Abstract
In response to stress stimuli, eukaryotic cells typically suppress protein synthesis. This leads to the release of mRNAs from polysomes, their condensation with RNA-binding proteins, and the formation of non-membrane-bound cytoplasmic compartments called stress granules (SGs). SGs contain 40S but generally lack 60S ribosomal subunits. It is known that cycloheximide, emetine, and anisomycin, the ribosome inhibitors that block the progression of 80S ribosomes along mRNA and stabilize polysomes, prevent SG assembly. Conversely, puromycin, which induces premature termination, releases mRNA from polysomes and stimulates the formation of SGs. The same effect is caused by some translation initiation inhibitors, which lead to polysome disassembly and the accumulation of mRNAs in the form of stalled 48S preinitiation complexes. Based on these and other data, it is believed that the trigger for SG formation is the presence of mRNA with extended ribosome-free segments, which tend to form condensates in the cell. In this study, we evaluated the ability of various small-molecule translation inhibitors to block or stimulate the assembly of SGs under conditions of severe oxidative stress induced by sodium arsenite. Contrary to expectations, we found that ribosome-targeting elongation inhibitors of a specific type, which arrest solitary 80S ribosomes at the beginning of the mRNA coding regions but do not interfere with all subsequent ribosomes in completing translation and leaving the transcripts (such as harringtonine, lactimidomycin, or T-2 toxin), completely prevent the formation of arsenite-induced SGs. These observations suggest that the presence of even a single 80S ribosome on mRNA is sufficient to prevent its recruitment into SGs, and the presence of extended ribosome-free regions of mRNA is not sufficient for SG formation. We propose that mRNA entry into SGs may be mediated by specific contacts between RNA-binding proteins and those regions on 40S subunits that remain inaccessible when ribosomes are associated.
Similar content being viewed by others
INTRODUCTION
The eukaryotic cell has the ability to promptly respond to sudden changes in environmental conditions through the launching a stress response program. A crucial component of this program is usually the rapid suppression of protein synthesis [1]. In many cases, the stress-induced suppression of translation is accompanied by the formation of specialized cytoplasmic ribonucleoprotein (RNP) condensates known as stress granules (SGs) [2-5]. It is believed that SGs contribute to cell survival under stress conditions by facilitating the optimal spatial distribution of mRNAs and translation machinery components while protein synthesis is inhibited and when it subsequently resumes, and also play a role in antiviral innate immunity [3, 5-8].
SGs contain untranslated mRNA molecules, 40S ribosomal subunits, and a large number of proteins: first of all, several translation initiation factors, participating in the 48S preinitiation complex formation under normal conditions [9], as well as some other translation factors (see [10] and references therein); they also include many mRNA-binding proteins (see e.g. [11-14]), proteins involved in intracellular signaling [15], and other components of translation machinery (reviewed in [1-5]). Some of them directly regulate SG formation, while others are involved through interaction with various components of the granules. The repertoire of proteins found in SG has expanded significantly over the past few years due to the use of proximal labeling approaches [16-20].
The mechanisms of SG formation are not fully understood. But a key principle underlying their formation is the liquid–liquid phase separation (LLPS) associated with inherent properties of biopolymers: RNA and RNA-binding proteins that have unstructured or repetitive regions prone to forming excessive contacts [21-30]. It is assumed that intermolecular base pairing of untranslated mRNAs leads to their aggregation, while interaction with RNA-binding proteins promotes the formation of RNP granules [22, 27, 31]. Protein–protein interactions involving internally disordered regions also play an important role in this process [4, 32]. In particular, the G3BP1/2 proteins, which are crucial for SG formation, under stress conditions are converted from a compact inactive state to a partially unfolded conformation that promotes cooperative RNA–protein interactions [33, 34]. This leads to the assembly of RNP clusters, wherein mRNA and protein molecules are combined into heterogeneous condensates.
Transcriptome analysis of SGs revealed that almost all cellular mRNA species are recruited into SGs, yet in varying degrees [35]. Inefficiently translated mRNAs as well as transcripts with long coding regions (CDSs) or 3′ untranslated regions (UTRs) are more abundant in SGs [35, 36]. This indicates a significant role of RNA–RNA interactions between long ribosome-free mRNA regions in SG formation. Interestingly, only a small portion (~10%) of cellular mRNA molecules are found in SGs at any given moment [35], being dynamically exchanged with the cytosolic pool of mRNA [37-39].
According to the classic model, it is untranslated mRNAs, which are not associated with ribosomes, that accumulate in SGs [9, 37, 40]. This is consistent with the fact that, despite the presence of the 40S ribosomal subunits, the 60S subunits are not found in SGs, as evidenced by immunofluorescence methods [9, 41, 42] and by analysis of 18S and 28S rRNA distribution [43]. However, recent evidence suggests that some actively translated mRNAs may be transiently associated with SGs [44] and even likely found inside SGs [45]. Finally, the statement about the complete absence of the 60S subunits in SGs is not unambiguous [45, 46]. However, there is still no doubt that mRNAs localized in SGs are largely not translated [45].
Anyway, the appearance of long mRNA regions devoid of ribosomes in the cell is considered a key requirement for SG formation in all existing models. Although this condition alone might not always trigger SG assembly, it typically promotes their formation. For instance, SG assembly can be induced by blocking translation initiation through various means. Numerous stress stimuli, leading to the inactivation of the translation initiation factor eIF2, which delivers Met-tRNAi to the pre-initiation complex [12], or small-molecule inhibitors of the RNA helicase eIF4A, involved in ribosomal scanning [47-50], serve as inducers for SG formation. SG assembly is also triggered by reducing the levels of eIF4B, eIF4H, or PABP, and to a lesser extent, by depleting the cap-binding protein eIF4E or preventing its incorporation into the eIF4F complex [51-53]. Remarkably, interference with the last step of translation initiation (60S subunit joining) using a small-molecule inhibitor MDMP or by depletion of the L28 protein does not induce SG formation or affect their assembly [51].
The formation of SG is also stimulated by the ribosome inhibitor puromycin, which induces premature termination and polysome disassembly [37]. In some cases, puromycin alone can even trigger SG assembly without any additional stimuli [37, 54]. In contrast, polysome stabilization by classic inhibitors of elongation such as cycloheximide, emetine, and anisomycin prevents SG assembly under stress conditions [14, 37-39, 51, 55].
The facts above fit into the model that emphasizes the role of extended ribosome-free mRNA regions as major RNA components of SGs that largely determine their formation. Here, we aimed to probe of this model. Thus, we analyzed the effects of various small-molecule ribosome inhibitors on SG formation triggered by sodium arsenite, the classical SG inducer. The diverse properties of these inhibitors [56] enable us to not only stabilize or completely disassemble polysomes but also ensure the presence of only a single 80S ribosome at the beginning of the mRNA CDS. We found that in this latter case, no SGs were formed. This suggests that mRNAs associated with only a solitary ribosome cannot be incorporated in SGs, despite having extended ribosome-free regions. Apparently, the 80S ribosome itself prevents the recruitment of mRNA into SG. These findings raise questions about the active role of the ribosome in excluding translated mRNAs from SGs and the universality of the classic model describing the mechanisms of SG assembly.
