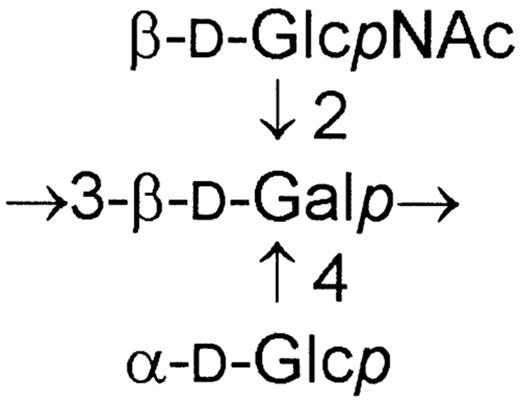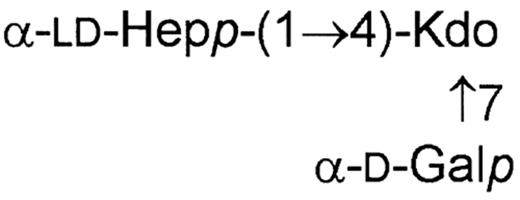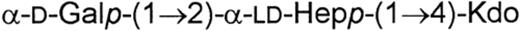-
PDF
- Split View
-
Views
-
Cite
Cite
Elżbieta Romanowska, Immunochemical aspects of Hafnia alvei O antigens, FEMS Immunology & Medical Microbiology, Volume 27, Issue 3, March 2000, Pages 219–225, https://doi.org/10.1111/j.1574-695X.2000.tb01433.x
Close - Share Icon Share
Abstract
In the article the composition and structure of the O-specific polysaccharide chains and core region of the O antigens isolated from over 20 H. alvei strains is overviewed. Moreover, the correlation between the structure and immunospecificity of the O antigens is presented and discussed.
1 Introduction
Hafnia forms a separate genus in the Enterobacteriaceae family. Although there are two genospecies within Hafnia[1], only a single species, Hafnia alvei, has been designated [2]. H. alvei occurs in natural environments such as soil, sewage and water, and in animals and birds. This organism seems to be an opportunistic pathogen since it is identified in hospital specimens (stool, sputum and urine).
The serology of cultures that were biochemically defined as members of the Hafnia genus was studied by Sakazaki [3] and Matsumoto [4]. Twenty-nine O groups and 23 H antigens were designated in the genus. However, when some test strains of this scheme were reexamined based on the current classification of the Enterobacteriaceae family, they proved to be no H. alvei. A new serotyping scheme of H. alvei including 39 O groups and 36 H antigens was announced by Baturo and Raginskaya [5].
Sakazaki [3], Matsumoto [6], Eveland and Faber [7] and Sedlak and Slajsova [8] demonstrated interrelationships between O antigens of H. alvei and those of certain Enterobacter cloacae, Escherichia coli, Citrobacter freundii and Salmonella.
O antigens (lipopolysaccharides), the cell wall components of Gram-negative bacteria, are responsible for their immunospecificity.
Chemical studies on lipopolysaccharides (LPSs) isolated from 33 strains of H. alvei started in 1988. SDS-PAGE analysis of the LPSs and their sugar composition were presented in [9]. In these preliminary studies it was found that all H. alvei LPSs contained glucose, glucosamine, heptose and 3-deoxy-octulosonic acid. Some of the tested LPSs also contained galactose, galactosamine, mannose, mannosamine, 6-deoxyhexose, sialic acid and unknown amino sugars.
2 Composition and structure of H. alvei O-specific polysaccharides
To obtain O-specific polysaccharide and core oligosaccharide in free form, the LPS was hydrolyzed with 1% acetic acid (100°C, 30–90 min) releasing the water-soluble carbohydrate portion that was then separated on BioGel P2 or Sephadex G50 column into O-specific polysaccharide and core oligosaccharide fractions. The fractions were then lyophilized.
