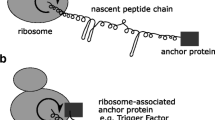
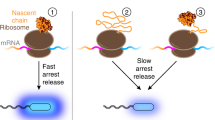
Abstract
The problem of linear polypeptide chain folding into a unique tertiary structure is one of the fundamental scientific challenges. The process of folding cannot be fully understood without its biological context, especially for big multidomain and multisubunit proteins. The principal features of biosynthetic folding are co-translational folding of growing nascent polypeptide chains and involvement of molecular chaperones in the process. The review summarizes available data on the early events of nascent chain folding, as well as on later advanced steps, including formation of elements of native structure. The relationship between the non-uniformity of translation rate and folding of the growing polypeptide is discussed. The results of studies on the effect of biosynthetic folding features on the parameters of folding as a physical process, its kinetics and mechanisms, are presented. Current understanding and hypotheses on the relationship of biosynthetic folding with the fundamental physical parameters and current views on polypeptide folding in the context of energy landscapes are discussed.
Similar content being viewed by others
References
Perrin, D. (1963) Complementation between products of the β-galactosidase structural gene of Escherichia coli, Cold Spring Harb. Symp. Quant. Biol., 28, 529-532, https://doi.org/10.1101/SQB.1963.028.01.070.
Fogel, Z., and Elson, D. (1964) The ribosomal beta-galactosidase of Escherichia coli, Biochim. Biophys. Acta, 80, 601-613, https://doi.org/10.1016/0926-6550(64)90305-6.
Kiho, Y., and Rich, A. (1964) Induced enzyme formed on bacterial polyribosomes, Proc. Natl. Acad. Sci. USA, 51, 111-118.
Lederberg, E. M., Cavalli-Sforza, L., and Lederberg, J. (1964) Interaction of streptomycin and a suppressor for galactose fermentation in E. coli K-12, Proc. Natl. Acad. Sci. USA, 51, 678-682, https://doi.org/10.1073/pnas.51.4.678.
Baldwin, R. L. (1975) Intermediates in protein folding reactions and the mechanism of protein folding, Ann. Rev. Biochem., 44, 453-475, https://doi.org/10.1146/annurev.bi.44.070175.002321.
Lim, V. I., and Spirin, A. S. (1986) Stereochemical analysis of ribosomal transpeptidation conformation of nascent peptide, J. Mol. Biol., 188, 565-574, https://doi.org/10.1016/S0022-2836(86)80006-7.
Jenner, L., Melnikov, S., de Loubresse, N. G., Ben-Shem, A., Iskakova, M., et al. (2012) Crystal structure of the 80S yeast ribosome, Curr. Opin. Struct. Biol., 22, 759-767, https://doi.org/10.1016/j.sbi.2012.07.013.
Lu, J., and Deutsch, C. (2005) Folding zones inside the ribosomal exit tunnel, Nat. Struct. Mol. Biol., 12, 1123-1129, https://doi.org/10.1038/nsmb1021.
Ziv, G., Haran, G., and Thirumalai, D. (2005) Ribosome exit tunnel can entropically stabilize α-helices, Proc. Natl. Acad. Sci. USA, 102, 18956-18961, https://doi.org/10.1073/pnas.0508234102.
Holtkamp, W., Kokic, G., Jäger, M., Mittelstaet, J., Komar, A. A., et al. (2015) Cotranslational protein folding on the ribosome monitored in real time, Science, 350, 1104-1107, https://doi.org/10.1126/science.aad0344.
Wruck, F., Katranidis, A., Nierhaus, K. H., Büldt, G., and Hegner, M. (2017) Translation and folding of single proteins in real time, Proc. Natl. Acad. Sci. USA, 114, E4399-E4407, https://doi.org/10.1073/pnas.1617873114.
Haase, N., Holtkamp, W., Lipowsky, R., Rodnina, M., and Rudorf, S. (2018) Decomposition of time-dependent fluorescence signals reveals codon-specific kinetics of protein synthesis, Nucleic Acids Res., 46, 12186-12187, https://doi.org/10.1093/nar/gky740.
Cassaignau, A. M. E., Launay, H. M. M., Karyadi, M.-E., Wang, X., Waudby, C. A., et al. (2016) A strategy for co-translational folding studies of ribosome-bound nascent chain complexes using NMR spectroscopy, Nat. Protoc., 11, 1492-1507, https://doi.org/10.1038/nprot.2016.101.
Waudby, C. A., Wlodarski, T., Karyadi, M.-E., Cassaignau, A. M. E., Chan, S. H. S., et al. (2018) Systematic mapping of free energy landscapes of a growing filamin domain during biosynthesis, Proc. Natl. Acad. Sci. USA, 115, 9744-9749, https://doi.org/10.1073/pnas.1716252115.