MATERIALS AND METHODS
Cell culture, stress induction and treatment with translation inhibitors. HeLa cells were grown in DMEM (Gibco, USA) with alanyl-glutamine (Paneco, Russia) supplemented with 10% FBS (HyClone, USA) in the presence of penicillin and streptomycin (Paneco) in a humidified 5% CO2 atmosphere at 37°C. The following translation inhibitors were used (manufacturers, concentrations of stock solutions, and solvents are indicated): cycloheximide (Sigma-Aldrich, USA, 10 mg/ml in water), cephaeline (Cayman Chemical, USA, 10 mM in ethanol), anisomycin (Sigma-Aldrich, 50 mM in DMSO), puromycin (Santa Cruz Biotechnology, USA, 100 mM in water), lactimidomycin (Sigma-Aldrich, 2 mM in DMSO), harringtonine (Santa Cruz Biotechnology, 10 mg/ml in DMSO), T-2 toxin (Cayman Chemical, 10 mM in DMSO), pactamycin (Sigma-Aldrich, 10 mM in DMSO), Torin-1 (Tocris Bioscience, UK, 10 mM in DMSO). Before use, stock solutions were diluted with water or PBS to appropriate concentrations (see below). To induce SG formation, cells were exposed to 100 μM sodium arsenite (NaH2AsO3, Sigma-Aldrich) for the indicated time.
mRNA transfection. One day before transfection, HeLa cells were transferred into the white FB/HB 96-well microplates (Greiner, Austria) in a volume of 75 μl per well, the space between the wells was filled with sterile water. m7G-capped polyadenylated mRNA encoding firefly luciferase (Fluc) and containing the 5′ UTR of the human β-actin mRNA and 3′ UTR of an SV40 virus mRNA, obtained according to the previously described protocol [57], was kindly provided by E. A. Panova (Moscow State University). The cells were transfected with reporter mRNAs using GenJect-U (Molecta, Russia) at a cell density of ~75% as described [58]. When preparing the transfection mixtures, D-luciferin (Promega, USA) was added into PBS, mRNA, and GenJect-U mix in such a way that its final concentration per microplate well would be 0.4 mM. To examine the effect of arsenite on mRNA translation, it was added to the indicated concentrations 1 h before transfection. Translation inhibitors were added into the wells in the form of 10× solutions immediately before transfection. Then the microplate was placed in the CLARIOstar multimodal plate reader (BMG Labtech, Germany) and luciferase activity was measured in living cells for 24 h as described [59]. For further analysis, values at the 10 h post-transfection were taken, approximately corresponding to the midpoint of the linear phase of the curve (except for sodium arsenite, for which the 2 h post-transfection time point was used). All transfections were performed at least twice on different cell passages, each in triplicate, and then the mean values and standard deviation were calculated.
Polysome analysis. HeLa cells were cultured to ~75% confluence on 10-cm dishes and then were treated with either the indicated concentrations of ribosome inhibitors followed by 30-min incubation, or with 100 μM sodium arsenite followed by 60-min incubation. The subsequent steps were performed according to the protocol described in [60] with the following minor changes. Cells were washed with ice-cold PBS and lysed in 200 µl of buffer containing 20 mM Tris-HCl (pH 7.5), 250 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 0.1 mg/ml cycloheximide, and 20 U/ml DNase TURBO (Ambion-Invitrogen, USA). Lysates were incubated on ice for 10 min, centrifuged at 16,000g for 10 min at 4°C, and the supernatant was carefully loaded onto a 10-60% (m/v) sucrose gradient (11 ml), containing 20 mM Tris–HCl (pH 7.5), 250 mM NaCl, 15 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, and 0.1 mg/ml cycloheximide, followed by centrifugation in an SW-41 rotor at 35,000 rpm for 3 h. The gradient was then manually fractionated into 300-µl aliquots and the A260 was measured in 96-well UV-Transparent microplates (Corning, USA) using the Infinite 200 PRO plate reader (TECAN, Switzerland). The obtained values were normalized to the sum of the A260 values for all aliquots from the same tube.
Immunocytochemistry and microscopy. For immunostaining, HeLa cells were cultured to ~70% confluence on coverslips and treated with ribosome inhibitors at the indicated concentrations for 30 min. Then oxidative stress was induced by exposing the cells to 100 μM sodium arsenite. After incubation for 1 h, the cells were fixed with anhydrous methanol at –20°C for 7 min, then post-fixed with freshly prepared 3% paraformaldehyde in PBS at 4°C for 30 min, and washed with PBS at room temperature twice for 15 min. This was followed by permeabilization with 0.5% Triton X-100 for 2 min, blocking by 3% BSA for 15 min, and washing with PBS for 15 min. Cells were then incubated in primary mouse anti-G3BP1 (H-10) antibodies (Santa Cruz Biotechnology) at room temperature for 1 h, followed by washing with PBS (3 × 5 min) and incubation with secondary anti-mouse IgG-specific antibodies conjugated with Alexa Fluor 488 (Invitrogen) under the same conditions, and similar washing. After that, the nuclei were stained with Hoechst 33342 (Thermo Fisher Scientific, USA) and coverslips were mounted using Aqua-Poly/Mount polyvinyl embedding medium (Polysciences Inc, USA). Microscopy was performed using a Zeiss LSM900 confocal microscope (provided by the Moscow State University Development Program). The figures represent the sum of all frames from the corresponding Z-stack.
RESULTS
Selection of small-molecule inhibitors. The following ribosome inhibitors were chosen for analysis: cycloheximide, cephaeline (an analog of emetine), anisomycin, puromycin, lactimidomycin, harringtonine, T-2 toxin, and pactamycin, as well as the mTOR kinase inhibitor torin-1 [56]. Their structures and essential features are summarized in the table.
Structural and functional properties of small-molecule translation inhibitors used in this study
Inhibitor (PubChem CID) | Structural formula | Target (binding site) | Mechanism of action | Effect on polysomes | Effect on SG assembly |
|---|---|---|---|---|---|
Cycloheximide (6197) |
| 60S (E site) | inhibits translocation by impeding the movement of the CCA end of deacylated tRNA to the E site | stabilizes | |
Cephaeline (442195), an analog of emetine |
| 40S (in front of E site) | inhibits translocation | stabilizes | |
Anisomycin (253602) |
| 60S (A site) | inhibits the peptidyl transferase center by destabilizing the binding of aminoacyl-tRNA in the A site | stabilizes | prevents assembly [55] |
Puromycin (439530) |
| 60S (A site) | induces premature termination by mimicking the aminoacylated CCA end of tRNA | disassembles | |
Pactamycin (5289124) |
| 40S (in front of E site) | inhibits translocation | disassembles | unknown |
Lactimidomycin (11669726) |
| 60S (E site) | inhibits translocation by impeding the movement of the CCA end of deacylated tRNA to the E site; can only bind to a vacant ribosome | disassembles | unknown |
Harringtonine (276389) |
| 60S (A site) | inhibits the peptidyl transferase center by preventing the accommodation of the aminoacyl residue of aminoacyl-tRNA in the A site; can only bind to a vacant ribosome | disassembles | unknown |
T-2 toxin (5284461) |
| 60S (A site) | inhibits the peptidyl transferase center by preventing the accommodation of the aminoacyl residue of aminoacyl-tRNA in the A site; can only bind to a vacant ribosome | disassembles | unknown |
Torin-1 (49836027) |
| mTOR (active site) | mTOR inactivation leads to dephosphorylation of 4E-BP1 and disruption of the interaction between the cap-binding protein eIF4E and eIF4G, thus inhibiting cap-dependent translation | disassembles |
Cycloheximide binds to the E site of the 60S ribosomal subunit, blocks translocation, and stabilizes polysomes. Cephaeline (6′-O-demethylemetine) also blocks translocation but interacts with the small subunit rather than the large one. Anisomycin binds to the A site of the 60S subunit and inhibits the peptidyl transferase reaction. Finally, puromycin mimics the aminoacylated CCA end of tRNA and, by entering the A site of the ribosome, induces premature termination, peptide release, and polysome disassembly. These compounds were chosen as controls due to their well-documented effects on SG formation: polysome stabilizing drugs (such as cycloheximide, emetine, and anisomycin) are expected to prevent SG assembly [14, 37-39, 51, 55], while puromycin, which disassembles polysomes, on the contrary, is expected to promote granule formation [37, 54].
