribonucleic acid in plasmolyzed cells of Escherichia coli. J Bacteriol 125:787–795.
- Selby CP and Sancar A (1997a) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A 94:11205–11209.
- Selby CP and Sancar A (1997b) Human transcription-repair coupling factor CSB / ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem 272:1885–1890.
- Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM Jr and Bohr VA (2002) Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res 30:782–793.
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L et al. (2013) Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110:3507–3512.
- Sin Y, Tanaka K and Saijo M (2016) The C-terminal region and SUMOylation of Cockayne syndrome group B protein play critical roles in transcription-coupled nucleotide excision repair. J Biol Chem 291:1387–1397.
- Spitsbergen JM and Kent ML (2003) The state of the art of the zebrafish model for toxicology and toxicologic pathology research - advantages and current limitations. Toxicol Pathol 31:62–87.
- Spivak G (2005) UV-sensitive syndrome. Mutat Res 577:162–169.
- Spivak G and Ganesan AK (2014) The complex choreography of transcription-coupled repair. DNA Repair (Amst) 19:64–70. Stadler J and Richly H (2017) Regulation of DNA repair mechanisms: How the chromatin environment regulates the DNA damage response. Int J Mol Sci 18:1–15.
- Stevnsner T, Muftuoglu M, Aamann MD and Bohr VA (2008) The role of Cockayne syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev 129:441–448.
- Stichel CC, Zhu XR, Bader V, Linnartz B, Schmidt S and Lübbert H (2007) Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum Mol Genet 16:2377–2393.
- Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC and Bohr VA (2005) Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res 33:3722–3732.
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker APM, Hanaoka F, Bootsma D and Hoeijmakers JHJ (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 2:223–232.
- Sunesen M, Selzer RR, Brosh Jr. RM, Balajee AS, Stevnsner T and Bohr VA (2000) Molecular characterization of an acidic region deletion mutant of Cockayne syndrome group B protein. Nucleic Acids Res 28:3151–3159.
- Taghdiri M, Dastsooz H, Fardaei M, Mohammadi S, Fard MAF and Faghihi MA (2017) A novel mutation in ERCC8 gene causing Cockayne syndrome. Front Pediatr 5:169.
- Tanaka K, Kawai K, Kumahara Y, Ikenaga M and Okada Y (1981) Genetic complementation groups in Cockayne syndrome. Somatic Cell Genet 7:445–455.
- Tantin D (1998) RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J Biol Chem 273:27794–27799.
- Tantin D, Kansal A and Carey M (1997) Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol 17:6803–6814.
- Tirode F, Busso D, Coin F and Egly JM (1999) Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell 3:87–95.
- Titus TA, Yan YL, Wilson C, Starks AM, Frohnmayer JD, Canestro C, Rodriguez-Mari A, He X and Postlethwait JH (2009) The Fanconi anemia/BRCA gene network in zebrafish: Embryonic expression and comparative genomics. Mutat Res 668:117–132.
- Tornaletti S (2005) Transcription arrest at DNA damage sites. Mutat Res 577:131–145.
- Trapp C, Reite K, Klungland A and Epe B (2007) Deficiency of the Cockayne syndrome B (CSB) gene aGG-NERavates the genomic instability caused by endogenous oxidative DNA base damage in mice. Oncogene 26:4044–4048.
- Troelstra C, Odijk LH, de Wit J, Westerveld A, Thompson LH, Bootsma D and Hoeijmakers JHJ (1990) Molecular cloning of the human DNA excision repair. Mol Cell Biol 10:5806–5813.
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D and Hoeijmakers JHJ (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71:939–953.
- Troelstra C, Hesen W, Bootsma D and Hoeijmakers JHJ (1993) Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res 21:419–426.
- Tuo J, Müftüoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Rodriguez H, Dizdaroglu M and Bohr VA (2001) The Cockayne syndrome group b gene product is involved in general genome base excision repair of 8-Hydroxyguanine in DNA. J Biol Chem 276:45772–45779.
- Tuo J, Chen C, Zeng X, Christiansen M and Bohr VA (2002a) Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 1:913–927.
- Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M and Bohr VA (2002b) The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem 277:30832–30837.
- Tuo J, Jaruga P, Rodriguez H, Bohr VA and Dizdaroglu M (2003) Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J 17:668–674.
