-
PDF
- Split View
-
Views
-
Cite
Cite
Chris J. Law, Graham J. Slater, Rita S. Mehta, Lineage Diversity and Size Disparity in Musteloidea: Testing Patterns of Adaptive Radiation Using Molecular and Fossil-Based Methods, Systematic Biology, Volume 67, Issue 1, January 2018, Pages 127–144, https://doi.org/10.1093/sysbio/syx047
Close - Share Icon Share
Abstract
Adaptive radiation is hypothesized to be a primary mechanism that drives the remarkable species diversity and morphological disparity across the Tree of Life. Tests for adaptive radiation in extant taxa are traditionally estimated from calibrated molecular phylogenies with little input from extinct taxa. With 85 putative species in 33 genera and over 400 described extinct species, the carnivoran superfamily Musteloidea is a prime candidate to investigate patterns of adaptive radiation using both extant- and fossil-based macroevolutionary methods. The species diversity and equally impressive ecological and phenotypic diversity found across Musteloidea is often attributed to two adaptive radiations coinciding with two major climate events, the Eocene–Oligocene transition and the Mid-Miocene Climate Transition. Here, we compiled a novel time-scaled phylogeny for 88% of extant musteloids and used it as a framework for testing the predictions of adaptive radiation hypotheses with respect to rates of lineage diversification and phenotypic evolution. Contrary to expectations, we found no evidence for rapid bursts of lineage diversification at the origin of Musteloidea, and further analyses of lineage diversification rates using molecular and fossil-based methods did not find associations between rates of lineage diversification and the Eocene–Oligocene transition or Mid-Miocene Climate Transition as previously hypothesized. Rather, we found support for decoupled diversification dynamics driven by increased clade carrying capacity in the branches leading to a subclade of elongate mustelids. Supporting decoupled diversification dynamics between the subclade of elongate mustelids and the ancestral musteloid regime is our finding of increased rates of body length evolution, but not body mass evolution, within the decoupled mustelid subclade. The lack of correspondence in rates of body mass and length evolution suggest that phenotypic evolutionary rates under a single morphological metric, even one as influential as mass, may not capture the evolution of diversity in clades that exhibit elongate body shapes. The discordance in evolutionary rates between body length and body mass along with evidence of decoupled diversification dynamics suggests that body elongation might be an innovation for the exploitation of novel Mid-Miocene resources, resulting in the radiation of some musteloids.
The remarkable disparity in both species richness and morphological diversity among clades represents one of the most salient macroevolutionary patterns across the Tree of Life. Since Darwin, many have asked why some clades exhibit low species diversity and morphological stasis for much of their evolutionary history whereas other clades diversified into numerous species and morphological forms. One mechanism theorized to drive species diversity and morphological disparity is adaptive radiation (Simpson 1955; Schluter 2000). In adaptive radiation theory, the occurrence of ecological opportunity and/or evolution of novel morphological innovation(s) create new adaptive zones. In turn, these new niches promote rapid increases in lineage diversification that coincides with increases in phenotypic disparity as colonizing lineages rapidly evolve adaptive traits that are highly correlated to their new resources (Schluter 2000). A key prediction of adaptive radiation theory is, therefore, that rapidly increasing rates of diversification and phenotypic evolution should be highest when ecology opportunity is greatest, typically early in the clade’s evolutionary history (Harmon et al. 2003; Rabosky and Lovette 2008a, 2008b; Gavrilets and Losos 2009; but see Mahler et al. 2013; Hopkins and Smith 2015; Slater 2015). These rates are subsequently expected to slow down as niche space fills to capacity (Harmon et al. 2003; Rabosky and Lovette 2008b).
Rates of diversification and phenotypic evolution through time can be estimated from calibrated molecular phylogenies of extant taxa (e.g., Harmon et al. 2003; Adams et al. 2009; Tran 2014; Colombo et al. 2015). A variety of methods have been developed to quantify the processes that have shaped the evolutionary history of a clade, assess rate variation among lineages, and, more recently, investigate rate variation through time (Rabosky 2006; Rabosky and Lovette 2008b; Alfaro et al. 2009; Stadler 2011; Etienne and Haegeman 2012; Rabosky et al. 2014). Interestingly, investigation of diversification and phenotypic evolutionary rates in some vertebrate clades hypothesized to have undergone adaptive radiations—for example, cetaceans (Slater et al. 2010), ovenbirds and woodcreepers (Derryberry et al. 2011), and New World monkeys (Aristide et al. 2015)—have revealed a lack of congruence between lineage diversification and phenotypic disparity: whereas rates of phenotypic evolution show evidence for declining rates through time, lineage diversification often does not exhibit an expected early burst (Slater et al. 2010; Derryberry et al. 2011; Aristide et al. 2015). A possible explanation for the lack of signal for an early burst of diversification is the absence of fossil data in these macroevolutionary analyses. Previous studies have shown that fossil data are important in understanding macroevolutionary processes (Quental and Marshall 2010; Liow et al. 2010); however, the majority of current macroevolutionary methods rely solely on time-calibrated phylogenies of extant taxa and do not account for extinct taxa in estimations of evolutionary rates. As a result, some researchers postulate the absence of extinct taxa in these analyses may have erased signatures of early rapid diversification that remain in patterns morphological disparity (Slater et al. 2010; Derryberry et al. 2011; Aristide et al. 2015). Although fossil-based methods for inferring diversification dynamics exist and have recently become more prominent in the evolutionary biology community (e.g., Alroy 2014; Silvestro et al. 2014a), many groups of organisms lack the adequate fossil record needed to assess diversification rates through time.
With approximately 85 putative extant species in 33 genera and over 400 described extinct species, the carnivoran superfamily Musteloidea is a prime candidate clade in which to investigate patterns of adaptive radiation using both extant- and fossil-based macroevolutionary methods. Musteloid habitats encompass several biomes including the arctic tundra, tropical and temperate forests, grasslands, deserts, and aquatic habitats such as inland waterways and coastal intertidal zones (Wilson and Mittermeier 2009). Equally impressive is the great ecomorphological diversity found across Musteloidea (Fabre et al. 2013, 2015; Dumont et al. 2015). Within each environment, musteloids exhibit diverse lifestyles, with taxa that are arboreal, fossorial, or aquatic as well as a variety of diets ranging from the generalist diets of raccoons, skunks, and badgers to the specialized diets of the herbivorous red panda, hypercarnivorous weasels, and piscivorous otters (Wilson and Mittermeier 2009). The incredible ecological and phenotypic diversity in Musteloidea is often attributed to adaptive radiation (Koepfli et al. 2008; Sato et al. 2009, 2012). Previous studies have postulated associations between the advent of ecological opportunity and rapid bursts of diversification during two distinct time periods in musteloid evolutionary history (Koepfli et al. 2008; Sato et al. 2009, 2012). The first putative burst of rapid diversification occurred 28–33 million years ago (Ma) right after the Eocene–Oligocene transition (33.5 Ma) and gave rise to Mephitidae (skunks and stink badgers), Ailuridae, (red panda), Procyonidae (raccoons and allies), and Mustelidae (badgers, martens, minks, otters, and weasels) (Sato et al. 2009, 2012). The second burst occurred near the root of extant Mustelidae (9.5–14 Ma), right after the Mid-Miocene Climate Transition, and gave rise to six of the eight extant subfamilies within Mustelidae (Koepfli et al. 2008). In both instances, rapid diversification closely followed major climatic events characterized by a sudden decrease in global temperatures (Zachos et al. 2008). These periods of abrupt cooling and drying resulted in a dramatic expansion of more open vegetation habitats such as grasslands and woodlands (Singh 1988; Estep et al. 2014; Leopold et al. 2014; Prothero and Berggren 2014), and, in turn, led to the diversification of rodent species (Finarelli and Badgley 2010; Calede et al. 2011; Fabre et al. 2012), which are the predominate prey of many mustelids (Wilson and Mittermeier 2009). The occurrence of ecological opportunity and rapid diversification at the roots of Musteloidea and Mustelidae led researchers to hypothesize that adaptive radiation may be the underlying process that promoted the ecological—and therefore phenotypic diversity—we observe within subsequent clades.
Although Musteloidea has qualitatively been described to exhibit two bursts of adaptive radiation (Koepfli et al. 2008; Sato et al. 2009, 2012), the quantitative evidence for the adaptive radiation hypotheses has yet to be investigated. Most importantly, incorporation of fossil data into current diversification analyses may help to link species with phenotypic diversification. The fossil record indicates that extinct musteloids were species-rich, and many of these groups are also hypothesized to have originated near major climate transitions; paleomustelids arose just after the Eocene–Oligocene transition (Baskin 1998; Finarelli 2008) and leptarctines originated near the Mid-Miocene Climate Transition (Bever and Zakrzewski 2009). Thus, how paleomustelids, leptarctinae, and other extinct musteloids may have contributed to patterns of lineage diversification remains unknown. In this study, we tested the hypothesis that patterns of lineage diversification and phenotypic evolution in Musteloidea are consistent with adaptive radiation theory. To accomplish our goals, we first constructed a novel time-calibrated phylogeny of 75 musteloid species (88%) that encompasses all 33 genera by compiling a 46 mitochondrial and nuclear gene dataset and then estimating divergence times using the fossilized birth–death (FBD) model with 74 musteloid fossils. We then tested for quantitative signatures of adaptive radiation within Musteloidea. We first examined decoupled diversification dynamics within Musteloidea using the program DDD. We then assessed patterns of lineage diversification using both molecular-based methods with extant taxa in the program BAMM as well as fossil-based methods with 453 extinct and extant musteloids (total of 2168 specimens) in the program PyRate. Lastly, to determine if ecological opportunity led to early bursts in rates of phenotypic evolution, we assessed evolutionary rates of body mass and body length in BAMM. We examined body size because it scales with many traits associated with the ecomorphology of species.
