Abstract
Background
Alzheimer’s disease (AD) is one of the most common neurodegenerative diseases, accompanied by cognitive and memory impairment, accounting for about 60% - 80% of dementia types. The pathogenesis of AD has not been clarified, and there is no effective therapy to prevent or treat AD. In this study, we aimed to identify the potential biomarkers involved in the brain immune microenvironment in AD.
Methods
AD datasets from GEO database were obtained to identify the differentially expressed disease-related genes (DEDRGs) in AD through weighted gene co-expression network analysis (WGCNA) and differential expression analysis. Functional Enrichment analysis was performed to explore the potential biological function of DEDRGs. The hub DEDRGs were identified through the protein-protein interaction (PPI) network. Furthermore, the CIBERSORT algorithm was employed to bulk gene expression profiles of AD to depict the immune microenvironment characteristics in AD. Pearson’s correlation analysis was utilized to depict the correlation between each of immune cells and hub DEDRGs.
Results
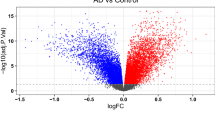
A total of 27 DEDRGs were identified through WGCNA and differential expression analysis. Functional enrichment analysis of 27 DEDRGs indicated that chemokine signaling pathway was the most significantly enriched KEGG pathway, response to biotic stimulus was the most significantly enriched GO term, and most of DEDRGs were enriched into urinary system cancer in DO analysis. 6 hub DEDRGs, ANGPT1, CCL2, CD44, CXCR4, GJA1 and VCAM1, were screened through PPI network and all of them were up-regulated in AD. Immune infiltration analysis revealed that there were higher infiltration levels of T cells CD4 memory activated, T cells gamma delta, NK cells resting and macrophages M0, and lower infiltration level of NK cell activated in AD, and macrophages M2 owned the highest positively association with VCAM1 and CXCR4, but VCAM1 was statistically and negatively correlated to T cells CD8.
Conclusion
Our study identified 6 hub DEDRGs, ANGPT1, CCL2, CD44, CXCR4, GJA1 and VCAM1, were statistically associated with immune infiltrating cells, and were significantly related to the pathological development of AD, which may provide a theoretical basis for developing potential biomarkers and implementing effective therapies against AD.





Similar content being viewed by others
Abbreviations
- Aβ:
-
amyloid β
- AD:
-
Alzheimer’s disease
- DEDRGs:
-
differentially expressed disease-related genes
- WGCNA:
-
weighted gene co-expression network analysis
- PPI:
-
protein-protein interaction
- NFTs:
-
neurofibrillary tangles
- SP:
-
senile plaque
- ADI:
-
Alzheimer’s Disease International
- CNS:
-
central nervous system
- TOM:
-
topological overlap matrix
- DEGs:
-
differentially expressed genes
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene Ontology
- BP:
-
biological process
- CC:
-
cellular component
- MF:
-
molecular functio
- DO:
-
disease ontology
- MCODE:
-
Molecular Complex Detection
- CMV:
-
cytomegalovirus
- APOE:
-
apolipoprotein E
References
Lane CA, Hardy J, Schott JM. Alzheimer’s disease. European journal of neurology. Jan 2018;25(1):59–70. doi:https://doi.org/10.1111/ene.13439
Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Primers. Oct 15 2015;1:15056. doi:https://doi.org/10.1038/nrdp.2015.56
Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO molecular medicine. Jun 2016;8(6):595–608. doi:https://doi.org/10.15252/emmm.201606210
Chen GF, Xu TH, Yan Y, et al. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol Sin. Sep 2017;38(9):1205–1235. doi:https://doi.org/10.1038/aps.2017.28
Spillantini MG, Goedert M. Tau protein pathology in neurodegenerative diseases. Trends in neurosciences. Oct 1998;21(10):428–33. doi:https://doi.org/10.1016/s0166-2236(98)01337-x
Bussian TJ, Aziz A, Meyer CF, Swenson BL, van Deursen JM, Baker DJ. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature. 2018/10/01 2018;562(7728):578–582. doi:https://doi.org/10.1038/s41586-018-0543-y
Gao Y, Tan L, Yu JT, Tan L. Tau in Alzheimer’s Disease: Mechanisms and Therapeutic Strategies. Current Alzheimer research. 2018;15(3):283–300. doi:https://doi.org/10.2174/1567205014666170417111859
Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. Nov 3 2006;314(5800):777–81. doi:https://doi.org/10.1126/science.1132814
Mayeux R, Stern YJNRN. Epidemiology of Alzheimer disease. 2011;7(3):137–52.