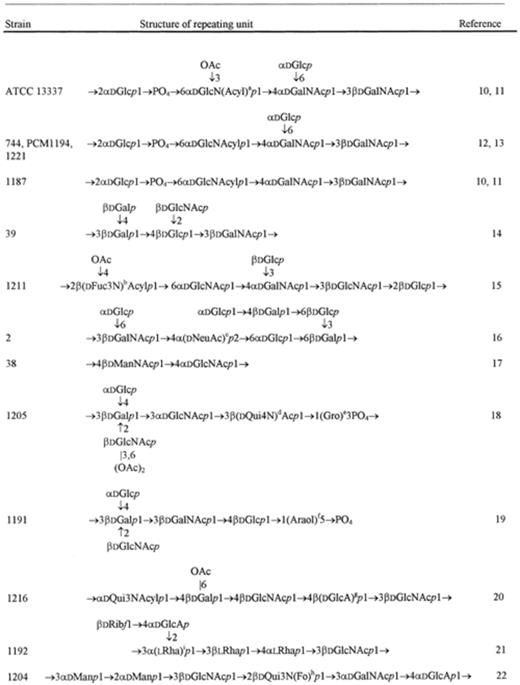
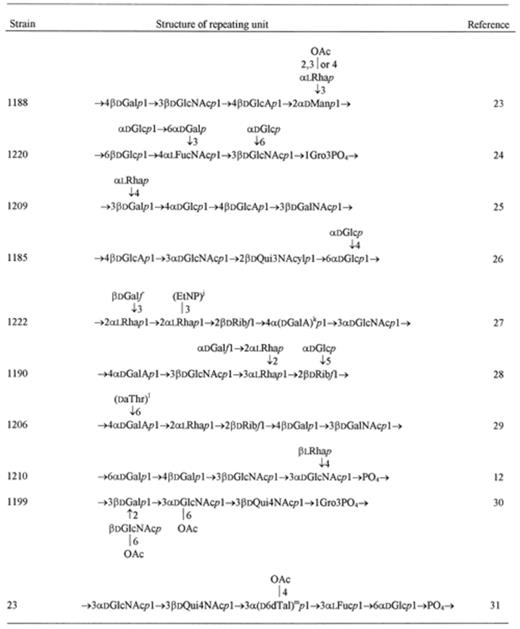
The characteristic feature of H. alvei O-specific polysaccharides is an unusual richness of sugar components forming their repeating units as well as a variety of nonsugar side groups (Table 1[10–31]). d-Glucose, d-galactose, d-glucosamine and d-galactosamine occur commonly in these polysaccharides. 6-Deoxy-sugars like l-fucose, l-rhamnose, N-acyl-d-fucosamine and N-acetyl-d-quinovosamine are often present in the examined polysaccharides. It is interesting to note that fucosamine as well as quinovosamine may have amino groups situated at position 2, 3 or 4 in alvei polysaccharides. Amino groups of hexosamines and of 6-deoxy-hexosamines are usually acetylated. However, in some cases they are acylated by d-3-hydroxybutyryl groups or in one case by a formyl group (H. alvei 1204 strain, [22]).
Uronic acids are also constituents of H. alvei polysaccharides: d-glucuronic acid occurs more often than d-galacturonic acid. d-Ribose was identified in the polysaccharides of four H. alvei strains [21,27–29]. Sialic (N-acetyl-neuraminic) acid was found in one strain (H. alvei 2 [16]). Moreover, a rare sugar, 6-deoxy-d-talose, was recognized as a constituent of H. alvei strain 23 [31].
O-Acetyl groups appear as the side groups in many O-specific polysaccharides. The microheterogeneity often observed in H. alvei polysaccharides is due to the incomplete substitution by these groups. As a nonsugar side group 2-aminoethyl phosphate was identified in the polysaccharide of strain PCM 1222 [27]. Amino acid allo-threonine amide-linked to d-galacturonic acid is present in the polysaccharide of strain PCM 1206 [29].
Based on sugar and methylation analyses, periodate oxidation, gas chromatography/mass spectrometry and NMR spectroscopy, the structures of the O-specific polysaccharides of H. alvei were established (Table 1). As seen in Table 1, the polysaccharides composed of di- to octasaccharide repeating units form O-specific chains of the lipopolysaccharides.
Among 24 H. alvei O-specific polysaccharides examined, 11 have a teichoic acid-like character (see Table 1). These polysaccharides contain phosphodiester linkages in their O-specific chains. Generally, glycerol participates in these linkages, except for strain 1191 where l-arabitol occurs instead.