Materials and Methods
Taxon Sampling
Previous musteloid phylogenetic studies primarily focused within individual families and genera, particularly within Mustelidae (e.g., Koepfli andWayne 1998; Koepfli et al. 2007; Fulton and Strobeck 2007; Koepfli et al. 2008; Harding and Smith 2009; Sato et al. 2009). Investigation of the phylogenetics of Musteloidea has focused on resolving relationships among the four families, and the largest musteloid phylogeny to date contains approximately 52% of extant musteloid species (Sato et al. 2012). To reconstruct a more comprehensive phylogeny, we downloaded GenBank sequence data from these previous studies, the majority of which came from Koepfli et al. (2008), Yu et al. (2011), and Sato et al. (2012) (see Supplementary Table S1 available at Dryad at http://dx.doi.org/10.5061/dryad.nj1bp for complete reference list). We obtained 46 genes (4 mitochondrial and 42 nuclear genes) from 75 of the 85 putative musteloid species (88.2% of musteloid species), representing all 33 musteloid genera (Wilson and Mittermeier 2009). Our dataset accounts for 8 of the 12 mephitids, 13 of the 14 procyonids, 53 of the 58 mustelids, and the single ailurid species (Supplementary Table S1 available on Dryad; see Supplementary Table S2 available on Dryad for missing species). The ursid Ursus arctos and phocid Phoca vitulina were chosen as outgroups based on previous studies indicating that Ursidae and Pinnipedia are the closest extant clades to Musteloidea (Flynn et al. 2005; Eizirik et al. 2010). The final 46-gene dataset consists of 34,857 base pairs (bps) with an average of 767 bps per gene (range 194–1419), and each gene was represented by an average of 40 species (range 15–76) (Supplementary Table S3 available on Dryad). Total percentage of missing genes was 48.7% (1680 of 3450).
Phylogenetic Analyses
We reconstructed the musteloid phylogeny using a supermatrix approach. Prior to phylogenetic analyses, the individual 46 gene datasets (Supplementary Table S3 available on Dryad) were aligned with MUSCLE 3.8 (Edgar 2004) under default settings and edited manually where needed using Mesquite (Maddison and Maddison 2011). Alignments were also trimmed to start and end with the first and third codon positions, and protein translations of protein coding sequences were checked for stop codons and frameshift mutations as well as the presence of nuclear mitochondrial DNA (numts) in the mitochondrial dataset. To ensure no mislabeled sequences, we constructed individual gene trees using MrBayes v. 3.2.2 (Ronquist et al. 2012) on the CIPRES (Cyberinfrastructure for Phylogenetic Research at the UC San Diego Supercomputer Center) Portal (Miller et al. 2010) using four runs of one chain, 10 million Markov Chain Monte Carlo (MCMC) generations sampled every 1000 generations, and the first 25% of samples discarded as burnin. We found no mislabeled sequences. We selected a 14 partition scheme using PartitionFinder (Lanfear et al. 2012) under the default settings with Bayesian information criterion scores and a greedy search and allowed each partition to have its own independent substitution model (Supplementary Table S4 available on Dryad).
The 46 individual gene datasets were concatenated and analyzed using maximum likelihood (ML) and Bayesian inference (BI) methods. ML analyses were performed in RAxML 8.0.9 (Stamatakis 2014) as a 14 partitioned dataset under the General Time Reversible |$+$| Gamma (GTR |$+$| G) model. Nonparametric bootstrap values were estimated using 1000 pseudo replicates under the same model conditions. BI analyses were conducted using MrBayes 3.2.2 (Ronquist et al. 2012) on the CIPRES Portal (Miller et al. 2010). Appropriate evolutionary models were applied to each respective partition (Supplementary Table S4 available on Dryad). The data partitions were unlinked for parameter estimations and ran on 4 parallel MCMC chains (one cold, three heated) for 2 independent sets at 10 million generations per set, sampling trees every 1000 generations. Convergence of these values and effective sample sizes was assessed in the program Tracer, 1.6 (Rambaut et al. 2014). Approximately 25% of sampled trees were discarded as burn-in, and the remaining trees were used to construct a majority rule consensus tree.
We also estimated species trees using supertree and multispecies coalescent methods. For the supertree analysis, a matrix representation with parsimony (MRP) matrix (Ragan 1992) was then compiled from the 46 Bayesian gene trees (from above) in the program Clann 3.2.3 (Creevey and McInerney 2005). With the MRP matrix, reconstruction of the supertree phylogeny was performed in PAUP* 4.0b10 (Swofford 2003) using a parsimony-based heuristic search with tree bisection and reconnection. For the coalescent analysis, we used ASTRAL-II v4.7.8 with 100 replicates of multilocus bootstrapping (Mirarab et al. 2014).
Estimation of Divergence Times
We estimated musteloid divergence times using the FBD tree process prior (Heath et al. 2014), extended to allow for sampled ancestors (Gavryushkina et al. 2014) as implemented in BEAST v2.3.2 (Bouckaert et al. 2014). The use of this model circumvents the need to define arbitrary age priors on nodes while maximizing the amount of fossil data that can be used. We compiled age ranges of 74 fossil musteloids from the literature (Appendix 1). Only fossil taxa that have been subject to formal phylogenetic analysis and could be robustly placed within a musteloid clade were included in our dataset. Our final dataset consisted of 4 stem arctoids, 4 stem musteloids, 9 stem mephitids, 8 ailurids, 16 procyonids, 7 oligobunines, 11 leptarctines, and 13 neomustelids. For each fossil, we also surveyed the literature to identify its stratigraphic range. Fossil ages were assigned a uniform distribution for their stratigraphic range, spanning age uncertainty in the case of taxa known from a single fossil or from first and last appearance dates in the case of taxa known from multiple localities.
We used an uncorrelated lognormal relaxed molecular clock model with exponential prior distributions on the mean and standard deviation of the clock rate. The same 14 partition scheme (Supplementary Table S4 available on Dryad) was used for the molecular data, and we allowed each partition to have its own independent substitution model. Three parameters are used in the MCMC optimization under the FBD model: net diversification rate, turnover rate, and fossil sampling rate. Preliminary diversification rate analyses suggested musteloids exhibit low-diversification rates; thus we used an exponential prior distribution with a mean of 1 for the net diversification rate parameter and uniform prior distributions on the range [0,1] for turnover rate and fossil sampling proportion. Lastly, we accounted for incomplete taxon sampling by setting the parameter |$\rho $| (Rho) to 0.88 for the 88% of extant species sampled.
Four separate MCMC analyses with 150 million generations sampled every 15,000 generations were run to provide independent posterior probability distribution estimations of model parameters and node ages, and a random starting tree was used for each analysis. Convergence of parameters and effective sample sizes were assessed in the program Tracer, 1.6 (Rambaut et al. 2014); all effective samples sizes were > 200 after discarding the first 35% of the combined samples as burn-in. Sampled trees from the four analyses were combined using LogCombiner v2.2.1 (http://beast.bio.ed.ac.uk/LogCombiner), pruned to remove all fossil taxa, and summarized as a maximum clade credibility (MCC) tree using TreeAnnotator v2.2.1 (http://beast.bio.ed.ac.uk/TreeAnnotator). The resulting posterior probabilities and corresponding 95% credible intervals for the mean divergence time estimates were visualized using the program FigTree v1.40 (http://beast.bio.ed.ac.uk/FigTree).
Estimation of Species Diversification
To test for temporally distinct adaptive radiations within Musteloidea, we employed the likelihood-based approach developed by (Etienne et al. 2012; Etienne and Haegeman 2012), implemented in the R library DDD v 3.2. We compared the fit of a suite of decoupled diversity dependent models against null models assuming either a time-homogenous constant rates birth–death process (CR0) or a diversity dependent process in which a single carrying capacity operated over musteloid history with diversity dependent speciation and constant extinction (CR1). The first set of alternative models assumes a shift in carrying capacity and/or rates operating across musteloid phylogeny at some time |$t$| in the past. We fit four variants of this process in which (i) only carrying capacities varied before and after |$t$| (SR1), (ii) carrying capacities and speciation rates varied before and after time |$t$| (SR2), (iii) carrying capacities and extinction rates varied before and after time |$t$| (SR3), and (iv) all parameters varied before and after time |$t$| (SR4). The shift point |$t$| was itself inferred from the data in all cases. The second set of alternative models that we considered tested for shifts in diversification parameters associated with clade-specific key innovations (KI models). Based on analyses of body length evolutionary rates (see below), we identified two putative branches within which key innovations may have evolved; the base of Mustelidae (KI_ M) and in the branch leading to the most recent common ancestor of Ictonychinae, Mustelinae and Lutrinae (KI_ IML). We further tested for decoupled dynamics in the branch leading to the most recent common ancestor of this clade plus Helictinae plus Martinae (KI_ M2), as this is the most inclusive clade exhibiting dramatic body elongation. Missing species were assigned as appropriate to the clade to which they belong for all analyses and model fitting was conditioned on nonextinction of the phylogeny (Supplementary Table S2 available on Dryad). We fit the same four variants of the KI models as for the SR models and assessed relative model support using Akaike weights (|$w_{A})$|.
Etienne et al. (2016) showed that the use of Akaike weights for model choice may be inappropriate when comparing the fit of diversity dependent and constant rates model as the likelihood ratio (LR) statistic is often not chi-square distributed. They suggested instead to use a parametric bootstrap procedure to determine the appropriate value for accepting or rejecting diversity dependence at a specified level of significance. Although such a procedure is not appropriate for decoupled models as they are not nested within either a constant rates or single diversity dependent process (R. Etienne pers. comm.), we can still use the parametric bootstrap procedure to more generally evaluate the reliability of model selection when comparing constant rates and diversity dependent models. Following Etienne et al. (2016), we used 1000 parametric bootstrap replicates to assess type I error, the appropriate critical value of the LR statistic at |$\alpha = 0.05$|, and power of the test for diversity dependence.
We also estimated the rates of diversification across the musteloid phylogeny using BAMM v2.5.0 (Rabosky et al. 2013; Rabosky 2014). BAMM uses a reversible jump Markov Chain Monte Carlo (RJMCMC) to explore candidate models of lineage diversification and trait evolution as well as quantify heterogeneity in evolutionary rates (Rabosky 2014). We performed 4 independent BAMM runs of 10 million generations on our MCC musteloid phylogeny from BEAST, sampling every 1000 generations and with priors chosen using the function setBAMMpriors (Rabosky et al. 2014). We assessed the convergence of the BAMM run using the R (R Core Team 2015) package BAMMtools v2.0 (Rabosky et al. 2014) and used the CODA library to check the effective sample sizes of log-likelihoods and the number of shift events present in each sample; all parameters had effective sample sizes > 1000. We accounted for incomplete taxon sampling using the analytical correction implemented in BAMM (Supplementary Table S2 available on Dryad). We note that some aspects of BAMM have recently been criticized (Moore et al. 2016, but see Rabosky et al. 2017); nevertheless, we cautiously retain our analyses here for comparative purposes, noting that they are one of several diversification rate analyses employed.