Briggs R, Kennelly SP, O’Neill D. Drug treatments in Alzheimer’s disease. Clinical medicine (London, England). Jun 2016;16(3):247–53. doi:https://doi.org/10.7861/clinmedicine.16-3-247
International AsD. World Alzheimer Report 2021-Journey through the diagnosis of dementia. 2022
Akiyama H, Barger S, Barnum S, et al. Inflammation and Alzheimer’s disease. Neurobiology of aging. May-Jun 2000;21(3):383–421. doi:https://doi.org/10.1016/s0197-4580(00)00124-x
Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer’s disease. The Lancet Neurology. Apr 2015;14(4):388–405. doi:https://doi.org/10.1016/s1474-422(15)70016-5
Griciuc A, Tanzi RE. The role of innate immune genes in Alzheimer’s disease. Current opinion in neurology. Apr 1 2021;34(2):228–236. doi:https://doi.org/10.1097/wco.0000000000000911
Griciuc A, Federico AN, Natasan J, et al. Gene therapy for Alzheimer’s disease targeting CD33 reduces amyloid beta accumulation and neuroinflammation. Human molecular genetics. Oct 10 2020;29(17):2920–2935. doi:https://doi.org/10.1093/hmg/ddaa179
Svensson M, Olsson G, Yang Y, et al. The effect of electroconvulsive therapy on neuroinflammation, behavior and amyloid plaques in the 5xFAD mouse model of Alzheimer’s disease. Scientific reports. Mar 1 2021;11(1):4910. doi:https://doi.org/10.1038/s41598-021-83998-0
Lu Y, Li K, Hu Y, Wang X. Expression of Immune Related Genes and Possible Regulatory Mechanisms in Alzheimer’s Disease. Frontiers in immunology. 2021;12:768966. doi:https://doi.org/10.3389/fimmu.2021.768966
Jevtic S, Sengar AS, Salter MW, McLaurin J. The role of the immune system in Alzheimer disease: Etiology and treatment. Ageing research reviews. Nov 2017;40:84–94. doi:https://doi.org/10.1016/j.arr.2017.08.005
Shi Y, Holtzman DM. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nature reviews Immunology. Dec 2018;18(12):759–772. doi:https://doi.org/10.1038/s41577-018-0051-1
Sarlus H, Heneka MT. Microglia in Alzheimer’s disease. The Journal of clinical investigation. Sep 1 2017;127(9):3240–3249. doi:https://doi.org/10.1172/jci90606
Streit WJ, Khoshbouei H, Bechmann I. The Role of Microglia in Sporadic Alzheimer’s Disease. Journal of Alzheimer’s disease: JAD. 2021;79(3):961–968. doi:https://doi.org/10.3233/jad-201248
Keren-Shaul H, Spinrad A, Weiner A, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. Jun 15 2017;169(7):1276–1290.e17. doi:https://doi.org/10.1016/j.cell.2017.05.018
Hanslik KL, Ulland TK. The Role of Microglia and the Nlrp3 Inflammasome in Alzheimer’s Disease. Frontiers in neurology. 2020;11:570711. doi:https://doi.org/10.3389/fneur.2020.570711
Fricker M, Tolkovsky AM, Borutaite V, Coleman M, Brown GC. Neuronal Cell Death. Physiological reviews. Apr 1 2018;98(2):813–880. doi:https://doi.org/10.1152/physrev.00011.2017
Cornell J, Salinas S, Huang HY, Zhou M. Microglia regulation of synaptic plasticity and learning and memory. Neural regeneration research. Apr 2022;17(4):705–716. doi:https://doi.org/10.4103/1673-5374.322423
Bisht K, Sharma K, Tremblay M. Chronic stress as a risk factor for Alzheimer’s disease: Roles of microglia-mediated synaptic remodeling, inflammation, and oxidative stress. Neurobiology of stress. Nov 2018;9:9–21. doi:https://doi.org/10.1016/j.ynstr.2018.05.003