In our study we aimed to uncover how the assembly of SGs is affected by “non-canonical” elongation inhibitors, which paradoxically lead to the disassembly of polysome [56]. For example, pactamycin, which binds to approximately the same site as emetine, has long been considered an inhibitor of translation initiation [64], but it is now known to actually block translocation, although its mechanism of action is still poorly understood [65, 66]. Lactimidomycin belongs to the same group of compounds (glutarimides) as cycloheximide [67] and interacts with the same site of the 60S subunit but, due to its larger size, cannot effectively compete with deacylated tRNA on an actively translating ribosome [68]. This is probably why it binds exclusively to the 60S subunit that has just recruited at the start codon during translation initiation and has a vacant E site. Therefore, it arrests only one (5′-proximal) ribosome across the entire CDS, resulting in disassembly of polysomes into monosomes rather than polysome stabilization [67, 69]. Harringtonine and T-2 toxin are inhibitors of the peptidyl transferase center, but their binding also shares the same characteristic as lactimidomycin: they selectively block newly initiated ribosomes. Therefore, their addition also leads to polysome disassembly, while only a solitary 80S ribosome remains bound to each mRNA molecule [64, 70-72].
In addition to the ribosome-targeting drugs, we also included torin-1, an inhibitor of the protein kinase mTOR that inactivates the cap-binding machinery of the cell [73], in our analysis. The reduced efficiency of translation initiation releases mRNA from polysomes and, according to the model described above, should stimulate SG assembly. However, torin-1 has previously been shown to slightly inhibit SG formation in two different models instead [61, 62], so we decided to investigate its effect in our system as well.
For the experiments, we chose a classical model widely used for studying SGs in cultured mammalian cells – severe oxidative stress induced by sodium arsenite (NaH2AsO3), whose effects on translation has been well studied [9, 58, 60, 74-76]. We also used a classic object, the human cervical cancer cell line HeLa.
Analysis of the effect of small-molecule inhibitors on translation in cultured human cells. Determining the effect of ribosome inhibitors on SG assembly implies complete translation arrest by adding the compounds in a certain excess. To determine the required drug concentrations, we analyzed their inhibitory effect on protein synthesis in our system. Translation suppression in living cells can be readily assessed using the fleeting mRNA transfection (FLERT), which we previously developed to study the effect of stress response [58, 77, 78] and small-molecule drugs [79, 80] on protein biosynthesis. A capped polyadenylated transcript containing the 5′ UTR of the human β-actin mRNA, the firefly luciferase CDS, and the 3′ UTR of an SV40 virus mRNA was used as a reporter since its translation level corresponds well to that of a typical cellular mRNA [57]. To obtain more reliable results, we used the method of real-time monitoring of luciferase activity in living cells [59, 79] and took the values at 10 h post-transfection as the final ones, although a similar pattern of suppression was observed at other time points as well (for details, see Materials and Methods).
Effects of selected small-molecule inhibitors on reporter mRNA translation in HeLa cells. Cells were transfected with a capped polyadenylated transcript containing the 5′ UTR of the human β-actin mRNA, the firefly luciferase CDS, and the 3′ UTR of an SV40 virus mRNA. Prior to transfection, the indicated compounds were added to the cells at different concentrations, then luciferase activity was measured in living cells in real-time for 24 h, and the values at 10 h post-transfection (2 h for sodium arsenite) were taken for analysis.
As expected, increasing the concentration of inhibitors resulted in a decrease in luciferase activity in the cells (Fig. 1). In all cases, except for torin-1, we were able to achieve complete suppression of translation of the reporter mRNA. The result obtained with torin-1 (Fig. 1i) was expected, as inactivation of the cap-binding protein eIF4E does not lead to complete block of the translation of most cellular mRNAs, but instead has differential effects on different transcripts [81, 82]. For further experiments, we used concentrations of inhibitors that were five times higher than the minimum concentrations at which complete suppression of luciferase production was observed in this experiment (for torin-1, the maximum concentration tested was used).
In a separate experiment, we examined the effect of sodium arsenite on luciferase production by the transfected cells (Fig. 1j) in order to assess the degree of translation suppression under the conditions we were going to use to analyze SG formation. Arsenite was added 1 h before transfection; in this case, the values presented in the figure correspond to the time point 2 h after transfection (since during the multi-hour incubation, the cells adapted to this type of stress and resumed translation, data not shown).
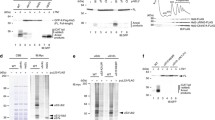
Analysis of the effects of small-molecule inhibitors on polysomes. To evaluate the effect of compounds on overall cellular translation and confirm that their effects on polysomes in our model are consistent with those described in the literature, we analyzed the polysome profile of HeLa cells treated with the inhibitors at selected concentrations for 30 min (Fig. 2). As expected, cycloheximide, emetine, and anisomycin caused the accumulation of the heavy fractions (indicating stabilization of polysomes), while puromycin, pactamycin, lactimidomycin, harringtonine, T-2 toxin, and torin-1 shifted the optical density towards the monosome and upper fractions (i.e., induced polysome disassembly). In the case of pactamycin and torin-1, complete polysome disassembly was not observed, but cells treated with puromycin, lactimidomycin, harringtonine, and T-2 toxin showed very similar changes in the polysome profile, indicating a complete transition of optical density from the polysome fractions to the monosome area. It is well known however (for review, see [56]) that puromycin produces monosomes that mainly consist of “empty” 80S ribosomes (containing no mRNA), whereas in the case of lactimidomycin, harringtonine, T-2 toxin, and pactamycin, the monosome peak mainly corresponds to single 80S ribosomes stopped at the mRNA start codons [56, 65, 69, 83, 84]. Thus, we were able to identify conditions in which mRNAs with extended ribosome-free regions accumulate in the cell, each carrying a solitary 80S ribosome.
Polysome profiles of HeLa cells treated with translation inhibitors. a) Scheme illustrating the distribution of monosomes and polysomes in sucrose gradient fractions. b) Polysome profile of untreated cells. c-k) Cells were treated for 30 min with an excess of translational inhibitors: 350 μM cycloheximide, 10 μM cephaeline, 10 μM anisomycin, 1 mM puromycin, 10 μM pactamycin, 10 μM lactimidomycin, 10 μM harringtonine, 10 μM T-2 toxin, or 1 μM torin-1. l) Cells were treated with 100 μM sodium arsenite for 1 h. 80S, monosome fraction; PS, polysome fraction.
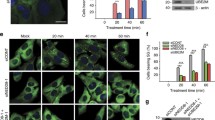
Analysis of the effects of ribosome inhibitors on the assembly of arsenite-induced stress granules. After identifying the appropriate conditions and ensuring that the inhibitors were exerting the expected effects, we proceeded to analyze their impacts on SG assembly. As mentioned earlier, sodium arsenite is a classic inducer of SG formation in cultured mammalian cells that causes severe oxidative stress [9, 74-76]. HeLa cells were incubated for 30 min with the same concentrations of translation inhibitors that we previously used in polysome profiling. Then 100 μM arsenite was applied and the cells were further incubated for 1 h, followed by fixation and staining (Fig. 3). Antibodies against the G3BP1 protein, a classic marker for SGs, were used to visualize the granules.