To incorporate fossil data in our analyses, we estimated lineage diversification rates from the fossil record using a birth–death MCMC (BDMCMC) analysis implemented in the program PyRate v. 0.604 (Silvestro et al. 2014a, 2014b). PyRate simultaneously estimates the speciation and extinction rates for each species using a birth-death process. The birth-death process uses each species’ temporal distribution in the fossil record to model the diversification dynamics by exploring alternative diversification models with different number of rate shifts that may underlie changes in diversity (Silvestro et al. 2014a, 2014b). We obtained musteloid fossil data from the Paleobiology Database, Fossilworks, and NOW Database (accessed 9/02/15) and included only fossils that were identified to the species level. Our complete fossil dataset comprised of 453 species (total of 2168 specimens), of which 404 are extinct (Supplementary Table S5 available on Dryad). We then used the R function extract.ages from the PyRate package to randomly resample the age of fossil occurrences 10 times. We assumed independent preservation rates for each epoch. Each replicate was analyzed independently for 15 million generations with the number of extant species set to 85 using Python 2.7.6 (python.org). Convergence of the BDMCMC sample and effective sample sizes was assessed in the program Tracer, 1.6 (Rambaut et al. 2014), and mean diversification rates through time were estimated after discarding the first 25% of the logged rate estimates as burn-in and combining the remaining samples across all 10 replicates. We also performed separate PyRate analyses for each of the major families: Mephitidae, Ailuridae, Procyonidae, and Neomustelidae. We did not include paleomustelids as the phylogenetic placement of the majority of these taxa is unknown. We used the same procedure as above.
We also plotted species richness of all putative extinct and extant musteloids using the same fossil dataset obtained from the Paleobiology Database, Fossilworks, and NOW Database as well as extant musteloids not represented in the fossil record. The combined database was pruned to exclude taxa that could not be placed within the taxonomic groups Mephitidae, Ailuridae, Procyonidae, Paleomustelidae, and Neomustelidae. Our complete dataset comprised of 447 putative musteloids (40 mephitids, 21 ailurids, 46 procyonids, 46 paleomustelids, and 294 neomustelids). We used the R package Paleotree (Bapst 2012) to calculate species richness within each taxonomic group over the past 36 Ma in 1 Ma intervals.
Lastly, we used PyRateContinuous to examine whether evolutionary (speciation and extinction) rates correlate with changes in clade diversity, suggesting diversity dependence, or with changes in global temperature. PyRateContinuous uses a birth–death model to quantify correlation parameters between time-varying evolutionary rates with a time continuous variable such as the clade’s own diversity or temperature (Silvestro et al. 2015). For each birth–death model—the diversity-dependent and temperature-dependent models—we discarded the first 25% of the logged rate estimates as burn-in combined the remaining samples across the 10 replicates, and ran each analysis independently for 1.25 million MCMC iterations with sampling frequency of 1000. For the temperature-dependent model, we used global temperature data derived from stable isotope proxies (Zachos et al. 2008) and rescaled for analyses in PyRate (Silvestro et al. 2015). We used Tracer, 1.6 (Rambaut et al. 2014) to examine the mean values and 95% HPD (highest posterior density) intervals of the posterior samples of the parameters. Following Silvestro et al. (2015), we considered the correlation to be statistically significant if 0 was not within the 95% HPD of the correlation parameter. We also used PyRateContinuous to examine if there was a shift in correlation parameters before and after the Mid-Miocene Climate Transition.
Estimation of Phenotypic Evolutionary Rates
It is a basic biological phenomenon that several ecological, physiological, and morphological traits scale with size (Schmidt-Nielsen 1984; LaBarbera 1989). Previous investigations of rates of phenotypic evolution in mammals have therefore used either body length or body mass as proxies for organismal size (Slater et al. 2010; Wilson et al. 2012; Aristide et al. 2015). Since both characters were readily attainable in the literature, we used BAMM to test for variation in rates of phenotypic evolution with both body size metrics as proxies for musteloid ecomorphology. We compiled the mean adult body length—excluding the tail length—and mean adult body mass for each species in our phylogeny and log-transformed the measurements prior to analysis (Supplementary Table S6 available on Dryad). Mean body sizes rather than maximum sizes were used to limit the effects of outliers and/or misreported measurements. Although most species of musteloids exhibit sexual dimorphism, we did not test body size rates between males and females because body size measurements based on sex are scarce in the literature. We performed 4 independent BAMM runs on our MCC musteloid phylogeny, with 10 million generations sampling every 1000 generations, and assessed convergence of the BAMM run as described above. Again, we accounted for incomplete taxon sampling using the analytical correction implemented in BAMM.
Results
Phylogenetic Analyses and Estimation of Divergence Times
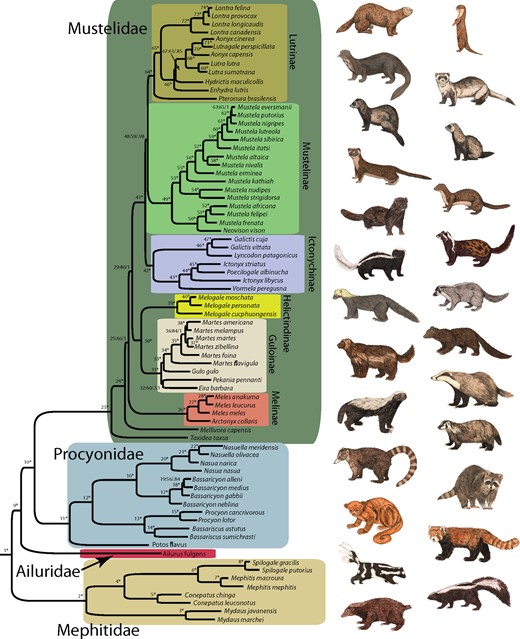
Supermatrix-based phylogenetic analyses with ML and BI methods resulted in identical topologies. We recovered a successive sister group relationship of Mephitidae (Node 1) and Ailuridae (Node 9), to a clade (Node 10) consisting of Procyonidae and Mustelidae (Fig. 1; Supplementary Text and Supplementary Table S7 available on Dryad). Species tree estimations using MRP and coalescent-based methods revealed similar topologies as the supermatrix approaches with the exception of the topological placement of some mustelid subfamilies (Appendix 2; Supplementary Fig. S1 available on Dryad).
Majority rule consensus trees of Musteloidea from Bayesian inference of combined molecular data. Nodes are numbered (1–74), followed by bootstrap support (BS) values from maximum likelihood and posterior probabilities (PP) from Bayesian inference. Support values are in the following order: BS/PP. An asterisk (*) indicates > 90% for BS and > 0.95 for PP for the node. Nodal support is also presented in Supplementary Table S7 available on Dryad. Outgroup taxa were pruned from the tree.
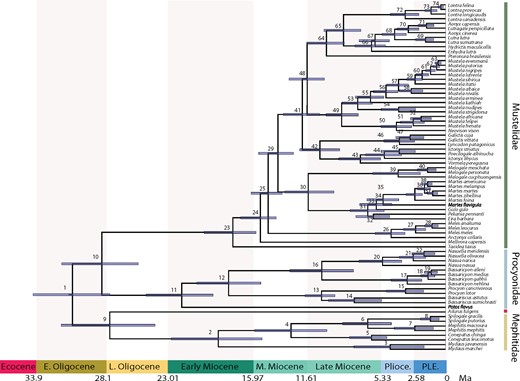
Our time-tree estimated using the FBD tree prior with 74 fossil taxa infers that the initial radiation of extant musteloid families (the basal-most nodes of the extant sample) occurred during a ~ 3 million year time interval (from 31.21 to 28.09 Ma) beginning in the Early Oligocene and continuing into the Late Oligocene (Fig. 2; Supplementary Fig. S2 and Supplementary Table S7 available on Dryad). When attempting to summarize our results as a MCC tree, we found that pruning all fossil taxa prior to identifying the MCC tree resulted in unconventional but weakly supported (posterior probability |$=0.69$|) higher level branching pattern, where Mephitidae and Ailuridae form a sister group, with Mephitidae diverging from Ailuridae around 28.09 Ma (node 9; 95% HPD, 24.30–32.15 Ma). Pruning fossils after finding the MCC tree resulted in the traditional higher level relationships of (Mephitidae, (Ailuridae, (Procyonidae, Mustelidae))); however, the placement of fossil taxa in our posterior sample of trees is somewhat arbitrary as we did not include morphological data in our analyses. We therefore focus our discussion on the results generated using the MCC tree recovered after pruning fossils from input trees, despite its unconventional higher level branching order. Reassuringly, diversification analyses using both time trees displayed identical results, suggesting that this minor topological distinction does not greatly impact macroevolutionary inferences. Regardless of the position of Ailuridae, we infer that Procyonidae and Mustelidae diverged from each other around 28.75 Ma (node 10; 95% HPD, 25.71–32.12 Ma) during the Late Oligocene.
Time-calibrated phylogenetic tree of Musteloidea. Mean divergence times estimated using a relaxed molecular clock model on the complete 46 gene dataset with 74 fossil priors. Blue bars across nodes indicate 95% HPD intervals around the mean divergence time estimates. Posterior estimates of mean and 95% HPD of divergence times are presented in Supplementary Table S7 available on Dryad. Nodes are numbered (1–74, same as Fig. 1). Outgroup taxa were pruned from the tree and geological time scale is shown below the tree.
Diversification Rates Through Time
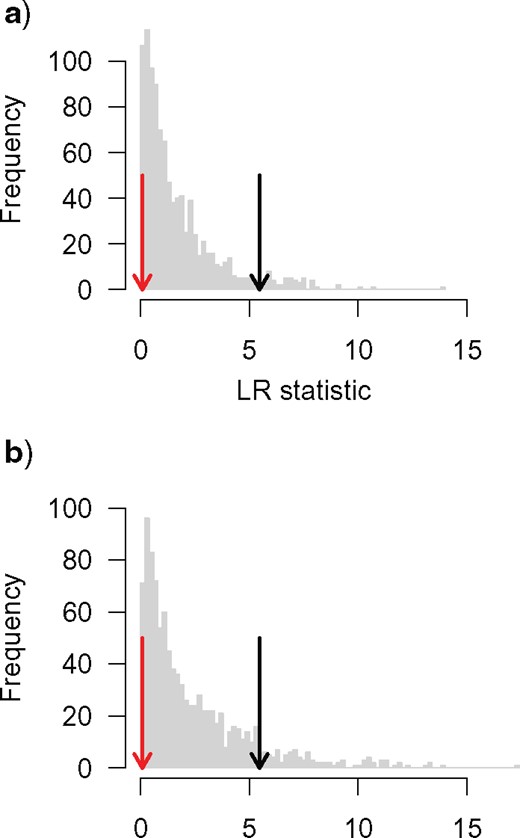
Comparison of constant rates and simple diversity dependent diversification models to those allowing for decoupled diversification dynamics yielded strong support (~ 85% of |$w_{A})$| for a decoupling of the diversification process along the branch leading to Helictidinae, Guloninae, Ictonychinae, Mustelinae and Lutrinae (KI_ M2 models; Table 1). The best-fitting models, receiving ~ 27 and ~ 31 % of weight, respectively, suggest a slightly larger carrying capacity for the decoupled sub-clade than for the main clade (56 vs. 32 species) with no change in intrinsic speciation or extinction rates, or else a much larger carrying capacity (~ 70 species) for the sub-clade coupled with a shift in extinction rates from 0.15 to 0.01. Key innovation models associated with crown Mustelidae (KI_ M) or Ictonychinae, Mustelinae and Lutrinae (KI_ IML) received negligible support (< 5% |$w_{A\thinspace })$|, as did simple homogeneous or clade-wide shift models. Diversity dependence with a single |$K$| is not significantly preferred over a constant rates process using a classical LR test based on the chi-square distribution with one degree of freedom (LR statistic |$=$| 0.088, |$P = 0.77$|) or AIC (Akaike information criterion) (|$w_{\mathrm{A}}$|[constant rates]) |$=$| 0.72). Parametric bootstrapping yields a critical value for significance at |$\alpha = 0.05$| level of 5.47, some 1.4 times larger than the critical value of 3.84 implied by a |$\chi^{\mathrm{2}}$| test (Fig. 3a). This increases the explanatory power of the constant rates process (|$P =$| 0.96) and suggests that model selection based on AIC may be inappropriate (type I error rate using classical (likelihood ratio test likelihood r |$=$| 0.11). However, parametric bootstrapping under the single |$K$| diversity dependent model also shows that power to detect a diversity dependent process of the magnitude implied by the musteloid data is very low (power |$=$| 0.1; Fig. 3b).