Sometimes phosphodiester bonds are formed between sugar components of the polysaccharide as in the standard strain ATCC 13337 and in some other H. alvei strains of close structure of their repeating units (strains 1187, 744 and PCM 1194).
The polysaccharide of strain 1222 has no teichoic acid-like structure, but it contains 2-aminoethyl phosphate as a side group and such rare components as d-galactofuranose and d-ribofuranose [27].
3 Immunospecificity of H. alvei lipopolysaccharides
LPSs of over 30 strains of H. alvei were tested serologically using homo- and heterologous antisera. For the evaluation of the cross-reactivity of H. alvei LPSs the passive hemagglutination test, rocket immunoelectrophoresis and immunoblotting were employed [13,32]. It is clearly visible from the results presented in Table 2 that the LPSs of strains ATCC 13337, 1221, 1187, 744 and PCM 1194 cross-reacted distinctly. As seen from Table 1, the common feature of these five strains is that the base chains of their O-specific polysaccharides are identical. Additional α-glucosyl and O-acetyl side groups occur in the standard strain ATCC 13337, but there are only α-glucosyl side residues in strains 1221, 744 and PCM 1194. Although LPS of strain 1187 has the base chain only, it reacted strongly with anti-ATCC 13337 and anti-1221 sera. On the other hand anti-1187 serum reacted with LPSs of strains ATCC 13337, 1221, 744 and PCM 1194. These facts proved evidence that α-glucosyl and O-acetyl side groups are not essential elements of epitopes of the LPSs mentioned above.
Passive hemagglutination (reciprocal titer) of H. alvei LPSs with homologous and heterologous antisera
| Anti-H. alvei serum | Lipopolysaccharide | ||||||||||||||||
| ATCC 13337 | 1221 | 744 | PCM1194 | 1187 | 39 | 1211 | 2 | 1205 | 1191 | 1216 | 1192 | 1188 | 1220 | 1185 | 1190 | 1199 | |
| Anti-ATCC 13337 | 10 240 | 81 900 | – | – | 20480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1221 | 5 120 | 5 120 | – | – | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1187 | 10 240 | 5 120 | 5 120 | 5 120 | 20 480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-39 | 0 | 0 | – | – | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1211 | 0 | 0 | – | – | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-2 | 0 | 0 | – | – | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1205 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
| Anti-1191 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1216 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 81 900 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1192 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 |
| Anti-1188 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 |
| Anti-1220 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 240 | 0 | 0 | 0 |
| Anti-1185 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 |
| Anti-1190 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 |
| Anti-1199 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 2 560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
| Anti-H. alvei serum | Lipopolysaccharide | ||||||||||||||||
| ATCC 13337 | 1221 | 744 | PCM1194 | 1187 | 39 | 1211 | 2 | 1205 | 1191 | 1216 | 1192 | 1188 | 1220 | 1185 | 1190 | 1199 | |
| Anti-ATCC 13337 | 10 240 | 81 900 | – | – | 20480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1221 | 5 120 | 5 120 | – | – | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1187 | 10 240 | 5 120 | 5 120 | 5 120 | 20 480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-39 | 0 | 0 | – | – | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1211 | 0 | 0 | – | – | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-2 | 0 | 0 | – | – | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1205 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
| Anti-1191 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1216 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 81 900 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1192 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 |
| Anti-1188 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 |
| Anti-1220 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 240 | 0 | 0 | 0 |
| Anti-1185 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 |
| Anti-1190 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 |
| Anti-1199 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 2 560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
–, not determined.