Effects of translational inhibitors on arsenite-induced stress granule formation. Confocal microscopy images of HeLa cells treated sequentially with the indicated ribosome inhibitors (CHX – 350 μM cycloheximide, CPH – 10 μM cephaeline, ANM – 10 μM anisomycin, PUR – 1 mM puromycin, PAC – 10 μM pactamycin, LTM – 10 μM lactimidomycin, HT – 10 μM harringtonine, T2T – 10 μM T-2 toxin, TOR – 1 μM torin-1) for 30 min, followed by 100 μM sodium arsenite (Ars) treatment for 60 min. The cells were fixed and stained with antibodies to G3BP1 (an SG marker, green channel) and Hoechst 33342 stain (nuclei, blue channel). Representative fields from several dozen analyzed samples are shown. All images were acquired at the same magnification (scale bar, 10 μm) and laser excitation power. Each panel represents the sum of all frames in the corresponding Z-stack.
As expected, in the absence of ribosome inhibitors, arsenite stress led to the appearance of microscopically visible SGs in cells. Similar granules were also observed in cells pretreated with puromycin, but were absent when cycloheximide, cephaeline, or anisomycin were added, which is fully consistent with the previously described effects of these drugs on SG assembly [14, 37-39, 51, 54, 55]. Pactamycin prevented granule formation; however, this result is difficult to interpret due to its dual effect on polysomes in our model (Fig. 2g). Treatment with torin-1 did not interfere with SG assembly, which generally corresponds to earlier observations [61, 62]; although this does not correlate well with the above-described model of SG assembly (see Introduction), in which extended mRNA regions devoid of ribosomes play a key role (as their quantity should obviously increase with partial polysome disassembly), this can be explained either by incomplete polysome disassembly (Fig. 2k) or by the important role of eIF4E-eIF4G interaction in SG formation [61].
Finally, in cells pre-incubated with lactimidomycin, harringtonine, or T-2 toxin (which also lead to disassembly of polysomes, but leave a single 80S ribosome on the mRNA in the coding region), arsenite did not induce SG formation. These data suggest that under conditions where the majority of mRNAs in the cell are released from polysomes but remain associated with solitary 80S ribosomes, SG formation is impossible. The simplest explanation for this fact is that mRNA-monosome complexes are unable to be recruited into SGs and therefore cannot contribute to LLPS-mediated condensation despite having extended ribosome-free regions.
DISCUSSION
Cytoplasmic RNA–protein condensates (SGs, P-bodies, germinal and neuronal granules, and other RNP-containing non-membrane-bound organelles and compartments) play important roles in RNA metabolism in eukaryotic cells [3, 5, 7, 85]. The physicochemical principles underlying their formation are associated with LLPS [21, 22, 24-26, 28-30], but the molecular mechanisms that drive these processes are still not fully understood.
It is widely believed that specific modifications of RNA-binding proteins (e.g., G3BP1/2) along with the appearance of extended ribosome-free mRNA in the cell serve as triggers for LLPS and subsequent SG assembly [27, 31, 33, 34]. The aggregation of these RNA regions can lead to the formation of condensates, similar to the aggregation of partially denatured proteins that accumulate in the cell during heat shock [86], while ribosomes, RNA helicases, and certain RNA-binding proteins act as RNA chaperones that regulate RNA–RNA interactions. With a sharp increase in mRNA regions available for base-pairing (as observed during massive mRNA release from polysomes), this machinery becomes overloaded, similar to how the amount of protein chaperones becomes insufficient to maintain proteostasis during heat shock [86].
In full accordance with this model, classic ribosome inhibitors that block elongation and stabilize polysomes (such as cycloheximide, emetine, and anisomycin) prevent SG assembly, while complete disassembly of polysomes triggered by puromycin, the premature termination inducer, on the contrary, promotes or even provokes their formation [10, 14, 37-39, 51, 55]. Here we further confirmed these findings.
However, in this study, we for the first time examined the effect of another type of elongation inhibitors: lactimidomycin, harringtonine, and T-2 toxin, on SG formation. They also target the ribosome (specifically the E site of the 60S subunit, similar to cycloheximide, or the peptidyl transferase center, similar to anisomycin); but due to some structural peculiarities [68], they are incapable of competing with tRNA on actively translating ribosomes and therefore only bind to vacant 60S subunits. As a result, on each mRNA they arrest only a single 80S ribosome that has just initiated elongation at the start of CDS (thoroughly discussed in the review [56]). The remaining mRNA-bound ribosomes continue elongation, successfully terminate, and eventually dissociate from the mRNA. In the case of harringtonine and lactimidomycin, this property is exploited for mapping translation start sites using ribosome profiling [69, 83, 84]. These drugs intrigued us because, although they produce an excess of extended ribosome-free mRNA regions in cells similarly to puromycin, they simultaneously lock an 80S ribosome at the 5′ proximal region of each polysome-released mRNA. At first glance, massive polysome disassembly and the appearance of long “naked” mRNA regions should promote SG formation. But instead, we unexpectedly observed an inhibition of their assembly. Thus, we have shown that polysome disassembly per se does not always promote SG formation. Moreover, our data suggest that if at least one 80S ribosome remains associated with mRNA, even when the rest of the CDS is free of ribosomes, the recruitment of this mRNA into SGs is impossible.
We could find no other explanation for our observation. Since all procedures for treating cells with different inhibitors were performed using the same protocol, we can hardly assume any artifacts of cell fixation. To be on the safe side, we repeated the microscopy experiments with some of the inhibitors in a live-cell format without fixation using the fusion protein PABP-mCherry, another SG marker, for granule visualization, and obtained the same results (data not shown). The chemicals we used were commercially available products of similar purity and exhibited the expected inhibitory effects on translation, making non-specific effects of “non-canonical” inhibitors highly unlikely. Furthermore, treatment with combination of “classic” inhibitors puromycin and cycloheximide, which also leads to the selective accumulation of solitary ribosomes at mRNA start codons, according to ribosome profiling data [87], had the same effect as lactimidomycin, harringtonine, or T-2 toxins (data not shown). Taking all these facts together, we consider the most plausible assumption that mRNA bound to a single 80S ribosome cannot be recruited into SGs.
This result is particularly interesting in the context of well-documented observations that SG formation can be induced by inhibiting the RNA helicase eIF4A by specific drugs like pateamine A, hippuristanol, and rocaglates such as silvestrol, that arrest 43S scanning ribosomal complexes within the mRNA 5′ leaders [47-50]. A similar effect was also described for edeine [88], which leads to the accumulation of 40S subunits on mRNA (however, it should be noted that this drug is unable to penetrate into mammalian cells under normal conditions [89] – in accordance with that, in our hands edeine did not inhibit translation in living cells, data not shown). Furthermore, inhibition of 60S subunit joining to the 48S preinitiation complex by another compound, MDMP, although did not induce SG formation, did not impede their assembly either [51]. Thus, the association of an mRNA with small ribosomal subunits, in contrast to 80S ribosomes, does not prevent its recruitment into SGs.