Cabrita, L. D., Cassaignau, A. M. E., Launay, H. M. M., Waudby, C. A., Wlodarski, T., et al. (2016) A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding, Nat. Struct. Mol. Biol., 23, 278-285, https://doi.org/10.1038/nsmb.3182.
Hoffmann, A., Becker, A. H., Zachmann-Brand, B., Deuerling, E., Bukau, B., et al. (2012) Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding, Mol. Cell, 48, 63-74, https://doi.org/10.1016/j.molcel.2012.07.018.
Nilsson, O. B., Nickson, A. A., Hollins, J. J., Wickles, S., Steward, A., et al. (2017) Cotranslational folding of spectrin domains via partially structured states, Nat. Struct. Mol. Biol., 24, 221-225, https://doi.org/10.1038/nsmb.3355.
Nilsson, O. B., Hedman, R., Marino, J., Wickles, S., Bischoff, L., et al. (2015) Cotranslational protein folding inside the ribosome exit tunnel, Cell Rep., 12, 1533-1540, https://doi.org/10.1016/j.celrep.2015.07.065.
Samelson, A. J., Bolin, E., Costello, S. M., Sharma, A. K., O’Brien, E. P., et al. (2018) Kinetic and structural comparison of a protein’s cotranslational folding and refolding pathways, Sci. Adv., 4, eaas9098, https://doi.org/10.1126/sciadv.aas9098.
Samelson, A. J., Jensen, M. K., Soto, R. A., Cate, J. H. D., and Marqusee, S. (2016) Quantitative determination of ribosome nascent chain stability, Proc. Natl. Acad. Sci. USA, 113, 13402-13407, https://doi.org/10.1073/pnas.1610272113.
Zhang, G., Hubalewska, M., and Ignatova, Z. (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding, Nat. Struct. Mol. Biol., 16, 274-280, https://doi.org/10.1038/nsmb.1554.
Deeng, J., Chan, K. Y., van der Sluis, E. O., Berninghausen, O., Han, W., et al. (2016) Dynamic behavior of trigger factor on the ribosome, J. Mol. Biol., 428, 3588-3602, https://doi.org/10.1016/j.jmb.2016.06.007.
Goldman, D. H., Kaiser, C. M., Milin, A., Righini, M., Tinoco, I., et al. (2015) Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo, Science, 348, 457-460, https://doi.org/10.1126/science.1261909.
Kaiser, C. M., Goldman, D. H., Chodera, J. D., Tinoco, I., and Bustamante, C. (2011) The ribosome modulates nascent protein folding, Science, 334, 1723-1727, https://doi.org/10.1126/science.1209740.
Liu, K., Maciuba, K., and Kaiser, C. M. (2019) The ribosome cooperates with a chaperone to guide multi-domain protein folding, Mol. Cell, 74, 310-319.e7, https://doi.org/10.1016/j.molcel.2019.01.043.
Tian, P., Steward, A., Kudva, R., Su, T., Shilling, P. J., Nickson, A. A., et al. (2018) Folding pathway of an Ig domain is conserved on and off the ribosome, Proc. Natl. Acad. Sci. USA, 115, E11284-E11293, https://doi.org/10.1073/pnas.1810523115.
Bhushan, S., Gartmann, M., Halic, M., Armache, J.-P., Jarasch, A., et al. (2010) alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel, Nat. Struct. Mol. Biol., 17, 313-317, https://doi.org/10.1038/nsmb.1756.
Farías-Rico, J. A., Selin, F. R., Myronidi, I., Frühauf, M., and von Heijne, G. (2018) Effects of protein size, thermodynamic stability, and net charge on cotranslational folding on the ribosome, Proc. Natl. Acad. Sci. USA, 115, E9280-E9287, https://doi.org/10.1073/pnas.1812756115.
Kudva, R., Tian, P., Pardo-Avila, F., Carroni, M., Best, R. B., et al. (2018) The shape of the bacterial ribosome exit tunnel affects cotranslational protein folding, ELife, 7, e36326, https://doi.org/10.7554/eLife.36326.
Zhou, H. X., and Dill, K. A. (2001) Stabilization of proteins in confined spaces, Biochemistry, 40, 11289-11293, https://doi.org/10.1021/bi0155504.
Deckert, A., Waudby, C. A., Wlodarski, T., Wentink, A. S., Wang, X., et al. (2016) Structural characterization of the interaction of α-synuclein nascent chains with the ribosomal surface and trigger factor, Proc. Natl. Acad. Sci. USA, 113, 5012-5017, https://doi.org/10.1073/pnas.1519124113.
