Abstract
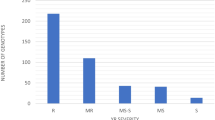
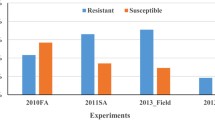
Powdery mildew caused by the fungal pathogen Blumeria graminis f. sp. tritici (Bgt) is an econo-mically important disease of common wheat T. aestivum L. One of the most effective and environmentally important ways of protection of wheat against Bgt is cultivation of the varieties with genetic resistance. The aim of this work was to study the genetic diversity of Russian spring wheat varieties for the powdery mildew resistance loci. Phytopathological evaluation of 97 wheat varieties showed that no more than 10% of the varieties have low level of susceptibility to the Bgt population specific to the Western Siberian region. Association mapping carried out on the basis of SNP genotyping and phytopathological testing during three environmental seasons identified eight loci in chromosomes 1AL, 1DS, 2BL, 5AS, 5DS, 6AL, 6DL, and 7AL. A high impact to the phenotypic manifestation of the trait was established for genetic factors localized in chromosomes 5AS, 6AL, and 6DL. The long arm of chromosome 6D contains the gene Pm6Ai=2, which was introduced from wheatgrass Thinopyrum intermedium and provides effective protection against the powdery mildew pathogen. On the basis of comparative analysis of the chromosomal localization of the known Pm resistance genes and loci mapped in this work, it was assumed that QTLs in chromosomes 1DS, 5AS, and 6AL are novel, not previously described resistance loci. The obtained results can be used in breeding programs for selection of target loci and for development of molecular markers specific to Bgt resistance loci.






Similar content being viewed by others
REFERENCES
Cunfer, B.M., Powdery mildew, in Bread Wheat: Improvement and Production, Italy: FAO, 2002, pp. 317—330.
Kiseleva, M.I., Kolomiets, T.M., Pakholkova, E.V., et al., The differentiation of winter wheat (Triticum aestivum L.) cultivars for resistance to the most harmful fungal pathogens, Agricultural Biol., 2016, vol. 51, no. 3, pp. 299—309. https://doi.org/10.15389/agrobiology.2016.3.299eng
Sanin, S.S. and Nazarova, A.N., Phytosanitary situation in wheat fields in the Russian Federation (1991—2008), Zashch. Karantin Rast., 2010, no. 2, pp. 70—78.
Teplyakova, O.I. and Teplyakov, V.I., Local monitoring of spring wheat leaf diseases in Siberia, Zashch. Karantin Rast., 2011, no. 6, pp. 39—41.
McIntosh, R.A., Yamazaki, Y., Dubcovsky, J., et al., Catalogue of Gene Symbols for Wheat, in The 12th International Wheat Genetics Symposium, Yokohama, 2013, suppl., pp. 2014—2017. http://www.shigen.nig.ac.jp/wheat/komugi/genes/.
Huang, X.O. and Röder, M.S., Molecular mapping of powdery mildew resistance genes in wheat: a review, Euphytica, 2004, vol. 137, pp. 203—223. https://doi.org/10.1023/B:EUPH.0000041576.74566.d7
Mohler, V., Hsam, S.L.K., Zeller, F.J., and Wenzel, G., An STS marker distinguishing the rye-derived powdery mildew resistance alleles at the Pm8/Pm17 locus of common wheat, Plant Breed., 2001, vol. 120, pp. 448—450. https://doi.org/10.1046/j.1439-0523.2001.00622.x
Spielmeyer, W., McIntosh, R.A., Kolmer, J., and Lagudah, E., Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust co-segregate at a locus on the short arm of chromosome 7D of wheat, Theor. Appl. Genet., 2005, vol. 111, pp. 731—735. https://doi.org/10.1007/s00122-005-2058-9
Lillemo, M., Asalf, B., Singh, R.P., et al., The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar, Theor. Appl. Genet., 2005, vol. 116, pp. 1155—1166. https://doi.org/10.1007/s00122-008-0743-1
Li, Z., Lan, C., He, Z., et al., Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat, Crop Sci., 2014, vol. 54, pp. 1907—1925. https://doi.org/10.2135/cropsci2014.02.0162
Alam, Md., Xue, F., Wang, C., and Ji, W., Powdery mildew resistance genes in wheat: identification and genetic analysis, J. Mol. Biol. Res., 2011, vol. 1, pp. 20—39. https://doi.org/10.5538/jmbr.v1n1p20
Shah, L., Rehman, S., Ali, A., et al., Genes responsible for powdery mildew resistance and improvement in wheat using molecular marker-assisted selection, J. Plant Dis. Protect., 2018, vol. 125, pp. 145—158. https://doi.org/10.1007/s41348-017-0132-6
Wang, S., Wong, D., Forrest, K., et al., Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array, Plant Biotechnol. J., 2014, vol. 12, pp. 787—796. https://doi.org/10.1111/pbi.12183
