Abstract
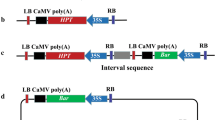
The cointegration rate into the aspen and birch genomes of foreign genes from a binary vector and a disarmed Ti plasmid pCBE21 carried by the same Agrobacterium tumefaciens strain was studied. The cotransformation rate for the genes within the Ti plasmid varied from 30 to 100%; while the transformation rate for the gene from TL region was twofold higher as compared with the TR region. On the average, the gene transfer from all three T-DNAs was recorded in 10.9% of the transgenic lines. For the vector pBI121, the cotransformation rates for the genes from both regions of pCBE21 T-DNA were higher as compared with the vector pGS. In addition, a concurrent transfer of the genes from the Ti plasmid TL and TR regions was recorded only after the transformation with the vector pBI121. These results can be used for constructing woody plants containing several genes.
Similar content being viewed by others
References
James, C., Global Status of Commercialized Biotech/GM Crops, no. 39 ISAAA Brief, Ithaca: ISAAA, 2008.
Lebedev, V.G., Skryabin, K.G., and Dolgov, S.V., Transgenic Pear Clonal Rootstocks Resistant to Herbicide “Basta”, Acta Hort., 2002, vol. 596, pp. 193–197.
Lyyra, S., Meagher, R.B., Kim, T., et al., Coupling Two Mercury Resistance Genes in Eastern Cottonwood Enhances the Processing of Organomercury, Plant Biotech. J., 2007, vol. 5, pp. 254–262.
An, G., Watson, B.D., Stachel, S., et al., New Cloning Vehicles for Transformation of Higher Plants, EMBO J., 1985, vol. 4, pp. 277–284.
Depicker, A., Herman, L., Jacobs, A., et al., Frequencies of Simultaneous Transformation with Different T-DNAs and Their Relevance to the Agrobacterium/Plant Cell Interaction, Mol. Gen. Genet., 1985, vol. 201, pp. 477–484.
Chen, L., Marmey, P., Taylor, N.J., et al., Expression and Inheritance of Multiple Transgenes in Rice Plants, Nat. Biotech., 1998, vol. 16, pp. 1060–1065.
McKnight, T.D., Lillis, M.T., and Simpson, R.B., Segregation of Genes Transferred to One Plant Cell from Two Separate Agrobacterium Strains, Plant. Mol. Biol., 1987, vol. 8, pp. 439–445.
De Buck, S., Jacobs, A., Van Montagu, M., and Depicker, A., Agrobacterium tumefaciens Transformation and Cotransformation Frequencies of Arabidopsis thaliana Root Explants and Tobacco Protoplasts, Mol. Plant Microbe Inter., 1998, vol. 11, pp. 449–457.
Radchuk, V.V., Thi Van, D., and Klocke, E., Multiple Gene Co-Integration in Arabidopsis thaliana Predominantly Occurs in the Same Genetic Locus after Simultaneous in planta Transformation with Distinct Agrobacterium tumefaciens Strains, Plant Sci., 2005, vol. 168, pp. 1515–1523.
De Buck, S., Podevin, N., Nolf, J., et al., The T-DNA Integration Pattern in Arabidopsis Transformants Is Highly Determined by the Transformed Target Cell, Plant J., 2009, vol. 60, pp. 134–145.
Jacob, S.S. and Veluthambi, K., Generation of Selection Marker-Free Transgenic Plants by Co-Transformation of a Co-Integrate Vector T-DNA and a Binary Vector T-DNA in One Agrobacterium tumefaciens Strain, Plant Sci., 2002, vol. 163, pp. 801–806.
Higgins, J.D., Newbury, H.J., Barbara, D.J., et al., The Production of Marker-Free Genetically Engineered Broccoli with Sense and Antisense ACC Synthase 1 and ACC Oxidases 1 and 2 to Extend Shelf Life, Mol. Breed., 2006, vol. 17, pp. 7–20.
Komari, T., Hiei, Y., Saito, Y., et al., Vectors Carrying Two Separate T-DNAs for Co-Transformation of Higher Plants Mediated by Agrobacterium tumefaciens and Segregation of Transformants Free from Selection Markers, Plant J., 1996, vol. 10, pp. 165–174.
Breitler, J., Meynard, D., Boxtel, J., et al., A Novel Two T-DNA Binary Vector Allows Efficient Generation of Marker-Free Transgenic Plants in Three Elite Cultivars of Rice (Oryza sativa L.), Trans. Res., 2004, vol. 13, pp. 271–278.
Li, L., Zhou, Y., Cheng, X., et al., Combinatorial Modification of Multiple Lignin Traits in Trees through Multigene Cotransformation, Proc. Natl. Acad. Sci. USA, 2003, vol. 100, pp. 4939–4944.
Lloyd, G. and McCown, B., Commercially Feasible Micropropagation of Mountain Laurel, Kalmia latifolia, by Use of Shoot-Tip Culture, Proc. Int. Plant Prop. Soc., 1981, vol. 30, pp. 421–427.
Jefferson, R.A., Assaying Chimeric Genes in Plants: The GUS Fusion System, Plant Mol. Biol. Rep., 1987, vol. 5, pp. 387–405.