Voss, N. R., Gerstein, M., Steitz, T. A., and Moore, P. B. (2006) The geometry of the ribosomal polypeptide exit tunnel, J. Mol. Biol., 360, 893-906, https://doi.org/10.1016/j.jmb.2006.05.023.
Kudlicki, W., Coffman, A., Kramer, G., and Hardesty, B. (1997) Ribosomes and ribosomal RNA as chaperones for folding of proteins, Folding Design, 2, 101-108, https://doi.org/10.1016/S1359-0278(97)00014-X.
Kramer, G., Ramachandiran, V., and Hardesty, B. (2001) Cotranslational folding – omnia mea mecum porto? Int. J. Biochem. Cell Biol., 33, 541-553, https://doi.org/10.1016/S1357-2725(01)00044-9.
Kramer, G., Kudlicki, W., McCarthy, D., Tsalkova, T., Simmons, D., et al. (1999) N-terminal and C-terminal modifications affect folding, release from the ribosomes and stability of in vitro synthesized proteins, Int. J. Biochem. Cell Biol., 31, 231-241, https://doi.org/10.1016/S1357-2725(98)00143-5.
Kudlicki, W., Odom, O. W., Merrill, G., Kramer, G., and Hardesty, B. (1995) Inhibition of the release factor-dependent termination reaction on ribosomes by DnaJ and the N-terminal peptide of rhodanese, J. Bacteriol., 177, 5517-5522, https://doi.org/10.1128/jb.177.19.5517-5522.1995.
Kudlicki, W., Coffman, A., Kramer, G., and Hardesty, B. (1997) Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing, J. Biol. Chem., 272, 32206-32210, https://doi.org/10.1074/jbc.272.51.32206.
Roder, H., and Colón, W. (1997) Kinetic role of early intermediates in protein folding, Curr. Opin. Struct. Biol., 7, 15-28, https://doi.org/10.1016/s0959-440x(97)80004-8.
Ptitsyn, O. B. (1995) Molten globule and protein folding, Adv. Prot. Chem., 47, 83-229, https://doi.org/10.1016/s0065-3233(08)60546-x.
Fedorov, A. N., and Baldwin, T. O. (1997) Cotranslational protein folding, J. Biol. Chem., 272, 32715-32718.
De Prat Gay, G., Ruiz-Sanz, J., Neira, J. L., Corrales, F. J., Otzen, D. E., et al. (1995) Conformational pathway of the polypeptide chain of chymotrypsin inhibitor-2 growing from its N terminus in vitro. Parallels with the protein folding pathway, J. Mol. Biol., 254, 968-979, https://doi.org/10.1006/jmbi.1995.0669.
De Prat Gay, G., Ruiz-Sanz, J., Neira, J. L., Itzhaki, L. S., and Fersht, A. R. (1995) Folding of a nascent polypeptide chain in vitro: cooperative formation of structure in a protein module, Proc. Natl. Acad. Sci. USA, 92, 3683-3686, https://doi.org/10.1073/pnas.92.9.3683.
Neira, J. L., and Fersht, A. R. (1999) Exploring the folding funnel of a polypeptide chain by biophysical studies on protein fragments, J. Mol. Biol., 285, 1309-1333, https://doi.org/10.1006/jmbi.1998.2249.
Hamlin, J., and Zabin, I. (1972) Galactosidase: immunological activity of ribosome-bound, growing polypeptide chains, Proc. Natl. Acad. Sci. USA, 69, 412-416, https://doi.org/10.1073/pnas.69.2.412.
Steinbacher, S., Miller, S., Baxa, U., Budisa, N., Weintraub, A., et al. (1997) Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage, J. Mol. Biol., 267, 865-880, https://doi.org/10.1006/jmbi.1997.0922.
Friguet, B., Djavadi-Ohaniance, L., King, J., and Goldberg, M. E. (1994) In vitro and ribosome-bound folding intermediates of P22 tailspike protein detected with monoclonal antibodies, J. Biol. Chem., 269, 15945-15949.
Tokatlidis, K., Friguet, B., Deville-Bonne, D., Baleux, F., Fedorov, A. N., et al. (1995) Nascent chains: folding and chaperone interaction during elongation on ribosomes, Philos. Trans. R. Soc. B Biol. Sci., 348, 89-95, https://doi.org/10.1098/rstb.1995.0049.
Clark, P. L., and King, J. (2001) A newly synthesized, ribosome-bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates, J. Biol. Chem., 276, 25411-25420, https://doi.org/10.1074/jbc.M008490200.
Jain, M., Evans, M. S., King, J., and Clark, P. L. (2005) Monoclonal antibody epitope mapping describes tailspike beta-helix folding and aggregation intermediates, J. Biol. Chem., 280, 23032-23040, https://doi.org/10.1074/jbc.M501963200.
