Abstract
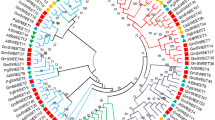
Transcriptome analysis of leaf and pitcher at different developmental stages in the carnivorous plant Nepenthes sp. identified 20 cDNAs of the SWEET gene family encoding sugar uniporters of classes I–IV. The structure of NSWEET proteins generally corresponded to the 3-1-3 scheme typical of SWEET proteins in eukaryotes. The variability in the expression of NSWEET genes in the mature leaf and in the three stages of pitcher development indicates the possible functional diversity of these genes. It has been suggested that class I transporters (NSWEET2d, 2f, and 2h) may participate in export of sugars from the leaf to the site of pitcher meristem initiation, while proteins NSWEET1, 2a, 2k (class I), 4a, 4b, 4d (II), and 12c (III) may participate in the delivery of sugars for the primary development of the pitcher. In subsequent stages, they can be replaced by NSWEET2b, 2c, 2e, 2i, 2j (class I), 4c (II), and 12b (III), delivering hexoses and sucrose to the growing pitcher. In the fully formed pitcher, proteins NSWEET12a (III), 2g (I), and 16 (IV) can export sugars from the digestive fluid to the leaf.




Similar content being viewed by others
REFERENCES
Lemoine, R., La Camera, S., Atanassova, R., et al., Source-to-sink transport of sugar and regulation by environmental factors, Front. Plant Sci., 2013, vol. 4, article 272. https://doi.org/10.3389/fpls.2013.00272
Chen, L.-Q., Cheung, L.S., Feng, L., et al., Transport of sugars, Annu. Rev. Biochem., 2015, vol. 84, pp. 865—894. https://doi.org/10.1146/annurev-biochem-060614-033904
Feng, C.Y., Han, J.X., Han, X.X., and Jiang, J., Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato, Gene, 2015, vol. 573, no. 2, pp. 261—272. https://doi.org/10.1016/j.gene.2015.07.055
Eom, J.S., Chen, L.Q., Sosso, D., et al., SWEETs, transporters for intracellular and intercellular sugar translocation, Curr. Opin. Plant Biol., 2015, vol. 25, pp. 53—62. https://doi.org/10.1016/j.pbi.2015.04.005
Hu, L.-P., Zhang, F., Song, S.-H., et al., Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber, J. Integr. Agric., 2017, vol. 16, no. 7, pp. 1486—1501. https://doi.org/10.1016/S2095-3119(16)61501-0
Chen, L.-Q., SWEET sugar transporters for phloem transport and pathogen nutrition, New Phytol., 2014, vol. 201, no. 4, pp. 1150—1155. https://doi.org/10.1111/nph.12445
Chen, L.-Q., Hou, B.-H., Lalonde, S., et al., Sugar transporters for intercellular exchange and nutrition of pathogens, Nature, 2010, no. 468, pp. 527—532.
Chen, L.-Q., Qu, X.-Q., Hou, B.-H., et al., Sucrose efflux mediated by SWEET proteins as a key step for phloem transport, Science, 2012, vol. 335, pp. 207—211. https://doi.org/10.1126/science.1213351
Yuan, M. and Wang, S., Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms, Mol. Plant, 2013, vol. 6, pp. 665—674. https://doi.org/10.1093/mp/sst035
Xuan, Y.H., Hu, Y.B., Chen, L.Q., et al., Functional role of oligomerization for bacterial and plant SWEET sugar transporter family, Proc. Natl. Acad. Sci. U.S.A., 2013, vol. 110, pp. E3685—E3694. https://doi.org/10.1073/pnas.1311244110
Patil, G., Valliyodan, B., Deshmukh, R., et al., Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis, BMC Genomics, 2015, vol. 16, article 520. https://doi.org/10.1186/s12864-015-1730-y
Gao, Y., Wang, Z.Y., Kumar, V., et al., Genome-wide identification of the SWEET gene family in wheat, Gene, 2018, vol. 642, pp. 284—292. https://doi.org/10.1016/j.gene.2017.11.044
Guo, C., Li, H., Xia, X., et al., Functional and evolution characterization of SWEET sugar transporters in Ananas comosus, Biochem. Biophys. Res. Commun., 2018, vol. 496, no. 2, pp. 407—414. https://doi.org/10.1016/j.bbrc.2018.01.024
Chandran, D., Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance, IUBMB Life, 2015, vol. 67, no. 7, pp. 461—471. https://doi.org/10.1002/iub.1394
Klemens, P.A., Patzke, K., Deitmer, J., et al., Over expression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis, Plant Physiol., 2013, vol. 163, pp. 1338—1352. https://doi.org/10.1104/pp.113.224972
Chardon, F., Bedu, M., Calenge, F., et al., Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis, Curr. Biol., 2013, vol. 23, pp. 697—702. https://doi.org/10.1016/j.cub.2013.03.021
Guo, W.J., Nagy, R., Chen, H.Y., et al., SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves, Plant Physiol., 2014, vol. 164, pp. 777—789. https://doi.org/10.1104/pp.113.232751
Sun, M.X., Huang, X.Y., Yang, J., et al., Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage, Plant Reprod., 2013, vol. 26, pp. 83—91. https://doi.org/10.1007/s00497-012-0208-1
Lin, I.W., Sosso, D., Chen, L.-Q., et al., Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9, Nature, 2014, vol. 508, pp. 546—549.