Parameter estimates (speciation rate [|$\lambda $|], extinction rate [|$\mu $|], clade carrying capacity [K]) and model support (log likelihood [lnL] values, |$\Delta $|AIC scores, Akaike weights [wA]) for constant rate (CR) and diversity-dependent models with shifts in clade-wide (SR) or clade-specific key innovation related (KI) carry capacities
| Model . | |$\lambda_{\mathrm{1}}$| . | |$\mu_{\mathrm{1}}$| . | K|$_{\mathrm{1}}$| . | |$\lambda_{\mathrm{2}}$| . | |$\mu_{\mathrm{2}}$| . | K|$_{\mathrm{2}}$| . | t|$_{\mathrm{shift}}$| . | LnL . | df . | AIC . | |$\Delta $|AIC . | |$w_{\mathrm{A}}$| . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR0 | 0.17 | 0.04 | NA | NA | NA | NA | NA | |$-218.39$| | 2 | 440.78 | 13.60 | < 0.01 |
| CR1 | 0.19 | 0.06 | 366.24 | NA | NA | NA | NA | |$-218.35$| | 3 | 442.7 | 15.52 | < 0.01 |
| SR1 | 0.24 | 0.04 | 37.17 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 178.95 | 6.51 | |$-216.16$| | 5 | 442.32 | 15.14 | < 0.01 |
| SR2 | 0.18 | 0.05 | 54.45 | 0.48 | |$\mu_{\mathrm{1}}$| | 96.38 | 6.51 | |$-213.42$| | 6 | 438.84 | 11.66 | < 0.01 |
| SR3 | 0.47 | 0.17 | 27.5 | 0.47 | 0.04 | 97.63 | 6.51 | |$-210.94$| | 6 | 433.88 | 6.70 | 0.01 |
| SR4 | 0.33 | 0.12 | 25.04 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 111.91 | 6.51 | |$-213.08$| | 7 | 440.16 | 12.98 | < 0.01 |
| KI1_ M | 0.52 | 0.16 | 25.71 | 0.52 | |$\mu_{\mathrm{1}}$| | 61.45 | 17.79 | |$-210.78$| | 5 | 431.56 | 4.38 | 0.03 |
| KI2_ M | 1.1 | 0.17 | 25.96 | |$\lambda_{\mathrm{1}}$| | 0.17 | 61.43 | 18.15 | |$-210.38$| | 6 | 432.76 | 5.58 | 0.02 |
| KI3_ M | 0.47 | 0.17 | 26.01 | 0.47 | 0.12 | 63.8 | 17.83 | |$-210.54$| | 6 | 433.08 | 5.90 | 0.02 |
| KI4_ M | 1.16 | 0.2 | 26.28 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 109.38 | 18.15 | |$-209.14$| | 7 | 432.28 | 5.10 | 0.02 |
| KI1_ M2 | 0.55 | 0.15 | 31.49 | 0.55 | |$\mu_{\mathrm{1}}$| | 55.89 | 14.02 | |$-208.73$| | 5 | 427.46 | 0.28 | 0.27 |
| KI2_ M2 | 0.38 | 0.14 | 32.27 | |$\lambda_{\mathrm{1}}$| | 0.14 | 55.66 | 14.03 | |$-208.33$| | 6 | 428.66 | 1.48 | 0.15 |
| KI3_ M2 | 0.38 | 0.15 | 32.56 | 0.38 | 0.01 | 70.16 | 14.02 | |$-207.59$| | 6 | 427.18 | 0.00 | 0.31 |
| KI4_ M2 | 0.46 | 0.17 | 32.27 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 73.32 | 14.02 | |$-207.47$| | 7 | 428.94 | 1.76 | 0.13 |
| KI1_ IML | 0.67 | 0.21 | 45.16 | 0.67 | |$\mu_{\mathrm{1}}$| | 38.39 | 11.98 | |$-212.56$| | 5 | 435.12 | 7.94 | 0.01 |
| KI2_ IML | 0.34 | 0.16 | 49.2 | |$\lambda_{\mathrm{1}}$| | 0.16 | 37.39 | 11.98 | |$-210.91$| | 6 | 433.82 | 6.64 | 0.01 |
| KI3_ IML | 0.39 | 0.19 | 47.64 | 0.39 | 0 | 47.76 | 11.98 | |$-210.21$| | 6 | 432.42 | 5.24 | 0.02 |
| KI4_ IML | 0.42 | 0.19 | 47.32 | 0.39 | 0 | 47.76 | 11.98 | |$-210.2$| | 7 | 434.4 | 7.22 | 0.01 |
| Model . | |$\lambda_{\mathrm{1}}$| . | |$\mu_{\mathrm{1}}$| . | K|$_{\mathrm{1}}$| . | |$\lambda_{\mathrm{2}}$| . | |$\mu_{\mathrm{2}}$| . | K|$_{\mathrm{2}}$| . | t|$_{\mathrm{shift}}$| . | LnL . | df . | AIC . | |$\Delta $|AIC . | |$w_{\mathrm{A}}$| . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR0 | 0.17 | 0.04 | NA | NA | NA | NA | NA | |$-218.39$| | 2 | 440.78 | 13.60 | < 0.01 |
| CR1 | 0.19 | 0.06 | 366.24 | NA | NA | NA | NA | |$-218.35$| | 3 | 442.7 | 15.52 | < 0.01 |
| SR1 | 0.24 | 0.04 | 37.17 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 178.95 | 6.51 | |$-216.16$| | 5 | 442.32 | 15.14 | < 0.01 |
| SR2 | 0.18 | 0.05 | 54.45 | 0.48 | |$\mu_{\mathrm{1}}$| | 96.38 | 6.51 | |$-213.42$| | 6 | 438.84 | 11.66 | < 0.01 |
| SR3 | 0.47 | 0.17 | 27.5 | 0.47 | 0.04 | 97.63 | 6.51 | |$-210.94$| | 6 | 433.88 | 6.70 | 0.01 |
| SR4 | 0.33 | 0.12 | 25.04 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 111.91 | 6.51 | |$-213.08$| | 7 | 440.16 | 12.98 | < 0.01 |
| KI1_ M | 0.52 | 0.16 | 25.71 | 0.52 | |$\mu_{\mathrm{1}}$| | 61.45 | 17.79 | |$-210.78$| | 5 | 431.56 | 4.38 | 0.03 |
| KI2_ M | 1.1 | 0.17 | 25.96 | |$\lambda_{\mathrm{1}}$| | 0.17 | 61.43 | 18.15 | |$-210.38$| | 6 | 432.76 | 5.58 | 0.02 |
| KI3_ M | 0.47 | 0.17 | 26.01 | 0.47 | 0.12 | 63.8 | 17.83 | |$-210.54$| | 6 | 433.08 | 5.90 | 0.02 |
| KI4_ M | 1.16 | 0.2 | 26.28 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 109.38 | 18.15 | |$-209.14$| | 7 | 432.28 | 5.10 | 0.02 |
| KI1_ M2 | 0.55 | 0.15 | 31.49 | 0.55 | |$\mu_{\mathrm{1}}$| | 55.89 | 14.02 | |$-208.73$| | 5 | 427.46 | 0.28 | 0.27 |
| KI2_ M2 | 0.38 | 0.14 | 32.27 | |$\lambda_{\mathrm{1}}$| | 0.14 | 55.66 | 14.03 | |$-208.33$| | 6 | 428.66 | 1.48 | 0.15 |
| KI3_ M2 | 0.38 | 0.15 | 32.56 | 0.38 | 0.01 | 70.16 | 14.02 | |$-207.59$| | 6 | 427.18 | 0.00 | 0.31 |
| KI4_ M2 | 0.46 | 0.17 | 32.27 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 73.32 | 14.02 | |$-207.47$| | 7 | 428.94 | 1.76 | 0.13 |
| KI1_ IML | 0.67 | 0.21 | 45.16 | 0.67 | |$\mu_{\mathrm{1}}$| | 38.39 | 11.98 | |$-212.56$| | 5 | 435.12 | 7.94 | 0.01 |
| KI2_ IML | 0.34 | 0.16 | 49.2 | |$\lambda_{\mathrm{1}}$| | 0.16 | 37.39 | 11.98 | |$-210.91$| | 6 | 433.82 | 6.64 | 0.01 |
| KI3_ IML | 0.39 | 0.19 | 47.64 | 0.39 | 0 | 47.76 | 11.98 | |$-210.21$| | 6 | 432.42 | 5.24 | 0.02 |
| KI4_ IML | 0.42 | 0.19 | 47.32 | 0.39 | 0 | 47.76 | 11.98 | |$-210.2$| | 7 | 434.4 | 7.22 | 0.01 |
Note: See text for model names.