Evans, M. S., Clarke, T. F., and Clark, P. L. (2005) Conformations of co-translational folding intermediates, Protein Pept. Lett., 12, 189-195, https://doi.org/10.2174/0929866053005908.
Clarke, T. F., and Clark, P. L. (2008) Rare codons cluster, PLoS One, 3, e3412, https://doi.org/10.1371/journal.pone.0003412.
Evans, M. S., Sander, I. M., and Clark, P. L. (2008) Cotranslational folding promotes beta-helix formation and avoids aggregation in vivo, J. Mol. Biol., 383, 683-692, https://doi.org/10.1016/j.jmb.2008.07.035.
Fedorov, A. N., Friguet, B., Djavadi-Ohaniance, L., Alakhov, Y. B., and Goldberg, M. E. (1992) Folding on the ribosome of Escherichia coli tryptophan synthase beta subunit nascent chains probed with a conformation-dependent monoclonal antibody, J. Mol. Biol., 228, 351-358, https://doi.org/10.1016/0022-2836(92)90825-5.
Friguet, B., Fedorov, A., Serganov, A. A., Navon, A., and Goldberg, M. E. (1993) A radioimmunoassay-based method for measuring the true affinity of a monoclonal antibody with trace amounts of radioactive antigen: illustration with the products of a cell-free protein synthesis system, Anal. Biochem., 210, 344-350, https://doi.org/10.1006/abio.1993.1206.
Bergman, L. W., and Kuehl, W. M. (1979) Formation of intermolecular disulfide bonds on nascent immunoglobulin polypeptides, J. Biol. Chem., 254, 5690-5694.
Land, A., Zonneveld, D., and Braakman, I. (2003) Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage, FASEB J., 17, 1058-1067, https://doi.org/10.1096/fj.02-0811com.
Maggioni, M. C., Liscaljet, I. M., and Braakman, I. (2005) A critical step in the folding of influenza virus HA determined with a novel folding assay, Nat. Struct. Mol. Biol., 12, 258-263, https://doi.org/10.1038/nsmb897.
Kim, J., Klein, P. G., and Mullet, J. E. (1991) Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1, J. Biol. Chem., 266, 14931-14938.
Mullet, J. E., Klein, P. G., and Klein, R. R. (1990) Chlorophyll regulates accumulation of the plastid-encoded chlorophyll apoproteins CP43 and D1 by increasing apoprotein stability, Proc. Natl. Acad. Sci. USA, 87, 4038-4042, https://doi.org/10.1073/pnas.87.11.4038.
Chen, W., Helenius, J., Braakman, I., and Helenius, A. (1995) Cotranslational folding and calnexin binding during glycoprotein synthesis, Proc. Natl. Acad. Sci. USA, 92, 6229-6233, https://doi.org/10.1073/pnas.92.14.6229.
Komar, A. A., Kommer, A., Krasheninnikov, I. A., and Spirin, A. S. (1997) Cotranslational folding of globin, J. Biol. Chem., 272, 10646-10651, https://doi.org/10.1074/jbc.272.16.10646.
Komar, A. A., Kommer, A., Krasheninnikov, I. A., and Spirin, A. S. (1993) Cotranslational heme binding to nascent globin chains, FEBS Lett., 326, 261-263, https://doi.org/10.1016/0014-5793(93)81803-8.
Khushoo, A., Yang, Z., Johnson, A. E., and Skach, W. R. (2011) Ligand-driven vectorial folding of ribosome-bound human CFTR NBD1, Mol. Cell, 41, 682-692, https://doi.org/10.1016/j.molcel.2011.02.027.
Peters, T., and Davidson, L. K. (1982) The biosynthesis of rat serum albumin. In vivo studies on the formation of the disulfide bonds, J. Biol. Chem., 257, 8847-8853.
Kudlicki, W., Chirgwin, J., Kramer, G., and Hardesty, B. (1995) Folding of an enzyme into an active conformation while bound as peptidyl-tRNA to the ribosome, Biochemistry, 34, 14284-14287, https://doi.org/10.1021/bi00044a003.
Makeyev, E. V., Kolb, V. A., and Spirin, A. S. (1996) Enzymatic activity of the ribosome-bound nascent polypeptide, FEBS Lett., 378, 166-170, https://doi.org/10.1016/0014-5793(95)01438-1.
Nicola, A. V., Chen, W., and Helenius, A. (1999) Co-translational folding of an alphavirus capsid protein in the cytosol of living cells, Nat. Cell Biol., 1, 341-345, https://doi.org/10.1038/14032.
Sánchez, I. E., Morillas, M., Zobeley, E., Kiefhaber, T., and Glockshuber, R. (2004) Fast folding of the two-domain semliki forest virus capsid protein explains co-translational proteolytic activity, J. Mol. Biol., 338, 159-167, https://doi.org/10.1016/j.jmb.2004.02.037.