There may be several explanations why the presence of 80S ribosome on mRNA interferes with its localization in SGs. This could be due to some properties of the 60S ribosomal subunits, which may be actively excluded from SGs. Alternatively, proteins recognizing stalled 80S ribosomes [90] could prevent the recruitment of such complexes into the granules. It is also possible that the topology of mRNA in the state associated with 80S ribosomes, but not with 40S subunits, does not favor RNA–RNA interactions or the incorporation of mRNA into condensates (for example, it is well known that translated mRNAs have a specific topology in both structural and functional terms – see discussion in [91, 92] and references therein). Lastly, perhaps the most intriguing explanation for our results is that mRNA recruitment into SGs may require direct interaction of some RNA-binding protein(s) with the intersubunit interface of the 40S subunit or some other regions that are inaccessible in the 80S ribosome. One such protein could be G3BP1/2, a key SG component, which has been previously shown to directly bind to the 40S ribosomal subunit [93, 94].
CONCLUSIONS
Thus, our data lead us to conclude that the presence of large amount of mRNA with extended regions devoid of ribosomes in the cell is insufficient for the formation of stress granules. In addition to that, it is required that at least a portion of the transcripts be free of 60S ribosomal subunits. This may be related to the association of RNA-binding proteins responsible for recruiting mRNA into stress granules with specific regions of the 40S subunit that are inaccessible within the 80S ribosome.
Abbreviations
- CDS:
-
coding region
- LLPS:
-
liquid–liquid phase separation
- RNP:
-
ribonucleoprotein
- SG:
-
stress granules
- UTR:
-
untranslated region
References
Bhatter, N., Dmitriev, S. E., and Ivanov, P. (2023) Cell death or survival: Insights into the role of mRNA translational control, Semin. Cell Dev. Biol., 154, 138-154, https://doi.org/10.1016/j.semcdb.2023.06.006.
Guzikowski, A. R., Chen, Y. S., and Zid, B. M. (2019) Stress-induced mRNP granules: Form and function of processing bodies and stress granules, Wiley Interdiscip. Rev. RNA, 10, e1524, https://doi.org/10.1002/wrna.1524.
Ivanov, P., Kedersha, N., and Anderson, P. (2019) Stress granules and processing bodies in translational control, Cold Spring Harb. Perspect. Biol., 11, a032813, https://doi.org/10.1101/cshperspect.a032813.
Protter, D. S., and Parker, R. (2016) Principles and properties of stress granules, Trends Cell Biol., 26, 668-679, https://doi.org/10.1016/j.tcb.2016.05.004.
Riggs, C. L., Kedersha, N., Ivanov, P., and Anderson, P. (2020) Mammalian stress granules and P bodies at a glance, J. Cell Sci., 133, jcs242487, https://doi.org/10.1242/jcs.242487.
Anderson, P., and Kedersha, N. (2008) Stress granules: the Tao of RNA triage, Trends Biochem. Sci., 33, 141-150, https://doi.org/10.1016/j.tibs.2007.12.003.
Lashkevich, K. A., and Dmitriev, S. E. (2021) mRNA targeting, transport and local translation in eukaryotic cells: from the classical view to a diversity of new concepts, Mol. Biol., 55, 507-537, https://doi.org/10.1134/S0026893321030080.
Sorokin, I. I., Vassilenko, K. S., Terenin, I. M., Kalinina, N. O., Agol, V. I., and Dmitriev, S. E. (2021) Non-canonical translation initiation mechanisms employed by eukaryotic viral mRNAs, Biochemistry (Moscow), 86, 1060-1094, https://doi.org/10.1134/S0006297921090042.
Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., and Anderson, P. (2002) Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules, Mol. Biol. Cell, 13, 195-210, https://doi.org/10.1091/mbc.01-05-0221.
Makeeva, D. S., Riggs, C. L., Burakov, A. V., Ivanov, P. A., Kushchenko, A. S., Bykov, D. A., Popenko, V. I., Prassolov, V. S., Ivanov, P. V., and Dmitriev, S. E. (2023) Relocalization of translation termination and ribosome recycling factors to stress granules coincides with elevated stop-codon readthrough and reinitiation rates upon oxidative stress, Cells, 12, 259, https://doi.org/10.3390/cells12020259.
Harvey, R., Dezi, V., Pizzinga, M., and Willis, A. E. (2017) Post-transcriptional control of gene expression following stress: the role of RNA-binding proteins, Biochem. Soc. Trans., 45, 1007-1014, https://doi.org/10.1042/BST20160364.
Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules, J. Cell Biol., 147, 1431-1442, https://doi.org/10.1083/jcb.147.7.1431.
Matsuki, H., Takahashi, M., Higuchi, M., Makokha, G. N., Oie, M., and Fujii, M. (2013) Both G3BP1 and G3BP2 contribute to stress granule formation, Genes Cells, 18, 135-146, https://doi.org/10.1111/gtc.12023.
Mazroui, R., Huot, M. E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E. W. (2002) Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression, Hum. Mol. Genet., 11, 3007-3017.
Kedersha, N., Ivanov, P., and Anderson, P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci., 38, 494-506, https://doi.org/10.1016/j.tibs.2013.07.004.
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016) ATPase-modulated stress granules contain a diverse proteome and substructure, Cell, 164, 487-498, https://doi.org/10.1016/j.cell.2015.12.038.
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., Bagci, H., Rathod, B., MacLeod, G., Eng, S. W. M., Angers, S., Morris, Q., Fabian, M., Cote, J. F., and Gingras, A. C. (2018) High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies, Mol. Cell, 69, 517-532.e511, https://doi.org/10.1016/j.molcel.2017.12.020.
Markmiller, S., Soltanieh, S., Server, K. L., Mak, R., Jin, W., Fang, M. Y., Luo, E. C., Krach, F., Yang, D., Sen, A., Fulzele, A., Wozniak, J. M., Gonzalez, D. J., Kankel, M. W., Gao, F. B., Bennett, E. J., Lecuyer, E., and Yeo, G. W. (2018) Context-dependent and disease-specific diversity in protein interactions within stress granules, Cell, 172, 590-604.e513, https://doi.org/10.1016/j.cell.2017.12.032.
Youn, J. Y., Dyakov, B. J. A., Zhang, J., Knight, J. D. R., Vernon, R. M., Forman-Kay, J. D., and Gingras, A. C. (2019) Properties of stress granule and P-body proteomes, Mol. Cell, 76, 286-294, https://doi.org/10.1016/j.molcel.2019.09.014.
Marmor-Kollet, H., Siany, A., Kedersha, N., Knafo, N., Rivkin, N., Danino, Y. M., Moens, T. G., Olender, T., Sheban, D., Cohen, N., Dadosh, T., Addadi, Y., Ravid, R., Eitan, C., Toth Cohen, B., Hofmann, S., Riggs, C. L., Advani, V. M., Higginbottom, A., Cooper-Knock, J., et al. (2020) Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis, Mol. Cell, 80, 876-891.e876, https://doi.org/10.1016/j.molcel.2020.10.032.
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017) Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev. Mol. Cell Biol., 18, 285-298, https://doi.org/10.1038/nrm.2017.7.
Jain, A., and Vale, R. D. (2017) RNA phase transitions in repeat expansion disorders, Nature, 546, 243-247, https://doi.org/10.1038/nature22386.
Alberti, S., Gladfelter, A., and Mittag, T. (2019) Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates, Cell, 176, 419-434, https://doi.org/10.1016/j.cell.2018.12.035.
Shin, Y., and Brangwynne, C. P. (2017) Liquid phase condensation in cell physiology and disease, Science, 357, https://doi.org/10.1126/science.aaf4382.