Hansen DV, Hanson JE, Sheng M. Microglia in Alzheimer’s disease. The Journal of cell biology. Feb 5 2018;217(2):459–472. doi:https://doi.org/10.1083/jcb.201709069
Cacabelos R, Torrellas C, Fernández-Novoa L, López-Muñoz F. Histamine and Immune Biomarkers in CNS Disorders. Mediators of inflammation. 2016;2016:1924603. doi:https://doi.org/10.1155/2016/1924603
Nava Catorce M, Acero G, Gevorkian G. Age- and sex-dependent alterations in the peripheral immune system in the 3xTg-AD mouse model of Alzheimer’s disease: Increased proportion of CD3+CD4-CD8- double-negative T cells in the blood. J Neuroimmunol. Nov 15 2021;360:577720. doi:https://doi.org/10.1016/j.jneuroim.2021.577720
Machhi J, Yeapuri P, Lu Y, et al. CD4+ effector T cells accelerate Alzheimer’s disease in mice. Journal of neuroinflammation. Nov 19 2021;18(1):272. doi:https://doi.org/10.1186/s12974-021-02308-7
Ferrer I, Boada Rovira M, Sánchez Guerra ML, Rey MJ, Costa-Jussá F. Neuropathology and pathogenesis of encephalitis following amyloid-beta immunization in Alzheimer’s disease. Brain pathology (Zurich, Switzerland). Jan 2004;14(1):11–20. doi:https://doi.org/10.1111/j.1750-3639.2004.tb00493.x
Lurain NS, Hanson BA, Martinson J, et al. Virological and immunological characteristics of human cytomegalovirus infection associated with Alzheimer disease. J Infect Dis. Aug 15 2013;208(4):564–72. doi:https://doi.org/10.1093/infdis/jit210
Fani L, Georgakis MK, Ikram MA, Ikram MK, Malik R, Dichgans M. Circulating biomarkers of immunity and inflammation, risk of Alzheimer’s disease, and hippocampal volume: a Mendelian randomization study. Translational psychiatry. May 17 2021;11(1):291. doi:https://doi.org/10.1038/s41398-021-01400-z
Gate D, Saligrama N, Leventhal O, et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature. Jan 2020;577(7790):399–404. doi:https://doi.org/10.1038/s41586-019-1895-7
Barrett T, Edgar R. Mining microarray data at NCBI’s Gene Expression Omnibus (GEO)*. Methods in molecular biology (Clifton, NJ). 2006;338:175–90. doi:https://doi.org/10.1385/1-59745-097-9:175
Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic acids research. Jan 2013;41(Database issue):D991–5. doi:https://doi.org/10.1093/nar/gks1193
Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. Apr 20 2015;43(7):e47. doi:https://doi.org/10.1093/nar/gkv007
Taminau J, Meganck S, Lazar C, et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics. Dec 24 2012;13:335. doi:https://doi.org/10.1186/1471-2105-13-335
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. Jan 2007;8(1):118–27. doi:https://doi.org/10.1093/biostatistics/kxj037
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. Dec 29 2008;9:559. doi:https://doi.org/10.1186/1471-2105-9-559
Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (New York, NY). Aug 28 2021;2(3):100141. doi:https://doi.org/10.1016/j.xinn.2021.100141
Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. May 2015;12(5):453–7. doi:https://doi.org/10.1038/nmeth.3337
Zeng D, Ye Z, Shen R, et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Frontiers in immunology. 2021;12:687975. doi:https://doi.org/10.3389/fimmu.2021.687975
Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. Jan 6 2023;51(D1):D638–d646. doi:https://doi.org/10.1093/nar/gkac1000