Parameter estimates (speciation rate [|$\lambda $|], extinction rate [|$\mu $|], clade carrying capacity [K]) and model support (log likelihood [lnL] values, |$\Delta $|AIC scores, Akaike weights [wA]) for constant rate (CR) and diversity-dependent models with shifts in clade-wide (SR) or clade-specific key innovation related (KI) carry capacities
| Model . | |$\lambda_{\mathrm{1}}$| . | |$\mu_{\mathrm{1}}$| . | K|$_{\mathrm{1}}$| . | |$\lambda_{\mathrm{2}}$| . | |$\mu_{\mathrm{2}}$| . | K|$_{\mathrm{2}}$| . | t|$_{\mathrm{shift}}$| . | LnL . | df . | AIC . | |$\Delta $|AIC . | |$w_{\mathrm{A}}$| . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR0 | 0.17 | 0.04 | NA | NA | NA | NA | NA | |$-218.39$| | 2 | 440.78 | 13.60 | < 0.01 |
| CR1 | 0.19 | 0.06 | 366.24 | NA | NA | NA | NA | |$-218.35$| | 3 | 442.7 | 15.52 | < 0.01 |
| SR1 | 0.24 | 0.04 | 37.17 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 178.95 | 6.51 | |$-216.16$| | 5 | 442.32 | 15.14 | < 0.01 |
| SR2 | 0.18 | 0.05 | 54.45 | 0.48 | |$\mu_{\mathrm{1}}$| | 96.38 | 6.51 | |$-213.42$| | 6 | 438.84 | 11.66 | < 0.01 |
| SR3 | 0.47 | 0.17 | 27.5 | 0.47 | 0.04 | 97.63 | 6.51 | |$-210.94$| | 6 | 433.88 | 6.70 | 0.01 |
| SR4 | 0.33 | 0.12 | 25.04 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 111.91 | 6.51 | |$-213.08$| | 7 | 440.16 | 12.98 | < 0.01 |
| KI1_ M | 0.52 | 0.16 | 25.71 | 0.52 | |$\mu_{\mathrm{1}}$| | 61.45 | 17.79 | |$-210.78$| | 5 | 431.56 | 4.38 | 0.03 |
| KI2_ M | 1.1 | 0.17 | 25.96 | |$\lambda_{\mathrm{1}}$| | 0.17 | 61.43 | 18.15 | |$-210.38$| | 6 | 432.76 | 5.58 | 0.02 |
| KI3_ M | 0.47 | 0.17 | 26.01 | 0.47 | 0.12 | 63.8 | 17.83 | |$-210.54$| | 6 | 433.08 | 5.90 | 0.02 |
| KI4_ M | 1.16 | 0.2 | 26.28 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 109.38 | 18.15 | |$-209.14$| | 7 | 432.28 | 5.10 | 0.02 |
| KI1_ M2 | 0.55 | 0.15 | 31.49 | 0.55 | |$\mu_{\mathrm{1}}$| | 55.89 | 14.02 | |$-208.73$| | 5 | 427.46 | 0.28 | 0.27 |
| KI2_ M2 | 0.38 | 0.14 | 32.27 | |$\lambda_{\mathrm{1}}$| | 0.14 | 55.66 | 14.03 | |$-208.33$| | 6 | 428.66 | 1.48 | 0.15 |
| KI3_ M2 | 0.38 | 0.15 | 32.56 | 0.38 | 0.01 | 70.16 | 14.02 | |$-207.59$| | 6 | 427.18 | 0.00 | 0.31 |
| KI4_ M2 | 0.46 | 0.17 | 32.27 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 73.32 | 14.02 | |$-207.47$| | 7 | 428.94 | 1.76 | 0.13 |
| KI1_ IML | 0.67 | 0.21 | 45.16 | 0.67 | |$\mu_{\mathrm{1}}$| | 38.39 | 11.98 | |$-212.56$| | 5 | 435.12 | 7.94 | 0.01 |
| KI2_ IML | 0.34 | 0.16 | 49.2 | |$\lambda_{\mathrm{1}}$| | 0.16 | 37.39 | 11.98 | |$-210.91$| | 6 | 433.82 | 6.64 | 0.01 |
| KI3_ IML | 0.39 | 0.19 | 47.64 | 0.39 | 0 | 47.76 | 11.98 | |$-210.21$| | 6 | 432.42 | 5.24 | 0.02 |
| KI4_ IML | 0.42 | 0.19 | 47.32 | 0.39 | 0 | 47.76 | 11.98 | |$-210.2$| | 7 | 434.4 | 7.22 | 0.01 |
| Model . | |$\lambda_{\mathrm{1}}$| . | |$\mu_{\mathrm{1}}$| . | K|$_{\mathrm{1}}$| . | |$\lambda_{\mathrm{2}}$| . | |$\mu_{\mathrm{2}}$| . | K|$_{\mathrm{2}}$| . | t|$_{\mathrm{shift}}$| . | LnL . | df . | AIC . | |$\Delta $|AIC . | |$w_{\mathrm{A}}$| . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR0 | 0.17 | 0.04 | NA | NA | NA | NA | NA | |$-218.39$| | 2 | 440.78 | 13.60 | < 0.01 |
| CR1 | 0.19 | 0.06 | 366.24 | NA | NA | NA | NA | |$-218.35$| | 3 | 442.7 | 15.52 | < 0.01 |
| SR1 | 0.24 | 0.04 | 37.17 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 178.95 | 6.51 | |$-216.16$| | 5 | 442.32 | 15.14 | < 0.01 |
| SR2 | 0.18 | 0.05 | 54.45 | 0.48 | |$\mu_{\mathrm{1}}$| | 96.38 | 6.51 | |$-213.42$| | 6 | 438.84 | 11.66 | < 0.01 |
| SR3 | 0.47 | 0.17 | 27.5 | 0.47 | 0.04 | 97.63 | 6.51 | |$-210.94$| | 6 | 433.88 | 6.70 | 0.01 |
| SR4 | 0.33 | 0.12 | 25.04 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 111.91 | 6.51 | |$-213.08$| | 7 | 440.16 | 12.98 | < 0.01 |
| KI1_ M | 0.52 | 0.16 | 25.71 | 0.52 | |$\mu_{\mathrm{1}}$| | 61.45 | 17.79 | |$-210.78$| | 5 | 431.56 | 4.38 | 0.03 |
| KI2_ M | 1.1 | 0.17 | 25.96 | |$\lambda_{\mathrm{1}}$| | 0.17 | 61.43 | 18.15 | |$-210.38$| | 6 | 432.76 | 5.58 | 0.02 |
| KI3_ M | 0.47 | 0.17 | 26.01 | 0.47 | 0.12 | 63.8 | 17.83 | |$-210.54$| | 6 | 433.08 | 5.90 | 0.02 |
| KI4_ M | 1.16 | 0.2 | 26.28 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 109.38 | 18.15 | |$-209.14$| | 7 | 432.28 | 5.10 | 0.02 |
| KI1_ M2 | 0.55 | 0.15 | 31.49 | 0.55 | |$\mu_{\mathrm{1}}$| | 55.89 | 14.02 | |$-208.73$| | 5 | 427.46 | 0.28 | 0.27 |
| KI2_ M2 | 0.38 | 0.14 | 32.27 | |$\lambda_{\mathrm{1}}$| | 0.14 | 55.66 | 14.03 | |$-208.33$| | 6 | 428.66 | 1.48 | 0.15 |
| KI3_ M2 | 0.38 | 0.15 | 32.56 | 0.38 | 0.01 | 70.16 | 14.02 | |$-207.59$| | 6 | 427.18 | 0.00 | 0.31 |
| KI4_ M2 | 0.46 | 0.17 | 32.27 | |$\lambda_{\mathrm{1}}$| | |$\mu_{\mathrm{1}}$| | 73.32 | 14.02 | |$-207.47$| | 7 | 428.94 | 1.76 | 0.13 |
| KI1_ IML | 0.67 | 0.21 | 45.16 | 0.67 | |$\mu_{\mathrm{1}}$| | 38.39 | 11.98 | |$-212.56$| | 5 | 435.12 | 7.94 | 0.01 |
| KI2_ IML | 0.34 | 0.16 | 49.2 | |$\lambda_{\mathrm{1}}$| | 0.16 | 37.39 | 11.98 | |$-210.91$| | 6 | 433.82 | 6.64 | 0.01 |
| KI3_ IML | 0.39 | 0.19 | 47.64 | 0.39 | 0 | 47.76 | 11.98 | |$-210.21$| | 6 | 432.42 | 5.24 | 0.02 |
| KI4_ IML | 0.42 | 0.19 | 47.32 | 0.39 | 0 | 47.76 | 11.98 | |$-210.2$| | 7 | 434.4 | 7.22 | 0.01 |
Note: See text for model names.
Parametric bootstrapping of diversification rate models. (a) The distribution of LR statistics comparing the fit of CR1 to CR0 when CR0 is the true model shows that the critical value for rejecting CR0 at the |$\alpha = 0.05$| level is larger than implied by a chi-squared distribution with one degree of freedom, and further increases confidence in rejecting a diversity dependent process. However, (b) shows that power to detect a true diversity dependent process is, in this case, also low. In both plots, the red/dark grey arrow shows the empirically derived LR statistic while the black arrow shows the critical value required to reject the null constant rates model derived from parametric bootstrapping.
Mean net diversification rates inferred from BAMM analyses are broadly congruent across families: Mephitidae |$=$| 0.138 lineages/lineage million years (95% HPD, 0.106–0.169 lineages/lineage million years), Procyonidae |$=$| 0.138 lineages/lineage million years (95% HPD, 0.106–0.169 lineages/lineage million years), and Mustelidae |$=$| 0.143 lineages/lineage million years (95% HPD, 0.114–0.175 lineages/lineage million years). The rate of diversification in Mustelidae is slightly higher than the background musteloid rate, but 99.3% of the samples from the posterior distribution are assigned to just one rate configuration with no indication of a rate shift (Fig. 4a). Contour plots of branch specific diversification rates and plots of clade specific rates through time indicate that diversification rates have tended to increase slightly through the past 31.2 Ma to the present, from 0.101 lineages/lineage million years to 0.145 lineages/lineage million years. Mean net diversification for all of Musteloidea is 0.132 lineages/lineage million years (95% HPD, 0.089–0.174 lineages/lineage million years). Speciation rate and extinction rate for all of Musteloidea are 0.15 lineages/lineage million years (95% HPD, 0.103–0.190 lineages/lineage million years) and 0.014 lineages/lineage million years (95% HPD, 0.001–0.043 lineages/lineage million years), respectively (Fig. 5a). These results are consistent across the pruned-before (Fig. 4) and pruned-after (Supplementary Fig. S3 available on Dryad) trees.