Lin, L., DeMartino, G. N., and Greene, W. C. (1998) Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome, Cell, 92, 819-828, https://doi.org/10.1016/s0092-8674(00)81409-9.
Lin, L., DeMartino, G. N., and Greene, W. C. (2000) Cotranslational dimerization of the Rel homology domain of NF-kappaB1 generates p50-p105 heterodimers and is required for effective p50 production, EMBO J., 19, 4712-4722, https://doi.org/10.1093/emboj/19.17.4712.
Sakahira, H., and Nagata, S. (2002) Co-translational folding of caspase-activated DNase with Hsp70, Hsp40, and inhibitor of caspase-activated DNase, J. Biol. Chem., 277, 3364-3370, https://doi.org/10.1074/jbc.M110071200.
Gilmore, R., Coffey, M. C., Leone, G., McLure, K., and Lee, P. W. (1996) Co-translational trimerization of the reovirus cell attachment protein, EMBO J., 15, 2651-2658.
Leone, G., Coffey, M. C., Gilmore, R., Duncan, R., Maybaum, L., et al. (1996) C-terminal trimerization, but not N-terminal trimerization, of the reovirus cell attachment protein is a posttranslational and Hsp70/ATP-dependent process, J. Biol. Chem., 271, 8466-8471, https://doi.org/10.1074/jbc.271.14.8466.
Kleizen, B., van Vlijmen, T., de Jonge, H. R., and Braakman, I. (2005) Folding of CFTR is predominantly cotranslational, Mol. Cell, 20, 277-287, https://doi.org/10.1016/j.molcel.2005.09.007.
Matthews, B. W. (2005) The structure of E. coli beta-galactosidase, C. R. Biol., 328, 549-556, https://doi.org/10.1016/j.crvi.2005.03.006.
Jacobson, R. H., Zhang, X. J., DuBose, R. F., and Matthews, B. W. (1994) Three-dimensional structure of beta-galactosidase from E. coli, Nature, 369, 761-766, https://doi.org/10.1038/369761a0.
Langley, K. E., Villarejo, M. R., Fowler, A. V., Zamenhof, P. J., and Zabin, I. (1975) Molecular basis of beta-galactosidase alpha-complementation, Proc. Natl. Acad. Sci. USA, 72, 1254-1257, https://doi.org/10.1073/pnas.72.4.1254.
Zeilstra-Ryalls, J. H., and Somerville, R. L. (1992) Protein-protein interaction in the alpha-complementation system of beta-galactosidase, Curr. Top. Cell. Reg., 33, 81-104, https://doi.org/10.1016/b978-0-12-152833-1.50011-3.
Klappa, P., Freedman, R. B., and Zimmermann, R. (1995) Protein disulphide isomerase and a lumenal cyclophilin-type peptidyl prolyl cis-trans isomerase are in transient contact with secretory proteins during late stages of translocation, Eur. J. Biochem., 232, 755-764.
Roth, R. A., and Pierce, S. B. (1987) In vivo cross-linking of protein disulfide isomerase to immunoglobulins, Biochemistry, 26, 4179-4182, https://doi.org/10.1021/bi00388a001.
Bulleid, N. J., and Freedman, R. B. (1988) Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes, Nature, 335, 649-651, https://doi.org/10.1038/335649a0.
Hesterkamp, T., Hauser, S., Lütcke, H., and Bukau, B. (1996) Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains, Proc. Natl. Acad. Sci. USA, 93, 4437-4441.
Valent, Q. A., Kendall, D. A., High, S., Kusters, R., Oudega, B., et al. (1995) Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides, EMBO J., 14, 5494-5505.
Komar, A. A., and Jaenicke, R. (1995) Kinetics of translation of gamma B crystallin and its circularly permutated variant in an in vitro cell-free system: possible relations to codon distribution and protein folding, FEBS Lett., 376, 195-198, https://doi.org/10.1016/0014-5793(95)01275-0.
Komar, A. A. (2009) A pause for thought along the co-translational folding pathway, Trends Biochem. Sci., 34, 16-24, https://doi.org/10.1016/j.tibs.2008.10.002.
Purvis, I. J., Bettany, A. J. E., Santiago, T. C., Coggins, J. R., Duncan, K., et al. (1987) The efficiency of folding of some proteins is increased by controlled rates of translation in vivo: A hypothesis, J. Mol. Biol., 193, 413-417, https://doi.org/10.1016/0022-2836(87)90230-0.
Thanaraj, T. A., and Argos, P. (1996) Protein secondary structural types are differentially coded on messenger RNA, Protein Sci., 5, 1973-1983, https://doi.org/10.1002/pro.5560051003.