Killcoyne S, Carter GW, Smith J, Boyle J. Cytoscape: a community-based framework for network modeling. Methods Mol Biol. 2009;563:219–39. doi:https://doi.org/10.1007/978-1-60761-175-2_12
Kiyota T, Gendelman HE, Weir RA, Higgins EE, Zhang G, Jain M. CCL2 affects β-amyloidosis and progressive neurocognitive dysfunction in a mouse model of Alzheimer’s disease. Neurobiol Aging. Apr 2013;34(4):1060–8. doi:https://doi.org/10.1016/j.neurobiolaging.2012.08.009
Liu Z, Condello C, Schain A, Harb R, Grutzendler J. CX3CR1 in microglia regulates brain amyloid deposition through selective protofibrillar amyloid-β phagocytosis. J Neurosci. Dec 15 2010;30(50):17091–101. doi:https://doi.org/10.1523/jneurosci.4403-10.2010
Li X, Zhang DF, Bi R, et al. Convergent transcriptomic and genomic evidence supporting a dysregulation of CXCL16 and CCL5 in Alzheimer’s disease. Alzheimers Res Ther. Jan 21 2023;15(1):17. doi:https://doi.org/10.1186/s13195-022-01159-5
Mangialasche F, Solomon A, Winblad B, Mecocci P, Kivipelto M. Alzheimer’s disease: clinical trials and drug development. The Lancet Neurology. Jul 2010;9(7):702–16. doi:https://doi.org/10.1016/s1474-4422(10)70119-8
Morimoto B. Drug Development for Neurodegenerative Diseases—Second Annual marcus evans Conference. Advances in drug development for NDD and expediting discovery through novel compounds and sound clinical trials. IDrugs: the investigational drugs journal. Jul 2010;13(7):440–3.
Srivastava S, Ahmad R, Khare SK. Alzheimer’s disease and its treatment by different approaches: A review. Eur J Med Chem. Apr 15 2021;216:113320. doi:https://doi.org/10.1016/j.ejmech.2021.113320
Hampel H, Mesulam MM, Cuello AC, et al. The cholinergic system in the pathophysiology and treatment of Alzheimer’s disease. Brain: a journal of neurology. Jul 1 2018;141(7):1917–1933. doi:https://doi.org/10.1093/brain/awy132
Liang D, Han G, Feng X, Sun J, Duan Y, Lei H. Concerted perturbation observed in a hub network in Alzheimer’s disease. PLoS One. 2012;7(7):e40498. doi:https://doi.org/10.1371/journal.pone.0040498
Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. Apr 25 2013;153(3):707–20. doi:https://doi.org/10.1016/j.cell.2013.03.030
Klein HU, Schäfer M, Bennett DA, Schwender H, De Jager PL. Bayesian integrative analysis of epigenomic and transcriptomic data identifies Alzheimer’s disease candidate genes and networks. PLoS Comput Biol. Apr 2020;16(4):e1007771. doi:https://doi.org/10.1371/journal.pcbi.1007771
Lin CX, Li HD, Deng C, et al. AlzCode: a platform for multiview analysis of genes related to Alzheimer’s disease. Bioinformatics. Mar 28 2022;38(7):2030–2032. doi:https://doi.org/10.1093/bioinformatics/btac033
Xu M, Zhang DF, Luo R, et al. A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer’s disease. Alzheimers Dement. Feb 2018;14(2):215–229. doi:https://doi.org/10.1016/j.jalz.2017.08.012
Sun Y, Lin J, Zhang L. The application of weighted gene co-expression network analysis in identifying key modules and hub genes associated with disease status in Alzheimer’s disease. Annals of translational medicine. Dec 2019;7(24):800. doi:https://doi.org/10.21037/atm.2019.12.59
Yu W, Yu W, Yang Y, Lü Y. Exploring the Key Genes and Identification of Potential Diagnosis Biomarkers in Alzheimer’s Disease Using Bioinformatics Analysis. Frontiers in aging neuroscience. 2021;13:602781. doi:https://doi.org/10.3389/fnagi.2021.602781