Zhang, H., Elbaum-Garfinkle, S., Langdon, E. M., Taylor, N., Occhipinti, P., Bridges, A. A., Brangwynne, C. P., and Gladfelter, A. S. (2015) RNA controls PolyQ protein phase transitions, Mol. Cell, 60, 220-230, https://doi.org/10.1016/j.molcel.2015.09.017.
Langdon, E. M., Qiu, Y., Ghanbari Niaki, A., McLaughlin, G. A., Weidmann, C. A., Gerbich, T. M., Smith, J. A., Crutchley, J. M., Termini, C. M., Weeks, K. M., Myong, S., and Gladfelter, A. S. (2018) mRNA structure determines specificity of a polyQ-driven phase separation, Science, 360, 922-927, https://doi.org/10.1126/science.aar7432.
Van Treeck, B., and Parker, R. (2018) Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies, Cell, 174, 791-802, https://doi.org/10.1016/j.cell.2018.07.023.
Kroschwald, S., Maharana, S., Mateju, D., Malinovska, L., Nuske, E., Poser, I., Richter, D., and Alberti, S. (2015) Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules, eLife, 4, e06807, https://doi.org/10.7554/eLife.06807.
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., Mittag, T., and Taylor, J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization, Cell, 163, 123-133, https://doi.org/10.1016/j.cell.2015.09.015.
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., Stoynov, S., Mahamid, J., Saha, S., Franzmann, T. M., Pozniakovski, A., Poser, I., Maghelli, N., Royer, L. A., Weigert, M., Myers, E. W., Grill, S., Drechsel, D., Hyman, A. A., and Alberti, S. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation, Cell, 162, 1066-1077, https://doi.org/10.1016/j.cell.2015.07.047.
Van Treeck, B., Protter, D. S. W., Matheny, T., Khong, A., Link, C. D., and Parker, R. (2018) RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome, Proc. Natl. Acad. Sci. USA, 115, 2734-2739, https://doi.org/10.1073/pnas.1800038115.
Panas, M. D., Ivanov, P., and Anderson, P. (2016) Mechanistic insights into mammalian stress granule dynamics, J. Cell Biol., 215, 313-323, https://doi.org/10.1083/jcb.201609081.
Guillen-Boixet, J., Kopach, A., Holehouse, A. S., Wittmann, S., Jahnel, M., Schlussler, R., Kim, K., Trussina, I., Wang, J., Mateju, D., Poser, I., Maharana, S., Ruer-Gruss, M., Richter, D., Zhang, X., Chang, Y. T., Guck, J., Honigmann, A., Mahamid, J., Hyman, A. A., et al. (2020) RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation, Cell, 181, 346-361.e317, https://doi.org/10.1016/j.cell.2020.03.049.
Yang, P., Mathieu, C., Kolaitis, R. M., Zhang, P., Messing, J., Yurtsever, U., Yang, Z., Wu, J., Li, Y., Pan, Q., Yu, J., Martin, E. W., Mittag, T., Kim, H. J., and Taylor, J. P. (2020) G3BP1 is a tunable switch that triggers phase separation to assemble stress granules, Cell, 181, 325-345 e328, https://doi.org/10.1016/j.cell.2020.03.046.
Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R., and Parker, R. (2017) The stress granule transcriptome reveals principles of mRNA accumulation in stress granules, Mol. Cell, 68, 808-820.e805, https://doi.org/10.1016/j.molcel.2017.10.015.
Namkoong, S., Ho, A., Woo, Y. M., Kwak, H., and Lee, J. H. (2018) Systematic characterization of stress-induced RNA granulation, Mol. Cell, 70, 175-187.e178, https://doi.org/10.1016/j.molcel.2018.02.025.
Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., Golan, D. E., and Anderson, P. (2000) Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules, J. Cell Biol., 151, 1257-1268, https://doi.org/10.1083/jcb.151.6.1257.
Mollet, S., Cougot, N., Wilczynska, A., Dautry, F., Kress, M., Bertrand, E., and Weil, D. (2008) Translationally repressed mRNA transiently cycles through stress granules during stress, Mol. Biol. Cell, 19, 4469-4479, https://doi.org/10.1091/mbc.e08-05-0499.
Brengues, M., Teixeira, D., and Parker, R. (2005) Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies, Science, 310, 486-489, https://doi.org/10.1126/science.1115791.
Kedersha, N., and Anderson, P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability, Biochem. Soc. Trans., 30, 963-969, https://doi.org/10.1042/bst0300963.
Reineke, L. C., Dougherty, J. D., Pierre, P., and Lloyd, R. E. (2012) Large G3BP-induced granules trigger eIF2alpha phosphorylation, Mol. Biol. Cell, 23, 3499-3510, https://doi.org/10.1091/mbc.E12-05-0385.
Kimball, S. R., Horetsky, R. L., Ron, D., Jefferson, L. S., and Harding, H. P. (2003) Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes, Am. J. Physiol. Cell Physiol., 284, C273-C284, https://doi.org/10.1152/ajpcell.00314.2002.
Souquere, S., Mollet, S., Kress, M., Dautry, F., Pierron, G., and Weil, D. (2009) Unravelling the ultrastructure of stress granules and associated P-bodies in human cells, J. Cell Sci., 122, 3619-3626, https://doi.org/10.1242/jcs.054437.
Moon, S. L., Morisaki, T., Khong, A., Lyon, K., Parker, R., and Stasevich, T. J. (2019) Multicolour single-molecule tracking of mRNA interactions with RNP granules, Nat. Cell Biol., 21, 162-168, https://doi.org/10.1038/s41556-018-0263-4.
Mateju, D., Eichenberger, B., Voigt, F., Eglinger, J., Roth, G., and Chao, J. A. (2020) Single-molecule imaging reveals translation of mRNAs localized to stress granules, Cell, 183, 1801-1812.e1813, https://doi.org/10.1016/j.cell.2020.11.010.
Seguin, S. J., Morelli, F. F., Vinet, J., Amore, D., De Biasi, S., Poletti, A., Rubinsztein, D. C., and Carra, S. (2014) Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly, Cell Death Differ., 21, 1838-1851, https://doi.org/10.1038/cdd.2014.103.
Dang, Y., Kedersha, N., Low, W. K., Romo, D., Gorospe, M., Kaufman, R., Anderson, P., and Liu, J. O. (2006) Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A, J. Biol. Chem., 281, 32870-32878, https://doi.org/10.1074/jbc.M606149200.
Mazroui, R., Sukarieh, R., Bordeleau, M. E., Kaufman, R. J., Northcote, P., Tanaka, J., Gallouzi, I., and Pelletier, J. (2006) Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation, Mol. Biol. Cell, 17, 4212-4219, https://doi.org/10.1091/mbc.e06-04-0318.
Cencic, R., Carrier, M., Galicia-Vazquez, G., Bordeleau, M. E., Sukarieh, R., Bourdeau, A., Brem, B., Teodoro, J. G., Greger, H., Tremblay, M. L., Porco, J. A., Jr., and Pelletier, J. (2009) Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol, PLoS One, 4, e5223, https://doi.org/10.1371/journal.pone.0005223.
Low, W. K., Dang, Y., Schneider-Poetsch, T., Shi, Z., Choi, N. S., Merrick, W. C., Romo, D., and Liu, J. O. (2005) Inhibition of eukaryotic translation initiation by the marine natural product pateamine A, Mol. Cell, 20, 709-722, https://doi.org/10.1016/j.molcel.2005.10.008.