Adzhubei, A. A., Adzhubei, I. A., Krasheninnikov, I. A., and Neidle, S. (1996) Non-random usage of ‘degenerate’ codons is related to protein three-dimensional structure, FEBS Lett., 399, 78-82, https://doi.org/10.1016/s0014-5793(96)01287-2.
Mukhopadhyay, P., Basak, S., and Ghosh, T. C. (2007) Synonymous codon usage in different protein secondary structural classes of human genes: implication for increased non-randomness of GC3 rich genes towards protein stability, J. Biosci., 32, 947-963, https://doi.org/10.1007/s12038-007-0095-z.
Oresic, M., and Shalloway, D. (1998) Specific correlations between relative synonymous codon usage and protein secondary structure, J. Mol. Biol., 281, 31-48, https://doi.org/10.1006/jmbi.1998.1921.
Xie, T., Ding, D., Tao, X., and Dafu, D. (1998) The relationship between synonymous codon usage and protein structure, FEBS Lett., 434, 93-96, https://doi.org/10.1016/s0014-5793(98)00955-7.
Gu, W., Zhou, T., Ma, J., Sun, X., and Lu, Z. (2003) Folding type specific secondary structure propensities of synonymous codons, IEEE Trans. NanoBiosci., 2, 150-157, https://doi.org/10.1109/TNB.2003.817024.
Thanaraj, T. A., and Argos, P. (1996) Ribosome-mediated translational pause and protein domain organization, Protein Sci., 5, 1594-1612, https://doi.org/10.1002/pro.5560050814.
Crombie, T., Swaffield, J. C., and Brown, A. J. (1992) Protein folding within the cell is influenced by controlled rates of polypeptide elongation, J. Mol. Biol., 228, 7-12, https://doi.org/10.1016/0022-2836(92)90486-4.
Crombie, T., Boyle, J. P., Coggins, J. R., and Brown, A. J. (1994) The folding of the bifunctional TRP3 protein in yeast is influenced by a translational pause which lies in a region of structural divergence with Escherichia coli indoleglycerol-phosphate synthase, Eur. J. Biochem., 226, 657-664, https://doi.org/10.1111/j.1432-1033.1994.tb20093.x.
Komar, A. A., Lesnik, T., and Reiss, C. (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation, FEBS Lett., 462, 387-391, https://doi.org/10.1016/s0014-5793(99)01566-5.
Cortazzo, P., Cerveñansky, C., Marín, M., Reiss, C., Ehrlich, R., et al. (2002) Silent mutations affect in vivo protein folding in Escherichia coli, Biochem. Biophys. Res. Commun., 293, 537-541, https://doi.org/10.1016/S0006-291X(02)00226-7.
Sharma, A. K., and O’Brien, E. P. (2018) Non-equilibrium coupling of protein structure and function to translation-elongation kinetics, Curr. Opin. Struct. Biol., 49, 94-103, https://doi.org/10.1016/j.sbi.2018.01.005.
Marin, M. (2008) Folding at the rhythm of the rare codon beat, Biotechnol. J., 3, 1047-1057, https://doi.org/10.1002/biot.200800089.
Angov, E., Hillier, C. J., Kincaid, R. L., and Lyon, J. A. (2008) Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host, PLoS One, 3, e2189, https://doi.org/10.1371/journal.pone.0002189.
Hatfield, G. W., and Roth, D. A. (2007) Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and translation engineering, Biotechnol. Ann. Rev., 13, 27-42, https://doi.org/10.1016/S1387-2656(07)13002-7.
Bergman, L. W., and Kuehl, W. M. (1979) Formation of an intrachain disulfide bond on nascent immunoglobulin light chains, J. Biol. Chem., 254, 8869-8876.
Bruckner, P., Eikenberry, E. F., and Prockop, D. J. (1981) Formation of the triple helix of type I procollagen in cellulo. A kinetic model based on cis-trans isomerization of peptide bonds, Eur. J. Biochem., 118, 607-613, https://doi.org/10.1111/j.1432-1033.1981.tb05562.x.
Berestovskaya, N. G., Shaloiko, L. A., Gorokhovatsky, A. Y., Bondar, V. S., Vysotski, E. S., et al. (1999) Cotranslational formation of active photoprotein obelin in a cell-free translation system: direct ultrahigh sensitive measure of the translation course, Anal. Biochem., 268, 72-78, https://doi.org/10.1006/abio.1998.3051.
Kolb, V. A., Makeyev, E. V., and Spirin, A. S. (1994) Folding of firefly luciferase during translation in a cell-free system, EMBO J., 13, 3631-3637.
Alexandrov, N. (1993) Structural argument for N-terminal initiation of protein folding, Protein Sci., 2, 1989-1991, https://doi.org/10.1002/pro.5560021121.