Serrano-Pozo A, Das S, Hyman BT. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. Jan 2021;20(1):68–80. doi:https://doi.org/10.1016/S1474-4422(20)30412-9
Lanni C, Masi M, Racchi M, Govoni S. Cancer and Alzheimer’s disease inverse relationship: an age-associated diverging derailment of shared pathways. Mol Psychiatry. Jan 2021;26(1):280–295. doi:https://doi.org/10.1038/s41380-020-0760-2
Kong DH, Kim YK, Kim MR, Jang JH, Lee S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. International journal of molecular sciences. Apr 2 2018;19(4)doi:https://doi.org/10.3390/ijms19041057
Kim CW, Oh ET, Park HJ. A strategy to prevent atherosclerosis via TNF receptor regulation. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. Mar 2021;35(3):e21391. doi:https://doi.org/10.1096/fj.202000764R
Koren Krajnc M, Hojs R, Holc I, Knez Ž, Pahor A. Accelerated atherosclerosis in premenopausal women with rheumatoid arthritis - 15-year follow-up. Bosnian journal of basic medical sciences. Aug 1 2021;21(4):477–483. doi:https://doi.org/10.17305/bjbms.2020.5176
Zhang L, Mao H. The Relationship Between Serum VCAM-1 and Alzheimer’s Disease in Patients with Type 2 Diabetes Mellitus. Diabetes, metabolic syndrome and obesity: targets and therapy. 2020;13:4661–4667. doi:https://doi.org/10.2147/dmso.S274232
Rangaraju S, Dammer EB, Raza SA, et al. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Molecular neurodegeneration. May 21 2018;13(1):24. doi:https://doi.org/10.1186/s13024-018-0254-8
Ou GY, Lin WW, Zhao WJ. Construction of Long Noncoding RNA-Associated ceRNA Networks Reveals Potential Biomarkers in Alzheimer’s Disease. Journal of Alzheimer’s disease: JAD. 2021;82(1):169–183. doi:https://doi.org/10.3233/jad-210068
Acknowledgments
We thank the GEO database, AlzData for providing free and public data sets, and we also thank other online platforms such as R software, ggplot2, Cytoscape software, Metascape, String, Sangerbox and Bioinformatics for facilitating data procession and visualization in our study.
Author information
Authors and Affiliations
Contributions
Author contributions: Data curation, Ni Zhang and Guan-yong Ou; Formal analysis, Fan Yang and Guan-yong Ou; Writing - original draft, Ni Zhang; Writing - review & editing, Fan Yang and Shuwen, Xu. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing financial interests: All authors declare no conflict of financial interests.
Ethical standards: GEO belongs to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open-source data, so there are no ethical issues and other conflicts of interest.
Supplementary Information for
42414_2024_5_MOESM1_ESM.docx
Integrated Bioinformatic Analysis and Validation Identifies Immune Microenvironment-Related Potential Biomarkers in Alzheimer’s Disease, approximately 7.69 MB.
Rights and permissions
About this article
Cite this article
Yang, F., Zhang, N., Ou, GY. et al. Integrated Bioinformatic Analysis and Validation Identifies Immune Microenvironment-Related Potential Biomarkers in Alzheimer’s Disease. J Prev Alzheimers Dis 11, 495–506 (2024). https://doi.org/10.14283/jpad.2024.5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.14283/jpad.2024.5