Mokas, S., Mills, J. R., Garreau, C., Fournier, M. J., Robert, F., Arya, P., Kaufman, R. J., Pelletier, J., and Mazroui, R. (2009) Uncoupling stress granule assembly and translation initiation inhibition, Mol. Biol. Cell, 20, 2673-2683, https://doi.org/10.1091/mbc.e08-10-1061.
Emara, M. M., Ivanov, P., Hickman, T., Dawra, N., Tisdale, S., Kedersha, N., Hu, G. F., and Anderson, P. (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly, J. Biol. Chem., 285, 10959-10968, https://doi.org/10.1074/jbc.M109.077560.
Fujimura, K., Sasaki, A. T., and Anderson, P. (2012) Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules, Nucleic Acids Res., 40, 8099-8110, https://doi.org/10.1093/nar/gks566.
Fukuda, T., Naiki, T., Saito, M., and Irie, K. (2009) hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions, Genes Cells, 14, 113-128, https://doi.org/10.1111/j.1365-2443.2008.01256.x.
Samir, P., Kesavardhana, S., Patmore, D. M., Gingras, S., Malireddi, R. K. S., Karki, R., Guy, C. S., Briard, B., Place, D. E., Bhattacharya, A., Sharma, B. R., Nourse, A., King, S. V., Pitre, A., Burton, A. R., Pelletier, S., Gilbertson, R. J., and Kanneganti, T. D. (2019) DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome, Nature, 573, 590-594, https://doi.org/10.1038/s41586-019-1551-2.
Dmitriev, S. E., Vladimirov, D. O., and Lashkevich, K. A. (2020) A quick guide to small-molecule inhibitors of eukaryotic protein synthesis, Biochemistry (Moscow), 85, 1389-1421, https://doi.org/10.1134/S0006297920110097.
Dmitriev, S. E., Andreev, D. E., Adyanova, Z. V., Terenin, I. M., and Shatsky, I. N. (2009) Efficient cap-dependent translation of mammalian mRNAs with long and highly structured 5′-untranslated regions in vitro and in vivo, Mol. Biol. (Mosk.), 43, 108-113, https://doi.org/10.1134/S0026893309010154.
Akulich, K. A., Andreev, D. E., Terenin, I. M., Smirnova, V. V., Anisimova, A. S., Makeeva, D. S., Arkhipova, V. I., Stolboushkina, E. A., Garber, M. B., Prokofjeva, M. M., Spirin, P. V., Prassolov, V. S., Shatsky, I. N., and Dmitriev, S. E. (2016) Four translation initiation pathways employed by the leaderless mRNA in eukaryotes, Sci. Rep., 6, 37905, https://doi.org/10.1038/srep37905.
Panova, E. A., Kleymenov, D. A., Shcheblyakov, D. V., Bykonia, E. N., Mazunina, E. P., Dzharullaeva, A. S., Zolotar, A. N., Derkaev, A. A., Esmagambetov, I. B., Sorokin, I. I., Usachev, E. V., Noskov, A. N., Ivanov, I. A., Zatsepin, T. S., Dmitriev, S. E., Gushchin, V. A., Naroditsky, B. S., Logunov, D. Y., and Gintsburg, A. L. (2023) Single-domain antibody delivery using an mRNA platform protects against lethal doses of botulinum neurotoxin A, Front. Immunol., 14, 1098302, https://doi.org/10.3389/fimmu.2023.1098302.
Andreev, D. E., O’Connor, P. B., Fahey, C., Kenny, E. M., Terenin, I. M., Dmitriev, S. E., Cormican, P., Morris, D. W., Shatsky, I. N., and Baranov, P. V. (2015) Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression, eLife, 4, e03971, https://doi.org/10.7554/eLife.03971.
Fournier, M. J., Coudert, L., Mellaoui, S., Adjibade, P., Gareau, C., Cote, M. F., Sonenberg, N., Gaudreault, R. C., and Mazroui, R. (2013) Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation, Mol. Cell. Biol., 33, 2285-2301, https://doi.org/10.1128/MCB.01517-12.
Ying, S., and Khaperskyy, D. A. (2020) UV damage induces G3BP1-dependent stress granule formation that is not driven by mTOR inhibition-mediated translation arrest, J. Cell Sci., 133, jcs248310, https://doi.org/10.1242/jcs.248310.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., and Bolton, E. E. (2023) PubChem 2023 update, Nucleic Acids Res., 51, D1373-D1380, https://doi.org/10.1093/nar/gkac956.
Tscherne, J. S., and Pestka, S. (1975) Inhibition of protein synthesis in intact HeLa cells, Antimicrob. Agents Chemother., 8, 479-487, https://doi.org/10.1128/AAC.8.4.479.
Akulich, K. A., Sinitcyn, P. G., Lomakin, I. B., Andreev, D. E., Terenin, I. M., Smirnova, V. V., Mironov, A. A., Shatsky, I. N., and Dmitriev, S. E. (2017) Peptidyl transferase inhibitors arrest the ribosome at specific amino acid codons: insights from an integrated approach, FEBS J., 284, 296-296, https://doi.org/10.1111/febs.14174.
Dinos, G., Wilson, D. N., Teraoka, Y., Szaflarski, W., Fucini, P., Kalpaxis, D., and Nierhaus, K. H. (2004) Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding, Mol. Cell, 13, 113-124, https://doi.org/10.1016/s1097-2765(04)00002-4.
Schneider-Poetsch, T., Ju, J., Eyler, D. E., Dang, Y., Bhat, S., Merrick, W. C., Green, R., Shen, B., and Liu, J. O. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin, Nat. Chem. Biol., 6, 209-217, https://doi.org/10.1038/nchembio.304.
Garreau de Loubresse, N., Prokhorova, I., Holtkamp, W., Rodnina, M. V., Yusupova, G., and Yusupov, M. (2014) Structural basis for the inhibition of the eukaryotic ribosome, Nature, 513, 517-522, https://doi.org/10.1038/nature13737.
Lee, S., Liu, B., Lee, S., Huang, S. X., Shen, B., and Qian, S. B. (2012) Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution, Proc. Natl. Acad. Sci. USA, 109, E2424-E2432, https://doi.org/10.1073/pnas.1207846109.
Fresno, M., Jimenez, A., and Vazquez, D. (1977) Inhibition of translation in eukaryotic systems by harringtonine, Eur. J. Biochem., 72, 323-330, https://doi.org/10.1111/j.1432-1033.1977.tb11256.x.
Cundliffe, E., Cannon, M., and Davies, J. (1974) Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins, Proc. Natl. Acad. Sci. USA, 71, 30-34, https://doi.org/10.1073/pnas.71.1.30.
Cannon, M., Smith, K. E., and Carter, C. J. (1976) Prevention, by ribosome-bound nascent polyphenylalanine chains, of the functional interaction of t-2 toxin with its receptor site, Biochem. J., 156, 289-294, https://doi.org/10.1042/bj1560289.
Thoreen, C. C., Kang, S. A., Chang, J. W., Liu, Q., Zhang, J., Gao, Y., Reichling, L. J., Sim, T., Sabatini, D. M., and Gray, N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1, J. Biol. Chem., 284, 8023-8032, https://doi.org/10.1074/jbc.M900301200.