Contreras Martínez, L. M., Martínez-Veracoechea, F. J., Pohkarel, P., Stroock, A. D., Escobedo, F. A., et al. (2006) Protein translocation through a tunnel induces changes in folding kinetics: a lattice model study, Biotechnol. Bioeng., 94, 105-117, https://doi.org/10.1002/bit.20832.
Huard, F. P. E., Deane, C. M., and Wood, G. R. (2006) Modelling sequential protein folding under kinetic control, Bioinformatics, 22, e203-e210, https://doi.org/10.1093/bioinformatics/btl248.
Deane, C. M., Dong, M., Huard, F. P. E., Lance, B. K., and Wood, G. R. (2007) Cotranslational protein folding – fact or fiction? Bioinformatics, 23, i142-i148, https://doi.org/10.1093/bioinformatics/btm175.
Taylor, W. R. (2006) Topological accessibility shows a distinct asymmetry in the folds of beta-alpha proteins, FEBS Lett., 580, 5263-5267, https://doi.org/10.1016/j.febslet.2006.08.070.
Lu, H.-M., and Liang, J. (2008) A model study of protein nascent chain and cotranslational folding using hydrophobic-polar residues, Proteins, 70, 442-449, https://doi.org/10.1002/prot.21575.
Fedorov, A. N., and Baldwin, T. O. (1995) Contribution of cotranslational folding to the rate of formation of native protein structure, Proc. Natl. Acad. Sci. USA, 92, 1227-1231, https://doi.org/10.1073/pnas.92.4.1227.
Fedorov, A. N., and Baldwin, T. O. (1999) Process of biosynthetic protein folding determines the rapid formation of native structure, J. Mol. Biol., 294, 579-586, https://doi.org/10.1006/jmbi.1999.3281.
Shieh, Y.-W., Minguez, P., Bork, P., Auburger, J. J., Guilbride, D. L., et al. (2015) Operon structure and cotranslational subunit association direct protein assembly in bacteria, Science, 350, 678-680, https://doi.org/10.1126/science.aac8171.
Kramer, G., Patzelt, H., Rauch, T., Kurz, T. A., Vorderwülbecke, S., et al. (2004) Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli, J. Biol. Chem., 279, 14165-14170, https://doi.org/10.1074/jbc.M313635200.
Baram, D., Pyetan, E., Sittner, A., Auerbach-Nevo, T., Bashan, A., et al. (2005) Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action, Proc. Natl. Acad. Sci. USA, 102, 12017-12022, https://doi.org/10.1073/pnas.0505581102.
Ferbitz, L., Maier, T., Patzelt, H., Bukau, B., Deuerling, E., et al. (2004) Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins, Nature, 431, 590-596, https://doi.org/10.1038/nature02899.
Morgado, L., Burmann, B. M., Sharpe, T., Mazur, A., and Hiller, S. (2017) The dynamic dimer structure of the chaperone Trigger Factor, Nat. Commun., 8, 1992, https://doi.org/10.1038/s41467-017-02196-7.
Saio, T., Kawagoe, S., Ishimori, K., and Kalodimos, C. G. (2018) Oligomerization of a molecular chaperone modulates its activity, ELife, 7, e35731, https://doi.org/10.7554/eLife.35731.
Kaiser, C. M., Chang, H.-C., Agashe, V. R., Lakshmipathy, S. K., Etchells, S. A., et al. (2006) Real-time observation of trigger factor function on translating ribosomes, Nature, 444, 455-460, https://doi.org/10.1038/nature05225.
Saio, T., Guan, X., Rossi, P., Economou, A., and Kalodimos, C. G. (2014) Structural basis for protein antiaggregation activity of the trigger factor chaperone, Science, 344, 1250494, https://doi.org/10.1126/science.1250494.
Beckmann, R. P., Mizzen, L. E., and Welch, W. J. (1990) Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly, Science, 248, 850-854, https://doi.org/10.1126/science.2188360.
Gaitanaris, G. A., Vysokanov, A., Hung, S. C., Gottesman, M. E., and Gragerov, A. (1994) Successive action of Escherichia coli chaperones in vivo, Mol. Microbiol., 14, 861-869, https://doi.org/10.1111/j.1365-2958.1994.tb01322.x.
Nelson, R. J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M., and Craig, E. A. (1992) The translation machinery and 70 kDa heat shock protein cooperate in protein synthesis, Cell, 71, 97-105, https://doi.org/10.1016/0092-8674(92)90269-i.
Kudlicki, W., Odom, O. W., Kramer, G., and Hardesty, B. (1994) Chaperone-dependent folding and activation of ribosome-bound nascent rhodanese. Analysis by fluorescence, J. Mol. Biol., 244, 319-331, https://doi.org/10.1006/jmbi.1994.1732.