Passive hemagglutination (reciprocal titer) of H. alvei LPSs with homologous and heterologous antisera
| Anti-H. alvei serum | Lipopolysaccharide | ||||||||||||||||
| ATCC 13337 | 1221 | 744 | PCM1194 | 1187 | 39 | 1211 | 2 | 1205 | 1191 | 1216 | 1192 | 1188 | 1220 | 1185 | 1190 | 1199 | |
| Anti-ATCC 13337 | 10 240 | 81 900 | – | – | 20480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1221 | 5 120 | 5 120 | – | – | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1187 | 10 240 | 5 120 | 5 120 | 5 120 | 20 480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-39 | 0 | 0 | – | – | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1211 | 0 | 0 | – | – | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-2 | 0 | 0 | – | – | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1205 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
| Anti-1191 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1216 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 81 900 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1192 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 |
| Anti-1188 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 |
| Anti-1220 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 240 | 0 | 0 | 0 |
| Anti-1185 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 |
| Anti-1190 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 |
| Anti-1199 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 2 560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
| Anti-H. alvei serum | Lipopolysaccharide | ||||||||||||||||
| ATCC 13337 | 1221 | 744 | PCM1194 | 1187 | 39 | 1211 | 2 | 1205 | 1191 | 1216 | 1192 | 1188 | 1220 | 1185 | 1190 | 1199 | |
| Anti-ATCC 13337 | 10 240 | 81 900 | – | – | 20480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1221 | 5 120 | 5 120 | – | – | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1187 | 10 240 | 5 120 | 5 120 | 5 120 | 20 480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-39 | 0 | 0 | – | – | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1211 | 0 | 0 | – | – | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-2 | 0 | 0 | – | – | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1205 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
| Anti-1191 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1216 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 81 900 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anti-1192 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 | 0 |
| Anti-1188 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 | 0 | 0 |
| Anti-1220 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 240 | 0 | 0 | 0 |
| Anti-1185 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 | 0 |
| Anti-1190 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 | 0 |
| Anti-1199 | 0 | 0 | – | – | 0 | 0 | 0 | 0 | 2 560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 120 |
–, not determined.
To study the serological relationship between strain ATCC 13337 and strain 1187 and also to characterize their LPS epitopes, a microprecipitin test was carried out [10]. The three preparations of ATCC 13337 polysaccharide (native, de-O-acetylated and re-N-acetylated after the removal of the d-3-hydroxybutyryl group) and two preparations of 1187 polysaccharide (native and re-N-acetylated) were used.
According to the serological results described above the antibodies of anti-ATCC 13337 and anti-1187 sera are directed predominantly to the base chains of both specific polysaccharides. However, d-3-hydroxybutyryl groups occurring in these antigens are also of some immunological importance because their replacement by N-acetyl groups resulted in a serious decrease of the serological activity of both polysaccharides [10].
d-3-Hydroxybutyryl groups are present in H. alvei LPSs of strains ATCC 13337, 1221, 1187, 744 and PCM 1194, but they were also identified in LPSs of strains 1211 and 1216. As seen from the serological results (Table 2) cross-reactivity occurs neither between LPSs 1211 and 1216 nor between these two and the LPSs of strains ATCC 13337, 1221 and 1187.
As explained above [10], d-3-hydroxybutyryl groups present in the LPSs are important for their serological activity, but they are not sufficient to cause cross-reactivity between the LPSs when the structures of their repeating units differ seriously. As seen from Table 1 the structures of the repeating units of LPSs isolated from strains ATCC 13337 and 1187, and from strains 1211 and 1216 have nothing in common except for the presence of d-3-hydroxybutyryl groups. As the consequence no serological relationship is observed.
The O-specific polysaccharides of H. alvei strains 1205 and 1199 have similar structures [30] (Table 1), which differ in the presence of a terminal α-d-glucosyl residue in the 1205 LPS only. The cross-reactivity between these two polysaccharides was proved in a quantitative immunoprecipitation test using anti-H. alvei 1205 and anti-H. alvei 1199 sera. The immunoblotting of the LPSs of these two strains also confirmed their serological affinity.
It is worth mentioning that O-specific polysaccharides of H. alvei strains 1191 and 1205 [18,19] are almost identical in some part of their repeating units:
In the case of strain 1205 the terminal β-d-GlcNAc of the fragment shown above is O-acetylated. However, this structural similarity was not expressed in their serological cross-reactivity as was proved by rocket immunoelectrophoresis and passive hemagglutination. This observation indicated that the epitopes of both O antigens also comprise those parts of their repeating units which differ.
As seen from Table 1 the majority of structures of the repeating units which occur in H. alvei LPSs are different except for the strains ATCC 13337, 1187, 1221, 744, PCM 1194, and strains 1205, 1199 mentioned above.
The results of passive hemagglutination of H. alvei LPSs with homologous and heterologous antisera, shown in Table 2, coincide with the structural data presented above. As can be seen, cross-reactivity between the LPSs and heterologous sera occurs rather rarely. LPSs of ATCC 13337, 1187, 1221, 744, PCM 1194 reacted distinctly with anti-1187 serum. The cross-reactions were also observed between LPSs 1205 and 1199 with their antisera.