Maccaferri, M., Ricci, A., Salvi, S., et al., A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding, Plant Biotechnol. J., 2015, vol. 13, pp. 648—663. https://doi.org/10.1111/pbi.12288
Gerard, G.S., Börner, A., Lohwasser, U., et al., Genome-wide association mapping of genetic factors controlling Septoria tritici blotch resistance and their association with plant height and heading date in wheat, Euphytica, 2017, vol. 213, pp. 27—41. https://doi.org/10.1007/s10681-016-1920-1
Kankwatsa, P., Singh, D., Thomson, P.C., et al., Characterization and genome-wide association mapping of resistance to leaf rust, stem rust and stripe rust in a geographically diverse collection of spring wheat landraces, Mol. Breed., 2017, vol. 37, pp. 113—136. https://doi.org/10.1007/s11032-017-0707-8
Tessmann, E.W. and Van Sanford, D.A., GWAS for Fusarium head blight related traits in winter wheat (Triticum aestivum L.) in an artificially warmed treatment, Agronomy, 2018, vol. 8, article no. 68. https://doi.org/10.3390/agronomy8050068
Liu, N., Bai, G., Lin, M., et al., Genome-wide association analysis of powdery mildew resistance in U.S. winter wheat, Sci. Rep., 2017, vol. 7, article no. 11743. https://doi.org/10.1038/s41598-017-11230-z
Ren, Y., Hou, W., Lan, C., et al., QTL analysis and nested association mapping for adult plant resistance to powdery mildew in two bread wheat populations, Front. Plant Sci., 2017, vol. 8, article no. 1212. https://doi.org/10.3389/fpls.2017.01212
Saari, E.E. and Prescott, J.M., A scale for appraising the foliar intensity of wheat diseases, Plant Dis. Rep., 1975, vol. 59, pp. 377—380.
Leonova, I.N., Dobrovol’skaya, O.B., Kaminskaya, L.N., et al., Molecular analysis of the triticale lines with different Vrn gene systems using microsatellite markers and hybridization in situ,Russ. J. Genet., 2005, vol. 41, no. 9, pp. 1014—1020. https://doi.org/10.1007/s11177-005-0193-7
Bradbury, P.J., Zhang, Z., Kroon, D.E., et al., TASSEL: software for association mapping of complex traits in diverse samples, Bioinformatics, 2007, vol. 23, pp. 2633—2635. https://doi.org/10.1093/bioinformatics/btm308
Pritchard, J., Stephens, M., and Donnelly, P., Inference of population structure using multilocus genotype data, Genetics, 2000, vol. 155, pp. 945—959.
Evanno, G., Regnaut, S., and Goudet, J., Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study, Mol. Ecol., 2005, vol. 14, pp. 2611—2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Earl D.A. and von Holdt, B.M., STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method, Conserv. Genet. Resour., 2012, vol. 4, pp. 359—361. https://doi.org/10.1007/s12686-011-9548-7
Hammer, Ø., Harper, D.A.T., and Ryan, P.D., PAST: paleontological statistics software package for education and data analysis, Palaeontol. Electron., 2001, vol. 4, pp. 1—9.
Lebedeva, T.V., Genetic diversity of common wheat Triticum aestivum L. for resistance to Blumeria graminis DC. f. sp. tritici Golovin, Vestnik VOGIS, 2008, vol. 12, pp. 685—690.
Lebedeva, T.V., Zuev, E.V., and Brykova, A.N., The expression of powdery mildew resistance in spring bread wheat cultivars from the VIR collection of plan genetic resources, Proceed. Applied Bot. Genet. Breed., 2018, vol. 179, no. 3, pp. 272—277. https://doi.org/10.30901/2227-8834-2018-3-272-277
Somers, D.J., Isaac, P., and Edwards, K., A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.), Theor. Appl. Genet., 2004, vol. 109, pp. 1105—1114. https://doi.org/10.1007/s00122-004-1740-7
Salina, E.A., Adonina, I.G., Badaeva, E.D., et al., A Thinopyrum intermedium chromosome in bread wheat cultivars as a source of genes conferring resistance to fungal diseases, Euphytica, 2015, vol. 204, pp. 91—101. https://doi.org/10.1007/s10681-014-1344-5
Leonova, I.N., Stasyuk, A.I., Skolotneva, E.S., and Salina, E.A., Enhancement of leaf rust resistance of Siberian winter wheat varieties by marker-assisted selection, Cereal Res. Commun., 2017, vol. 45, pp. 621—632. https://doi.org/10.1556/0806.45.2017.048
Li, H., Chen, X., Xin, Z.Y., et al., Development and identification of wheat—Haynaldia villosa 6DL.6VS chromosome translocation lines conferring resistance to powdery mildew, Plant Breed., 2005, vol. 124, pp. 203—205. https://doi.org/10.1111/j1439-0523.2004.01062.x
Qi, L.L., Chen, P.D., Liu, D.J., et al., The gene Pm21—a new source of resistance to wheat powdery mildew, Acta Agric. Sin., 1995, vol. 21, pp. 257—261.