Svetlov, M. S., Kommer, A., Kolb, V. A., and Spirin, A. S. (2006) Effective cotranslational folding of firefly luciferase without chaperones of the Hsp70 family, Protein Sci., 15, 242-247, https://doi.org/10.1110/ps.051752506.
Frydman, J., Nimmesgern, E., Ohtsuka, K., and Hartl, F. U. (1994) Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones, Nature, 370, 111-117, https://doi.org/10.1038/370111a0.
Frydman, J., and Hartl, F. U. (1996) Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms, Science, 272, 1497-1502, https://doi.org/10.1126/science.272.5267.1497.
Lorimer, G. H. (1996) A quantitative assessment of the role of the chaperonin proteins in protein folding in vivo, FASEB J., 10, 5-9, https://doi.org/10.1096/fasebj.10.1.8566548.
Reid, B. G., and Flynn, G. C. (1996) GroEL binds to and unfolds rhodanese posttranslationally, J. Biol. Chem., 271, 7212-7217, https://doi.org/10.1074/jbc.271.12.7212.
Schatz, G. (1996) The protein import system of mitochondria, J. Biol. Chem., 271, 31763-31766, https://doi.org/10.1074/jbc.271.50.31763.
Ying, B.-W., Taguchi, H., and Ueda, T. (2006) Co-translational binding of GroEL to nascent polypeptides is followed by post-translational encapsulation by GroES to mediate protein folding, J. Biol. Chem., 281, 21813-21819, https://doi.org/10.1074/jbc.M603091200.
Ying, B.-W., Taguchi, H., Kondo, M., and Ueda, T. (2005) Co-translational involvement of the chaperonin GroEL in the folding of newly translated polypeptides, J. Biol. Chem., 280, 12035-12040, https://doi.org/10.1074/jbc.M500364200.
Shimizu, Y., Kuruma, Y., Ying, B.-W., Umekage, S., and Ueda, T. (2006) Cell-free translation systems for protein engineering, FEBS J., 273, 4133-4140, https://doi.org/10.1111/j.1742-4658.2006.05431.x.
Baker, D., and Agard, D. A. (1994) Kinetics versus thermodynamics in protein folding, Biochemistry, 33, 7505-7509, https://doi.org/10.1021/bi00190a002.
Eder, J., Rheinnecker, M., and Fersht, A. R. (1993) Folding of subtilisin BPN’: characterization of a folding intermediate, Biochemistry, 32, 18-26, https://doi.org/10.1021/bi00052a004.
Shinde, U. P., Liu, J. J., and Inouye, M. (1997) Protein memory through altered folding mediated by intramolecular chaperones, Nature, 389, 520-522, https://doi.org/10.1038/39097.
Weissman, J. S., and Kim, P. S. (1992) The pro region of BPTI facilitates folding, Cell, 71, 841-851, https://doi.org/10.1016/0092-8674(92)90559-u.
Dill, K. A., Bromberg, S., Yue, K., Fiebig, K. M., Yee, D. P., et al. (1995) Principles of protein folding--a perspective from simple exact models, Protein Sci., 4, 561-602.
Onuchic, J. N., Wolynes, P. G., Luthey-Schulten, Z., and Socci, N. D. (1995) Toward an outline of the topography of a realistic protein-folding funnel, Proc. Natl. Acad. Sci. USA, 92, 3626-3630, https://doi.org/10.1073/pnas.92.8.3626.
Waudby, C. A., Dobson, C. M., and Christodoulou, J. (2019) Nature and regulation of protein folding on the ribosome, Trends Biochem. Sci., 44, 914-926, https://doi.org/10.1016/j.tibs.2019.06.008.
Fiaux, J., Horst, J., Scior, A., Preissler, S., Koplin, A., et al. (2010) Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction, J. Biol. Chem., 285, 3227-3234, https://doi.org/10.1074/jbc.M109.075804.
Fedorov, A. N., and Baldwin, T. O. (1997) GroE modulates kinetic partitioning of folding intermediates between alternative states to maximize the yield of biologically active protein, J. Mol. Biol., 268, 712-723, https://doi.org/10.1006/jmbi.1997.1007.
Acknowledgments
The author dedicates this review to his mentors – Lev P. Ovchinnikov and Alexander S. Spirin.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares no conflicts of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by the author.
Additional information
Translated from Uspekhi Biologicheskoi Khimii, 2022, Vol. 62, pp. 279-318.
Rights and permissions
About this article
Cite this article
Fedorov, A.N. Biosynthetic Protein Folding and Molecular Chaperons. Biochemistry Moscow 87 (Suppl 1), S128–S145 (2022). https://doi.org/10.1134/S0006297922140115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297922140115