Revenkova, E.V., Kraev, A.S. and Skryabin, K.G., Construction of a Disarmed Derivative of the Supervirulent Ti Plasmid pTiBo542, in Biotechnology and Molecular Biology, Moscow, 1993, pp. 67–76.
Murashige, T. and Skoog, F., A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Culture, Phys. Plant., 1962, vol. 15, pp. 473–497.
Rogers, S.O. and Bendich, A.J., Extraction of Total Cellular DNA from Plants, Algae and Fungi, Plant Mol. Biol. Manual, Gelvin, S.B. and Schilperoort, R.A., Eds., Kluwer, 1994, pp. 1–8.
Sripriya, R., Raghupathy, V., and Veluthambi, K., Generation of Selectable Marker-Free Sheath Blight Resistant Transgenic Rice Plants by Efficient Co-Transformation of a Cointegrate Vector T-DNA and a Binary Vector T-DNA in One Agrobacterium tumefaciens Strain, Plant Cell Rep., 2008, vol. 27, pp. 1635–1644.
Ream, W., Production of a Mobile T-DNA by Agrobacterium tumefaciens, Agrobacterium: From Biology to Biotechnology, Tzfira, T., Citovsky, V., Eds., 2008, pp. 279–313.
Gulina, I.V., Shul’ga, O.A., Mironov, V.N., et al., The Expression of Partly Modified d-Endotoxin Gene from B. thuringiensis var. tenebrionis in Potato Transgenic Plants, Mol. Biol., 1994, vol. 28, pp. 1166–1175.
Lebedev, V.G., Lavrova, N., Lunin, V.G., et al., Plant-Defensin Genes Introduction for Improvement of Pear Phytopathogene Resistance, Acta Hort., 2002, vol. 596, pp. 167–172.
Schestibratov, K.A. and Dolgov, S.V., Transgenic Strawberry Plants Expressing a Thaumatin II Gene Demonstrate Enhanced Resistance to Botrytis cinerea, Sci. Hort., 2005, vol. 106, pp. 177–189.
Palanichelvam, K., Oger, P., Clough, S.J., et al., A Second T-Region of the Soybean-Supervirulent Chrysopine-Type Ti Plasmid pTiChry5, and Construction of a Fully Disarmed vir Helper Plasmid, Mol. Plant Microbe Inter., 2000, vol. 13, pp. 1081–1091.
Ko, T.-S., Lee, S., Farrand, S.K., and Korban, S.S., A Partially Disarmed vir Helper Plasmid, pKYRT1, in Conjunction with 2.4-Dichlorophenoxyactic Acid Promotes Emergence of Regenerable Transgenic Somatic Embryos from Immature Cotyledons of Soybean, Planta, 2004, vol. 218, pp. 536–554.
Hooykaas, P.J.J. and Schilperoort, R.A., Agrobacterium and Plant Genetic Engineering, Plant. Mol. Biol., 1992, vol. 19, pp. 15–38.
McCormac, A.C., Fowler, M.R., Chen, D.-F., and Elliott, M.C., Efficient Co-Transformation of Nicotiana tabacum by Two Independent T-DNAs, the Effect of T-DNA Size and Implications for Genetic Separation, Trans. Res., 2001, vol. 10, pp. 143–155.
Miller, M., Tagliani, L., Wang, N., et al., High Efficiency Transgene Segregation in Co-Trans-Formed Maize Plants Using an Agrobacterium tumefaciens 2 T-DNA Binary System, Trans. Res., 2002, vol. 11, pp. 381–396.
Windels, P., De Buck, S., and Depicker, A., Agrobacterium tumefaciens-Mediated Transformation: Patterns of T-DNA Integration into the Host Genome, Agrobacterium: From Biology to Biotechnology, Tzfira, T. and Citovsky, V., Eds., 2008, pp. 441–481.
Simpson, R.B., Spielmann, A., Margossian, L., and McKnight, T.D., A Disarmed Binary Vector from Agrobacterium tumefaciens Functions in Agrobacterium rhizogenes, Plant. Mol. Biol., 1986, vol. 6, pp. 403–415.
Lebedev, V.G., Shestibratov, K.A., and Dolgov, S.V., Efficiency of Cointegration of Heterologous DNA Sequences from T-Area of Different Agrobacterial Replicons in Plant Nuclear Genome, Tezisy dokladov Otchetnoi konferentsii FIBKh RAN za 2001 g (Abstracts of Summary Conference of the Institute of Physiology and Biochemistry RAS 2001), Pushchino, 2001.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © V.G. Lebedev, K.A. Schestibratov, T.E. Shadrina, I.V. Bulatova, D.G. Abramochkin, A.I. Miroshnikov, 2010, published in Genetika, 2010, Vol. 46, No. 11, pp. 1458–1466.
Rights and permissions
About this article
Cite this article
Lebedev, V.G., Schestibratov, K.A., Shadrina, T.E. et al. Cotransformation of aspen and birch with three T-DNA regions from two different replicons in one Agrobacterium tumefaciens strain. Russ J Genet 46, 1282–1289 (2010). https://doi.org/10.1134/S1022795410110025
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795410110025