From the results of structural and serological studies on the O-specific polysaccharides of H. alvei it can be accepted that there is one group (A) of the LPSs with a common structure of their repeating units: ATCC 13337, 1221, 744, PCM 1194 and 1187, and the second group (B) of the LPSs of strains 1205 and 1199. The remaining LPSs of 17 H. alvei strains are characterized (1) by the different structure of their repeating units and (2) by the lack of cross-reactivity between them.
These two features should allow us to assume that the remaining H. alvei strains form 17 separate serotypes.
4 Structure and serological characterization of H. alvei LPS core region
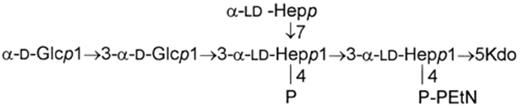
The core oligosaccharides isolated from LPSs of nine H. alvei strains proved to have identical hexasaccharide skeletons according to the results of sugar and methylation analysis with the use of gas chromatography/mass spectrometry and 1H NMR spectroscopy [21,33]. The structure of the H. alvei core is presented below: where Kdo=3-deoxy-oct-2-ulosonic acid; P-PEtN=diphosphorylethanolamine.
The core hexasaccharide is composed of two d-glucoses, three ld-heptoses and one 3-deoxy-oct-2-ulosonic acid. The chain heptose residues of the core hexasaccharide are phosphorylated in two positions: (1) by phosphoryl and (2) by diphosphorylethanolamine groups.
Serological characterization of the H. alvei core oligosaccharides was performed using anti-H. alvei core serum [34]. These antibodies were produced against covalent glycoconjugate containing LPS core of H. alvei standard strain ATCC 13337 and tetanus toxoid. This serum reacted positively with core oligosaccharides isolated from over 30 H. alvei strains. The serum was tested in rocket immunoelectrophoresis, immunoblotting and ELISA. The results indicated that the core region present in standard strain ATCC 13337 is a prevailing structure in the entire Hafnia genus.
However, chemical analysis of core oligosaccharides of H. alvei strains 23, 1222 and 39 has shown that their sugar composition is different from the typical H. alvei core. The core oligosaccharides of strains 23 and 1222 contained three d-galactoses, two d-glucoses and three ld-heptoses. The core oligosaccharide of strain 39 contained two d-galactoses, two d-glucoses, one N-acetyl-d-glucosamine and nearly three ld-heptoses.
On the basis of sugar and methylation analyses, 1H NMR spectra and MALDI-TOF mass spectrometry it was found that the core oligosaccharides of strains 23 and 1222 have the same structure as the Escherichia coli R4 core region, while the core oligosaccharide of strain 39 has the structure of the Salmonella Ra core. The data of serological analysis (passive hemagglutination, ELISA and immunoblotting) in which anti-E. coli R4 and anti-Salmonella Ra conjugate sera were used, confirmed the structural results [35].
5 Core trisaccharides of H. alvei
In the LPSs of some H. alvei strains (2, 1211, 32 and 1192) an additional core fragment, besides the presence of the typical core region, was identified [36,37]. The structure of the trisaccharide was determined to be as follows:
The position of this trisaccharide in the LPS molecule is not clear. It may originate from a non-reducing terminal Kdo III residue substituted at carbons 4 and 7 by ld-heptose and d-galactose units respectively, or may be directly linked to lipid A.
A trisaccharide of the same composition, but of a different structure was found in LPSs of H. alvei strains 1188 and 1196 [38]. The linear oligosaccharide of the core origin is built as follows:
6 Final remark
Although the numerous carbohydrate structures presented above do not cover entirely the lipopolysaccharides of the Hafnia genus, it seems advantageous for all interested in the immunochemistry of O antigens of Enterobacteriaceae to have access to the results obtained hitherto. So far parts of the structural results on the carbohydrate moiety of H. alvei lipopolysaccharides presented above have been reviewed in Polish only [39].
Acknowledgements
I gratefully thank Prof. Czeslaw Lugowski for providing the samples of H. alvei lipopolysaccharides of strains 744 and PCM 1194.
References
Author notes
E-mail: romanows@immuno.iitd.pan.wroc.pl