Brostrom, C. O., Prostko, C. R., Kaufman, R. J., and Brostrom, M. A. (1996) Inhibition of translational initiation by activators of the glucose-regulated stress protein and heat shock protein stress response systems. Role of the interferon-inducible double-stranded RNA-activated eukaryotic initiation factor 2alpha kinase, J. Biol. Chem., 271, 24995-25002, https://doi.org/10.1074/jbc.271.40.24995.
Bernstam, L., and Nriagu, J. (2000) Molecular aspects of arsenic stress, J. Toxicol. Environ. Health. Part B Crit. Rev., 3, 293-322, https://doi.org/10.1080/109374000436355.
Ruiz-Ramos, R., Lopez-Carrillo, L., Rios-Perez, A. D., De Vizcaya-Ruiz, A., and Cebrian, M. E. (2009) Sodium arsenite induces ROS generation, DNA oxidative damage, HO-1 and c-Myc proteins, NF-kappaB activation and cell proliferation in human breast cancer MCF-7 cells, Mutat. Res., 674, 109-115, https://doi.org/10.1016/j.mrgentox.2008.09.021.
Akulich, K. A., Sinitcyn, P. G., Makeeva, D. S., Andreev, D. E., Terenin, I. M., Anisimova, A. S., Shatsky, I. N., and Dmitriev, S. E. (2019) A novel uORF-based regulatory mechanism controls translation of the human MDM2 and eIF2D mRNAs during stress, Biochimie, 157, 92-101, https://doi.org/10.1016/j.biochi.2018.11.005.
Terenin, I. M., Dmitriev, S. E., Andreev, D. E., and Shatsky, I. N. (2008) Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2, Nat. Struct. Mol. Biol., 15, 836-841, https://doi.org/10.1038/nsmb.1445.
Osterman, I. A., Wieland, M., Maviza, T. P., Lashkevich, K. A., Lukianov, D. A., Komarova, E. S., Zakalyukina, Y. V., Buschauer, R., Shiriaev, D. I., Leyn, S. A., Zlamal, J. E., Biryukov, M. V., Skvortsov, D. A., Tashlitsky, V. N., Polshakov, V. I., Cheng, J., Polikanov, Y. S., Bogdanov, A. A., Osterman, A. L., Dmitriev, S. E., et al. (2020) Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel, Nat. Chem. Biol., 16, 1071-1077, https://doi.org/10.1038/s41589-020-0578-x.
Prokhorova, I. V., Akulich, K. A., Makeeva, D. S., Osterman, I. A., Skvortsov, D. A., Sergiev, P. V., Dontsova, O. A., Yusupova, G., Yusupov, M. M., and Dmitriev, S. E. (2016) Amicoumacin A induces cancer cell death by targeting the eukaryotic ribosome, Sci. Rep., 6, 27720, https://doi.org/10.1038/srep27720.
Andreev, D. E., Dmitriev, S. E., Terenin, I. M., Prassolov, V. S., Merrick, W. C., and Shatsky, I. N. (2009) Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs, Nucleic Acids Res., 37, 6135-6147, https://doi.org/10.1093/nar/gkp665.
Thoreen, C. C., Chantranupong, L., Keys, H. R., Wang, T., Gray, N. S., and Sabatini, D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation, Nature, 485, 109-113, https://doi.org/10.1038/nature11083.
Ingolia, N. T., Lareau, L. F., and Weissman, J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes, Cell, 147, 789-802, https://doi.org/10.1016/j.cell.2011.10.002.
Gerashchenko, M. V., Peterfi, Z., Yim, S. H., and Gladyshev, V. N. (2021) Translation elongation rate varies among organs and decreases with age, Nucleic Acids Res., 49, e9, https://doi.org/10.1093/nar/gkaa1103.
Anderson, P., and Kedersha, N. (2006) RNA granules, J. Cell Biol., 172, 803-808, https://doi.org/10.1083/jcb.200512082.
Ripin, N., and Parker, R. (2022) Are stress granules the RNA analogs of misfolded protein aggregates? RNA, 28, 67-75, https://doi.org/10.1261/rna.079000.121.
Fritsch, C., Herrmann, A., Nothnagel, M., Szafranski, K., Huse, K., Schumann, F., Schreiber, S., Platzer, M., Krawczak, M., Hampe, J., and Brosch, M. (2012) Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting, Genome Res., 22, 2208-2218, https://doi.org/10.1101/gr.139568.112.
Thomas, M. G., Martinez Tosar, L. J., Loschi, M., Pasquini, J. M., Correale, J., Kindler, S., and Boccaccio, G. L. (2005) Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes, Mol. Biol. Cell, 16, 405-420, https://doi.org/10.1091/mbc.e04-06-0516.
Contreras, A., and Carrasco, L. (1979) Selective inhibition of protein synthesis in virus-infected mammalian cells, J. Virol., 29, 114-122, https://doi.org/10.1128/JVI.29.1.114-122.1979.
Schuller, A. P., and Green, R. (2018) Roadblocks and resolutions in eukaryotic translation, Nat. Rev. Mol. Cell Biol., 19, 526-541, https://doi.org/10.1038/s41580-018-0011-4.
Alekhina, O. M., Terenin, I. M., Dmitriev, S. E., and Vassilenko, K. S. (2020) Functional cyclization of eukaryotic mRNAs, Int. J. Mol. Sci., 21, 1677, https://doi.org/10.3390/ijms21051677.
Baymukhametov, T. N., Lyabin, D. N., Chesnokov, Y. M., Sorokin, I. I., Pechnikova, E. V., Vasiliev, A. L., and Afonina, Z. A. (2023) Polyribosomes of circular topology are prevalent in mammalian cells, Nucleic Acids Res., 51, 908-918, https://doi.org/10.1093/nar/gkac1208.
Kedersha, N., Panas, M. D., Achorn, C. A., Lyons, S., Tisdale, S., Hickman, T., Thomas, M., Lieberman, J., McInerney, G. M., Ivanov, P., and Anderson, P. (2016) G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits, J. Cell Biol., 212, 845-860, https://doi.org/10.1083/jcb.201508028.
Meyer, C., Garzia, A., Morozov, P., Molina, H., and Tuschl, T. (2020) The G3BP1-family-USP10 deubiquitinase complex rescues ubiquitinated 40S subunits of ribosomes stalled in translation from lysosomal degradation, Mol. Cell, 77, 1193-1205.e1195, https://doi.org/10.1016/j.molcel.2019.12.024.
Acknowledgments
We are grateful to Eugenia A. Panova (Moscow State University) for the Actin-Fluc mRNA and the MSU Development Program for providing access to the confocal microscope Zeiss LSM900
Funding
The study was supported by the Russian Science Foundation, grant no. 18-14-00291.
Author information
Authors and Affiliations
Contributions
A.G.F., A.V.B., I.M.T., D.A.B., K.A.L., V.I.P., N.E.M., I.I.S. performing the experiments; D.A.B., S.E.D. research concept; I.M.T., P.V.I., S.E.D. discussion of the results; V.S.P., S.E.D. providing infrastructure and project administration; A.G.F., A.V.B., I.M.T., A.P.S., S.E.D. creating the figures, writing, editing, and translation of the manuscript.
Corresponding author
Ethics declarations
The authors declare no conflict of interest. This paper does not contain a description of any research involving humans or animals.
Rights and permissions
Open access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fedorovskiy, A.G., Burakov, A.V., Terenin, I.M. et al. A Solitary Stalled 80S Ribosome Prevents mRNA Recruitment to Stress Granules. Biochemistry Moscow 88, 1786–1799 (2023). https://doi.org/10.1134/S000629792311010X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S000629792311010X