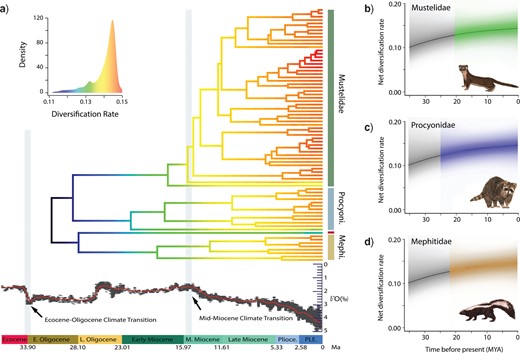
Lineage diversification rates through time. a) Phylorate plot of lineage diversification based on the “pruned-before” MCC phylogeny. Colors at each point in time along the branches of the phylorate plot denote instantaneous rate of diversification. Warmer/lighter colors (red) indicate faster rates and cooler/darker colors (blue) indicate slower rates. Below the phylorate plot is the global deep-sea oxygen isotope records (modified from Zachos et al. 2008). These records indicate a rapid decrease in global temperatures following the Eocene–Oligocene Transition and the Mid-Miocene Climate Transition (Zachos et al. 2008), giving rise to more open vegetation habitats such as grasslands and woodlands (Singh 1988; Prothero and Berggren 2014; Leopold et al. 2014). However, the lack of rate shifts suggests that there are no significant increases in diversification rates after these climate events. b–d) Diversification-through-time plots depicting family-specific net diversification trajectories computed from the joint posterior density of macroevolutionary rate parameters in BAMM. The black lines denote the background diversification rate (the rate of all musteloids minus the rate of each respective family). Shading intensity of the colored lines indicate the 5% through 95% Bayesian credible regions on the distribution of rates at any point in time for Mustelidae (b), Procyonidae (c), and Mephitidae (d). The black lines denote the mean background diversification rate-through-time (the rate of all musteloids minus the rate of each respective family), and the grayscale shading illustrates the 95% credible interval of the distribution of background rates through time.
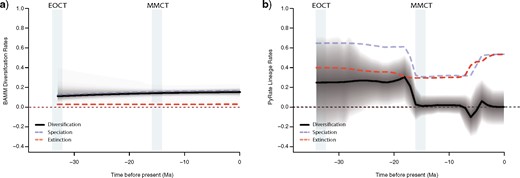
Rate-through-time plot in extant musteloids using BAMM (a) and in fossil musteloids using PyRate (b). Net diversification rates are shown in black, speciation rates in blue/light grey, and extinction rates in red/dark grey. The grayscale shading illustrates the 95% credible interval for net diversification. EOCT|$=$| Eocene–Oligocene Climate Transition; MMCT |$=$| Mid-Miocene Climate Transition.
Paleontological estimates of speciation and extinction rates derived from 2168 fossil occurrences indicate a relatively fast net diversification rate for musteloids throughout the Oligocene and the earliest Miocene due to a speciation rate (0.713 lineages/lineage million years, 95% HSD, 0.506–1.001 lineages/lineage million years) that is almost double the extinction rate (0.417 lineages/lineage million years, 95% HSD, 0.259–0.696 lineages/lineage million years) (Fig. 5b). At approximately 19–16 Ma, net diversification rate dramatically decreased to almost 0 lineages/lineage million years (0.020 lineages/lineage million years, 95% HSD, -0.094–0.153 lineages/lineage million years) due to a sudden slowdown in speciation rate just before the onset of the Mid-Miocene Climate Transition. Most of the remaining Miocene is characterized by relatively stable and balanced speciation and extinction rates. Both speciation and extinction rates rapidly increased during the Late Miocene and into the Pliocene, resulting in a brief decrease in net diversification rate. By the late Pliocene, speciation and extinction rates stabilized and net diversification rate returned to near 0 lineages/lineage million years.
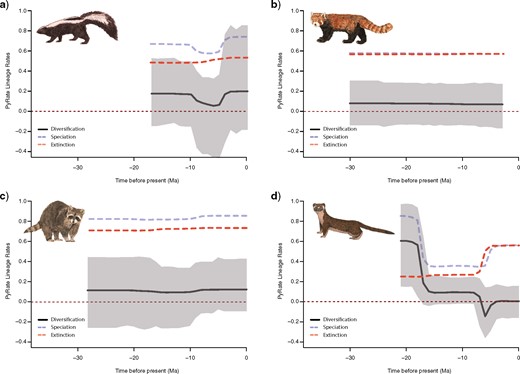
Similar to the BAMM results, mean net diversification rates inferred from our PyRate analyses using fossil data are broadly congruent across families: Mephitidae |$=$| 0.153 lineages/lineage million years (95% HPD, |$-$|0.241 to 0.545 lineages/lineage million years), Ailuridae |$=$| 0.009 lineages/lineage million years (95% HPD, |$-$|0.223 to 0.232 lineages/lineage million years), Procyonidae |$=$| 0.112 lineages/lineage million years (95% HPD, |$-$|0.205 to 0.413 lineages/lineage million years), and Neomustelidae |$=$| 0.152 lineages/lineage million years (95% HPD, |$-$|0.066 to 0.373 lineages/lineage million years). Diversification rates for Mephitidae, Ailuridae, and Procyonidae were relatively constant through time (Fig. 6). Neomustelids, in contrast, exhibited similar diversification rate patterns when compared to diversification patterns of the whole musteloid clade. Net diversification rate dramatically decreased to 0.041 lineages/lineage million years (95% HSD, |$-$|0.115 to 0.209 lineages/lineage million years) around 19–16 Ma. Speciation and extinction rates rapidly increased during the Late Miocene and into the Pliocene, resulting in a brief decrease in net diversification rate. Net diversification rate returned to near 0 lineages/lineage million years by the late Pliocene.
Rate-through-time plot in fossil mephitids (a), ailurids (b), procyonids (c), and mustelids (d) from PyRate. Net diversification rates are shown in black, speciation rates in blue/light grey, and extinction rates in red/dark grey. The grayscale shading illustrates the 95% credible interval for net diversification.
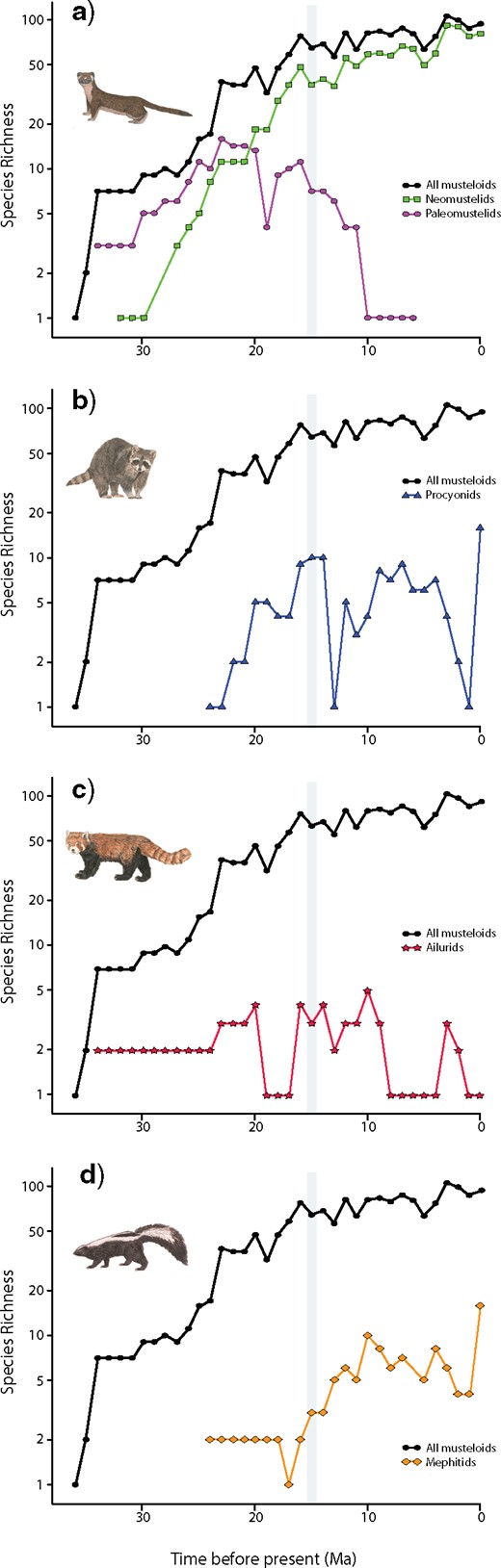
Total musteloid species richness continued to increase over the past 37 Ma based on plots of fossil diversity through time (Fig. 7). Closer examination of individual musteloid clades reveals that neomustelids have dominated total musteloid species richness since ~ 20 Ma, when they replaced paleomustelids as the most numerically abundant clade (Fig. 7a). Procyonid richness has been relatively stable since the Mid-Miocene Climate Transition and mephitid richness leveled off around 10 Ma (Fig. 7b,d). Furthermore, ailurid richness decreased (Fig. 7c) whereas paleomustelids became extinct by 6 Ma (Fig. 7a).
Species richness of Mustelidae (a), Procyonidae (b), Ailuridae (c), and Mephitidae (d) across the past 37 Ma. The black curve in each of plot denotes total musteloid richness. The grey bar indicates the Mid-Miocene Climate Transition. Colored curves denote species richness of each respective clade: green squares |$=$| neomustelids; pink circles |$=$| paleomustelids; blue triangles |$=$| procyonids; red stars |$=$| ailurids; orange diamonds |$=$| mephitids.
Clade diversity is negatively correlated with speciation rate (|$-$|0.010, 95% HPD |$-$|0.016 to |$-$|0.004) but is not significantly correlated with extinction rate (0.005, 95% HPD |$-$|0.001 to 0.011). Global temperature was also negatively correlated with speciation rate (|$-$|0.0781, 95% HPD |$-$|0.122 to 0.041) and with extinction rate (|$-$|0.129, 95% HPD |$-$|0.170 to |$-$|0.088). Correlations with global temperature are driven by the last 13.65 Ma: from the root age to 13.65 Ma, global temperature is not correlated with speciation rate (|$-$|0.585, 95% HPD |$-$|2.230 to |$-$|0.818) or extinction rate (|$-$|0.965, 95% HPD |$-$|2.571 to |$-$|0.782), whereas from 13.65 Ma to the present, global temperature is negatively correlated with speciation rate (|$-$|2.722, 95% HPD |$-$|4.135 to |$-$|1.526) and extinction rate (|$-$|2.371, 95% HPD |$-$|3.746 to |$-$|1.130). The negative correlation found between clade diversity and speciation rate remains the same in both time periods.