- van den Boom V, Citterio E, Hoogstraten D, Zotter A, Egly JM, van Cappellen WA, Hoeijmakers JHJ, Houtsmuller AB and Vermeulen W (2004) DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J Cell Biol 166:27–36.
- van der Horst GTJ, van Steeg H, Berg RJW, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR et al. (1997) Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer pre-disposition. Cell 89:425–435.
- van der Horst GTJ, Meira L, Gorgels TG, de Wit J, Velasco-Miguel S, Richardson JA, Kamp Y, Vreeswijk MPG, Smit B, Bootsma D et al. (2002) UVB radiation-induced cancer pre-disposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair (Amst) 1:143–157.
- van der Pluijm I, Garinis GA, Brandt RMC, Gorgels TG, Wijnhoven SW, Diderich KEM, De Wit J, Mitchell JR, van Oostrom C, Beems R et al. (2007) Impaired genome maintenance suppresses the growth hormone-insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5:0023–0038.
- van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D and Hoeijmakers JH (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J 16:5955–5965.
- van Hoffen A, Natarajan AT, Mayne LV, Zeeland AA Van, Mullendersl LHF and Venemal J (1993) Deficient repair of the transcribed strand of active in Cockayne’s syndrome cells. Nucleic Acids Res 21:5890–5895.
- van Houten B, Hunter SE and Meyer JN (2016) Mitochondrial DNA damage induced autophagy, cell death, and disease. Front Biosci 21:42–54.
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA and Mayne LV (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci U S A 87:4707–4711.
- Vermeij WP, Dollé MET, Reiling E, Jaarsma D, Payan-Gomez C, Bombardieri CR, Wu H, Roks AJM, Botter SM, van der Eerden BC et al. (2016) Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 537:427–431.
- Vessoni AT, Herai RH, Karpiak JV, Leal AMS, Trujillo CA, Quinet A, Lima LFA, Menck CFM and Muotri AR (2016) Cockayne Syndrome-derived neurons display reduced synapse density and altered neural network synchrony. Hum Mol Genet 25:1271–1280.
- Wallace DC (2012) Mitochondria and cancer. Nat Rev Cancer 12:685–698.
- Wang W, Xu J, Chong J and Wang D (2018) Structural basis of DNA lesion recognition for eukaryotic transcription-coupled nucleotide excision repair. DNA Repair (Amst) 71:43–55.
- Wang Y, Chakravarty P, Ranes M, Kelly G, Brooks PJ, Neilan E, Stewart A, Schiavo G and Svejstrup JQ (2014) Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease. Proc Natl Acad SciUSA 111:14454–14459.
- Weeda G, van Ham RCA, Vermeulen W, Bootsma D, van der Eb AJ and Hoeijmakers JHJ (1990) A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders Xeroderma pigmentosum and Cockayne’s syndrome. Cell 62:777–791.
- Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D and Hoeijmakers JHJ (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol 7:427–439.
- Wei L, Nakajima S, Böhm S, Bernstein KA, Shen Z and Tsang M (2015) DNA damage during the G0 / G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc Natl Acad Sci U S A 112:E3495–E3504.
- Weidenheim KM, Dickson DW and Rapin I (2009) Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech Ageing Dev 130:619–636.
- Wilson BT, Stark Z, Sutton RE, Danda S, Ekbote AV, Elsayed SM, Gibson L, Goodship JA, Jackson AP, Keng WT et al. (2015) The Cockayne Syndrome Natural History (CoSyNH) study: Clinical findings in 102 individuals and recommendations for care. Genet Med 18:483–493.
- Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA and Wilson DM (2007) Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res 35:4103–4113.
- Wu M, Wei W, Chen J, Cong R, Shi T, Li J, Wong J and Du JX (2017) Acidic domains differentially read histone H3 lysine 4 methylation status and are widely present in chromatin-associated proteins. Sci China Life Sci 60:138–151.
- Xu C and Min J (2011) Structure and function of WD40 domain proteins. Protein Cell 2:202–214.
- Yu S, Chen L, Ye L, Fei L, Tang W and Tian Y (2014) Identification of two missense mutations of ERCC6 in three chinese sisters with Cockayne syndrome by Whole Exome Sequencing. PloS One 9:1–11.
- Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T et al. (2012) Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 44:593–597.
Supplementary material
The following online material is available for this article:
Edited by
Publication Dates
-
Publication in this collection
20 May 2020 -
Date of issue
2020
History
-
Received
18 Mar 2019 -
Accepted
15 Jan 2020