Hao, M., Liu, M., Luo, J., et al., Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat, Front. Plant Sci., 2018, vol. 9, article no. 1040. https://doi.org/10.3389/fpls.2018.01040
Keller, M., Keller, B., Schachermayr, G., et al., Quantitative trait loci for resistance against powdery mildew in a segregating wheat x spelt population, Theor. Appl. Genet., 1999, vol. 98, pp. 903—912. https://doi.org/10.1007/s001220051149
Jakobson, I., Peusha, H., Timofejeva, L., and Jarve, K., Adult plant and seedling resistance to powdery mildew in a Triticum aestivum × Triticum militinae hybrid line, Theor. Appl. Genet., 2006, vol. 112, pp. 760—769. https://doi.org/10.1007/s00122-005-0181-2
Miranda, L.M., Murphy, J.P., Marshall, D., et al., Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.), Theor. Appl. Genet., 2006, vol. 113, pp. 1497—1504. https://doi.org/10.1007/s00122-006-0397-9
Zhang, R., Sun, B., Chen, J., et al., Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat, Theor. Appl. Genet., 2016, vol. 129, pp. 1975—1985. https://doi.org/10.1007/s00122-016-2753-8
Xue, F., Wang, C., Li, C., et al., Molecular mapping of a powdery mildew resistance gene in common wheat landrace Baihulu and its allelism with Pm24,Theor. Appl. Genet., 2012, vol. 125, pp. 1425—1432. https://doi.org/10.1007/s00122-012-1923-6
Lan, C., Liang, S., Wang, Z., et al., Quantitative trait loci mapping for adult-plant resistance to powdery mildew in Chinese wheat cultivar Bainong 64, Phytopathology, 2009, vol. 99, pp. 1121—1126. https://doi.org/10.1094/PHYTO-99-10-1121
Zhu, Z., Zhou, R., Kong, X., et al., Microsatellite markers linked to 2 powdery mildew resistance genes introgressed from Triticum carthlicum accession PS5 into common wheat, Genome, 2005, vol. 48, pp. 585—590. https://doi.org/10.1139/g05-016
Zhan, H., Li, G., Zhang, X., et al., Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat—Thinopyrum ponticum introgression line, PLoS One, 2014, vol. 9. e113455. https://doi.org/10.1371/journal.pone.0113455
Liu, W., Koo, D.H., Xia, Q., et al., Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat, Theor. Appl. Genet., 2017, vol. 130, pp. 841—848. https://doi.org/10.1007/s00122-017-2855-y
Zhao, Z., Sun, Y., Somg, W., et al., Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99, Theor. Appl. Genet., 2013, vol. 126, pp. 3081—3089. https://doi.org/10.1007/s00122-013-2194-6
Neu, C., Stein, N., and Keller, B., Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat, Genome, 2002, vol. 45, pp. 737—744. https://doi.org/10.1139/g02-040
Aoun, M., Breiland, B., Turner, M.K., et al., Genome-wide association mapping of leaf rust response in a durum wheat worldwide germplasm collection, Plant Genome, 2016, vol. 9, no. 3, pp. 1—24. https://doi.org/10.3835/plantgenome2016.01.0008
Buloichik, A.A. and Borzyak, V.S., The occurrence of virulence genes in the Belarusian population of Blumeria graminis f. sp. tritici,Mikol. Fitopatol., 2013, vol. 47, pp. 405—409.
Cowger, C., Mehra, L., Arellano, C., et al., Virulence differences in Blumeria graminis f. sp. tritici from the Central and Eastern United States, Phytopathology, 2018, vol. 108, pp. 402—411. https://doi.org/10.1094/PHYTO-06-17-0211-R
Sochalova, L.P. and Likhenko, I.E., The study of wheat resistance to leaf pathogens in Western Siberia, Sib. Vestn. S.-kh. Nauki, 2011, no. 1, pp. 18—25.
Sochalova, L.P. and Piskarev, V.V., Resistance of varieties of spring soft wheat to agents of infections under conditions the changing climate of Western Siberia, Achivements Sci. Technol. AIC, 2017, vol. 31, no. 2, pp. 21—25.
Funding
This work was supported by the grant of the Russian Science Foundation 16-16-00011-P. The collection of spring wheat varieties was reproduced and cultivated within the framework of the project of the Ministry of Education and Science of the Russian Federation no. 0324-2019-0039.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any research using animals as an object. This article does not contain any research involving people as an object.
Additional information
Translated by K. Lazarev
Rights and permissions
About this article
Cite this article
Leonova, I.N. Genome-Wide Association Study of Powdery Mildew Resistance in Russian Spring Wheat (T. aestivum L.) Varieties. Russ J Genet 55, 1360–1374 (2019). https://doi.org/10.1134/S1022795419110085
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795419110085