Phenotypic Evolution Rate Through Time
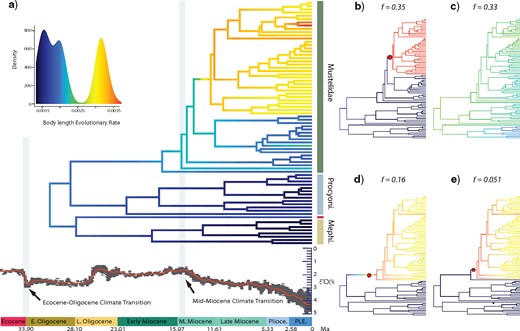
Using BAMM, we recover seven distinct rate shift configurations for body length, of which three configurations account for the majority (|$f = 0.84$|) of the posterior probability of the data (Fig. 8b–d). The first shift configuration (|$f = 0.35$|) suggests a single rate shift at the node leading to the diversification of Ictonychinae, Mustelinae, and Lutrinae (node 41; Fig. 8b), the second shift configuration (|$f = 0.33$|) suggests that there are no significant shifts in phenotypic evolutionary rate across the entire phylogeny (Fig. 8c), and the third shift configuration (|$f = 0.16$|) suggests a single shift of increased phenotypic evolutionary rate at the root of Mustelidae (node 23; Fig. 8d). Overall, our BAMM analyses inferred a general pattern of increased rates of body length evolution within Mustelidae after the Mid-Miocene Climate Transition followed by a slowdown in evolutionary rates (Figs. 8 and 9a; Supplementary Fig. S4 available on Dryad). Mephitidae and Procyonidae also exhibited a slowdown in evolutionary rates toward to the present but did not exhibit an inferred rate shift.
Phylorate plot of phenotypic (body length) evolution rates through time a) Phylorate plot of the mean phenotypic evolutionary rate of body length across all shift configurations based the MCC phylogeny. b–d) Phylorate plots of four distinct shift configurations with the highest posterior probability. Rate shifts, shown as red circles with sizes proportional to the marginal probability of the shift, demonstrate significant increase in evolutionary rate. Three distinct shift configurations account for the majority of the posterior probability of the data but result in conflicting rate configurations. The most frequent shift configuration (f = 0.35) signifies no rate shift (b) whereas the second most frequent shift configuration (f = 0.33) indicates an increase in evolutionary rate at the node leading to the divergence of Ictonychinae, Mustelinae, and Lutrinae (c). Conversely, the thirdmost frequent shift configuration (f =0.16) reveals a rate shift at the root of Mustelidae (d).
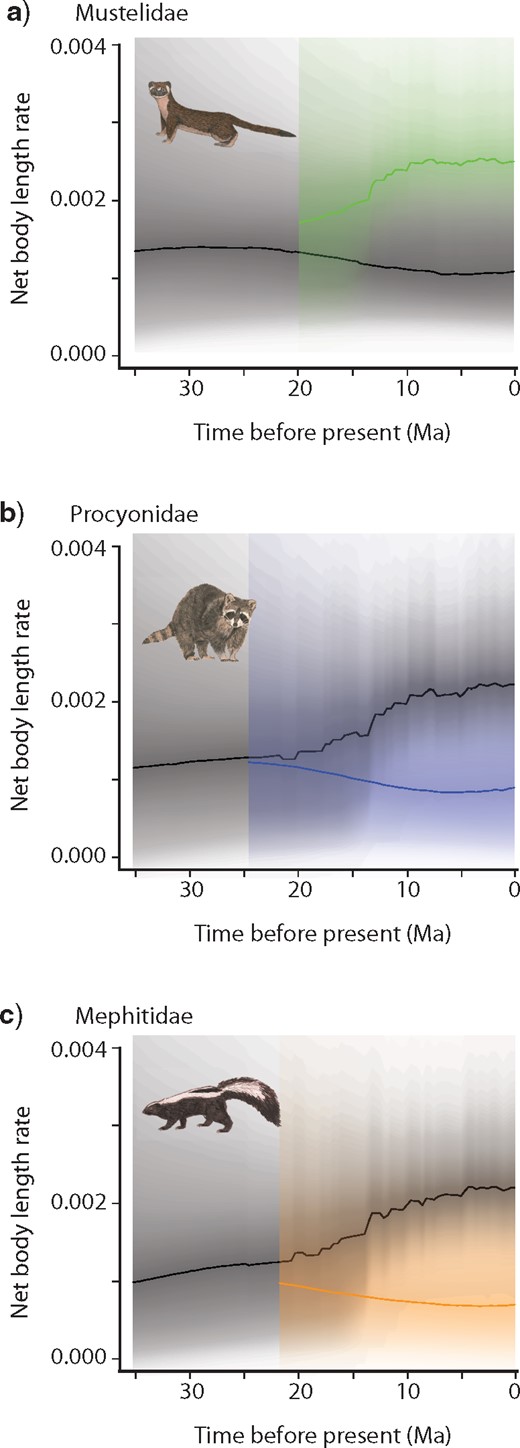
Rate-through-time plots depicting net body length evolutionary rate in Mustelidae (a), Procyonidae (b), and Mephitidae (c). Shading intensity of the colored lines indicates the 5% through 95% Bayesian credible regions on the distribution of rates at any point in time for Mustelidae (a), Procyonidae (b), and Mephitidae (c). The black curve in each plot denotes the mean background diversification rate-throughtime (the rate of all musteloids minus the rate of each respective family), and the grayscale shading illustrates the 95% credible interval of the distribution of background rates through time. Mustelids (mean rate, 0.0023; 95% HPD, 0.0016–0.0033) exhibit greater body length evolutionary rates than procyonids (mean rate, 0.0011; 95% HPD, 0.0004–0.0023) and mephitids (mean rate, 0.0011; 95% HPD, 0.0004–0.0023).
We found no shifts in rates of body mass evolution (Supplementary Fig. S5 available on Dryad). We did, however, observe a general slowdown in body mass evolutionary rate through time.
Discussion
We compiled a novel time-scaled phylogeny for 88% of extant musteloids and a database of occurrences for fossil musteloids to examine if rates of lineage diversification and phenotypic evolution in this clade followed patterns predicted under adaptive radiation theory. Contrary to expectations linking diversification to ecological opportunity, we found no association between changes in rates of lineage diversification and the Eocene–Oligocene transition or Mid-Miocene Climate Transition as previously hypothesized by Koepfli et al. (2008) and Sato et al. (2009, 2012). Nevertheless, analyses of phenotypic evolutionary rates help to shed some light on musteloid diversification dynamics through time, and explicit modeling of decoupled diversity-dependent dynamics informed by these analyses results in a more nuanced picture of how ecological opportunity has shaped extant diversity and phylogenetic structure of this diverse carnivoran clade. Specifically, our results suggest that body elongation may have served as an innovation that allowed a subclade of mustelids to escape niche competition and rapidly diversify after the onset of ecological opportunity.
Diversification of Musteloidea
The inference of decoupled diversification dynamics with respect to carrying capacity within Mustelidae suggest that ecological opportunity after the onset of the Mid-Miocene Climate Transition played a key role in musteloid diversification, but was phylogenetically restricted in its effects. The DDD analyses, which specifically test for clade-specific shifts in carrying capacity in addition to diversification rates (Etienne and Haegeman 2012), provide support for decoupled dynamics between the mustelid subclade consisting of Helictidinae, Guloninae, Ictonychinae, Mustelinae, and Lutrinae and the remaining musteloid clade (Table 1). This pattern seems to be largely driven by a carrying capacity for the mustelid subclade that is nearly twice as large as that of the main clade, though associated increases in base speciation rate and/or decreases in base extinction rate are also implied. It is important to note that these decoupled diversity-dependent dynamics necessarily imply a pulse of increased diversification in the decoupled clade at its origin, even if there is no increase in basal speciation rate, as speciation rates decline as a function of diversity and the decoupling effectively “resets” them (Sepkoski 1996; Etienne and Haegeman 2012). Within this framework, “early bursts” of net diversification are therefore necessarily implied both at the base of Musteloidea and the decoupled clade, regardless of their magnitude or whether the molecular phylogeny retains a signal. Our ability to reject a null, constant-rates process may be compromised here by the poor performance of AIC as a model selection tool (Etienne et al. 2016) and our parametric bootstraps suggest caution should be taken in interpreting these findings due to slightly elevated type I error rates (Fig. 3a). However, at least heuristically, our parametric bootstrap results are consistent with decoupled dynamics. Etienne et al. (2016) found that power to detect diversity dependent processes declines as extinction rate increases or as intrinsic speciation rate decreases or, alternatively, when clade diversity is low relative to |$K$| (Liow et al. 2010). Because decoupled dynamics necessarily imply pulses of increased diversification toward the tips of ultrametric phylogenies, we might expect to infer high-carrying capacities, relative to actual clade diversity, when attempting to explain such data under a common diversity dependent regime framework. In our case, the inferred |$K$| of ~ 366 species for the CR1 model is an order of magnitude larger than the background |$K$| in our preferred decoupled models (Table 1) and several times larger than the largest |$K$| inferred for a decoupled clade. While it is clear that further work is needed to understand and develop appropriate model selection tools for decoupled diversity dependent diversification models, as has recently begun with trait evolutionary models (Ho and Ané 2014), taken at face value our results are consistent with the hypothesis that an elongate mustelid subclade was able to escape niche competition and diversify rapidly after the onset of ecological opportunity in a new niche that could support more species than the ancestral one (Etienne and Haegeman 2012).