Fukushima, K., Fang, X., Alvarez-Ponce, D., et al., Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory, Nat. Ecol. Evol., 2017, vol. 1, no. 3, article 59. https://doi.org/10.1038/s41559-016-0059
Fukushima, K., Fujita, H., Yamaguchi, T., et al., Oriented cell division shapes carnivorous pitcher leaves of Sarracenia purpurea, Nat. Commun., 2015, no. 6, article 6450. https://doi.org/10.1038/ncomms7450
Frazier, C.K., The enduring controversies concerning the process of protein digestion in Nepenthes (Nepenthaceae), Carnivorous Plant Newslett., 2000, vol. 29, pp. 56—61.
Rottloff, S., Miguel, S., Biteau, F., et al., Proteome analysis of digestive fluids in Nepenthes pitchers, Ann. Bot., 2016, vol. 117, pp. 479—495. https://doi.org/10.1093/aob/mcw001
Saganová, M., Bokor, B., Stolárik, T., and Pavlovič, A., Regulation of enzyme activities in carnivorous pitcher plants of the genus Nepenthes, Planta, 2018. https://doi.org/10.1007/s00425-018-2917-7
Ravin, N.V., Gruzdev, E.V., Beletsky, A.V., et al., The loss of photosynthetic pathways in the plastid and nuclear genomes of the non-photosynthetic mycoheterotrophic eudicot Monotropa hypopitys, BMC Plant Biol., 2016, vol. 16, suppl. 3, pp. 153—161. https://doi.org/10.1186/s12870-016-0929-7
Bailey, T.L. and Elkan, C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers, in Proc. Sec. Int. Conf. Intelligent Syst. Mol. Biol., 1994, pp. 28—36.
Kumar, S., Stecher, G., and Tamura, K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, no. 7, pp. 1870—1874. https://doi.org/10.1093/molbev/msw054
Wang, L., Yao, L., Hao, X., et al., Tea plant SWEET transporters: expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis, Plant Mol. Biol., 2018, vol. 96, no. 6, pp. 577—592. https://doi.org/10.1007/s11103-018-0716-y
Miguel, S., Hehn, A., and Bourgaud, F., Nepenthes: state of the art of an inspiring plant for biotechnologists, J. Biotechnol., 2018, vol. 265, pp. 109—115. https://doi.org/10.1016/j.jbiotec.2017.11.014
Filyushin, M.A., Reshetnikova, N.M., Kochieva, E.Z., and Skryabin, K.G., Intraspecific variability of ITS sequences in the parasitic plant Monotropa hypopitys L. from the European Russian populations, Russ. J. Genet., 2015, no. 51, no. 11, pp. 1149—1152. https://doi.org/10.1134/S102279541511006X
Shulga, O.A., Shchennikova, A.V., Beletsky, A.V., et al., Transcriptome-wide characterization of the MADS-box family in pinesap Monotropa hypopitys reveals flowering conservation in non-photosynthetic myco-heterotrophs, J. Plant Growth Regul., 2017. https://doi.org/10.1007/s00344-017-9772-9
Shchennikova, A.V., Slugina, M.A., Beletsky, A.V., et al., The YABBY genes of leaf and leaf-like organ polarity in leafless plant Monotropa hypopitys, Int. J. Genomics, 2018, vol. 15, article ID 7203469. https://doi.org/10.1155/2018/7203469
ACKNOWLEDGMENTS
This work was performed using the experimental climate control facility (Institute of Bioengineering, Research Center of Biotechnology, Russian Academy of Sciences).
Funding
This work was supported in part by the grant of the Russian Science Foundation (no. 14-24-00175).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by K. Lazarev
Rights and permissions
About this article
Cite this article
Filyushin, M.A., Kochieva, E.Z., Shchennikova, A.V. et al. SWEET Uniporter Gene Family Expression Profile in the Pitcher Development in the Carnivorous Plant Nepenthes sp.. Russ J Genet 55, 692–700 (2019). https://doi.org/10.1134/S1022795419050089
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795419050089