Theories linking ecological opportunity to adaptive radiation often invoke the evolution of key innovations as a mechanism to facilitate the exploitation of novel niches (Simpson 1944, 1955; Schluter 2000). Interestingly, many members of the decoupled subclade have been observed to exhibit relatively elongated body plans compared to other carnivorans and even to other musteloids (Brown and Lasiewski 1972; King 1989). Thus, one possible innovation facilitating decoupled diversification dynamics via increased carrying capacity is body elongation, where body lengths are drastically increased relative to body depth. Body elongation in mustelids is often hypothesized to have evolved as a response to the Late Miocene diversification of rodents, permitting mustelid species to enter burrows and confined spaces to capture prey (Brown and Lasiewski 1972; King 1989). In addition, body elongation may have served as a preadaptation that facilitated the exploitation of aquatic habitats by streamlining the body profile and reducing total body drag and energetic demands during high speed swimming (Fish 1996), and semi-aquatic mustelids therefore have a distinct advantage for high speed swimming compared to other nonelongated mammals of similar sizes (Williams 1983, 1989; Fish 1994). A number of authors have noted that mustelid guilds are often particularly diverse in comparison to other carnivore guilds (e.g., Powell et al. 1983), and size-based character displacement appears common (e.g., Dayan et al. 1989; Dayan and Simberloff 1994) though the effects on resource use may be strongest between sexes, rather than among species (McDonald 2002). Significantly, Dayan and Simberloff (1994) found that equal-size ratios among British mustelids were only found when the Eurasian badger (Meles meles) was excluded. The remaining members of the British mustelid guild (Martes martes, Mustela erminea, M. nivalis, M. putorius) all belong to the elongate clade identified in our DDD analyses, suggesting that these taxa may indeed form part of an ecologically discrete community. In line with the observation that many mustelids exhibit relatively elongated bodies for a given mass (Brown and Lasiewski 1972; King 1989; Fish 1994), we found a lack of correspondence in patterns of phenotypic evolutionary rates when examining body mass and length independently (Figs. 7 and 8a; Supplementary Figs. S4 and S5 available on Dryad). This is consistent with Gans (1975) definition of body elongation in which elongate organisms exhibit an increased in relative body length for a given mass. Although not observed in helictidines and gulonines, the discordance of body length and body mass evolutionary rates in ictonychines, mustelines, and lutrines nonetheless suggested that body shape variation may have been partitioned rapidly after the origin of ecological opportunity. Thus, elongate mustelids may have been ideally suited to postMid-Miocene grassland environments and accompanying prey resources, resulting in their diversification whereas other musteloids steadily declined. While this may ultimately suggest that body elongation served as an adaptive trait for this clade, additional investigation is needed to quantify body shape to determine whether body elongation truly conferred adaptation to the ecological opportunity presented by the onset of the Mid-Miocene Climate Transition as well as to further support the decoupling diversification dynamics we found within Mustelidae.
Results from our analyses of diversification rates through time using molecular- and fossil-based methods lead to slightly different interpretations of musteloid diversification dynamics (Fig. 5). Using both BAMM and PyRate, we found no evidence for rapid bursts of musteloid diversification rates during times of hypothesized ecological opportunity near the Eocene–Oligocene transition and Mid-Miocene Climate Transition, contrary to expectations derived from classic adaptive radiation theory (Simpson 1955; Schluter 2000; Harmon et al. 2003; Rabosky and Lovette 2008a). Nevertheless, relative magnitudes of diversification rates throughout the Eocene, Oligocene, and much of the Early Miocene differed substantially between the two sets of analyses (Fig. 5). We inferred low-net diversification rates from the molecular phylogeny throughout this time period, likely due to a lack of extant clades originating at this time (Fig. 5a). In contrast, we found diversification rates inferred from the fossil record were relatively high until just before the Mid-Miocene Climate Transition, when a sudden decrease in origination resulted in net diversification rate of almost 0 lineages/lineage million years for the rest of the Middle Miocene. This pattern is strongly suggestive of equilibrium dynamics (Sepkoski 1978; Rabosky and Lovette 2008b; Etienne et al. 2012; Etienne and Haegeman 2012); indeed, speciation rate correlated negatively with musteloid paleodiversity over the entire interval examined. Though net diversification rate remained relatively constant, fluctuations in the absolute magnitudes of speciation and extinction rates from the Middle/Late Miocene boundary to the present appear to correlate with global temperature change. The increase in extinction rate observed for the Late Miocene is unsurprising considering that this period is often characterized as a period of high extinction of mammalian taxa hypothesized to be driven by changes in climate and environments (e.g., the Vallesian Crisis; Agusti and Moya-Sola 1990; Agustí et al. 2013; Fortelius et al. 2014). Concomittant increases in speciation rates further suggest that musetelids experienced high turnover during the Neogene as a result of climate driven environmental change (Baskin 1998).
Similar patterns of sustained diversification have been documented in other clades that were at one time hypothesized to have undergone adaptive radiations, such as Neotropical ovenbirds and woodcreepers (Derryberry et al. 2011), African Synodontis catfish (Day et al. 2013), and Heliconius butterflies (Kozak et al. 2015). These multiple results, including this present study, suggest that the “early burst” model of lineage diversification may be inappropriate for many ecologically diverse clades that otherwise fit within the adaptive radiation paradigm. It is possible that many clades are simply unable to rapidly exploit all suitable niches as soon as they arise, especially across complex and/or large environments such as entire continents (Derryberry et al. 2011; Day et al. 2013; Kozak et al. 2015; Liedtke et al. 2016). Because mustelids are globally distributed and, presumably, good dispersers, it is possible that ecological opportunity may be less of a limiting factor than it would be in more restrictive settings, such as islands, or for less mobile taxa. It is similarly tempting to infer that the signature of sustained diversification rates recovered from our analyses is an indication that musteloid niche space is not yet fully exploited.
The inconsistency between the BAMM and PyRate results provide a compelling reminder of how extinction can erase the signal of past diversification history in the structure of a molecular phylogeny (Rabosky and Lovette 2008a; Quental and Marshall 2009; Liow et al. 2010) and stress the importance of combining paleontological and neontological data in a unified framework (Slater and Harmon 2013). Such methods are in their infancy at present, but the FBD variant of the skyline model shows some promise in incorporating both datasets in diversification analyses by allowing stepwise rate changes across a FBD tree with extant and extinct species (Stadler et al. 2013; Gavryushkina et al. 2014). Using the FBD skyline model implemented in BEAST on a fixed, time-scaled tree including fossil tips, we found little support for distinct diversification rates before and after 19 Ma (see Appendix 3 for full methods and results). However, we found strong support for higher net diversification rate after 13.65 Ma (Mid-Miocene Climate Transition), which corroborates with our findings that global temperature and diversification rates correlate during this timeframe. Nevertheless, the reduced fossil dataset used to date our FBD tree may affect signals of shifts in evolutionary rates; of the 453 fossils used in our PyRate analyses, only 74 of these fossil species had sufficient information required to conservatively place them within a phylogenetic context for the FBD analyses (Appendix 1). Therefore, while the addition of a few fossil taxa to a phylogeny may improve inferences regarding trait evolutionary rates or mode (Slater et al. 2012), more complete sampling may also be necessary for comparable improvements in diversification rate studies.
Body Length or Body Mass?
While the size of an organism has important functional implications for how it utilizes its environment (Schmidt-Nielsen 1984), the question arises of which size metric most thoroughly describes “body size”? Both body length and body mass are readily obtainable in the literature and thus are commonly used as proxies to quantify rates of phenotypic evolution (Gonzalez-Voyer et al. 2009; Slater et al. 2010; Derryberry et al. 2011; Wilson et al. 2012; Aristide et al. 2015). Because mass and length are normally highly correlated (Schmidt-Nielsen 1984), the majority of studies use just one body size metric, commonly body mass, as a proxy for ecomorphological diversity to test patterns of phenotypic evolution. In this present study, the decoupling of these two body size metrics within Mustelidae illustrates the possibility that rates of phenotypic evolution under a single body size metric may mislead our interpretation of patterns of diversification. Therefore, using both body length and body mass may be a more robust approach for testing patterns of ecomorphological disparity in taxa with derived body plans such as elongation and deeper and/or flatter shapes. Alternatively, future studies interested in characterizing elongation may consider using recent metrics such as elongation ratio (ER) (Ward and Azizi 2004) or the vertebrate shape index (VSI) (Collar et al. 2013). While these metrics require the measurement of character traits that are not readily found in the literature, such as body width or depth for ER or vertebral measurements for VSI, they will provide a more comprehensive view of how clades are changing their body shape. Nevertheless, we recognize that, for several clades, comprehensive ecomorphological datasets are unavailable and we are usually left with body length and/or body mass. We therefore emphasize the importance of using both body size metrics when testing phenotypic evolutionary rates when other more rigorous metrics are unavailable.
Conclusion
Adaptive radiation theory predicts that ecological opportunity promotes rapid lineage diversification coinciding with increases in phenotypic disparity as colonizing lineages rapidly evolve adaptive traits that are highly correlated to their new resources (Simpson 1955; Schluter 2000). Qualitative inferences derived from time calibrated phylogenies have suggested that Musteloidea, an ecomorphological diverse clade of carnivorans, exhibits two pulses of adaptive radiation, once after the Eocene–Oligocene transition and a second radiation after the Mid-Miocene Climate Transition. Using quantitative phylogenetic methods with both extant and fossil musteloids and a comprehensive time-calibrated phylogeny, we found no evidence an early burst in the rate of lineage diversification throughout the entire phylogeny, including during these climate transitions. Nevertheless, using DDD to specifically test for the effects of key innovations on diversification, we found some support for decoupled diversification dynamics driven by increased clade carrying capacity in the branches leading to a subclade of elongate mustelids. Supporting decoupled diversification dynamics between the mustelid subclade and the remaining musteloids is our finding that there is a lack of correspondence in patterns of body length and body mass evolutionary rates within the decoupled mustelid subclade. The increase in the rate of body length evolution but not body mass evolution suggested that body elongation might be a key innovation for the exploitation of novel Mid-Miocene habitats and resources and subsequent diversification in some musteloids. Additional studies quantifying body shape of both extant and extinct musteloids will further elucidate patterns of ecomorphological diversification within this clade.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.nj1bp.
Author’s Contributions
CJL designed the study, collected the sequence, body size, and fossil data, performed the majority of data analyses, and drafted the manuscript. G.J.S helped compile fossil data to date the phylogeny and performed the DDD analyses. R.S.M and G.J.S helped develop the approach, provided crucial insights, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a University of California Santa Cruz Regents Fellowship; a Rebecca and Steve Sooy Graduate Fellowship for Marine Mammals; a Rosemary Grant Graduate Student Research Award from the Society for the Study of Evolution; and a National Foundation of Sciences Graduate Research Fellowship to C.J.L and a Peter Buck postdoctoral fellowship through the National Museum of Natural History to G.J.S.
Acknowledgements
We are grateful to Nancy Hung and Shohei Burns for assisting in the collection of GenBank sequences, Giacomo Bernardi for discussion about phylogenetic methods, Remco Bouckaert and Beth Shapiro for advice with BEAST, Dan Rabosky and Mike Grundler for assistance with BAMM, Daniele Silvestro for advice with PyRate, David Bapst for assistance with Paleotree, Rampal Etienne for advice with DDD, Sasha Gavryushkina for advice with the FBD skyline model, Alan Shabel for insight on Aonyx, and Vikram Baliga for helpful discussions. Lastly, we thank Editor Frank Anderson, Associate Editor Tanja Stadler, Michael Alfaro, Rampal Etienne, and 1 anonymous reviewer for valuable comments that greatly improved previous versions of this manuscript.
Appendix 1. Fossils used to calibrate divergence times estimations using the fossilized birth–death model implemented in BEAST.
Appendix 2. Results of phylogenetic analyses and divergence time estimations.
Appendix 3. Methods and results for FBD skyline model.
References
Author notes
Associate Editor: Tanja Stadler